Abstract
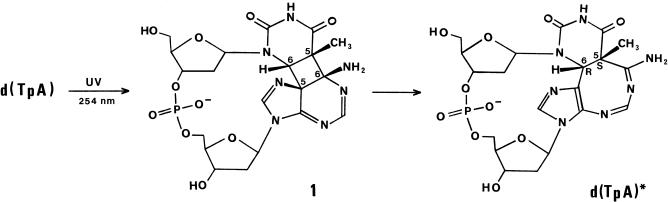
A high-resolution crystal structure is reported for d(TpA)*, the intramolecular thymine–adenine photoadduct that is produced by direct ultraviolet excitation of the dinucleoside monophosphate d(TpA). It confirms the presence of a central 1,3-diazacyclooctatriene ring linking the remnants of the T and A bases, as previously deduced from heteronuclear NMR measurements by Zhao et al. (The structure of d(TpA)*, the major photoproduct of thymidylyl-(3′-5′)-deoxyadenosine. Nucleic Acids Res., 1996, 24, 1554–1560). Within the crystal, the d(TpA)* molecules exist as zwitterions with a protonated amidine fragment of the eight-membered ring neutralizing the charge of the internucleotide phosphate monoanion. The absolute configuration at the original thymine C5 and C6 atoms is determined as 5S,6R. This is consistent with d(TpA)* arising by valence isomerization of a precursor cyclobutane photoproduct with cis–syn stereochemistry that is generated by [2 + 2] photoaddition of the thymine 5,6-double bond across the C6 and C5 positions of adenine. This mode of photoaddition should be favoured by the stacked conformation of adjacent T and A bases in B-form DNA. It is probable that the primary photoreaction is mechanistically analogous to pyrimidine dimerization despite having a much lower quantum yield.
INTRODUCTION
Despite its strong absorption at ultraviolet (UV) wavelengths, native DNA is a relatively photostable molecule because it dissipates electronic excitation energy very rapidly, primarily through internal conversion processes (1). Nonetheless, a variety of intramolecular photoproducts can be formed with low quantum efficiency when cellular DNA is exposed to UV radiation which, if not repaired, may have mutagenic or lethal consequences (2–4). The photoproducts induced by direct UV excitation arise almost exclusively from photoaddition reactions between adjacent nucleobases on the same strand of DNA. The pyrimidine bases are much more reactive than their purine counterparts in this respect so that photodamage is predominantly targeted to the thymine, cytosine and (where applicable) 5-methylcytosine bases. The resulting bipyrimidine photoproducts comprise either cyclobutane photodimers or the less abundant pyrimidine (6–4) pyrimidone photoadducts and their Dewar valence isomers. There is now a wealth of experimental evidence demonstrating the mutagenic properties of these species and implicating them as the major lesions responsible for non-melanoma skin cancers induced by sunlight (5). Reflecting their biological importance, the dimeric pyrimidine photoproducts have been the subject of very detailed physical and chemical characterization and their individual yields have been determined in both isolated and cellular DNA (6). Progress in defining the role of the purines adenine and guanine in the photochemistry and photobiology of DNA has been much more limited, not least because of their marked resistance to photomodification both as monomers and at the polynucleotide level. As yet, no dimeric photoproducts derived from guanine have been detected in DNA although a number of cross-links identified in UV-irradiated 16 S ribosomal RNA are indicative of their formation (7). Adenine, however, is known to undergo photoaddition reactions with adjacent adenine or 5′-thymine bases in duplex DNA (8–12) but with quantum yields (12,13) that are approximately 100-fold lower than for pyrimidine dimerization. The individual photoproducts can be prepared by irradiating the dinucleoside monophosphates d(ApA) and d(TpA) in aqueous solution at 254 nm (9,10,14). In the case of d(ApA), two structurally distinct photoadducts are formed (10) but d(TpA) gives a single photoproduct d(TpA)* whose base moiety is denoted TA*.
