Abstract
Altritol-modified nucleic acids (ANAs) support RNA-like A-form structures when included in oligonucleotide duplexes. Thus altritol residues seem suitable as candidates for the chemical modification of siRNAs. Here we report that ANA-modified siRNAs targeting the MDR1 gene can exhibit improved efficacy as compared to unmodified controls. This was particularly true of ANA modifications at or near the 3′ end of the sense or antisense strands, while modification at the 5′ end of the antisense strand resulted in complete loss of activity. Multiple ANA modifications within the sense strand were also well tolerated. Duplexes with ANA modifications at appropriate positions in both strands were generally more effective than duplexes with one modified and one unmodified strand. Initial evidence suggests that the loss of activity associated with ANA modification of the 5′-antisense strand may be due to reduced phosphorylation at this site by cellular kinases. Treatment of drug resistant cells with MDR1-targeted siRNAs resulted in reduction of P-glycoprotein (Pgp) expression, parallel reduction in MDR1 message levels, increased accumulation of the Pgp substrate rhodamine 123, and reduced resistance to anti-tumor drugs. Interestingly, the duration of action of some of the ANA-modified siRNAs was substantially greater than that of unmodified controls. These observations suggest that altritol modifications may be helpful in developing siRNAs with enhanced pharmacological effectiveness.
INTRODUCTION
There has recently been substantial interest in potential therapeutic exploitation of gene regulation based on RNA interference (RNAi). The endogenous regulatory mechanisms involved in the physiological role of RNAi (1) can be activated via the delivery of short double-stranded RNAs (siRNA) to the cell interior (2). The mechanism of targeted mRNA degradation by siRNA is highly complex and not fully understood. It involves the formation of an RNA-induced silencing complex (RISC) that contains the argonaute 2 protein, while the dicer RNase and other proteins may also be important in loading of siRNA on to the RISC complex (3). In order to exploit RNAi therapeutically in mammals it will be important to develop siRNAs that are specific, potent, efficacious and persistent in their action. Enhancing the delivery of siRNA in the in vivo setting will also be a key problem. Many of these issues are being addressed through various chemical modification approaches.
A variety of strategies for siRNA modification have been pursued including alterations in the backbone chemistry, 2′-sugar modifications, nucleobase modifications and others, as recently reviewed (4,5). These efforts have, in some cases, resulted in significant improvements in the biological effectiveness of siRNA and have provided important insights into how chemical modification affects function. In terms of overall design, understanding of the biochemical mechanism of RNA interference has provided important guidelines; first, the siRNA must maintain an A-form (RNA-like) duplex, second, the 5′-position on the antisense strand must be able to be phosphorylated, third, to be effective siRNAs should have low thermodynamic stability in the 5′ antisense region (1,5,6). These general design principles can then be further evolved through the use of selected chemical modifications.
Sulfurization of the backbone to form phosphorothioate oligonucleotides (PS) has been a popular modification for both antisense and siRNA. The PS modification enhances nuclease stability but at the cost of reduced melting temperature and increased non-specific binding to proteins (7). In general, PS modifications of siRNA are well-tolerated although some reports indicate increased toxicity and reduced efficacy (8,9). Boranophosphates are another interesting backbone modification that have been reported to be compatible with high siRNA activity (10). Modifications of the 2′ sugar residue have also often been well tolerated. Compounds with 2′ fluoro modifications have increased thermodynamic stability and increased resistance to nucleases; several reports indicate that extensive modification of this type in either sense or antisense strand seems compatible with retention of siRNA activity (4). The 2′-O-Me modification is also consistent with retention of an A-form helix; however the impact of this substitution is somewhat controversial with some reports indicating substantial reduction in siRNA activity with extensive (but not limited) substitution, in a position specific manner (9,11). On the other hand, one report indicates that extensive 2′-O-Me modification can be made in the sense strand without any loss of activity (12). Bicyclic-locked nucleic acids (LNAs) that confer high binding affinity can be introduced selectively, particularly when the central region of the siRNA duplex is avoided (13). Relatively few nucleobase modifications have been studied thoroughly for effects on siRNA activity (4). An interesting recent example is ribo-fluorotoluyl nucleotides that display increases in nuclease stability along with maintained siRNA effects (14). Thus a variety of chemical modifications are consistent with maintenance of siRNA activity; however, there are few examples of substantial improvement in the efficacy of RNA interference through such modifications.
Most of the modifications described above were originally developed for use with antisense deoxy-oligonucleotides; it seems worthwhile then to also examine chemical modifications that are particularly suited to RNA-type oligonucleotides. One such modification is the use of the 6-carbon sugar, altritol, in place of the usual pentose ring. d-altritol oligonucleotides (ANA) have been synthesized as mimics of RNA with a conformationally rigid sugar moiety (15–17). The main difference with most rigid conformation RNA mimics is that a free hydroxyl group is present at the carbon atom adjacent to the base moiety. ANA is able to form very stable duplexes with RNA in a sequence-specific manner, due to its pre-organization in a A-type helix conformation (18). ANA is stable against enzymatic degradation in serum and has been shown to be an excellent template for non-enzymatic synthesis of complementary RNA's (19). Although the structural properties of ANA have been described, ANAs have not been evaluated in a biological system as RNA mimic until now.
MDR1, the gene chosen as the target for this study, codes for the P-glycoprotein, a transmembrane ATPase that serves as an efflux pump for a wide range of drug molecules. MDR1 is often over-expressed in tumor cells thus contributing to the multi-drug resistant phenotype commonly seen in cancer (20). The fact that MDR1 mRNA and P-glycoprotein are highly expressed in multi-drug resistant cells, while P-glycoprotein has a long turnover time, makes this target very refractory to inhibition with antisense or siRNA oligonucleotides (21). Thus we have evaluated ANA siRNAs in the context of a challenging test system.
