Abstract
Expansions of (GAA)n repeats within the first intron of the frataxin gene reduce its expression, resulting in a hereditary neurodegenerative disorder, Friedreich's ataxia. While it is generally believed that expanded (GAA)n repeats block transcription elongation, fine mechanisms responsible for gene repression are not fully understood. To follow the effects of (GAA)n·(TTC)n repeats on gene expression, we have chosen E. coli as a convenient model system. (GAA)n·(TTC)n repeats were cloned into bacterial plasmids in both orientations relative to a promoter, and their effects on transcription and RNA stability were evaluated both in vitro and in vivo. Expanded (GAA)n repeats in the sense strand for transcription caused a significant decrease in the mRNA levels in vitro and in vivo. This decrease was likely due to the tardiness of the RNA polymerase within expanded (GAA)n runs but was not accompanied by the enzyme's dissociation and premature transcription termination. Unexpectedly, positioning of normal- and carrier-size (TTC)n repeats into the sense strand for transcription led to the appearance of RNA transcripts that were truncated within those repetitive runs in vivo. We have determined that these RNA truncations are consistent with cleavage of the full-sized mRNAs at (UUC)n runs by the E. coli degradosome.
INTRODUCTION
Friedreich's ataxia (FRDA) is the most commonly inherited ataxia, occurring in 1–2 of every 50 000 people (1,2). It is characterized by muscle weakness along with progressive gait and limb ataxia and is often accompanied by vision impairment, skeletal abnormalities, heart disease and diabetes (3). FRDA is a recessive disease that is predominantly caused by expansions of (GAA)n repeats within the first intron of the human frataxin gene X25 (4). These expansions lead to the lowered level of the frataxin protein, which is responsible for iron metabolism in mitochondria, resulting in disease (5). Short (GAA)n repeats, ranging from 5 to 34 units, are always present in normal frataxin alleles and were apparently originated by the primordial insertion of an Alu element (6). In FRDA alleles, (GAA)n stretches range from 66 to 1700 units (7). It is believed that these expanded stretches affect transcription of the X25 gene since FRDA patients have low levels of the frataxin mRNA (8,9). Further support for transcription inhibition by expanded (GAA)n stretches was obtained in transient transfection experiments with cultured mammalian cells (10).
The exact mechanisms responsible for transcription inhibition by long (GAA)n repeats are not entirely clear. Several groups have demonstrated that transcription elongation in vitro was disrupted by (GAA)n runs (9,11,12). On linear templates, (GAA)n repeats efficiently trapped T7 RNA polymerase leading to the reduced amounts of the full-length transcripts, but premature termination was not detected (11). In contrast, on supercoiled templates, transcripts prematurely terminated at the 3′ ends of repeats (11). Transcription inhibition in vitro was linked to the repeat's ability to form triple-helical H-DNA or a composite triplex structure, called sticky DNA (11,12). Intramolecular triplexes formed by (GAA)n·(TTC)n can exist in either H-y or H-r conformations depending on the nature of the third DNA strand (13,14). The H-r conformation is more likely to form during transcription in vitro, as it is stabilized by the presence of magnesium cations presented in transcription mixtures. The H-r triplex is more favorable in superhelical DNA than in linear DNA and could be additionally stabilized by the homopurine RNA tail, formed after RNA polymerase has passed the repeat (15).
The interpretation of the data on transcriptional repression in vivo is somewhat less obvious. Formation of triplex-like structures was suggested to cause transcription inhibition at expanded repeats in transient transfection experiments (10). An alternative explanation could be a manifold binding of nuclear protein(s) to the expanded repeat. (GAA)n-binding proteins were indeed identified in HeLa nuclear extracts (16). Another possibility could be an alteration in RNA splicing, processing or stability caused by the expanded repeat. Finally, expansions of (GAA)n repeats could lead to the formation of heterochromatin, thus silencing the frataxin gene. Supporting this, long (GAA)n stretches led to the expression variegation of an adjacent reporter in mice (17). The reversal of the frataxin gene silencing in FRDA lymphocytes by the synthetic histone deacetylase inhibitors further supports the role of heterochromatin formation in the disease (18).
In this study, we looked at the effects of (GAA)n·(TTC)n repeats on RNA synthesis by E. coli RNA polymerase in vitro and in vivo. This system, where formation of heterochromatin or RNA splicing are out of the question, allowed us to concentrate on the effects of FRDA repeats on transcription and RNA stability. We have found that expanded (GAA)n repeats, when in the sense strand for transcription, impair RNA polymerase progression. While elongating RNA polymerase is severely slowed by the repeat, it does not dissociate from the template. Surprisingly, in an opposite orientation of the repeat, i.e. when the (TTC)n run is in the sense strand for transcription, truncated RNA species accumulate even for normal-size repeats. We found that cleavage of full-length mRNAs within (UUC)n runs by the E. coli degradosome is responsible for these truncations.