Initial characterization of d(TpA)* suggested that it was a cyclobutane derivative (of uncertain stereochemistry) generated by [2 + 2] photoaddition of the 5,6-double bond of thymine across the C6 and C5 positions of adenine (11,15). A subsequent investigation of its solution structure by multi-dimensional 1H NMR and molecular modelling methods (14) led to the conclusion that it was a trans–syn cyclobutane adduct akin to structure (1) in Figure 1 (which is drawn with cis–syn stereochemistry). However, this assignment was later shown to be incompatible with the 13C NMR spectrum by Zhao et al. (16) who deduced that d(TpA)* results from strain-relieving ring expansion of the precursor cyclobutane adduct to give the more stable valence isomer incorporating an 8-membered heterocyclic ring that is depicted in Figure 1. In this structure, the configuration at the original thymine C5 and C6 carbon atoms is diagnostic of the stereochemistry of the initial cyclobutane adduct. Thus, the 5S,6R isomer of d(TpA)* shown in Figure 1 will be derived from the cis–syn adduct (1) in which both the thymidine and 2′-deoxyadenosine moieties are in the anti conformation about their glycosidic bonds. Conversely, the 5R,6S isomer will be formed from the originally proposed trans–syn cyclobutane adduct (14) where thymidine is in the syn orientation.
Figure 1.
Structure of d(TpA)* and the precursor cyclobutane photoadduct (1).
Zhao et al. (16) attempted to distinguish between these two possibilities by modelling the structure of d(TpA)* using molecular mechanics calculations informed by NMR-derived torsion angle and distance constraints. The results were inconclusive, however, because both stereoisomers could be reconciled with the available data. But, as these authors emphasized, the compatibility with 5S,6R stereochemistry allows a strong case to be made for cis–syn photoaddition because it should be favoured by the stacked geometry of TpA doublets in duplex DNA.
In this article, we report the X-ray crystal structure of d(TpA)* in a zwitterionic form which fully confirms the covalent structure of the photoproduct advanced by Zhao et al. (16). Significantly, the configuration at the central ring junction is unambiguously established as 5S,6R thus implying that the primary photoreaction generates a cyclobutane adduct (1) having cis–syn stereochemistry.
MATERIALS AND METHODS
Preparation of d(TpA)*
Ten-milligram batches of d(TpA) ammonium salt (from Sigma-Aldrich) were dissolved in deionized water (110 ml) and then adjusted to pH 7 by addition of very dilute aqueous ammonia. The solution was irradiated at 254 nm under nitrogen in a quartz tube as described earlier (15) until its absorbance at 260 nm had fallen to 55% of the initial value. The irradiated solution was reduced to a small volume by rotary evaporation at room temperature and finally lyophilized in a vacuum concentrator. The residue was redissolved in water (∼300 μl) and fractionated (in 50 μl aliquots) by reversed-phase HPLC on a phenomenex Jupiter 5 μ C18 column (250 × 10 mm) using essentially the same operating conditions as described previously (14). Isocratic elution with water was sufficient to elute the photoproduct d(TpA)* and a subsequent methanol gradient was used to free the column of unchanged d(TpA) and minor contaminants. The fractions containing d(TpA)* were pooled, lyophilized, and re-purified by repeating the HPLC separation once more. This gave chromatographically homogeneous material, with the expected UV absorption profile (11), which was used for crystallization purposes.
Crystallization and structure determination
Initial studies showed that needle-shaped microcrystals of d(TpA)* could be produced simply by allowing concentrated aqueous solutions (∼10 mg/ml) to evaporate at room temperature on a glass slide. However, while X-ray diffraction analysis of a crystal thus obtained revealed the connectivity of all the non-hydrogen atoms in the molecule, the crystals were invariably too small to permit complete definition of the structure. This difficulty was overcome when crystals were grown by the hanging-droplet method.
Drops containing 2 mg/ml d(TpA)* in water were equilibrated against a reservoir of 10% v/v MPD (2-methyl-2,4-pentanediol) solution in water at 20°C. Crystals grew as long rods after ∼1 month. These were up to 1.25 mm in length and 0.06 × 0.06 mm in cross section. Crystals are in the space group P212121, with cell dimensions of a = 8.327, b = 10.732, c = 33.128 Å. Diffraction data were collected at 100 K on an Oxford Diffraction Gemini-S-Ultra instrument using Cu-Kα radiation, to a resolution of 0.87 Å. The 16145 reflections measured merged with an Rmerge of 3.2%, and had an average redundancy of 5.9. A total of 99% of the available reflections in the range 33.0–0.87 Å were observed, compared with 94% in the highest resolution range of 0.90–0.87 Å. This high-resolution set had an Rmerge of 7.1%.