MATERIAL AND METHODS
The synthesis of the ANA oligonucleotides has been described previously (16). The techniques used to evaluate the impact of siRNAs on MDR1 expression and function have been fully described in our previous publications (21–23); a brief recapitulation is given below.
Treatment of cells with siRNA oligonucleotides
Multi-drug-resistant NIH 3T3-MDR cells were cultured as described (21). Hybridization of the siRNA strands was done in Dharmacon universal buffer by heating the solutions to 90°C in a Perkin Elmer PCR machine, then gradually cooling to 30°C for 30 min. Complexation of siRNA with the cationic lipid lipofectamine 2000 (Invitrogen) was done according to the manufacturer's standard procedure. The oligonucleotides bound to lipofectamine 2000 were mixed in 10% FBS/DMEM-H and incubated with cells at 37°C for 4 h; media was then removed and replaced with 2% FBS/DMEM-H and the cells incubated for an additional 68–72 h. Most of the experiments were done with an MDR1 siRNA sequence termed ORF1 (antisense sequence 3′-dTdTCAUAACUGUCGAUAAGCUU-5′) while a limited number of experiments were done with a second sequence termed ORF2 that targets a distinct region on the MDR1 mRNA (antisense sequence 3′-dTdT CUUGAGAAUCGCAUACGUU-5′).
Cytotoxicity
NIH 3T3-MDR cells were treated with lipofectamine 2000 complexes of standard or ANA-modified siRNA oligos as described above. The cells were then replated onto new 12-well plates and incubated overnight followed by the addition of various concentrations of doxorubicin for 24 h. Evaluation of drug-induced cytotoxicity was performed as described by monitoring changes in cell number (22).
Immunostaining of P-glycoprotein
The level of cell surface expression of P-glycoprotein (Pgp) was quantitated by immunostaining using a flow cytometry assay. After treating NIH 3T3-MDR cells with standard or ANA-modified siRNA complexes as described above, the cells were washed with PBS, trypsinized, resuspended in complete medium and washed again with PBS after centrifugation. The levels of cell surface Pgp were quantitated using 17F9 R-Phycoerythrin conjugated mouse anti human P-glycoprotein monoclonal antibody (BD-Pharmingen) as described previously (22). Analysis was done on a Becton Dickinson flow cytometer using Cicero software (Cytomation, Fort Collins, CO). Our previous studies have shown that P-glycoprotein levels are not affected by non-targeted siRNAs.
Rhodamine 123 accumulation
The fluorophore rhodamine 123 is a substrate for the P-glycoprotein efflux pump; thus rhodamine 123 accumulation is often used as a surrogate for anti-tumor drug uptake/retention (24). The accumulation of Rh123 by siRNA-treated NIH 3T3-MDR cells was measured via a flow cytometry assay as described (22).
RNA extraction and real-time RT-PCR
Total RNA was isolated using a Tri Reagent kit (Molecular Research Center, Inc.), and cDNA was synthesized from total RNA using an oligo-dT primer. Real-time PCR was performed using the ABI PRISM 7900 sequence detection system (Applied Biosystems, Foster City, CA) as previously described (22). Primers (Oligonucleotide Synthesis Core Facility, University of North Carolina) and probes (Integrated DNA Technologies, Santa Clara, CA) were designed using Primer 4 software and were designed to span exon–intron junctions. The MDR1 signal was normalized using human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) by dividing the copies of MDR1 by the copies of human GAPDH.
Nuclease stability
For nuclease stability experiments, unmodified or ANA-modified siRNA duplexes were incubated with partially purified nucleases or with calf serum. Thereafter the material was analyzed on 3% agarose/ethidium bromide gels in BPB/XC loading buffer, electrophoresed at 100 V for 45 min, and residual duplexes imaged by ultraviolet illumination.
Thermal transitions
Melting points (Tm) for conventional and modified ORF1 duplexes were measured using a Shimadszu thermal melt apparatus.
RESULTS
Altritol nucleic acids (ANAs) are novel RNA analogs that have a six membered d-altritol ring with a nucleobase at the 2-(S)- position (Figure 1A). They can be considered as hexitol nucleic acid (25) analogs that have an additional hydroxy group at the 3′ position. In duplex RNA the hydroxy group is directed into the minor groove and contributes to helix stability. A diagram of an ANA nucleotide and of an ANA strand duplexed with conventional RNA is shown in Figure 1B. A series of ANA-modified oligonucleotides were synthesized with modifications at various positions in the sense or antisense strands. These are indicated in Table 1. The sequences ORF1 and ORF2 target two distinct regions of the MDR1 open reading frame.
Figure 1.