MATERIALS AND METHODS
Bacteria
E. coli JM109 strain (Promega) was used for maintaining the p185ΔCTT10 and p185ΔCTT20 plasmids. Strains carrying these plasmids were grown at 37°C in LB media containing 10 μg/ml tetracycline. Longer (TTC)n repeats were maintained in a JM109 derivative carrying an extra pTrc99A plasmid to prevent leakage of the trc-promoter in repeat-containing plasmids. Expanded GAA repeats were maintained in an N3433 (lacZ43(Fs), relA1, spoT1, thi-1)-derived strain also carrying an extra pTrc99A plasmid. Plasmid-bearing strains were grown at 37°C in minimal M9 media supplemented with 1 μg/ml vitamin B1, 40 μg/ml amino acids mix, 10 μg/ml tetracycline and 50 μg/ml ampicillin. For RNA isolation, they were grown until A600 ∼ 0.2, then IPTG (Sigma) was added to 2 mM and cells were incubated for another 1 h.
Strain N3431 (lacZ43(Fs), λ−, rne-3071(ts), relA1, spoT1, thi-1), carrying the temperature-sensitive mutation in the RNase E gene, was obtained courtesy of the E. coli Genetic Stock Center. For RNA isolation, this strain was grown at 30°C up to A600 ∼ 0.4, then transferred to 43°C for 1 h. Transcription was induced by the addition of 2 mM IPTG for 40 min prior to cell harvesting. For the experiments with protein synthesis inhibition, N3431 cells were grown at 30°C until the early logarithmic stage (A600 = 0.2), followed by the addition of chloramphenicol (170 μg/ml) and subsequent incubation for 2 h. Cultures were then transferred to 43°C for 1 h. Transcription was induced by the addition of 2 mM IPTG for 40 min prior to cell harvesting.
Yeast
Yeast transcription experiments were performed in Saccharomyces cerevisiae CH1585 (MATa leu2Δ1 trpΔ63 ura3-52 his3-200) strain (ATCC 96098) transformed with pYES2-derived plasmids carrying (GAA)n·(TTC)n repeats, as described earlier (19).
Plasmids for transcription studies in vivo
The p185Δ vector for cloning of various (GAA)n·(TTC)n repeats was obtained in two steps. First, the p185 plasmid was made by ligating the PstI–BspHI fragment, carrying the cat gene from the pTrcCAT/Pst plasmid (20), with the XmnI–BsaAI fragment of the pACYC184 plasmid (New England Biolabs), carrying the p15A replication origin and tet gene. To improve stability of the cat mRNA, a BsrDI–XmnI fragment from the cat gene was deleted from the resultant plasmid.
To obtain the p185ΔGAA57 and p185ΔCTT57 plasmids, an EcoRI–HindIII fragment of the pBluescript-GAA57 (19), carrying the (GAA)57·(TTC)57 repeat, was cloned into the Kpn2I site of the p185Δ in both orientations. To obtain the p185ΔGAA114 plasmid, a (GAA)114·(TTC)114 repeat was excised upon the BsgI digestion from the pYES–GAA114 (19) and re-cloned into the Kpn2I site of the p185Δ. Other repeat lengths were generated as a result of spontaneous deletions of the (GAA)114·(TTC)114 repeat during the cultivation of the p185ΔGAA114 plasmid.
Plasmids with the (TTC)15 repeat and its mutative derivatives were obtained by cloning pre-annealed pairs of oligonucleotides into the Kpn2I site of the p185Δ. 5′-CCGG(TTC)15TGT-3′ and 5′-CCGGAC(AAG)15A-3′ were used to generate the p185CTT15 plasmid; 5′-CCGG(TTC)8TCT(TTC)2TCTTT(CTT)2GT-3′ and 5′-CCGGAC(AAG)2AAAGA(GAA)2AGA(GAA)8-3′ were used to create the p185CTT15-M1 plasmid; 5′-CCGG(TTC)2TTTCT(CTT)2TCT(CTT)8GT-3′ and 5′-CCGGAC(AAG)8AGA(AAG)2AGAAA(GAA)2-3′ were used for obtaining the p185CTT15-M2 plasmid; and 5′-CCGG(TTC)2TTTCT(CTT)2TCTCTTCTCT(TTC)2TCTTT(CTT)2GT-3′ and 5′-CCGGAC(AAG)2AAAGA(GAA)2AGAGAAGAGA(AAG)2AGAAA(GAA)2-3′ were used for generating the p185CTT15-M12 plasmid.