The structure was solved by direct methods using the SHELX program (17), and refined by full-matrix least-squares techniques. All non-hydrogen atoms were refined with anisotropic temperature factors. Hydrogen atom positions were located from difference Fourier maps, and were included in subsequent refinement cycles, with isotropic temperature factors being varied. Solvent atoms were also located by difference maps. Their identification as water oxygen atoms, ammonium or sodium ions was verified by systematic evaluation of each in turn, monitoring changes in R and Rfree values together with considerations of hydrogen bonding to the d(TpA)* molecule and to other solvent atoms. The final R value is 2.83% for the 4560 unique reflections; Rfree is 7%. The value of the Flack x parameter at the end of the refinement was 0.00, indicating that the assigned absolute configuration is correct (this is an independent crystallographic verification of the absolute stereochemistry, which is also defined by the absolute stereochemistry of the two 2-deoxyribose sugar rings). Final coordinates and structure factors have been deposited with the Nucleic Acid Database as ID number UD0072.
RESULTS
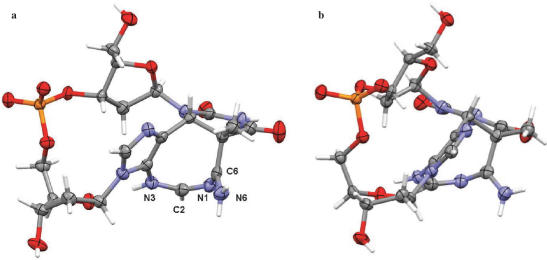
The crystal structure of d(TpA)* shows that, as predicted by Zhao et al. (16), the photoreaction between the T and A bases in d(TpA) has resulted in a tricyclic ring system in which a central diazacyclooctatriene ring is fused to the original pyrimidine ring of thymine and to the imidazole ring of adenine. As can be seen from Figure 2, the stereochemistry at the thymine C5 and C6 positions is established as 5S,6R.
Figure 2.
Two views (a and b) of the d(TpA)* structure, showing thermal ellipsoids for the non-hydrogen atoms. Drawn with the MERCURY program (18).
The availability of highly diffracting, well-ordered crystals has enabled the crystal structure to be determined to atomic resolution, so that the positions of hydrogen atoms have been unequivocally defined and individual solvent atoms assigned correctly. It was initially assumed that one of the solvent molecules was actually an ammonium ion (or less likely a sodium ion), in order to preserve charge neutrality with the phosphate group. A total of six solvent non-hydrogen atoms were located, all of which were well-ordered. They were initially refined as water oxygen atoms, at which point each in turn was changed to an ammonium nitrogen atom, and re-refined. In every case the refinement resulted in higher R and Rfree values. Difference Fourier maps showed that each of the six solvent atoms has two bound hydrogen atoms, which are fully in accord with the hydrogen-bonding network (detail not shown), and therefore with all the solvent molecules being assigned as waters.
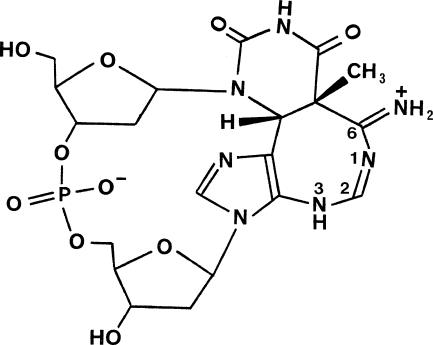
This therefore implies that the d(TpA)* molecule itself is protonated and exists as an internal salt (zwitterion). Examination of the geometry of the diazacyclooctatriene ring and its substituents has enabled the site of protonation to be unequivocally identified, and thus the tautomeric state of d(TpA)* to be determined. The C6–N6 bond (Figure 2a) is short (1.285(3) Å), compared to the consensus length in adenine itself of 1.337 Å (19), consistent with the double bond character of an imino bond (Figure 3), as is the in-plane geometry of the two hydrogen atoms attached to N6. Atoms N1, C2, N3 and N6 are closely co-planar. Bond lengths N1–C2 and N1–C6 of 1.284(3) Å and 1.356(3) Å are indicative of double bond and sp2–sp2 single bond character respectively, while bond C2–N3 (1.344(3) Å) also conforms to the latter category. These assignments, together with the location and satisfactory refinement of hydrogen atoms on C2, N3 and N6, define the tautomerism to be as shown in Figure 3.