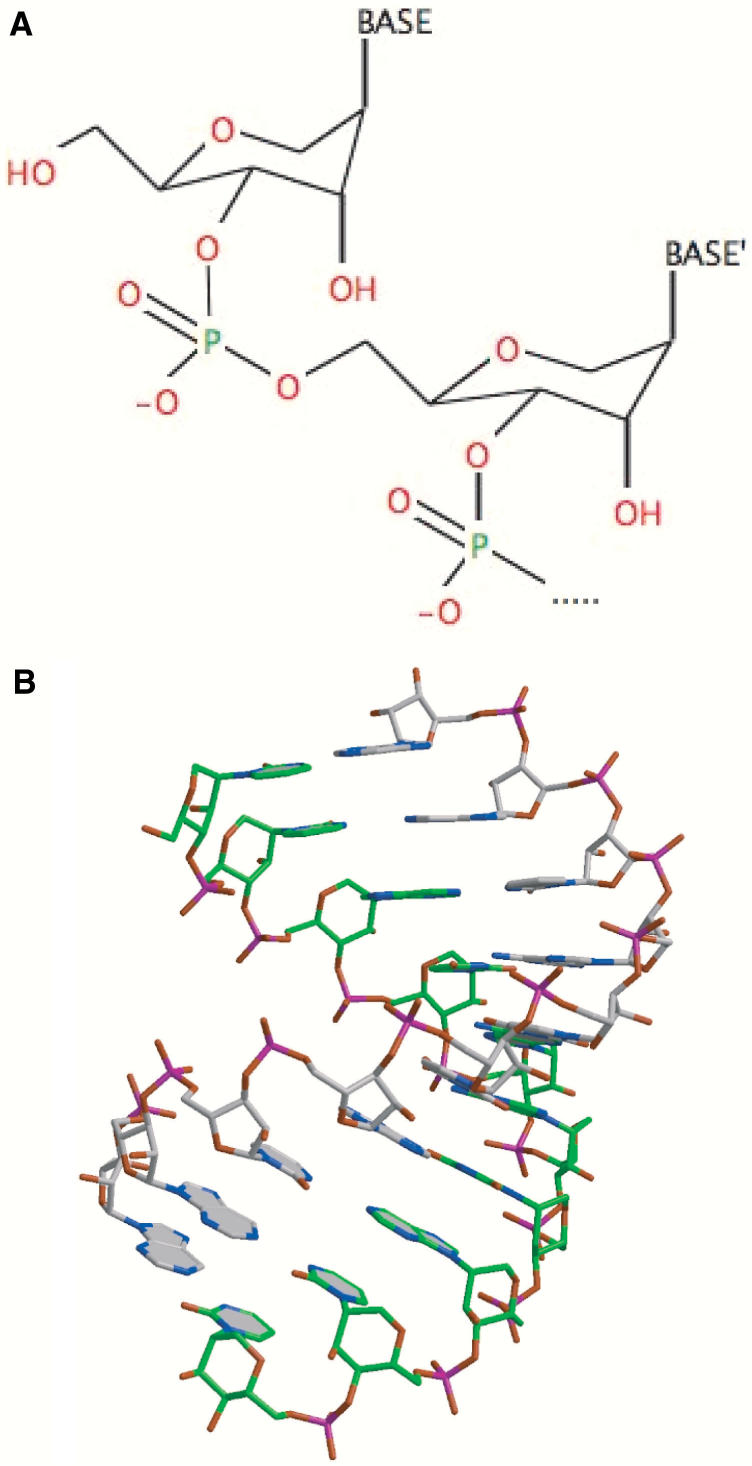
The chemistry and structure of altritol-modified oligonucleotides. (A) The chemical structure of ANA oligonucleotides. (B) Model of an ANA–RNA duplex; the green strand represents the ANA–modified oligonucleotide.
Table 1.
ANA(*)-modified RNAs that were used in the siRNA experiment. (Note: ORF1 and ORF2 refer to two distinct target regions of the MDR1 mRNA)
| ORF1 | |||||||
| Sense strand (5′ → 3′) | |||||||
| 2191 | GUA* | UUG | A*CA | GCU | AUU | CGA | AdTdT |
| 2192 | GUA | UUG | ACA | GCU | AUU | CGA* | AdTdT |
| 2307 | GUA | UUG | ACA | GCU | AUU* | CGA | AdTdT |
| 2308 | GUA | UUG | ACA | GCU* | AUU | CGA | AdTdT |
| 2309 | GUA | UU*G | ACA | GCU | AUU | CGA | AdTdT |
| 2310 | GU*A | UUG | ACA | GCU | AUU | CGA | AdTdT |
| 2311 | GUA | UU*G | ACA | GCU* | AU*U* | CGA | AdTdT |
| 2312 | GUA* | UU*G | ACA* | GCU* | AUU* | CGA* | AdTdT |
| 2313 | GU*A* | U*U*G | A*CA* | GCU* | A*U*U* | CGA* | A*dTdT |
| 2357 | GUA | UU*G | ACA* | GC*U | AU*U | CG*A | AdTdT |
| 2358 | G*U*A | UUG | ACA | GCU | AUU | CGA* | A*dTdT |
| 2359 | GUA | UUG | ACA* | G*C*U | AUU | CGA | AdTdT |
| 2385 | GUA | UUG | ACA | GCU | AUU | CGA* | A*dTdT |
| 2360 | GUA | U*U*G* | A*C*A* | G*C*U* | A*U*U* | C* CA | AdTdT |
| 2471 | G*UA | UUG | ACA | GCU | AUU | CGA | AdTdT |
| 2472 | G*U*A | UUG | ACA | GCU | AUU | CGA | AdTdT |
| Antisense strand (5′ → 3′) | |||||||
| 2193 | UUC | GAA | UAG | CUG | UCA | AUA* | CdTdT |
| 2194 | UUC | GAA | UAG | CUG | UCA | A*UA | CdTdT |
| 2314 | UUC | GAA | UAG | CUG | UCA | AU*A | CdTdT |
| 2315 | UUC | GAA | UAG | CUG | U*CA | AUA | CdTdT |
| 2316 | UUC | GAA | UAG | CU*G | UCA | AUA | CdTdT |
| 2317 | UUC | GAA | U*AG | CUG | UCA | AUA | CdTdT |
| 2318 | UU*C | GAA | UAG | CUG | UCA | AUA | CdTdT |
| 2361 | U*U*C | GAA | UAG | CUG | UCA | AUA | CdTdT |
| 2362 | UUC | GAA | UAG* | C*U*G | UCA | AUA | CdTdT |
| 2363 | U*U*C | GAA | UAG | CUG | UCA | AUA* | C*dTdT |
| 2364 | UUC | GA*A | UA*G | CU*G | UC*A | AU*A | CdTdT |
| 2365 | UUC | G*A*A* | U*A*G* | C*U*G* | U*C*A* | A*UA* | CdTdT |
| 2469 | UUC | GAA | UAG | CUG | UCA | AUA | C*dTdT |
| 2470 | UUC | GAA | UAG | CUG | UCA | AUA* | C*dTdT |
| ORF2 | |||||||
| Sense strand (5′ → 3′) | |||||||
| 2485 | GAA | CUC | UUA | GCG | UAU | GCA* | A*dTdT |
| 2486 | G*A*A | CUC | UUA | GCG | UAU | GCA* | A*dTdT |
| Antisense strand (5′ → 3′) | |||||||
| 2487 | UUG | CAU | ACG | CUA | AGA | GUU* | C*dTdT |
| 2488 | U*U*G | CAU | ACG | CUA | AGA | GUU | CdTdT |
Screening and dose–response relationships
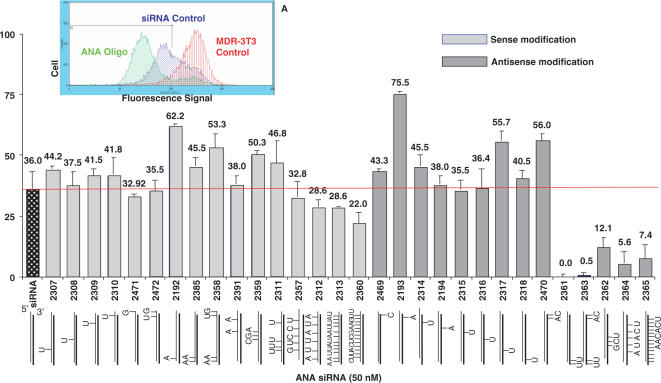
ANA-modified strands of the ORF1 sequence were complexed with conventional RNA to form siRNA duplexes and screened for activity in NIH-3T3-MDR cells by measuring the reduction in cell surface expression of P-glycoprotein, the MDR1 gene product. As seen in Figure 2, a wide range of efficacy was observed. Modifications at the 5′ end of the antisense strand abolished activity, while extensive modification within the central region of the antisense strand also lowered activity. By