Yeast plasmids pYES–Control and pYES–TTC57 for yeast transcription studies were described earlier (19). Plasmids pYES–TTC35 and pYES–TTC20 were obtained by cloning the EcoRI–Kpn2I repeat-containing fragments from the p185ΔTTC20 and p185ΔTTC35 plasmids, respectively, into the XhoI site of the pYES+ plasmid (19).
Plasmids for transcription studies in vitro
The plasmid pYES–GAA57TspR1 was obtained as follows. A 6608-bp fragment was generated upon partial digestion of the pYES–GAA57 plasmid (19) with BsgI. This fragment was then digested with KpnI and ligated to the pre-annealed oligonucleotides 5′-CGGCACTGGGCGGCCGCTCGAAGCTTGATGA-3′ and 5′-ATCAAGCTTCGAGCGGCCGCCCAGTGCCGGTAC-3′.
The plasmid pYES–TTC57TspR1 was obtained by ligating pre-annealed oligonucleotides 5′-CATCGATGGCACTGGGC-3′ and 5′-GGCCGCCCAGTGCATCGATGGTAC-3′ to the pYES–TTC57 plasmid (19) digested with the NotI and KpnI endonucleases.
The plasmid YEp24TspR1 was obtained by ligating pre-annealed oligonucleotides 5′-CCGGCACTGGGCGGCCGCTCGAAGCTTGATGCATG-3′ and 5′-CATCAAGCTTCGAGCGGCCGCCCAGTG-3′ to the YEp24 vector (New England Biolabs) digested with SphI and Kpn21 endonucleases.
For transcription studies in vitro, preformed initiation complexes were ligated to one of the following fragments: (i) a 208-bp TspR1–AlwN1 fragment of the pYES–GAA57TspR1 plasmid; (ii) a 214-bp TspR1–EcoRI fragment of the pYES–TTC57TspR1 plasmid; or (iii) a 220-bp TspR1–BamH1 fragment of the Yep24TspR1 plasmid.
Analysis of RNA synthesis and stability in vivo
RNA from E. coli cells was isolated by a modified method of Dennis and Nomura as described earlier (21). Northern blot hybridization was performed according to standard protocol (22). RNA samples were denatured in 2.2 M formaldehyde, 1 × MOPS, 50% v/v deionized formamide followed by the separation in a 1.5% agarose gel containing 2.2 M formaldehyde in a 1 × MOPS buffer. The gel was capillary-blotted onto a Hybond N+ membrane (Amersham). Membranes were hybridized either with the 279-bp EcoRI–KpnI fragment of the p185, corresponding to 5′ part of the reporter's mRNA, or with an oligonucleotide 5′-AAAACATTGCATACGGAATTCCGG-3′ that corresponds to the sequence immediately downstream from the (TTC)n stretch. All fragments were radioactively labeled using the Hexalabel DNA labeling kit (Fermentas). Oligonucleotides for hybridization were labeled with T4 polynucleotide kinase (Invitrogen).
To analyze RNA stability in vivo, E. coli cultures were grown up to OD = 0.3, 2 mM IPTG was added, and cell growth was continued for 20 min. After addition of rifampicin (0.2 mg/ml) to block transcription, cell culture aliquots were taken at 0, 7, 12 and 17 min. These aliquots were instantly mixed with ice-cold phenol–ethanol, pelleted and frozen at −70°C. RNA was isolated, and equal amounts of total mRNA were analyzed by northern blot hybridization as described above.
Yeast RNA was isolated from 10 ml of cultures exponentially growing in the complete URA-synthetic medium with galactose using the RNeasy Mini Kit (Qiagen). Northern blot hybridization was performed according to the standard protocol (22). RNA samples were denatured in 2.2 M formaldehyde, 1 × MOPS, 50% (v/v) deionized formamide and separated in the 1.5% agarose gel with 2.2 M formaldehyde in the 1 × MOPS buffer. The gel was capillary-blotted onto a Hybond N+ membrane (Amersham). The membrane was hybridized with either the 112-bp PvuII–XhoI fragment or the 473-bp BamHI–EcoRI fragment of the pYES+, corresponding to the 5′- or 3′-part, respectively, of the analyzed mRNA.