Figure 3.
Zwitterionic form of the d(TpA)* molecules in the crystals grown for structure determination.
The values of the d(TpA)* dinucleoside monophosphate backbone angles are given in Table 1. These show that the effect of the cross-linking has been to produce a highly asymmetric dinucleoside phosphate, with in particular the values for the two δ angles differing by almost 130°, and the two glycosidic angles by over 80°. Another consequence of the cross-linking is the large change in backbone angle ε(T) from its value in duplex polynucleotides, which has resulted in the thymidine deoxyribose ring being twisted to an almost perpendicular orientation compared to the deoxyadenosine sugar (Figure 2b). The latter has a C2′-endo pucker whereas that of the thymidine sugar is in the C3′-endo range. Other backbone torsion angles (Table 1) are closer to those of A- than B-form duplex DNA. Molecular modelling has been used to assess the effect of the TA* photoadduct on polynucleotide structure, by inserting it into a B-DNA helix. The resulting structure, after molecular dynamics simulation, shows remarkably little distortion beyond the immediacy of the cross-linked region.
Table 1.
Backbone torsion angles (in °) in d(TpA)*, compared to standard nucleic acid values (19). Esds for the d(TpA)* values average 0.4°
| Angle | d(TpA)* | A-DNA | B-DNA |
|---|---|---|---|
| δ(T) | 176.8 | 79 | 143 |
| ε(T) | 86.5 | −148 | −141 |
| ζ(T) | −106.8 | −75 | −161 |
| α(A) | − 59.9 | −52 | −30 |
| β(A) | 176.4 | 175 | 136 |
| γ(A) | 44.7 | 42 | 31 |
| δ(A) | 49.7 | ||
| χ(T) | −126.9 | −157 | −98 |
| χ(A) | −42.9 |
DISCUSSION
The determination of the crystal structure of d(TpA)* resolves important questions regarding its chemical identity and the nature of the photochemical reaction leading to its formation. It also provides a molecular basis for researching the structural and mutagenic consequences of thymine–adenine photoaddition in DNA. X-ray crystallographic studies of the major dimeric pyrimidine photoproducts have advanced to the point where structures are available for oligonucleotide duplexes incorporating a cis–syn thymine dimer both per se (20) and when complexed with DNA repair (21,22) or polymerase enzymes (23). A crystal structure has also been reported for the (6–4) photoadduct of d(TpT) bound to an antibody fragment (24). The present work, however, represents the first example where the covalent structure of a dimeric DNA photoproduct containing a purine base has been determined at atomic resolution. In the context of RNA photochemistry, a crystal structure has been reported for a model photocrosslink compound formed between 4-thiothymidine and adenosine (25).
The most striking structural feature of d(TpA)* is the 1,3-diazacyclooctatriene ring which contains a conjugated amidine grouping that imparts basic character to the molecule. Due to its presence, the pK for protonation is shifted (15) from 3.5 in d(TpA) to 5.5 in d(TpA)*. This satisfactorily explains why d(TpA)* crystallizes from unbuffered aqueous solution as the zwitterion shown in Figure 3 in which the negative charge of the phosphate monoanion is neutralized by the positive charge on the protonated amidino moiety. By contrast, it is reasonably certain that the NMR studies of its solution structure will have been carried out on the monoanionic form of d(TpA)*, shown in Figure 1, where the 8-membered heterocyclic ring is uncharged. Thus, the measurements of Koning et al. (14) were made in a phosphate buffer at pD 6.55. Although Zhao et al. (16) do not specify a particular NMR solvent, they prepared d(TpA)* by HPLC elution with a phosphate buffer at pH 6.8 and the 1H NMR spectrum was stated to be the same as that reported by Koning et al. (14). Given the difference in charge state and tautomeric form, it is not surprising that some conformational features of the crystal structure are at variance with those of the solution structure. Thus, while the diazacyclooctatriene ring is inferred to adopt a tub shape in solution, its protonation leads to atoms N1, C2, N3 and N6 being essentially co-planar in the crystal structure (Figure 2a). There are also significant differences in the pucker of both deoxyribose rings and the backbone torsion angles δ(T) and ε(T) between the crystal and solution structures (compare data in Table 1 and reference 16).