contrast, modifications at the 3′ end of either the sense or antisense strand resulted in efficacies that were substantially greater than that of unmodified siRNA (for example, oligonucleotides 2392, 2193). Multiple modifications within the sense strand were also tolerated with only modest reductions in siRNA effectiveness (e.g. oligonucleotides 2312, 2313). The activity of the various modified oligonucleotides was not correlated in any way with their thermal stability; for example, multiply modified oligonucleotide 2365 had the highest Tm when complexed with unmodified RNA (73°C versus 67°C for conventional ORF1 siRNA) but had little activity (Tm data not shown).
Figure 2.
Screening of ANA-modified siRNAs for activity versus MDR1. NIH 3T3-MDR cells were treated with 50 nM ORF1 sequence siRNA complexed to lipofectamine 2000 for 4 h at 37°C in complete medium. Thereafter the cells were placed in fresh medium (2% FBS/DMEM-H) and cultured for an additional 72 h. Cell surface P-glycoprotein levels were quantitated by immunostaining and flow cytometery as described in methods. Results are expressed as the percentage reduction in Pgp expression compared to untreated control cells. The positions of the ANA modifications are indicated in the figure. Results are means of three determinations. Figure 2 inset: a typical flow cytometry analysis is shown comparing Pgp levels in untreated control cells with cells treated with ANA-modified or control (unmodified) siRNAs. In this and subsequent figures unmodified anti-MDR1 siRNA is designated simply as ‘siRNA’ or ‘siRNA control’.
We also examined the effects of ANA-modified siRNAs targeted to a distinct region of the MDR1 mRNA. These siRNAs, termed ORF2 ANA sequences, also displayed enhanced activity as compared to conventional siRNA targeting the ORF2 sequence (Table 2). Thus the beneficial effects of ANA modification are not limited to a particular sequence within the MDR1 message. As with the ORF1 sequences, modifications at the 5′ end of the antisense strand for ORF2 were not well tolerated.
Table 2.
Effects of ORF2 siRNAs on P-glycoprotein expression (means and standard errors of 6 determinations)
| Oligonucleotide | Percentage reduction in P-glycoprotein |
|---|---|
| siRNA control | 30.7+/−6.3 |
| 2485/2487 | 50.8+/−4.1 |
| 2485/2488 | 4.1+/−1.4 |
| 2511/2487 | 47.8+/−5.9 |
| 2511/2488 | 2.9+/−1.6 |
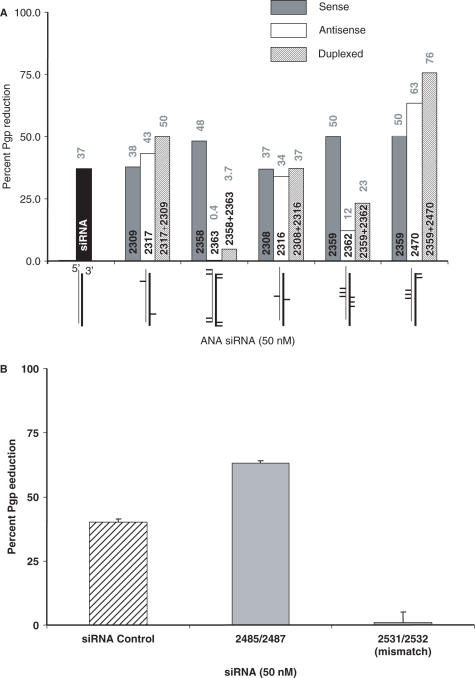
Use of two ANA-modified strands in a duplex usually provided additional advantages in terms of effectiveness (Figure 3A). For example, the modified sense strand 2359 and the modified antisense strand 2470 were both effective when duplexed with an unmodified partner; however, the 2359/2470 duplex was even more effective. Interestingly, duplexing a poorly effective antisense strand with an effective sense strand often partially ‘rescued’ siRNA activity; this can be seen in the 2359/2362 duplex in Figure 3A, suggesting that the overall characteristics of the duplex are important in RISC loading and mRNA degrading activity.
