Abstract
Base excision repair (BER) is the major pathway for the repair of simple, non-bulky lesions in DNA that is initiated by a damage-specific DNA glycosylase. Several human DNA glycosylases exist that efficiently excise numerous types of lesions, although the close proximity of a single strand break (SSB) to a DNA adduct can have a profound effect on both BER and SSB repair. We recently reported that DNA lesions located as a second nucleotide 5′-upstream to a DNA SSB are resistant to DNA glycosylase activity and this study further examines the processing of these ‘complex’ lesions. We first demonstrated that the damaged base should be excised before SSB repair can occur, since it impaired processing of the SSB by the BER enzymes, DNA ligase IIIα and DNA polymerase β. Using human whole cell extracts, we next isolated the major activity against DNA lesions located as a second nucleotide 5′-upstream to a DNA SSB and identified it as DNA polymerase δ (Pol δ). Using recombinant protein we confirmed that the 3′-5′-exonuclease activity of Pol δ can efficiently remove these DNA lesions. Furthermore, we demonstrated that mouse embryonic fibroblasts, deficient in the exonuclease activity of Pol δ are partially deficient in the repair of these ‘complex’ lesions, demonstrating the importance of Pol δ during the repair of DNA lesions in close proximity to a DNA SSB, typical of those induced by ionizing radiation.
INTRODUCTION
Endogenous oxidative metabolism and exogenous agents, such as ionizing radiation, can generate reactive oxygen species (ROS) that attack DNA to produce a plethora of lesions including oxidized bases, strand breaks and deamination of DNA bases, such as the formation of uracil and hypoxanthine (1,2). Since ROS can be produced at high local concentrations through the deposition of ionizing radiation energy in small volumes of nanometer dimensions, base lesions and DNA single strand breaks (SSBs) may arise in close proximity to each other and may affect each other during repair (3). Such ‘complex’ lesions may delay repair and generate DNA double strand breaks and mutations (4–6).
The majority of these ‘complex’ lesions are repaired by proteins of the base excision repair (BER) pathway (7–10). The repair process involves removal of the damaged base, excision of the abasic sugar, filling the created gap with an undamagerd nucleotide and sealing the DNA strands. Processing of different alkylated/methylated bases is initiated by N-methylpurine DNA glycosylase (MPG), that also excises hypoxanthine lesions derived from the oxidative or radiation-induced deamination of adenine (11). The effect of a nearby SSB on MPG activity has not been investigated previously. Two major DNA glycosylases are involved in recognition and excision of oxidative base lesions in human cells, namely 8-oxoguanine-DNA glycosylase (OGG1) and the endonuclease III homologue (NTH1) (12–15). However, the endonuclease VIII (Nei)-like proteins (NEIL) that possess a broad substrate specificity have recently been identified and were thought to act as a backup repair system to OGG1 and NTH1 (16,17). Although these DNA glycosylases cover removal of the majority of oxidative DNA lesions, recent reports have demonstrated that NTH1 and OGG1 have limited activity on oxidative base lesions located at the 3′-termini of SSBs and repair of such lesions is carried out by other BER enzymes. For example, the major activity against 3′-terminal 8-oxoguanine in human cell extracts was shown to be AP endonuclease-1 (APE1) (18). We have also recently shown that NEIL1 is involved in the repair of 5-hydroxyuracil and 8-oxoguanine located in close proximity to the 3′-end of a DNA SSB that are not excised by OGG1 and NTH1 (19).
However, some base lesions located in close proximity to SSBs are resistant to removal by BER enzymes. In our previous study, we demonstrated that DNA base lesions located as a second nucleotide 5′- to the SSB end are resistant to cleavage by human DNA glycosylases and to APE1 (19). In the present work, we have extended these findings and isolated the major activity against 5-hydroxyuracil (5-OHU) and hypoxanthine (Hx) located as a second nucleotide 5′- to a SSB in human cells using fractionated human cell extracts and oligonucleotide duplexes. Using this approach, we discovered that the 3′-5′-exonuclease activity of DNA polymerase δ (Pol δ) is the major activity involved in repair of these lesions.
MATERIALS AND METHODS
Materials
Synthetic oligodeoxyribonucleotides were purchased from Eurogentec and gel purified on a 20% polyacrylamide gel. [γ-32P]ATP (3000 Ci/mmol) was purchased from PerkinElmer Life Sciences. Histidine-tagged human DNA polymerase β, PCNA, FEN-1 and DNA ligase IIIα were purified on a Ni2+-charged His-Bind resin (Novagen) as described by the manufacturer. DNA ligase I was a gift from A. Tomkinson and Pol δ purified as described (20) was kindly provided by V. Podust. MPG was purified as described (21). Antibodies against human Pol δ were raised in rabbit and affinity purified as described (22) and antibodies against XPF were purchased from Abcam. Wild-type and DNA polymerase δ exonuclease deficient (D400A) mouse embryonic fibroblasts were grown as described previously (23).
Substrate labelling
Oligonucleotides were 5′-end labelled with [γ-32P]ATP using T4 polynucleotide kinase and unincorporated label was removed on a BioSpin P-6 spin column (BioRad). To prepare the substrates used in the repair assays, the labelled oligonucleotides were annealed to the relevant oligonucleotides shown in Figure 1 at 90°C for 3–5 min followed by slow cooling to room temperature.
Figure 1.
Structures of oligonucleotides used. Oligonucleotides (20-mer) containing 5-OHU or Hx (designated X) were 5′-end labelled and a 24-mer adjacent oligonucleotide added and annealed to the corresponding complementary strand (with base Y corresponding to guanine and thymine opposite to 5-OHU and Hx, respectively) to generate substrates containing DNA lesions located 1–4 nucleotides apart from a DNA single strand break. Substrates containing 5′-end labelled isolated DNA lesions were also used as a control. N refers to a normal base.