As pointed out by Zhao et al. (16), the diazacyclooctatriene ring of d(TpA)* can arise by electrocyclic or acid-catalysed valence tautomerization of an intermediate cyclobutane adduct. An important consequence of this process is that the configuration of the C5 and C6 atoms at the junction with the original thymine ring will be determined by the stereochemistry of the precursor cyclobutane photoproduct and thus provide insight into its mechanism of formation. The crystal structure of d(TpA)* allows the configuration at these carbons to be defined as 5S,6R relative to the known absolute configuration of the d-deoxyribose moieties. This can only result from ring expansion of the cis–syn cyclobutane adduct (1) shown in Figure 1. It is incompatible with valence isomerization of a trans–syn structure as this would lead to the 5R,6S configuration.
For photoaddition between the thymine and adenine bases of d(TpA) to generate the cis–syn photoproduct (1) it is necessary for both of the 2′-deoxyribonucleoside units to adopt an anti orientation about their respective glycosidic bonds. This mode of cycloaddition should therefore be favoured by TpA doublets in native B-form DNA where all the nucleotide residues are constrained in the anti conformation. The initial formation of a cis–syn cyclobutane photoadduct by d(TpA) formally resembles the [2 + 2] photodimerization of adjacent pyrimidine bases in DNA that leads almost exclusively to products with cis–syn stereochemistry (6).
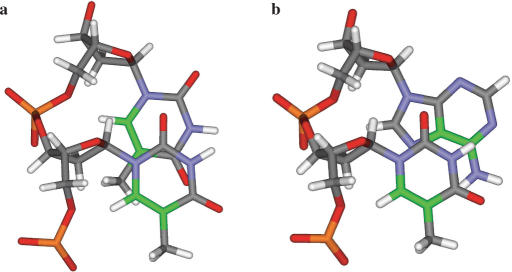
There is, however, a very marked difference in the efficiency of these photoreactions when double stranded DNA is irradiated at 254 nm. Thus, while the quantum yield for production of thymine–thymine photodimers is ∼0.02 mol einstein−1 (2), the yield for formation of TA* is estimated as ∼1 × 10−4 mol einstein−1 (12). The reason(s) for this greatly reduced yield is not clear but it may have its origin in altered excited state dynamics of the stacked bases (1,26,27) and/or stereoelectronic factors. In the latter regard, it is noteworthy that the photoreactive C5–C6 bond of a T base in canonical B-form DNA is separated by 4.4 Å from the C5–C6 bond of an adjacent T, but by a distance of 4.9 Å from the C6–C5 bond of a 3′-A (Figure 4). The greater conformational freedom of single polynucleotide strands should facilitate closer interaction of the photoreactive centres and might thus account for the approximately 4-fold increase in quantum yield observed with denatured DNA (12). Significant formation of TA* has been noted to accompany and compete with pyrimidine dimerization in some single-stranded oligonucleotides such as d(GTATTATG) (28–30). It has also been demonstrated to occur in short hairpin sequences (31), and in a duplex dodecamer incorporating a TATA box where the yield of TA* is suppressed by interaction with the TATA-binding protein, TBP (32).
Figure 4.
Views of (a) the TpT and (b) the TpA steps in canonical B-DNA, looking down the helix axis. The C5–C6 atoms and bonds have been coloured green.
By incorporating a single TA* photolesion into a 49-mer oligonucleotide (28) and thence into an M13mp18-derived phage construct, Zhao and Taylor (33) were able to assess the mutagenicity of TA* in transfected Escherichia coli cells. In the absence of the SOS response, the main effect of TA* was to block DNA replication. However, when SOS was activated, TA* was highly mutagenic and generated single base substitutions (mainly of A for T opposite the 3′-A) as well as tandem mutations. Rare instances of base sequence changes that map to TA doublets, and may therefore arise from TA* formation, have been observed in several studies of UV-induced mutagenesis conducted with both bacterial (34,35) and mammalian (36–38) cells. Again, there is a strong tendency for A.T → T.A transversions to occur as would be consistent with a modified 3′-A base in TA* coding for the incorporation of A during DNA replication. This scenario is, however, at odds with the tautomeric form of TA* found in its crystal structure (Figure 3). The protonated amidino fragment of the 8-membered ring could nominally base pair with T or G, but not with C or A. This lends support to the inference of Zhao and Taylor (33) that the predominant incorporation of A opposite the 3′-base moiety of TA* cannot be rationalized simply in terms of hydrogen bonding specificity.