Transcription in vitro
Promoter-independent assembly of elongation complexes and their ligation to the repeat-containing and control fragments was done as described earlier (23). Elongation complexes were immobilized on Ni-NTA agarose via a hexahistidine tag on the largest subunit of RNA polymerase. Prior to the assembly, the RNA was labeled at its 5′ end by the T4 polynucleotide kinase in the presence of 500 μCi of γ-32P ATP (MP Biomedicals, Irvine, CA). For transcription analyses, the assembled complexes were moved beyond the ligation junction by incubation with 100 μM each of ATP, GTP and CTP to form an elongation complex with 45-nt RNA (EC45). EC45 was chased with 1 mM of all four NTPs (Roche Biosciences, Palo Alto, CA).
RESULTS
Effects of expanded (GAA)n repeats on transcription in vivo
To study transcription through (GAA)n repeats of increasing lengths, they were cloned into a pACYC184-derived plasmid p185Δ downstream from an artificial trc promoter inside the cat gene (Figure 1A). This promoter is a hybrid between the 5′ part of the trp promoter and the 3′ part of the lacUV5 promoter, i.e. it is inducible by IPTG but is not a subject for catabolic repression (24). A strong terminator from the E. coli rrnB gene contributes to the stability of the cat mRNA. For the purpose of this study, we have deleted a 3′ portion of the cat gene, making the transcript approximately 500 nt shorter. To clone the longest repeats, it was crucial to achieve complete repression of the trc promoter, as the stability of the repeats appeared to decrease in transcribed areas (25). For this purpose, repeat-containing plasmids were co-transformed with the compatible pTrc99A plasmid (24) carrying lacIq repressor gene.
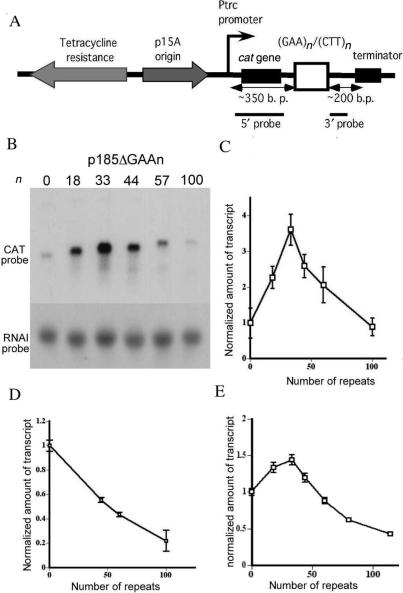
Figure 1.
Effect of expanded (GAA)n repeats on RNA synthesis in E. coli. (A) Structure of the p185Δ plasmid, carrying (GAA)n·(TTC)n repeats in different orientations. (B) Northern blot analysis of GAA-containing mRNAs from the wild-type E. coli cells. Total RNA was hybridized with a probe, corresponding to either the 5′ part of the cat gene (upper panel) or RNAI, corresponding to the p15A origin (lower panel). (C) Quantitative analysis of the above data. (D) Normalized amounts of GAA-containing mRNAs from cells treated with chloramphenicol. (E) Normalized amount of the GAA-containing mRNAs from the rne(ts) mutant strain upon its cultivation at the non-permissive temperature.
Total RNA from cells containing plasmids with different (GAA)n repeats in the sense strand for transcription were separated in the denaturing agarose gel, followed by the detection of repeat-containing transcripts via northern hybridization with the probe to the 5′ part of the cat gene (Figure 1A). Since long (GAA)n repeats are known to affect plasmid replication (26), normalizing repeat-containing transcripts to the plasmid copy number was necessary. For this purpose, the same gel was hybridized with the probe specific to the repressor RNAI for the pACYC184 replication origin, the amount of which directly reflects the plasmid copy number. Our primary results are presented in Figure 1B and their quantitative analysis is shown in Figure 1C. Rather unexpectedly, lengthening of (GAA)n repeats first leads to an increase in the amount of the full-sized transcript, reaching 3-fold at 30 repeats, followed by its gradual decrease down to the control level at 100 repeats.
On the one hand, these results are consistent with the earlier observations (8,9) that an increase in the repeat's length from 30 to 100 units results in a 3-fold decrease in the amount of transcript. On the other hand, this inhibitory effect is masked in our experiment by the fact that short repeats amplify the amount of transcript relative to the control, repeat-free RNA. The latter could be due to either RNA stabilization or transcription stimulation somehow mediated by short repeats. Both processes are likely to depend on protein binding to the repeat in either RNA or DNA form. We thus decided to evaluate transcription of repeat-containing plasmids in cells that were extensively incubated with the protein synthesis inhibitor chloramphenicol.