Fractionation of cell extracts
HeLa cell pellets were purchased from Paragon, USA. WCE were prepared by the method of Manley et al. (24) dialysed overnight against buffer containing 25 mM HEPES–KOH, pH 7.9, 100 mM KCl, 12 mM MgCl2, 0.1 mM EDTA, 17% glycerol and 2 mM DTT and aliquots frozen at −80°C. An aliquot of this extract (250 mg protein) was then fractionated by phosphocellulose chromatography using a step elution of 150 mM KCl (PC-FI) and 1 M KCl (PC-FII) as previously described (25,26). Proteins (50 mg) from the PC-FII fraction were further separated by gel filtration on a Superose-12 column (Amersham) in a buffer containing 50 mM HEPES (pH 7.9), 150 mM KCl, 1 mM EDTA, 1 mM DTT and 1 mM PMSF and 0.5 ml fractions collected. Active fractions were pooled, dialysed against buffer containing 50 mM HEPES (pH 7.9), 50 mM KCl, 1 mM EDTA, 1 mM DTT and further separated on a 1 ml Mono-Q column (Amersham) using a 20 ml gradient elution of 50–1000 mM KCl, collecting 0.5 ml fractions. Activity assays were performed as described below.
Repair assays
Assays using purified proteins contained 300 fmol oligonucleotide per reaction in 10 μl reaction buffer containing 50 mM HEPES-KOH, pH 7.8, 50 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, 1.5 mM DTT, 8.5% glycerol and 100 μg/ml BSA. Primer extension reactions also included 3.5 pmol PCNA and 20 μM of each dCTP, dATP, dGTP and TTP. Ligation reactions were supplemented with 2 mM ATP and full reconstitution reactions contained all the above. Repair assays using fractionated cell extract (2 μl) included 0.25 mM NAD+ and 1 μg of carrier DNA (single stranded 30-mer oligonucleotide). All reactions were incubated for 20 min at 37°C and 10 μl formamide loading dye (95% formamide, 0.02% xylene cyanole, 0.02% bromophenol blue) added and the samples heated to 95°C for 5 min. Products were subsequently analysed by 20% denaturing polyacrylamide gel electrophoresis and gels exposed to storage phosphor screens at 4°C prior to analysis by phosphorimaging.
Immunodepletion of Pol δ
Protein A Sepharose CL-4B was allowed to swell for 1 h in PBS and following washes with PBS a 50% suspension was prepared. To 200 μl suspension was added 5 μl Pol δ antibodies or 5 μl pre-immune serum and incubation was carried out for 2 h at 4°C with gentle shaking. The anti Pol δ -Sepharose was washed five times with 0.5 ml PBS and resuspended in 100 µl PBS. Fractions purified from Mono-Q chromatography (50 μl) were mixed with 50 μl anti Pol δ -Sepharose and incubated for 2 h at 4°C with gentle shaking. The mixture was filtered through Spin-X columns at 4°C and aliquots taken and SDS-PAGE loading dye (25 mM Tris-HCl, pH 6.8, 2.5% mercaptoethanol, 1% SDS, 5% glycerol, 1 mM EDTA, 0.15 mg/ml bromophenol blue) added. The samples were heated to 90°C for 3 min prior to loading on a 10% SDS-polyacrylamide gel followed by transfer to a PVDF membrane and immunoblot analysis with the indicated antibodies. Aliquots were also taken for repair assays as described above.
Host cell reactivation assay
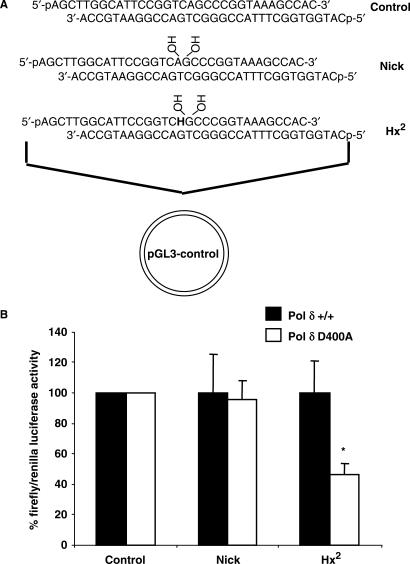
The pGL3-control luciferase reporter vector (Promega) was digested with Hind III and Nco I restriction enzymes to remove a sequence of 33 bp between the promoter and luciferase gene. The plasmid was electrophoresed on a 0.7% agarose gel and the linear plasmid DNA isolated from the gel using the QIAquick gel extraction kit (Qiagen). Oligonucleotides were 5′-phosphorylated using T4 polynucleotide kinase and duplexes were constructed (Figure 8A) by heating to 90°C for 3–5 min followed by slow cooling to room temperature. A 200 μl ligation reaction containing 1 μg (300 fmol) linear plasmid DNA, 1.5 pmol oligonucleotide duplex and 40 U T4 DNA ligase in buffer containing 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM ATP, 10 mM DTT, 25 μg/ml BSA was incubated overnight at 4°C. Analysis by agarose gel electrophoresis demonstrated that approximately 10% of the plasmid DNA was circularized with oligonucleotide duplex during the ligation reaction. The DNA was concentrated using Amicon-30 columns (Millipore) and buffer exchanged by washing twice with 1 mM Tris-HCl (pH 8.0), prior to concentration to 10 μl. Pol δ D400A exonuclease deficient and the corresponding wild-type cell lines were seeded at 1 × 105 cells/well in a volume of 500 μl in a 24-well plate and cultured overnight at 37°C in 5% CO2. The crude DNA ligation reactions, in combination with 0.2 μg pRL-TK renilla luciferase reporter vector, used as an internal control, were transfected into the cells using Lipofectamine 2000 reagent (Invitrogen) as recommended by the manufacturer using a 1:1 ratio of plasmid (μg) to reagent (μl) in a volume of 100 μl. The cells were incubated further for 8 h at 37°C in 5% CO2 and cell extracts prepared by washing the cells in PBS prior to the addition of 100 μl passive lysis buffer (Promega) and incubation at room temperature for 15 min with agitation. The extracts were collected and frozen at −70°C until required. For measurement of firefly and renilla luciferase expression, the dual-luciferase reporter assay system (Promega) and a luminoskan ascent luminometer (Thermo) were used as described by the manufacturers. The results were expressed as a ratio of firefly to renilla luciferase expression and an average ratio was obtained in each experiment from duplicate transfections. These values were then expressed as a percentage against the average ratio for the undamaged sequence and then normalized to that obtained with the wild-type cell line that was set to 100%. At least five transfection experiments were performed for each substrate prepared.