Figure 3.
(A) Effects of ANA modifications in both strands of the duplex. NIH 3T3-MDR cells were treated with 50 nM siRNA and monitored for Pgp expression by immunostaining and flow cytometry as in Figure 2. A comparison is shown between siRNAs with ANA modifications in one strand (gray bars = modified in sense strand, white = modified in antisense strand) duplexed with a conventional RNA complementary strand, or siRNAs with ANA modifications in both strands (hatched bars). Results are the means of triplicate determinations. (B) Effects of mismatches. ANA-modified ORF2 siRNAs (50 nM) with (2531/2532) or without (2485/2487) 4 mismatches to the target sequence were tested for inhibition of Pgp expression versus control siRNA. The 2531/2532 (sense/antisense) mismatch duplex has ANAs in exactly the same positions as 2485/2487 (2532-uuc gUa uag GuC ucU aua* c*dtdt; mismatches indicated by capital letters, positions of ANAs indicated by *). Results are the means and standard errors of triplicate determinations.
In control experiments, we confirmed that variants of active ANA-modified oligonucleotides that have several mismatches in their sequences are completely inactive (Figure 3B); this is similar to the situation for conventional siRNAs targeting MDR1, as we have previously shown (21).
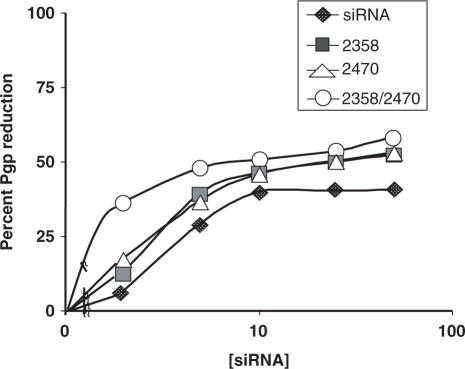
Some of the more promising duplexes with modifications in one or both strands were tested for inhibition of P-glycoprotein expression over a range of siRNA concentrations. As seen in Figure 4, the enhanced activities of the ANA-modified versions were present over the entire concentration range. The profiles indicate both increased potency with effects at lower concentration and increased efficacy with greater responses at the plateau.
Figure 4.
Dose–response curves for ANA-modified siRNAs. NIH 3T3-MDR cells were treated with various concentrations of siRNA complexed with lipofectamine 2000 for 4 h at 37°C in complete medium; thereafter the cells were placed in fresh medium (2% FBS/DMEM-H) and cultured for an additional 72 h. Cells were monitored for Pgp expression by immunostaining and flow cytometry as in Figure 2. Ordinate: percentage reduction in Pgp expression versus untreated control cells. Abscissa: concentration of siRNA (nM). Results are the means of triplicate determinations.
Role of 5′ Phosphorylation
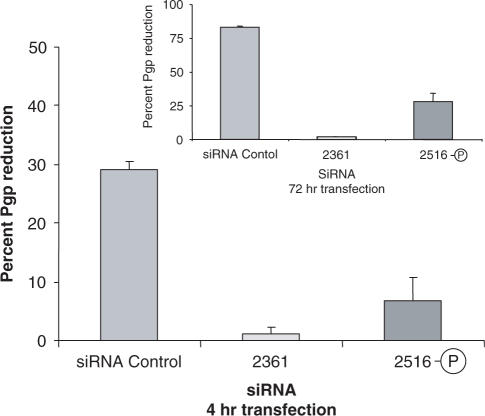
Antisense strands with ANA modifications at the 5′ position were completely inactive, but the reason for this was not immediately apparent. There is a stabilizing effect of the ANA residue, thus potentially undermining the requirement for local instability at the 5′ antisense position (6); however, the same stabilizing effect would occur with modifications near the 3′ end of the sense strand, but these are well tolerated. Another possibility is that the 5′ ANA residue interferes with the required 5′ phosphorylation of the antisense strand of siRNA by endogenous kinases (5). To examine this possibility, we synthesized a version of an inactive oligonucleotide (2361) that has a 5′ phosphate group (termed 2516-P). As seen in Figure 5, the presence of a 5′-phosphate group partially rescues the effectiveness of the ANA-modified antisense strand. Oligonucleotides 2361 and 2516-P have exactly the same ANA modification at the 5′ position, but 2516-P was phosphorylated during synthesis. The observed ‘rescue’ of the ‘knockdown’ effect is not large but it is consistently observed. The degree of rescue was not affected by the presence or absence of serum during the transfection process (data not shown), suggesting that the relatively modest rescue is not because of the possible dephosphorylation of 2516-P by serum phosphatases, but rather that dephosphorylation and inactivation of 2516-P may take place within the cell. Interestingly, when we extended the period of transfection beyond the 4 h usually used, the effect of the 5′ phosphorylated ANA antisense was much greater, while the unphosphorylated ANA antisense remained inactive (Figure 5 inset). This suggests that maintaining a persistent external pool of 5′ phosphorylated siRNA, and thus continuous delivery, may allow adequate loading of the RISC complex even in the face of rapid dephosphorylation by cellular phosphatases.
Figure 5.
Rescue of siRNA effectiveness by 5′ phosphorylation. Antisense oligonucleotide 2361 which has ANA modifications at its 5′end (and is therefore inactive) was synthesized including a 5′-terminal phosphate group and now termed 2516-P. NIH 3T3 MDR cells were treated with 50 nM siRNAs for 4 h, washed, and then analyzed for Pgp expression after 72 h. Results are expressed as the percentage reduction in Pgp expression compared to untreated control cells (means and standard errors of 3 determinations). (Inset) Assays after long exposures. The analysis was exactly the same as above except that the siRNAs and transfection agent were left in contact with the cells for the entire 76-h period.