Protein synthesis de novo is not required for the replication of our plasmids. Consequently, they rapidly amplify in the presence of chloramphenicol while the cellular protein content remains stagnant. Thus, if a protein binding to (GAA)n repeats is responsible for their stimulatory effects on transcription, one would expect this effect to diminish or disappear altogether upon chloramphenicol treatment. Our experimental data (Figure 1D) show that transcriptional stimulation, caused by short (GAA)n repeats, disappears in the presence of chloramphenicol. At the same time, transcription inhibition caused by longer repeats becomes profound, resulting in a roughly 5-fold inhibition of the RNA synthesis by the 100-repeat stretch. No truncated products were observed (data not shown), indicating that either RNA synthesis at long (GAA)n repeats was arrested rather than terminated, or was in fact terminated but the truncated products rapidly degraded.
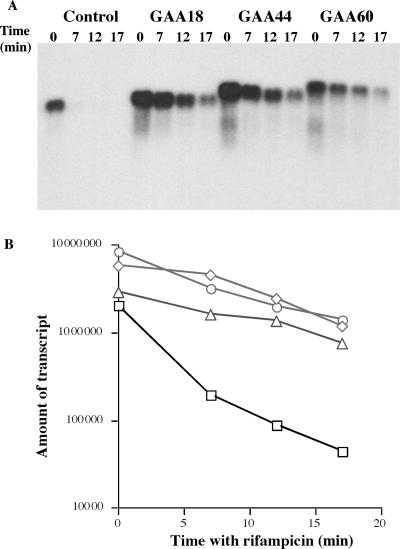
Two groups of data indicate that the elevated level of RNAs carrying short (GAA)n repeats is due to their increased stability. First, stabilities of repeat-containing and control RNAs were directly compared in conditions where transcription was arrested by rifampicin by measuring the levels of full-length messages as a function of time upon antibiotic addition. Figure 2A shows the primary experimental data and Figure 2B presents their quantitative analysis. One can clearly see that repeat-free RNA rapidly degrades, decaying by an order of magnitude within the first 7 min and as much as 50-fold after 17 min post transcription blockage. At the same time, RNA containing 18, 44, or 60 (GAA)n repeats are significantly more stable: there is only a 4- to 6-fold decay after 17 min in the presence of rifampicin. Interestingly, this stabilization does not differ significantly between 18 and 60 repeats, indicating that normal-size (GAA)n repeats are already sufficient to stabilize RNA in E. coli cells.
Figure 2.
Measuring RNA stability in rifampicin-treated cells. (A) Northern blot analysis of the control and GAA-containing RNAs at different time points upon rifampicin addition. (B) Quantitative analysis of the northern blot data. Squares—control RNA, diamonds—(GAA)18-containing RNA, circles—(GAA)44-containing RNA, triangles—(GAA)60-containing RNA.
Second, we compared control and repeat-containing transcripts in the RNase E mutant. RNase E is the key component of degradosome, which is the major player in the endonucleolytic degradation of bacterial RNA. Figure 1E shows the results of transcription experiments performed in the N3431 strain carrying a temperature-sensitive rne-3071 mutation. At the non-permissive temperature, short (GAA)n repeats lead to only a marginal increase in the amount of transcripts, indicating that amplification of the corresponding transcripts in the wild-type cells was due to the RNA stabilization. Long repeats, such as (GAA)114, cause a significant (3-fold) decrease in transcription relative to the control, apparently due to unmasking RNA stabilization in the RNase E mutant.
Altogether, our data indicate that short (GAA)n repeats stabilize RNA transcripts in E. coli whereas long repeats lead to fairly strong, albeit incomplete, transcription inhibition. Since this inhibition is particularly evident in chloramphenicol-treated cells, it is likely caused by the repeat structure rather than by protein binding to it.
Transcription through (GAA)n·(TTC)n repeats in vitro
If the interpretation of transcription data in vivo is correct, one would expect a strong inhibitory effect of (GAA)n runs on E. coli RNA polymerase in vitro. Note that the only data on the effects of (GAA)n repeats on the activity of purified transcription complexes were obtained for the T7 RNA polymerase (11,12). Since E. coli RNA polymerase belongs to the same family as mammalian RNA polymerase II, analysis of its progression through (GAA)n·(TTC)n repeats seems more relevant to human disease than that of a phage polymerase.
For these studies, we used the promoter-independent initiation with the immobilized transcription system (23). Briefly, the elongation complexes assembled on a synthetic DNA template with a 9-nt RNA primer, labeled at its 5′ end, were ligated in vitro to the DNA fragments containing (GAA)n·(TTC)n or a control sequence of a comparable length. The RNA polymerase was then ‘walked’ to the first UTP in position +46 in the presence of ATP, GTP and CTP. The resulting elongation complex was stalled just a few bases upstream from the repeat. Addition of all four NTPs allowed transcription run-off with three possible outcomes: (i) productive transcription through the repeat; (ii) transcription arrest within the repeat without RNA polymerase dissociation; or (iii) premature transcription termination inside the repeat (Figure 3). To access the transcription efficiency though the repeat, the amount of the 45-nt RNA (before chase) was compared with the amount of the run-off RNA from the complexes that successfully transcribed through the repeat.