Figure 8.
Repair ability of mouse embryonic fibroblasts deficient in Pol δ exonuclease against Hx located as a second nucleotide 5′- to a DNA single strand break. Plasmid constructs used in the experiments were prepared by ligation of the shown oligonucleotide duplexes into a luciferase reporter vector (A). Plasmids were transfected into either wild-type (Pol δ +/+) or mutant (Pol δ D400A) cells and after 8 h cell extracts were prepared and luciferase activity was measured. The average and standard error are shown graphically (B). Asterix (*) represents a statistical difference P < 0.02 compared to the wild-type cell line using t test.
RESULTS
Excision of DNA lesions located in close proximity to the 3′-end of a DNA single strand break
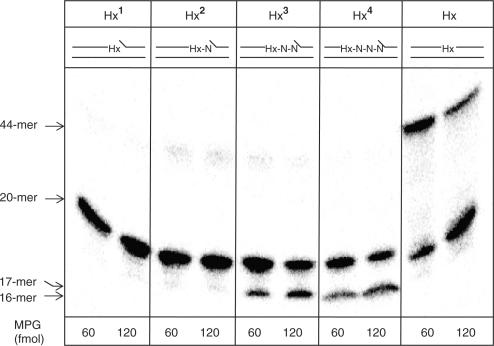
Recently, we have shown that when an oxidative base lesion is situated as a second nucleotide 5′-upstream to a SSB, then it is resistant to excision by the major DNA glycosylases, including NTH1, OGG1 and NEIL1 (19). To understand whether this observation can be extended to other types of base lesions, we have examined the excision of Hx by MPG when placed in close proximity to the 3′-end of a DNA SSB. Excision of the modified base was monitored by incubation of the 5′-end labelled oligonucleotide substrate (Figure 1) with purified MPG and subsequent excision of the AP site with piperidine. Similar to our observation of the inability of NTH1 and OGG1 to remove 3′-proximal oxidative lesions (18,19), we observed that Hx located immediately on the 3′-terminus (Hx1) or as a second nucleotide 5′-upstream to the SSB (Hx2) is not excised by MPG (Figure 2). However, we found that human AP endonuclease (APE1), that we recently identified as the major 3′-phosphoglycolate and 3′-8-oxoguanine activity in human cell extracts (18,26) is also able to excise 3′-terminal Hx (data not shown). In contrast, we found that APE1, NEIL1 or NEIL2 can not process Hx (or 5-OHU) located as a second nucleotide 5′-upstream to the SSB (data not shown). Furthermore, 5-OHU in this position is also not excised by UNG2 and SMUG1 (data not shown) that have previously been demonstrated to contain 5-OHU activity (27,28). We thus conclude that DNA base lesions of a broad chemical nature located as a second nucleotide 5′-upstream to the strand break are resistant to repair by DNA glycosylases and other enzymes should be involved in their processing.
Figure 2.
Excision of Hx in close proximity to the 3′-end of a DNA single strand break by MPG. Oligonucleotide substrates (0.3 pmol), shown at the top of each panel, were incubated with MPG (60 or 120 fmol) for 20 min at 37°C prior to treatment with 5% piperidine for 15 min at 95°C. The samples were subsequently dried, resuspended in formamide loading dye and an aliquot was analysed by 20% denaturing polyacrylamide gel electrophoresis and phosphorimaging.
DNA single strand breaks containing 3′-end proximal lesions are poorly processed by DNA ligase and DNA polymerase
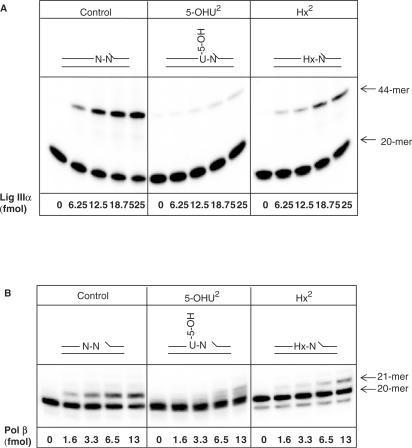
To exclude the possibility that DNA lesions located as a second nucleotide 5′-upstream to the SSB are sealed by a DNA ligase and then subsequently repaired by a DNA glycosylase, we investigated the effect of Hx and 5-OHU on DNA ligase IIIα, the major ligase employed during BER. We found a significant reduction in the amount of substrate, containing either 5-OHU or Hx in close proximity to the SSB, which was ligated by DNA ligase IIIα in comparison to the control duplex oligonucleotide (Figure 3A). The most dramatic reduction was observed in the case of 5-OHU2 that caused an approximate 10-fold inhibition in ligation, with Hx2 causing an approximate 3-fold inhibition.
Figure 3.
DNA lesions located as a second nucleotide 5′- to a SSB inhibit DNA ligase IIIα and Pol β activities. Oligonucleotide substrates (0.3 pmol), shown at the top of the panel, were incubated with increasing concentrations of DNA ligase IIIα (A) or Pol β (B) for 20 min at 37°C prior to the addition of formamide loading dye. An aliquot was analysed by 20% denaturing polyacrylamide gel electrophoresis and phosphorimaging.
Since after incorporation of even one nucleotide to the 3′-end of the strand break, the base lesion may become removable by a DNA glycosylase, we also examined the effect of neighbouring lesions on DNA polymerase β (Pol β). Similar to DNA ligase IIIα, we observed a defect in one nucleotide addition by Pol β, with both 5-OHU2 and Hx2 substrates exhibiting an approximate 4-fold decrease (Figure 3B). We thus conclude that Pol β and DNA ligase IIIα-XRCC1 heterodimer, that are the major DNA polymerase and DNA ligase activities involved in BER and SSB repair, operate with very low efficiency on such ‘complex’ SSBs. We hypothesized that the damaged base should be removed prior to SSB repair and that a specific activity involved in the excision of damaged bases located as a second nucleotide 5′-upstream to a SSB is apparent in human cells.