RT-PCR and functional correlates
We examined whether the observed siRNA-mediated reductions in P-glycoprotein expression were accompanied by similar changes at the message level and by functional changes in drug transport. As seen in Figure 6, reductions MDR1 mRNA were observed that paralleled reduced expression of the protein product, suggesting that the action of the ANA-modified siRNAs is at the level of message degradation.
Figure 6.
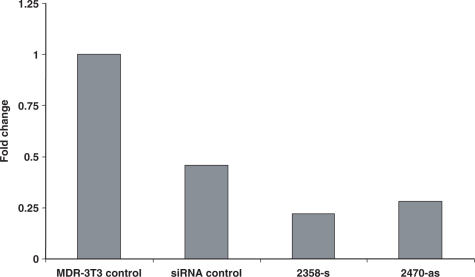
Analysis of effects of ANA-modified siRNA by real-time PCR. NIH 3T3 MDR cells were treated with 50 nM siRNAs as in Figure 2 and cellular mRNA levels were analyzed by RT-PCR as described in methods. Ordinate: fraction of MDR1 mRNA as compared to untreated control cells. Results are means of three determinations.
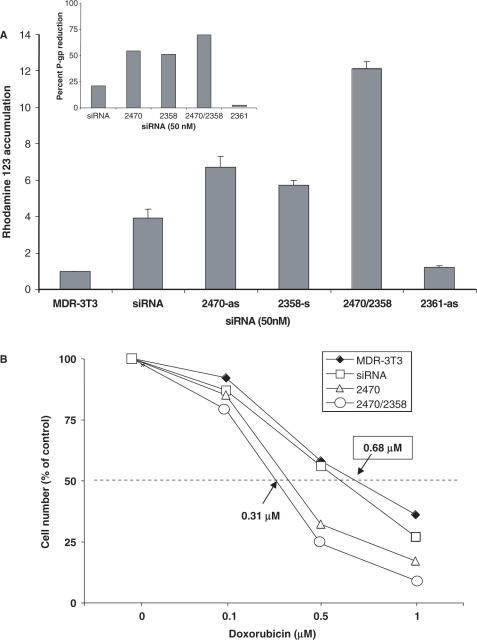
The observed reductions in P-glycoprotein levels were also accompanied by increases in the intracellular accumulation of P-glycoprotein substrates. Thus Figure 7A illustrates the effect of conventional or ANA-modified siRNAs on cell levels of the lipophilic dye Rh123. Notably a duplex with modifications in both strands (2470/2358) was significantly more effective than either modified strand complexed with unmodified RNA, which in turn were superior to conventional siRNA. A strand that was inactive in terms of P-glycoprotein reduction (2361) was also inactive in this assay. The changes in Rh123 accumulation were closely paralleled by changes in cell surface P-glycoprotein expression in the same experiment, as indicated in the inset in Figure 7A. Treatments with conventional or modified siRNA also resulted in changes in cellular response to cytotoxic drugs. Thus as seen in Figure 7B, the dose versus toxicity curve for the anti-tumor drug doxorubicin was left-shifted by treatment of NIH-3T3-MDR cells with siRNAs that reduced P-glycoprotein expression. Once again the most robust effects were attained with duplexes having ANA modifications in both strands. The maximal observed change in doxorubicin ID50 was slightly more than 2-fold, consistent with the 50–75% reductions in P-glycoprotein expression observed.
Figure 7.
(A) Effects of siRNAs on Rh123 accumulation. NIH 3T3-MDR cells were treated with 50 nM ANA-modified or conventional siRNAs as in Figure 2. After 72 h the cells were exposed to 1 μg/ml rhodamine 123 for 1 h. The cells were washed, harvested and analyzed for Rh123 levels by flow cytometry, as described in methods. Results are normalized based on a value of 1 for untreated cells. The data represents means and standard errors for three determinations. The inset in 7A illustrates cell surface Pgp levels in the cells used in one of the Rh123 experiments. (B) Effects of siRNAs on doxorubicin toxicity. NIH 3T3-MDR cells were treated with 50 nM ANA-modified or conventional siRNAs as in Figure 2. After 72 h the cells were exposed for 24 h to various concentrations of doxorubicin. After a further 48 h, cells were harvested and cell numbers were determined using a particle counter as described in methods. Results are expressed as the percentage of the cell number in controls not treated with siRNA. Ordinate: cell number as percentage of control. Abscissa: doxorubicin concentration (nM). Results are means of triplicate determinations.
Persistence of effects
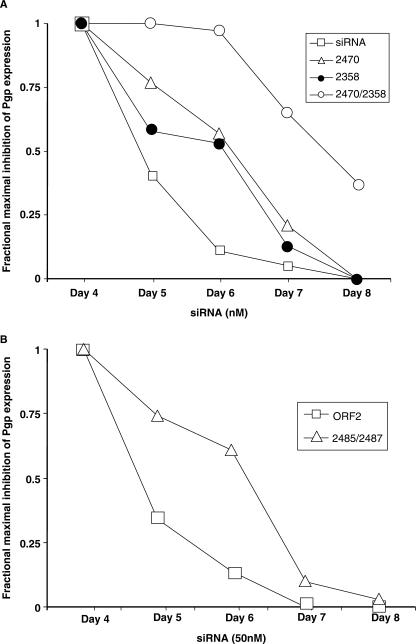
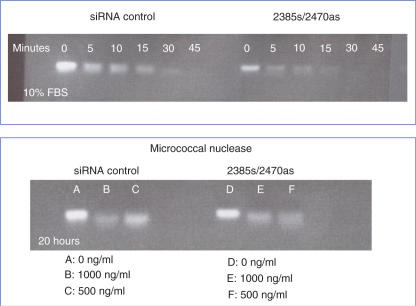
An important aspect of siRNA pharmacology is to attain persistent ‘knockdown’ of the target. Many factors will contribute to this, including the overall stability of the siRNA to nucleases, cellular uptake and import processes, and biochemical interactions with the RISC complex. We compared the duration of P-glycoprotein reduction in cells treated with conventional and ANA-modified siRNAs. As shown in Figure 8A, with conventional ORF1 siRNA almost all of the ‘knockdown’ effect seen at 4 days post-siRNA treatment was lost by day 6 as mRNA and protein levels returned to normal. By contrast, using ORF1 sequence siRNAs with ANA modifications in a single strand, 50% of the day 4 effect was maintained to day 6, while with modifications in both strands essentially all of the ‘knockdown’ persisted to day 6 and substantial effects were still present at day 8. A similar but slightly more modest effect on duration of action was seen with ANA-modified ORF2 oligonucleotides (Figure 8B). This remarkable enhancement of persistence of action may be an important pharmacological advantage for ANA siRNAs. While many factors can contribute to persistent biological effects of siRNA, one of them may well be nuclease stability. Thus we tested the stability of conventional and ANA-modified siRNAs to nucleases in an in vitro assay. However, as seen in Figure 9, when we compared a standard siRNA to one of the more potent ANA-modified siRNA duplexes (2385/2470), we did not observe any significant increase in stability to micrococcal nuclease or to the nuclease activity in serum. Thus, factors other than nuclease stability may contribute to the long duration of the ANA siRNA effects, although we cannot as yet rule out the possibility that ANA modification may increase stability to endogenous cellular nucleases.