Given that the measured pK of 5.5 (15) for protonation of d(TpA)* is close to neutrality, there is necessarily some uncertainty as to the tautomeric state adopted by TA* photoadducts in native DNA under physiological conditions. Although the unprotonated form shown in Figure 1 might be expected to predominate, some contribution from the protonated species (Figure 3) cannot be discounted. The presence of either tautomer will inevitably cause considerable distortion of the double helix at the sites of photoreaction. Nonetheless, a limited molecular modelling study based on the crystal structure coordinates indicates that longer range effects on the polynucleotide structure of B-DNA will be minimal.
The local structural perturbations induced by TA* formation are bound to impact on the protein–DNA recognition processes that, inter alia, govern replication and transcription. They may also serve as a trigger to initiate DNA repair. Reflecting its low abundance in UV-irradiated DNA, nothing is yet known about the capacity of TA* to act as a substrate for nucleotide excision or other forms of cellular DNA repair. Its behaviour in this regard is evidently a key determinant of its overall biological significance and requires investigation. Knowledge of the exact molecular structure of the photoadduct in d(TpA)* provides a secure foundation for studies of this type as well as being essential for a definitive description of the photochemical mechanism that leads to its formation.
ACKNOWLEDGEMENTS
Studies at the School of Pharmacy were supported by Cancer Research UK (Programme Grant No. C129/A4489). Y.G. acknowledges an ORSAS award and Overseas Postgraduate Studentship from the University of Reading. The Gemini diffractometer was funded by EPSRC Grant No. EP/C533526/1 to CJC. Funding to pay the Open Access Publication charge was provided by Queen's University Belfast.
Conflict of interest statement. None declared.
REFERENCES
- 1.Crespo-Hernández CE, Cohen B, Hare PM, Kohler B. Ultrafast excited-state dynamics in nucleic acids. Chem. Rev. 2004;104:1977–2019. doi: 10.1021/cr0206770. [DOI] [PubMed] [Google Scholar]
- 2.Cadet J, Vigny P. The photochemistry of nucleic acids. In: Morrison H, editor. Bioorganic Photochemistry. Vol. 1. New York: John Wiley and Sons; 1990. pp. 1–272. [Google Scholar]
- 3.Davies RJH. Ultraviolet radiation damage in DNA. Biochem. Soc. Trans. 1995;23:407–418. doi: 10.1042/bst0230407. [DOI] [PubMed] [Google Scholar]
- 4.Ravanat J-L, Douki T, Cadet J. Direct and indirect effects of UV radiation on DNA and its components. J. Photochem. Photobiol. B: Biol. 2001;63:88–102. doi: 10.1016/s1011-1344(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 5.Sage E, Drouin R, Rouabhia R, editors. Comprehensive Series in Photochemical and Photobiological Sciences. Vol. 5. UK: RSC Publishing, Cambridge; 2005. From DNA photolesions to mutations, skin cancer and cell death. [Google Scholar]
- 6.Douki T, Cadet J. Individual determination of the yield of the main UV-induced dimeric pyrimidine photoproducts in DNA suggests a high mutagenicity of CC photolesions. Biochemistry. 2001;40:2495–2501. doi: 10.1021/bi0022543. [DOI] [PubMed] [Google Scholar]
- 7.Huggins W, Ghosh SK, Nanda K, Wollenzien P. Internucleotide movements during formation of 16 S rRNA-rRNA photocrosslinks and their connection to the 30 S subunit conformational dynamics. J. Mol. Biol. 2005;354:358–374. doi: 10.1016/j.jmb.2005.09.060. [DOI] [PubMed] [Google Scholar]