Isolation of enzyme activity against 5-OHU in close proximity to a single strand break
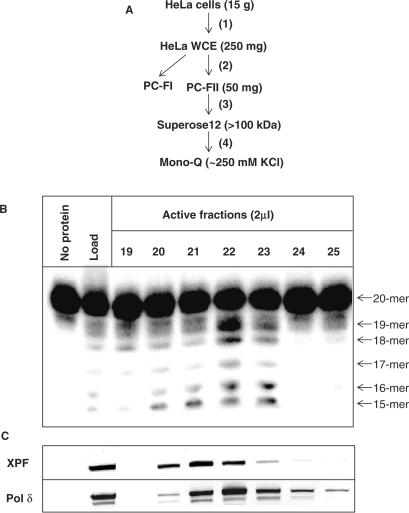
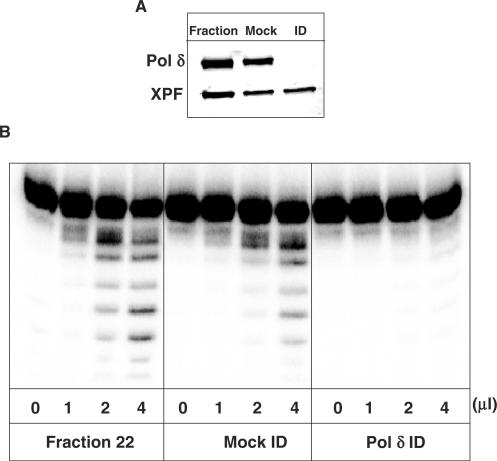
To isolate the major excision activity against DNA lesions located as a second nucleotide 5′-upstream to a SSB, a protein purification scheme was designed using the 5-OHU2 substrate to monitor excision activity (Figure 4A). Briefly, HeLa WCE was generated by the method of Manley et al. (24) and proteins subsequently separated by phosphocellulose chromatography using a step elution of buffers containing 0.15 M (PC-FI) and 1 M KCl (PC-FII). An excision activity against 5-OHU2 was detected in PC-FII that was subsequently fractionated by gel filtration chromatography on a Superose 12 column. The major peak of activity corresponded to a protein of molecular weight greater than 100 kDa using protein standards (data not shown). The peak activity fractions were pooled, dialysed and separated on a Mono-Q column using an elution gradient of 50–1000 mM KCl. Fractions containing excision activity against the 5-OHU2 substrate were subsequently detected, eluting at approximately 250 mM KCl (Figure 4B). What was apparent from the activity profile was that the enzyme may be an exonuclease due to the cleavage of at least five sequential nucleotides. The major known exonucleases in human cells include DNA polymerase δ (Pol δ), DNA polymerase ɛ, Werner (WRN) protein and MRE11 (29). However, recent evidence has suggested that xeroderma pigmentosum group F complementing protein (XPF) may be involved in the removal of 3′-blocked termini from DNA strand breaks (30). We therefore probed active fractions for XPF protein and found that it co-purified, although not completely aligned, with the activity against the 5-OHU2 substrate (Figure 4C). However, immunodepletion of active fractions using XPF antibodies or analysing the activity of purified ERCC1–XPF complex on the 5-OHU2 substrate, known to be active on a stem-loop containing substrate, did not ablate the activity (data not shown). As the 3′-5′ exonuclease activity of WRN protein has been shown to be inhibited by the presence of certain oxidative modifications (31) we eliminated this protein as the major activity against this substrate. We switched our attention to Pol δ and using western blotting and antibodies specific to this protein, similar to XPF protein, we observed Pol δ in active fractions (Figure 4C). Immunodepletion of Pol δ from the peak active fraction and subsequent analysis by western blotting revealed that the levels of Pol δ are only slightly reduced by mock immunodepletion compared to the original fraction, while the Pol δ specific antibodies completely removed the Pol δ protein from the fraction (Figure 5A). However, we also show that XPF remains in the mock- and Pol δ− immunodepleted fraction indicating the antibody specificity (Figure 5A). We further demonstrated that the mock-immunodepleted fraction has a slightly reduced excision activity against 5-OHU2 (Figure 5B), in accordance with the slightly reduced levels of Pol δ compared to the original fraction caused by the mock-immunodepletion protocol. Furthermore, immunodepletion using Pol δ specific antibodies ablates the activity observed in the purified Mono-Q fraction indicating that Pol δ is the major activity in human cell extracts eliminating 5-OHU located as a second nucleotide 5′-upstream to a SSB (Figure 5B).
Figure 4.
Purification of the major activity against 5-OHU located as a second nucleotide 5′- to a DNA single strand break in human cells. A protein purification scheme was designed using HeLa cells (A) and involved generating whole cell extracts (step 1), separation of proteins by phosphocellulose chromatography using a step elution of 0.15 M KCl (PC-FI) and 1 M KCl (PC-FII; step 2), separation of proteins in PC-FII by Superose 12 gel-filtration chromatography (step 3), followed by separation of proteins (>100 kDa) using a Mono-Q column and a gradient elution of 0.05–1 M KCl (step 4). During each stage, the activity against a 5′-labelled 5-OHU lesion located as a second nucleotide 5′- to a DNA single strand break (5-OHU2) was measured and the corresponding active fractions pooled, dialysed if necessary and used in the following step. Activity against 5-OHU2 from fractions eluted from the final Mono-Q stage are shown (B) and these were further analysed by western blotting using XPF and Pol δ specific antibodies.
Figure 5.