Figure 8.
Persistent effects of ANA-modified siRNAs. (A) Control and ANA-modified ORF1 oligonucleotides. NIH 3T3-MDR cells were treated with 50 nM ANA-modified or conventional ORF1 siRNAs as in Figure 2. Cells were recovered on days 4–8 after siRNA treatment and monitored for P-glycoprotein expression by immunostaining and flow cytometry. The percentage reduction in Pgp expression on day 4 was taken as 1 and the results for subsequent days were expressed as a fraction of that value. Thus a decline in the value of the ordinate represents an increase in the level of cell surface Pgp. Results are means of triplicate determinations. (B) Control and ANA-modified ORF2 oligonucleotides. The experiment was the same as in (A) except that we compared conventional ORF2 siRNA to an ANA-modified ORF2 oligonucleotide duplex (2485/2487).
Figure 9.
Nuclease stability of ANA-modified siRNAs. Standard MDR1 siRNA or the 2385/2470 ANA-modified siRNA were exposed to nuclease activity and the degradation of the siRNAs monitored by agarose gel electophoresis with ethidium bromide staining. Upper panel: exposure to 10% fetal calf serum for various times. Lower panel: exposure to various doses of micrococcal nuclease.
DISCUSSION
Based on the results reported here it seems that ANA-modified oligonucleotides have some desirable features for use as siRNAs including increased efficacy and prolonged duration of action. The effects of ANA modifications were screened via quantification of the reduction in expression of P-glycoprotein and were further examined at the mRNA level and via functional assays.
ANA modifications at the 5′ position of the antisense strand were not tolerated; however, the reason for this is not entirely clear. While there is a stabilizing effect of the ANA residue, thus opposing the requirement for local thermodynamic instability at the 5′ antisense position (6), the same would be true of modifications near the 3′ end of the sense strand, but these are well tolerated. One likely possibility is that the presence of an ANA modification hinders phosphorylation of the 5′-OH by cellular kinases. This is supported by our observations that a 5′ ANA antisense strand synthesized with a 5′ phosphate moiety was more effective than the equivalent unphosphorylated strand. Chemical modification of the 5′ position of the antisense strand often results is loss of activity (5); at least in the case of ANAs, reduced susceptibility to phosphorylation seems to be an important aspect of this phenomenon.
Excellent effects were attained with ANA modifications of the 3′ regions of the sense or antisense strands with efficacies as much as 2-fold greater than conventional siRNA. Once again the basis for this is uncertain, but it does not seem to be due to increased stability to nucleases. Although fully modified ANA oligonucleotides seem to be more resistant to serum nucleases, this was not the case for the partially modified compounds tested here. Extensive modification of the central region of the sense strand is also compatible with maintenance of siRNA activity. In some respects this is surprising based on the finding that the sense strand usually is degraded as the RISC complex becomes functional; however, an alternate pathway that does not require sense strand degradation has also been noted (26). Our observations are consistent with other reports indicating that certain modifications (e.g. 2′-O-Me) are well tolerated on the sense strand (12). As interesting aspect of our findings is that, in general, use of ANA modifications in both strands increases effectiveness while increasing the number of ANA residues on one strand does not. Thus the greatest improvements in efficacy were attained with duplexes having 3′ ANA modifications in both strands. The mechanistic basis of this effect is not yet understood.
The reductions in P-glycoprotein expression observed subsequent to siRNA treatment were paralleled by reduction in MDR1 mRNA levels and by changes in drug accumulation and drug resistance. In general there was good quantitative agreement among all of these parameters, with certain siRNAs having ANA modifications on both strands demonstrating the greatest activity in each of these assays. It is interesting that the ANA modifications often were not only compatible with retention of siRNA activity, but in many cases led to enhanced activity. Particularly noteworthy is the substantially increased duration of action of some of the ANA-modified siRNAs. This can be a significant advantage for therapeutic applications where persistent effects may be desirable. A recent publication has indicated that 2′-deoxy-2′-fluoro-β-d-arabinonucleotide(FANA)-modified siRNAs also displayed increased persistence of action, an effect that was attributed to nuclease stability (27). The increased biological activity and persistence of action of ANA-modified siRNAs may be due to other factors, including a better recognition by the RISC machinery, or greater accumulation/duration within cells. In future studies, we will investigate these possible factors including the recognition of ANA by the PAZ and PIWI domains of the RISC complex. Another interesting issue is that even single modifications can increase effectiveness. This might be due to small conformational changes of the duplex introduced by the modified nucleotide. The conformational change introduced by a rigid nucleotide is normally transferred to neighboring nucleotides so that the change is extended over several base pairs.