- 8.Pörschke D. Analysis of a specific photoreaction in oligo- and polydeoxyadenylic acids. J. Am. Chem. Soc. 1973;95:8440–8446. doi: 10.1021/ja00806a040. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S, Sharma ND, Davies RJH, Phillipson DW, McCloskey JA. The isolation and characterisation of a new type of dimeric adenine photoproduct in UV-irradiated deoxyadenylates. Nucleic Acids Res. 1987;15:1199–1216. doi: 10.1093/nar/15.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S, Joshi PC, Sharma ND, Bose SN, Davies RJH, Takeda N, McCloskey JA. Adenine photodimerization in deoxyadenylate sequences: elucidation of the mechanism through structural studies of a major d(ApA) photoproduct. Nucleic Acids Res. 1991;19:2841–2847. doi: 10.1093/nar/19.11.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bose SN, Davies RJH, Sethi SK, McCloskey JA. Formation of an adenine-thymine photoadduct in the deoxydinucleoside monophosphate d(TpA) and in DNA. Science. 1983;220:723–725. doi: 10.1126/science.6836308. [DOI] [PubMed] [Google Scholar]
- 12.Bose SN, Davies RJH. The photoreactivity of T-A sequences in oligodeoxyribonucleotides and DNA. Nucleic Acids Res. 1984;12:7903–7914. doi: 10.1093/nar/12.20.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clingen PH, Davies RJH. Quantum yields of adenine photodimerization in poly(deoxyadenylic acid) and DNA. J. Photochem. Photobiol. B: Biol. 1997;38:81–87. [Google Scholar]
- 14.Koning TMG, Davies RJH, Kaptein R. The solution structure of the intramolecular photoproduct of d(TpA) derived with the use of NMR and a combination of distance geometry and molecular dynamics. Nucleic Acids Res. 1990;18:277–284. doi: 10.1093/nar/18.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bose SN, Kumar S, Davies RJH, Sethi SK, McCloskey JA. The photochemistry of d(T-A) in aqueous solution and in ice. Nucleic Acids Res. 1984;12:7929–7947. doi: 10.1093/nar/12.20.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao X, Nadji S, Kao JL-F, Taylor J-S. The structure of d(TpA)*, the major photoproduct of thymidylyl-(3′-5′)-deoxyadenosine. Nucleic Acids Res. 1996;24:1554–1560. doi: 10.1093/nar/24.8.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheldrick GM. SHELX 97-2. Germany: University of Göttingen; 1998. [Google Scholar]
- 18.Macrae CF, Edgington PR, McCabe P, Pidcock E, Shields GP, Taylor R, Towler M, van de Streek J. Mercury: visualization and analysis of crystal structures. J. Appl. Cryst. 2006;39:453–457. [Google Scholar]
- 19.Neidle S. Nucleic Acid Structure and Recognition. Oxford: Oxford University Press; 2002. [Google Scholar]
- 20.Park H, Zhang K, Ren Y, Nadji S, Sinha N, Taylor J-S, Kang C. Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15965–15970. doi: 10.1073/pnas.242422699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vassylyev DG, Kashiwagi T, Mikami Y, Ariyoshi M, Iwai S, Ohtsuka E, Morikawa K. Atomic model of a pyrimidine dimer excision-repair enzyme complexed with a DNA substrate – structural basis for damaged DNA recognition. Cell. 1995;83:773–782. doi: 10.1016/0092-8674(95)90190-6. [DOI] [PubMed] [Google Scholar]
- 22.Mees A, Klar T, Gnau P, Hennecke U, Eker APM, Carell T, Essen L-O. Crystal structure of a photolyase bound to a CPD-like lesion after in situ repair. Science. 2004;306:1789–1793. doi: 10.1126/science.1101598. [DOI] [PubMed] [Google Scholar]
- 23.Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W. Replication of a cis-syn thymine dimer at atomic resolution. Nature. 2003;424:1083–1087. doi: 10.1038/nature01919. [DOI] [PubMed] [Google Scholar]
- 24.Yokoyama H, Mizutani R, Satow Y, Komatsu Y, Ohtsuka E, Nikaido O. Crystal structure of the 64M-2 antibody Fab fragment in complex with a DNA dT(6-4)T photoproduct formed by ultraviolet radiation. J. Mol. Biol. 2000;299:711–723. doi: 10.1006/jmbi.2000.3772. [DOI] [PubMed] [Google Scholar]