Immunodepletion of Pol δ from purified human whole cell extract fractions containing activity directed against 5-OHU2. Active fraction (fraction 22) from Mono-Q chromatography containing 5-OHU2 activity was mock-immunodepleted and immunodepleted using Pol δ specific antibodies and samples analysed by SDS-polyacrylamide gel electrophoresis and western blotting using antibodies against XPF and Pol δ. (A) The original fraction, mock-immunodepleted and Pol δ immunodepleted fraction was tested for 5-OHU2 activity using 300 fmol of duplex oligonucleotide (B) Samples were incubated for 20 min at 37°C prior to the addition of formamide loading dye and analysis by 20% denaturing polyacrylamide gel electrophoresis and phosphorimaging.
Characterization of recombinant Pol δ activity against DNA lesions in close proximity to a SSB
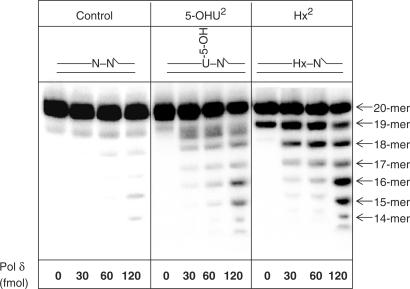
As we demonstrated that the major excision activity in human cell extracts against 5-OHU2 is Pol δ, we analysed the activity of recombinant Pol δ against 5-OHU and Hx located as a second nucleotide 5′-upstream to a SSB. We confirmed that purified recombinant Pol δ, albeit with different efficiencies, is able to excise 5-OHU and Hx in close proximity to a SSB using its associated 3′-5′-exonuclease activity in comparison to the control oligonucleotide substrate, at the enzyme concentrations tested (Figure 6). The exonuclease activity of Pol δ was markedly greater on the Hx-containing substrate in comparison to the 5-OHU-containing substrate, although very little exonuclease products were observed with the control substrate indicating substrate specificity. Although PCNA notably stimulated the DNA polymerase activity of Pol δ, it only moderately (1.2 fold) stimulated the 3′-5′-exonuclease activity (data not shown). These results imply that under the conditions of the exonuclease assay Pol δ is able to excise DNA lesions in close proximity to a SSB.
Figure 6.
The 3′-5′-exonuclease activity of recombinant Pol δ against DNA lesions located as a second nucleotide 5′- to a DNA single strand break. Oligonucleotide substrates (0.3 pmol), shown at the top of each panel, were incubated with increasing concentrations of Pol δ (0–240 fmol) in reaction mixture without dNTPs for 20 min at 37°C prior to the addition of formamide loading dye. An aliquot was analysed by 20% denaturing polyacrylamide gel electrophoresis and phosphorimaging.
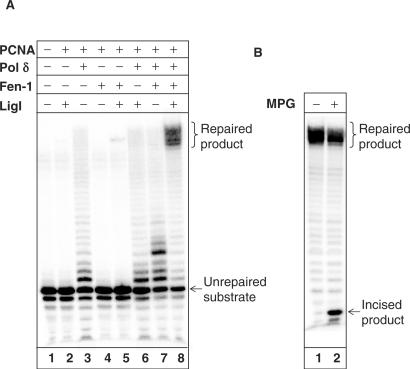
Subsequently, we reconstituted the repair of Hx2, an efficient substrate for the 3′-5′-exonuclease activity of Pol δ, using purified BER proteins (Figure 7A). DNA ligase I, in the presence of the processivity factor PCNA that was added to all enzyme reactions (lanes 2–8), was unable to ligate the substrate DNA directly to generate the fully repaired 44-mer product at the concentration tested (lane 2). Exonuclease and polymerase extension intermediates were observed on incubation with Pol δ only (lane 3). Interestingly, the addition of FEN-1 appeared to slightly stimulate the activities of both DNA ligase I (compare lanes 5 and 2), forming a minor amount of full length 44-mer product, and Pol δ (compare lanes 7 and 3), increasing the level of nucleotide incorporation, although FEN-1 alone had no effect on the Hx2 substrate (lane 4). Incubation with both Pol δ and DNA ligase I was unable to generate full length product (lane 6), however a repaired product was effectively observed on incubation of the full complementation reaction consisting of PCNA, Pol δ, DNA ligase I and FEN-1 (lane 8).
Figure 7.
Reconstitution of repair of Hx located as a second nucleotide 5′- to a DNA single strand break using purified proteins. Hx2 oligonucleotide substrate (0.3 pmol) was incubated with the indicated amount of either PCNA (3.5 pmol), Pol δ (240 fmol), FEN-1 (230 fmol) or DNA ligase I (10 fmol) in the presence of dNTPs for 20 min at 37°C prior to the addition of formamide loading dye. An aliquot was analysed by 20% denaturing polyacrylamide gel electrophoresis and phosphorimaging (A). The full complementation reaction (lane 8) was subsequently purified by phenol–chloroform extraction and Biospin P-30 gel filtration columns, incubated with MPG (600 fmol) and APE1 (600 fmol) for 20 min at 37°C prior to analysis by 20% denaturing polyacrylamide gel electrophoresis and phosphorimaging (B).
To examine whether Pol δ is either simply extending from the SSB or using its associated exonuclease activity prior to gap filling and repair, we subsequently purified the full length product obtained from the full complementation reaction (Figure 7A, lane 8) and further incubated with an excess of MPG and APE1 known to cleave 100% of full length product (see Figure 2). We found that on average 65% of the repaired product was resistant to excision by MPG (Figure 7B) demonstrating that this proportion of substrate is repaired using the 3′-5′-exonuclease activity of Pol δ.