Figure 3.
Schematic representation of the transcription experiment in vitro. Immobilized RNA polymerase binds to the RNA primer (arrow) annealed to a single-strand DNA oligonucleotide. A complementary non-template DNA is then added to the complex, creating a 3′ overhang for the repeat-containing fragment ligation. The repeat-containing fragment is excised from the plasmid and ligated to the elongation complex. After addition of ATP, CTP and GTP, the elongation complex is stalled upstream from the repeat. Three scenarios are possible after this stalled complex is released in the presence of all four NTPs: (i) successful transcription through the repeat; (ii) transcription arrest within the repeat; and (iii) transcription termination within the repeat.
Figure 4A shows that the (GAA)57 repeat in the sense strand severely inhibited transcription under these experimental conditions. While roughly 40% of starting EC45s formed run-off products upon chase with all four NTPs on the control and (TTC)n-encoding templates (Figure 4A (lanes 2 and 6) and B), only 7% of those complexes were able to complete transcription through the repetitive stretch on (GAA)n-encoding templates (Figure 4A (lane 4) and B). Overexposure of the gel revealed an array of shorter RNAs spanning the entire repetitive run (Figure 4A, lane 7). Therefore, the repeats caused either transcription arrest or termination.
Figure 4.
RNA polymerase progression is blocked by the (GAA)n repeats in vitro. (A) The immobilized elongation complexes were ligated to the linear fragment containing the repeat (GAA or TTC in the sense strand) or the control sequence. The 45-nt RNA in the starting EC and the 300-nt run-off (RO) product are indicated by arrows. (B) Quantitative analyses of data shown in (A); the fraction of the starting elongation complexes that reached the end of the template reflects the transcription efficiency. (C) The ECs obtained as in (A) were chased for 5 min with 1 mM NTPs, the KCl concentration was adjusted to 1 M, and the complexes were incubated in high salt for an additional 5 min. Then, the supernatant was removed (S, odd lanes) and loaded side-by-side with the unfractionated reaction mix (T, even lanes). The run-off products are indicated by arrows; the range of the transcripts from the TECs blocked by the repeat is shown by a red line.
It was possible to distinguish between the two alternatives because transcription was performed with the immobilized elongation complexes. In this system, transcription termination results in the release of the RNA in the supernatant. In Figure 4C, the products released in the supernatant (lanes 1, 3 and 5) were analyzed side-by-side with the unfractionated reaction mixtures (lanes 2, 4 and 6). One can see that the transcripts shorter than the run-off product, which evidently belong to the elongation complexes stopped within the (GAA)n repeat (a smear indicated by a vertical bar in Figure 4A, lane C), have not been released to the supernatant (lane 1) but remained associated with the polymerase (lane 2), thus suggesting that the (GAA)n repeat presents a block for the transcription elongation, but does not cause its premature termination. In contrast, the run-off product was completely released in supernatant, indicating that the disruption of the elongation complex results in dissociation of the transcript from the immobilized RNA polymerase.
Altogether, our in vitro data show RNA polymerase stalling by (GAA)n repeats in the sense strand for transcription and the lack of such stalling by (TTC)n repeats.
Effect of normal-size (TTC)n repeats on RNA stability in vivo
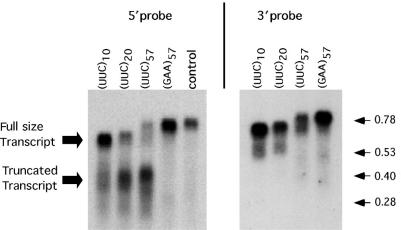
Based on the above in vitro data, one should not expect transcription inhibition by (TTC)n repeats in the sense strand for transcription. Strikingly, however, we observed that transcription of plasmids carrying normal-size (TTC)n repeats in the sense strand resulted in the appearance of specifically truncated mRNAs (Figure 5). The lengths of the shortened RNA species indicated that truncations occurred around (UUC)n repeats. Furthermore, the fraction of truncated products rapidly increased with an increase in the repeat's length whereas the fraction of the full-size transcripts concurrently decreased. This length dependence strongly suggested that (TTC)n repeats in the sense strand were indeed responsible for the RNA truncation. Hybridization of the same blots with the probe corresponding to the 3′ end of the cat mRNA (Figure 5B) revealed only full-size RNA transcripts but not the 3′ truncations. This could either be due to the lack of transcription beyond short (TTC)n runs or an endonucleolytic cleavage of the repeat-containing transcripts followed by the rapid degradation of their 3′ halves.