- 25.Saintomé C, Clivio P, Favre A, Fourrey J-L, Riche C. RNA photolabeling mechanistic studies: X-ray crystal structure of the photoproduct formed between 4-thiothymidine and adenosine upon near UV irradiation. J. Am. Chem. Soc. 1996;118:8142–8143. [Google Scholar]
- 26.Crespo-Hernández CE, Cohen B, Kohler B. Base stacking controls excited-state dynamics in A.T DNA. Nature. 2005;436:1141–1144. doi: 10.1038/nature03933. [DOI] [PubMed] [Google Scholar]
- 27.Markovitsi D, Onidas D, Gustavsson T, Talbot F, Lazzarotto E. Collective behavior of Franck-Condon excited states and energy transfer in DNA double helices. J. Am. Chem. Soc. 2005;127:17130–17131. doi: 10.1021/ja054955z. [DOI] [PubMed] [Google Scholar]
- 28.Zhao X, Kao JL-F, Taylor J-S. Preparation and characterization of a deoxyoligonucleotide 49-mer containing a site-specific thymidylyl-(3′-5′)-deoxyadenosine photoproduct. Biochemistry. 1995;34:1386–1392. doi: 10.1021/bi00004a033. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Taylor J-S, Gross ML. Isolation and mass spectrometric characterization of dimeric adenine photoproducts in oligodeoxynucleotides. Chem. Res. Toxicol. 2001;14:738–745. doi: 10.1021/tx010009u. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Taylor J-S, Gross ML. Fragmentation of photomodified oligodeoxynucleotides adducted with metal ions in an electrospray-ionization ion-trap mass spectrometer. J. Am. Mass Spectrom. 2001;12:1174–1179. doi: 10.1016/S1044-0305(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 31.Kundu LM, Linne U, Marahiel M, Carell T. RNA is more UV resistant than DNA: The formation of UV-induced DNA lesions is strongly sequence and conformation dependent. Chem. Eur. J. 2004;10:5697–5705. doi: 10.1002/chem.200305731. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Gross ML, Taylor J-S. Use of a combined enzymatic digestion/ESI mass spectrometry assay to study the effect of TATA-binding protein on photoproduct formation in a TATA box. Biochemistry. 2001;40:11785–11793. doi: 10.1021/bi0111552. [DOI] [PubMed] [Google Scholar]
- 33.Zhao X, Taylor J-S. Mutation spectra of TA*, the major photoproduct of thymidylyl-(3′-5′)-deoxyadenosine, in Escherichia coli under SOS conditions. Nucleic Acids Res. 1996;24:1561–1566. doi: 10.1093/nar/24.8.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Youderian P, Bouvier S, Susskind MM. Sequence determinants of promoter activity. Cell. 1982;30:843–853. doi: 10.1016/0092-8674(82)90289-6. [DOI] [PubMed] [Google Scholar]
- 35.LeClerc J, Istock NL, Saran BR, Allen R., Jr Sequence analysis of ultraviolet-induced mutations in M13lacZ hybrid phage DNA. J. Mol. Biol. 1984;180:217–237. doi: 10.1016/s0022-2836(84)80001-7. [DOI] [PubMed] [Google Scholar]
- 36.Basic-Zaninovic T, Meschini R, Calcagnile AS, Palombo F, D’Errico M, Proietti-De Sanctis L, Dogliotti E. Strand bias of ultraviolet light-induced mutations in a transcriptionally active gene in human cells. Mol. Carcinogenesis. 1995;14:214–225. doi: 10.1002/mc.2940140311. [DOI] [PubMed] [Google Scholar]
- 37.Otoshi E, Yagi T, Mori T, Matsunaga T, Nikaido O, Kim S-T, Hitomi K, Ikenaga M, Todo T. Respective roles of cyclobutane pyrimidine dimers, (6–4) photoproducts, and minor photoproducts in ultraviolet mutagenesis of repair-deficient xeroderma pigmentosum A cells. Cancer Res. 2000;60:1729–1735. [PubMed] [Google Scholar]
- 38.You Y-H, Lee D-H, Yoon J-H, Nakajima S, Yasui A, Pfeifer GP. Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J. Biol. Chem. 2001;276:44688–44694. doi: 10.1074/jbc.M107696200. [DOI] [PubMed] [Google Scholar]