Repair of Hx in close proximity to a SSB is impaired in cells deficient in Pol δ exonuclease activity
As we observed that Pol δ efficiently processes Hx lesions located as a second nucleotide 5′-upstream to a SSB in vitro, we examined specifically whether the exonuclease activity of Pol δ is required for the repair of this lesion in vivo using cultured cells derived from mice containing a mutant form of Pol δ (D400A) that is deficient in exonuclease activity (23), in combination with a host cell reactivation assay. This assay uses a firefly luciferase reporter vector that on transfection into cultured cells will cause expression of the luciferase gene. We replaced part of the original plasmid sequence with oligonucleotide duplexes containing either a control sequence (no damage), a SSB or a SSB with Hx located as a second nucleotide 5′-upstream to the SSB (Figure 8A). The oligonucleotide duplexes were introduced into the vector between the promoter and the luciferase gene, such that only repair of the DNA damage will cause luciferase expression. Following 8 h of transfection into either wild-type or Pol δ exonuclease-deficient cultured cell lines, we measured the ratio of firefly and renilla (internal transfection control) luciferase activities in both cell lines. Using the nick-containing plasmid, we observed a similar level of luciferase expression in both the wild-type and Pol δ exonuclease mutant cell lines, indicating that SSBs are efficiently repaired in both cell lines (Figure 8B). However, using a substrate containing Hx located as a second nucleotide 5′-upstream to the SSB (Hx2), we observed a significant decrease (∼50%) in the levels of expression in the mutant in comparison to the wild-type cell line indicating that this substrate partially requires the exonuclease activity of Pol δ for complete repair.
DISCUSSION
Endogenous oxidative metabolism or exogenous agents, such as ionizing radiation may generate DNA lesions, such as 5-OHU and Hx, in close proximity to a DNA SSB. Some of these lesions may impair repair by DNA polymerases and ligases and should be promptly removed, since delay in SSB repair results in genome instability and presents a threat to cell survival. The major human DNA glycosylases OGG1, NTH1 and NEIL1 excise 8-oxoG and oxidized pyrimidines from DNA in human cells, including some of the lesions located in close proximity to the SSB (6,12–17,19,32). However, we recently demonstrated that oxidative DNA lesions located as a second nucleotide 5′- to the SSB are not excised by these major human DNA glycosylases or by APE1 (19). In the present study, we extended these observations and have also shown that MPG is also unable to excise Hx in the same position. These data suggest that the position of the lesion, rather than their chemical nature, determines its repairability by DNA glycosylases. We also demonstrated that some of these ‘complex’ lesions efficiently block Pol β and DNA ligase IIIα thus preventing an alternative pathway for repair. We therefore pursued isolation of the major activity against DNA lesions positioned as a second nucleotide 5′- to the SSB using HeLa whole cell extracts and conventional protein purification methods.
Our results demonstrated that the enzyme involved was an exonuclease and not a specific activity directed at the DNA lesion. The major known 3′-5′-exonucleases in human cells include DNA polymerase δ (Pol δ), DNA polymerase ɛ, Werner (WRN) protein and MRE11 (29) indicating these as potential enzymes involved in the repair of DNA lesions located as a second nucleotide 5′- to the SSB. However, under our experimental conditions we did not observe any activity of ERCC1–XPF complex on these substrates, although it has been recently shown that this complex may be involved in the removal of 3′-blocked termini from DNA strand breaks (30). Furthermore, the 3′-5′ exonuclease activity of WRN protein has been shown to be inhibited by the presence of certain oxidative modifications (31). Exonuclease I, the major human 5′-3′-exonuclease that also possesses 3′-5′-exonuclease activity was found in the low-salt elution fraction (PC-FI) from phosphocellulose chromatography and did not co-purify with the major activity isolated (data not shown). We therefore focused on Pol δ and demonstrated that in fact the 3′-5′-exonuclease activity of Pol δ is the major activity that excises DNA lesions located as a second nucleotide 5′- to the SSB in human cell extracts. We were also able to demonstrate using an in vitro reconstituted system that efficient repair requires the presence of Pol δ, PCNA, DNA ligase I and FEN-1. However, the ability of Pol δ to process different ‘complex’ lesions varies. For example, Pol δ is very active on Hx-containing lesions, but much less active on 5-OHU-containing substrates and also very low activity was observed against 8-oxoG located as a second nucleotide 5′- to the SSB (data not shown). Therefore, one can expect that the role of Pol δ in cellular processing of these lesions may also vary and may be more important for some lesions than for others.
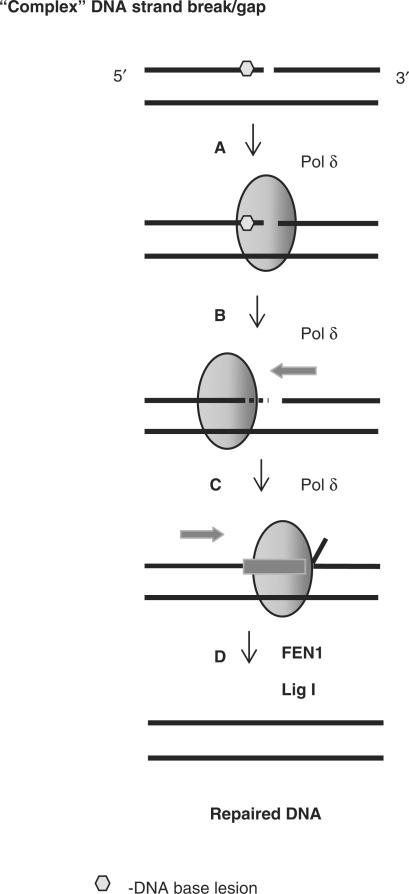
Pol δ is one of the major replicative polymerases in chromosomal DNA synthesis although it is also known to be involved in BER (33). The ‘long-patch’ BER pathway involves the incorporation of several nucleotides into the repair gap by Pol δ that cause strand displacement of the adjacent strand creating a flap structure that is excised by the flap endonuclease FEN1 (34,35). This reaction occurs in a PCNA-dependent manner (35) and the fidelity of nucleotide insertion during ‘long-patch’ BER and DNA replication is enhanced by the associated 3′-5′-exonuclease activity of the polymerases (36). However, ‘long-patch’ BER usually occurs as a result of the inability of Pol β to remove the 5′-deoxyribose phosphate moiety following apurinic/apyrimidinic site incision due to modification of the 5′-end by oxidation or reduction. Our study suggests a new important role of Pol δ in the repair of ‘complex’ SSBs. In this scenario the inability of both DNA ligase to seal the DNA strands and of Pol β to ‘move’ the SSB further away from the damaged base by DNA repair synthesis should initiate a switch of repair synthesis to Pol δ. However, damage removal by Pol δ exonuclease activity should precede DNA synthesis (Figure 9).