Figure 5.
Effect of normal-size (TTC)n repeats on RNA synthesis in E. coli cells. Northern blot analysis of UUC-containing mRNAs from the wild-type E. coli cells. Hybridization was carried out with either the 5′ or 3′ part (Figure 1A) of the repeat-containing transcript.
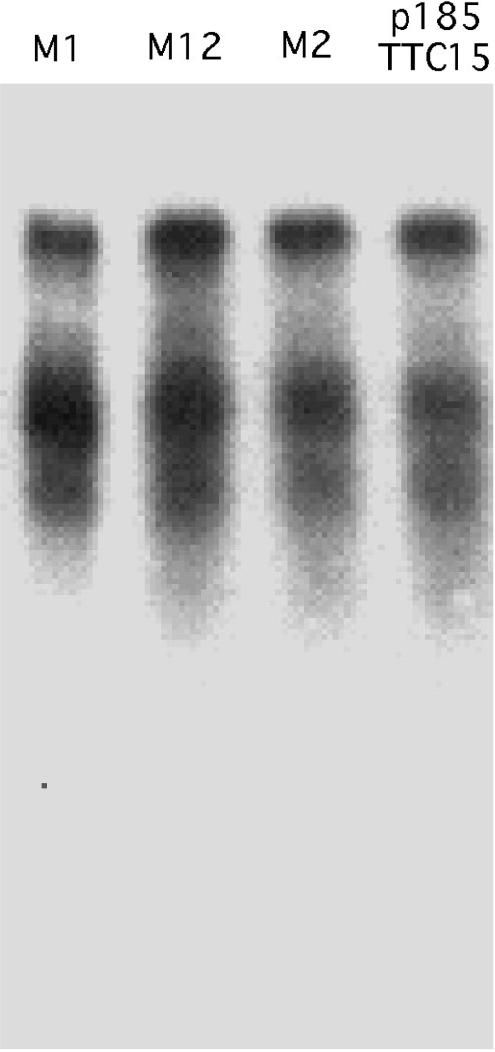
Since (GAA)n·(TTC)n repeats are capable of forming triplex DNA structures, we first analyzed whether transcript truncations in the (TTC)n orientation were due to the formation of intramolecular triplexes (H-DNA). To check this hypothesis, we used an approach based on the fact that intramolecular triplexes require the mirror symmetry from a homopurine–homopyrimidine repeat. We generated mutants M1 and M2 that carried two C-to-T transitions in either the 5′ or 3′ half of the (TTC)15 repeat, respectively, thus destroying its mirror symmetry without changing the CT content. We have also generated a mutant M12, which combined substitutions from the M1 and M2 mutants together, restoring the mirror symmetry of the repeat. Northern blot analysis of transcripts from the resultant plasmids revealed that RNA truncations at TC-rich stretches remained the same for all three mutants and was indistinguishable from that in the non-interrupted (TTC)15 stretch (Figure 6). We therefore concluded that RNA truncations did not depend on the repeat's ability to form triplex structures. Since the RNA pattern did not change even in the M12 mutant where the (TTC)n repeat was significantly disturbed, we concluded that the perfect repetitive nature of the sequence is not required for transcript truncations.
Figure 6.
RNA truncations caused by (TTC)n repeats are not due to the H-DNA formation. RNA from a plasmid with the (TTC)15 repeat and three mutant derivatives of this repeat, M1, M2 and M12, was analyzed by northern blot hybridization with the 5′ probe shown in Figure 1A.
Since we did not observe transcription inhibition in vitro for the (TTC)n-encoding template but saw a potent truncation of the UUC-containing transcripts in vivo, we strongly suspected that these truncations could be due to RNA cleavage at or within (UUC)n repeats. To study whether (UUC)n repeats within mRNAs could lead to their degradation, we have chosen a genetic approach based on the use of various RNase mutants in E. coli. Specifically, we analyzed (UUC)n-containing mRNA synthesis in strains that carried mutations in the genes encoding RNase I, RNase III, RNase H, RNase P and RNase E. Mutant and isogenic control strains were transformed with the plasmids carrying (TTC)20 repeats in the sense strand for transcription, and RNA products were analyzed by northern blot hybridization. Mutations in RNase I, RNase III, RNase H and RNase P did not have any effect on the synthesis of repeat-containing mRNA and/or accumulation of truncated RNA fragments (data not shown).