In summary, the altritol modification provides an RNA-like oligonucleotide that is well suited for siRNA applications. A number of questions remain as to the basis of the enhanced effectiveness and persistence of ANA siRNAs; these will be explored in future work as will the behavior and utility of ANA siRNAs in the in vivo setting.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by NIH grant PO1 GM59299 (to RLJ) and by KULeuven (GOA) (to PH). Funding to pay the Open Access publication charge was provided by GM59299.
REFERENCES
- 1.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 2.Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 3.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 4.Nawrot B, Sipa K. Chemical and structural diversity of siRNA molecules. Curr. Top. Med. Chem. 2006;6:913–925. doi: 10.2174/156802606777303658. [DOI] [PubMed] [Google Scholar]
- 5.Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 7.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 8.Harborth J, Elbashir SM, Vandenburgh K, Manninga H, Scaringe SA, Weber K, Tuschl T. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 9.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall AH, Wan J, Spesock A, Sergueeva Z, Shaw BR, Alexander KA. High potency silencing by single-stranded boranophosphate siRNA. Nucleic Acids Res. 2006;34:2773–2781. doi: 10.1093/nar/gkl339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czauderna F, Fechtner M, Dames S, Aygun H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraynack BA, Baker BF. Small interfering RNAs containing full 2'-O-methylribonucleotide-modified sense strands display Argonaute2/eIF2C2-dependent activity. RNA. 2006;12:163–176. doi: 10.1261/rna.2150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 14.Xia J, Noronha A, Toudjarska I, Li F, Akinc A, Braich R, Frank-Kamenetsky M, Rajeev KG, Egli M, Manoharan M. Gene silencing activity of siRNAs with a ribo-difluorotoluyl nucleotide. ACS Chem. Biol. 2006;1:176–183. doi: 10.1021/cb600063p. [DOI] [PubMed] [Google Scholar]
- 15.Allart B, Busson R, Rozenski J, Van Aerschot A, Herdewijn P. Synthesis of protected D-altriol nucleosides as building blocks for oligonucleotide synthesis. Tetrahedron. 1999;55:6527–6546. [Google Scholar]
- 16.Allart B, Khan K, Rosemeyer H, Schepers G, Hendrix C, Rothenbacher K, Seela F, Van Aerschot A, Herdewijn P. Properties of oligonucleotides with six membered carbohydrate mimics and a 1,4-relationship between the base moiety and the hydroxymethyl group. Chem. Eur. J. 1999;5:2424–2431. [Google Scholar]
- 17.Abramov M, Marchand A, Calleja-Marchand A, Herdewijn P. Synthesis of d-altritol nucleosides with a 3'-O-tert-butyldimethylsilyl protecting group. Nucleosides Nucleotides Nucleic Acids. 2004;23:439–455. doi: 10.1081/ncn-120028338. [DOI] [PubMed] [Google Scholar]
- 18.Froeyen M, Wroblowski B, Esnouf R, De Winter H, Allart B, Lescrinier E, Herdewijn P. Molecular dynamics studies of single stranded hexitol, altritol, mannitol, and ribose nucleic acides (HNA, MNA, ANA, and RNA, resp.) and of the stability of HND, RNA, ANA-RNA and MNA-RNA duplexes. Helv. Chim. Acta. 2000;83:2153–2182. [Google Scholar]
- 19.Kozlov IA, Zielinski M, Allart B, Kerremans L, Van Aerschot A, Busson R, Herdewijn P, Orgel LE. Nonenzymatic template-directed reactions on altritol oligomers, preorganized analogues of oligonucleotides. Chemistry. 2000;6:151–155. doi: 10.1002/(sici)1521-3765(20000103)6:1<151::aid-chem151>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 20.Ambudkar SV, Dey S, Hrycyna CA, Ramachandra M, Pastan I, Gottesman MM. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu. Rev. Pharmacol. Toxicol. 1999;39:361–398. doi: 10.1146/annurev.pharmtox.39.1.361. [DOI] [PubMed] [Google Scholar]
- 21.Xu D, Kang H, Fisher M, Juliano RL. Strategies for inhibition of MDR1 gene expression. Mol. Pharmacol. 2004;66:268–275. doi: 10.1124/mol.66.2.268. [DOI] [PubMed] [Google Scholar]
- 22.Xu D, McCarty D, Fernandes A, Fisher M, Samulski RJ, Juliano RL. Delivery of MDR1 small interfering RNA by self-complementary recombinant adeno-associated virus vector. Mol. Ther. 2005;11:523–530. doi: 10.1016/j.ymthe.2004.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang H, Fisher MH, Xu D, Miyamoto YJ, Marchand A, Van Aerschot A, Herdewijn P, Juliano RL. Inhibition of MDR1 gene expression by chimeric HNA antisense oligonucleotides. Nucleic Acids Res. 2004;32:4411–4419. doi: 10.1093/nar/gkh775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broxterman HJ, Schuurhuis GJ. Transport proteins in drug resistance: detection and prognostic significance in acute myeloid leukemia. J. Intern. Med. Suppl. 1997;740:147–151. [PubMed] [Google Scholar]
- 25.Declercq R, Van Aerschot A, Read RJ, Herdewijn P, Van Meervelt L. Crystal structure of double helical hexitol nucleic acids. J. Am. Chem. Soc. 2002;124:928–933. doi: 10.1021/ja016570w. [DOI] [PubMed] [Google Scholar]
- 26.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 27.Dowler T, Bergeron D, Tedeschi AL, Paquet L, Ferrari N, Damha MJ. Improvements in siRNA properties mediated by 2'-deoxy-2'-fluoro-beta-D-arabinonucleic acid (FANA) Nucleic Acids Res. 2006;34:1669–1675. doi: 10.1093/nar/gkl033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.