Figure 9.
Proposed mechanism of repair of DNA lesions located in close proximity to the 3′-end of a DNA single strand break by Pol δ. Complex DNA strand breaks containing 3′-proximal lesions that are resistant to the major BER enzymes are recognized by Pol δ (step A) that excises the lesion through its associated 3′-5′-exonuclease activity (step B). Pol δ is then able to insert the correct nucleotides into the gap causing strand displacement of the adjacent strand (step C). The subsequent 5′-flap generated is removed by FEN1 and DNA ligase I seals the nick (step D). PCNA can notably stimulate steps C and D.
The observation that Pol δ is able to excise modified DNA bases through its 3′-5′-exonuclease activity is a novel finding, as it is regarded as a proofreading activity to remove misincorporated bases, improving polymerase fidelity. Interestingly, mice deficient in the 3′-5′-exonuclease activity of Pol δ, but still retaining DNA polymerase activity, develop a high incidence of epithelial cancers, demonstrating the importance of this proofreading activity in genome stability (23). Using embryonic fibroblasts generated from these mutant mice, we demonstrated that a luciferase reporter vector containing Hx, located as a second nucleotide 5′-upstream to a SSB, are inefficiently repaired (50%) when transfected into the cells, demonstrating the requirement for Pol δ exonuclease activity during the repair of these ‘complex’ lesions. The remainder of these lesions are presumably repaired by either Pol β or DNA ligase IIIα as we only observed a 3- and 4-fold decrease, respectively, in the activities of both proteins in vitro. Also in vitro reconstitution experiments demonstrated that approximately 35% of Hx2 is repaired by Pol δ through simple extension of the SSB causing strand displacement that makes it amenable to MPG excision. Interestingly, a mammalian homologue of Escherchia. coli endonuclease V has recently been characterized that incises DNA at the second phosphodiester bond 3′ to Hx and uracil-containing nucleotides and thus would generate the Hx2 substrate used in our study that was predicted to be repaired by an exonuclease, that we now demonstrate is Pol δ (37,38).
In summary, our data suggest that Pol δ plays an important role in the repair of DNA damage in close proximity to a SSB that is not efficiently repaired by other proteins of the BER pathway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
ACKNOWLEDGEMENTS
We would like to acknowledge R.D. Wood for supplying us with ERCC1–XPF complex, A.E. Tomkinson for providing DNA ligase I, V. Podust for supplying Pol δ protein, B. Woodhouse for sequencing of the Pol δ gene and I. Dianova for assistance with the luciferase assay. Funding to pay the Open Access publication charge was provided by the Medical Research Council, Beckman Research Institute of the City of Hope and the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ponnamperuma C, Lemmon RM, Bennett EL, Calvin M. Deamination of adenine by ionizing radiation. Science. 1961;134:113. doi: 10.1126/science.134.3472.113. [DOI] [PubMed] [Google Scholar]
- 2.An Q, Robins P, Lindahl T, Barnes DE. C –> T mutagenesis and gamma-radiation sensitivity due to deficiency in the Smug1 and Ung DNA glycosylases. EMBO J. 2005;24:2205–2213. doi: 10.1038/sj.emboj.7600689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward JF. Radiation mutagenesis: the initial DNA lesions responsible. Radiat. Res. 1995;142:362–368. [PubMed] [Google Scholar]
- 4.Dianov GL, Timchenko TV, Sinitsina OI, Kuzminov AV, Medvedev OA, Salganik RI. Repair of uracil residues closely spaced on the opposite strands of plasmid DNA results in double-strand break and deletion formation. Mol. Gen. Genet. 1991;225:448–452. doi: 10.1007/BF00261686. [DOI] [PubMed] [Google Scholar]
- 5.Yang N, Chaudhry MA, Wallace SS. Base excision repair by hNTH1 and hOGG1: a two edged sword in the processing of DNA damage in gamma-irradiated human cells. DNA Repair (Amst) 2006;5:43–51. doi: 10.1016/j.dnarep.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Eot-Houllier G, Eon-Marchais S, Gasparutto D, Sage E. Processing of a complex multiply damaged DNA site by human cell extracts and purified repair proteins. Nucleic Acids Res. 2005;33:260–271. doi: 10.1093/nar/gki165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dianov GL, O'Neill P, Goodhead DT. Securing genome stability by orchestrating DNA repair: removal of radiation-induced clustered lesions in DNA. BioEssays. 2001;23:745–749. doi: 10.1002/bies.1104. [DOI] [PubMed] [Google Scholar]
- 8.Weinfeld M, Rasouli-Nia A, Chaudhry MA, Britten RA. Response of base excision repair enzymes to complex DNA lesions. Radiat. Res. 2001;156:584–589. doi: 10.1667/0033-7587(2001)156[0584:robere]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 9.Blaisdell JO, Harrison L, Wallace SS. Base excision repair processing of radiation-induced clustered DNA lesions. Radiat. Prot. Dosimetry. 2001;97:25–31. doi: 10.1093/oxfordjournals.rpd.a006634. [DOI] [PubMed] [Google Scholar]
- 10.Budworth H, Matthewman G, O'Neill P, Dianov GL. Repair of tandem base lesions in DNA by human cell extracts generates persisting single-strand breaks. J. Mol. Biol. 2005;351:1020–1029. doi: 10.1016/j.jmb.2005.06.069. [DOI] [PubMed] [Google Scholar]
- 11.Saparbaev M, Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc. Natl. Acad. Sci. U.S.A. 1994;91:5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeda S, Biswas T, Roy R, Izumi T, Boldogh I, Kurosky A, Sarker AH, Seki S, Mitra S. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III - Direct identification of Lys-212 as the active nucleophilic residue. J. Biol. Chem. 1998;273:21585–21593. doi: 10.1074/jbc.273.34.21585. [DOI] [PubMed] [Google Scholar]
- 13.Rosenquist TA, Zharkov DO, Grollman AP. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarker AH, Ikeda S, Nakano H, Terato H, Ide H, Imai K, Akiyama K, Tsutsui K, Bo Z, et al. Cloning and characterization of a mouse homologue (mNthl1) of Escherichia coli endonuclease III. J. Mol. Biol. 1998;282:761–774. doi: 10.1006/jmbi.1998.2042. [DOI] [PubMed] [Google Scholar]
- 16.Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, van der Horst GT, Yasui A. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J. Biol. Chem. 2002;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- 17.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parsons JL, Dianova II, Dianov GL. APE1-dependent repair of DNA single-strand breaks containing 3′-end 8-oxoguanine. Nucleic Acids Res. 2005;33:2204–2209. doi: 10.1093/nar/gki518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsons JL, Zharkov DO, Dianov GL. NEIL1 excises 3′ end proximal oxidative DNA lesions resistant to cleavage by NTH1 and OGG1. Nucleic Acids Res. 2005;33:4849–4856. doi: 10.1093/nar/gki816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Podust VN, Chang L-S, Ott R, Dianov GL, Fanning E. Reconstitution of human DNA polymerase δ using recombinant baculoviruses. J. Biol. Chem. 2002;277:3894–3901. doi: 10.1074/jbc.M109684200. [DOI] [PubMed] [Google Scholar]
- 21.O'Connor TR. Purification and characterization of human 3-methyladenine-DNA glycosylase. Nucleic Acids Res. 1993;21:5561–5569. doi: 10.1093/nar/21.24.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dianova II, Bohr VA, Dianov GL. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry. 2001;40:12639–12644. doi: 10.1021/bi011117i. [DOI] [PubMed] [Google Scholar]
- 23.Goldsby RE, Hays LE, Chen X, Olmsted EA, Slayton WB, Spangrude GJ, Preston BD. High incidence of epithelial cancers in mice deficient for DNA polymerase delta proofreading. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15560–15565. doi: 10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manley JL, Fire A, Samuels M, Sharp PA. In vitro transcription: whole-cell extract. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- 25.Dianov GL, Jensen BR, Kenny MK, Bohr VA. Replication protein A stimulates proliferating cell nuclear antigen- dependent repair of abasic sites in DNA by human cell extracts. Biochemistry. 1999;38:11021–11025. doi: 10.1021/bi9908890. [DOI] [PubMed] [Google Scholar]
- 26.Parsons JL, Dianova II, Dianov GL. APE1 is the major 3′-phosphoglycolate activity in human cell extracts. Nucleic Acids Res. 2004;32:3531–3536. doi: 10.1093/nar/gkh676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dizdaroglu M, Karakaya A, Jaruga P, Slupphaug G, Krokan HE. Novel activities of human uracil DNA N-glycosylase for cytosine-derived products of oxidative DNA damage. Nucleic Acids Res. 1996;24:418–422. doi: 10.1093/nar/24.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masaoka A, Matsubara M, Hasegawa R, Tanaka T, Kurisu S, Terato H, Ohyama Y, Karino N, Matsuda A, et al. Mammalian 5-formyluracil-DNA glycosylase. 2. Role of SMUG1 uracil-DNA glycosylase in repair of 5-formyluracil and other oxidized and deaminated base lesions. Biochemistry. 2003;42:5003–5012. doi: 10.1021/bi0273213. [DOI] [PubMed] [Google Scholar]
- 29.Shevelev IV, Hubscher U. The 3′ 5′ exonucleases. Nat. Rev. Mol. Cell Biol. 2002;3:364–376. doi: 10.1038/nrm804. [DOI] [PubMed] [Google Scholar]
- 30.Guzder SN, Torres-Ramos C, Johnson RE, Haracska L, Prakash L, Prakash S. Requirement of yeast Rad1-Rad10 nuclease for the removal of 3′-blocked termini from DNA strand breaks induced by reactive oxygen species. Genes Dev. 2004;18:2283–2291. doi: 10.1101/gad.1232804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machwe A, Ganunis R, Bohr VA, Orren DK. Selective blockage of the 3′–>5′ exonuclease activity of WRN protein by certain oxidative modifications and bulky lesions in DNA. Nucleic Acids Res. 2000;28:2762–2770. doi: 10.1093/nar/28.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.David-Cordonnier MH, Boiteux S, O'Neill P. Efficiency of excision of 8-oxo-guanine within DNA clustered damage by XRS5 nuclear extracts and purified human OGG1 protein. Biochemistry. 2001;40:11811–11818. doi: 10.1021/bi0112356. [DOI] [PubMed] [Google Scholar]
- 33.Stucki M, Pascucci B, Parlanti E, Fortini P, Wilson SH, Hubscher U, Dogliotti E. Mammalian base excision repair by DNA polymerases delta and epsilon. Oncogene. 1998;17:835–843. doi: 10.1038/sj.onc.1202001. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto Y, Kim K, Hurwitz J, Gary R, Levin DS, Tomkinson AE, Park MS. Reconstitution of proliferating cell nuclear antigen-dependent repair of apurinic/apyrimidinic sites with purified human proteins. J. Biol. Chem. 1999;274:33703–33708. doi: 10.1074/jbc.274.47.33703. [DOI] [PubMed] [Google Scholar]
- 35.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J. Biol. Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 36.Albertson TM, Preston BD. DNA replication fidelity: proofreading in trans. Curr. Biol. 2006;16:211. doi: 10.1016/j.cub.2006.02.031. [DOI] [PubMed] [Google Scholar]
- 37.Moe A, Ringvoll J, Nordstrand LM, Eide L, Bjoras M, Seeberg E, Rognes T, Klungland A. Incision at hypoxanthine residues in DNA by a mammalian homologue of the Escherichia coli antimutator enzyme endonuclease V. Nucleic Acids Res. 2003;31:3893–3900. doi: 10.1093/nar/gkg472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kow YW. Repair of deaminated bases in DNA. Free Radic. Biol. Med. 2002;33:886–893. doi: 10.1016/s0891-5849(02)00902-4. [DOI] [PubMed] [Google Scholar]