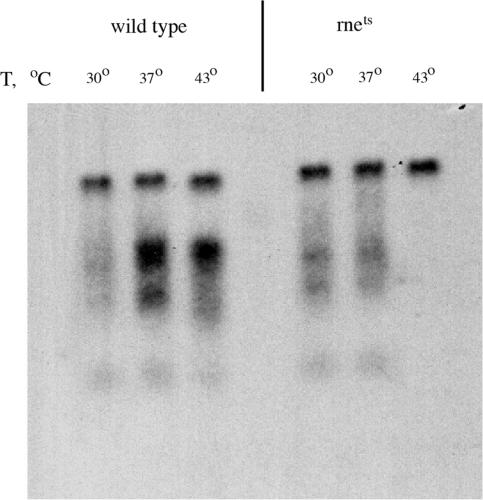
RNase E, to the contrary, appeared to be crucial for the repeat-caused RNA cleavage. Since RNase E is an essential protein, we used a temperature-sensitive rne-3071(ts) mutant and followed the fate of (UUC)n-containing RNAs upon shifting the logarithmic cells to different temperatures. Figure 7 shows a clear-cut decrease in the amount of truncated RNA fragments at the non-permissive compared to permissive temperatures. RNase E is the key component of the so-called degradosome, which is involved in the endonucleolytic degradation of bacterial mRNA. We believe, therefore, that RNA cleavage within (UUC)n repeats by the bacterial degradosome is responsible for the accumulation of truncated RNAs, which could be mistaken for the products of premature transcription attenuation.
Figure 7.
RNase E is responsible for the cleavage of (UUC)n-containing RNAs. Northern blot analysis of repeat-containing RNA isolated from the JM109 (control) and N3431 (rnets) cells carrying the p185ΔCTT20 plasmid. Prior to RNA isolation, cells were incubated at permissive (30°C), semi-permissive (37°C) or non-permissive (43°C) temperatures for 1 h.
DISCUSSION
The reduced amount of transcripts carrying expanded (GAA)n repeats in the sense strand for transcription was detected in FRDA patients (8,9) and in transient transfection experiments with cultured mammalian cells (10). At the same time, expanded (GAA)n runs did not strongly affect transcription in yeast plasmids (19) or in the FRDA knock-in mice (27). The reasons for these discrepancies are unclear. Furthermore, while it is generally believed that expanded repeats block transcription elongation (9,10), other mechanisms such as changes in the chromatin structure (17) or RNA splicing can be considered.
To gain a better insight into the mechanisms of gene repression caused by FRDA repeats, we analyzed the effects of (GAA)n·(TTC)n repeats of varying lengths and orientations within an E. coli plasmid on transcription. The advantage of this system is that in the lack of chromatin structure and RNA splicing, the results are easier to interpret in terms of transcription and RNA stability.
Rather unexpectedly, we have found that progressive lengthening of (GAA)n runs in the sense strand for transcription first leads to an increase followed by a decrease in the amount of the repeat-containing transcript (Figure 1C). The initial increase was completely eliminated upon chloramphenicol treatment of E. coli cells (Figure 1D), indicating that it was protein mediated. Furthermore, a much more modest increase was observed in the RNase E mutant (Figure 1E). These data suggested that short (18 < n < 50) (GAA)n repeats stabilize RNA transcripts in E. coli cells. We have proven this suggestion by measuring stabilities of control and repeat-containing RNA in cells treated with the transcription inhibitor rifampicin (Figure 2). While the reasons of this stabilization remain to be unraveled, an increase in the mRNA levels at carrier-size repeat lengths was also reported for other expansion diseases (28–30).
Expanded (n > 50) (GAA)n repeats in the sense strand for transcription significantly decreased transcription efficiency. This effect was particularly pronounced in cells treated with chloramphenicol or in the RNase E mutant (Figure 1D and E). Furthermore, the strength of transcription inhibition increased proportionally to the repeat's length. These results are highly indicative that the repeat's secondary structure, rather than protein binding to it, is responsible for the transcription inhibition.
This assumption is further supported by our in vitro studies of the E. coli RNA polymerase. Transcription was notably delayed within (GAA)n runs in linear templates. At the same time, premature termination within a repetitive run did not occur in vitro or in vivo, suggesting that the polymerase's progression was arrested within the repeat but not aborted altogether. Note that transcription inhibition in vitro was significantly stronger than that observed in vivo: the same (GAA)57 repeat inhibits transcription 6-fold in vitro but only 2-fold in chloramphenicol-treated cells. While the reasons for this discrepancy are unknown, we believe that accessory transcription factors facilitate the RNA polymerase progression through repetitive DNA.
By-and-large, our data on the effects of (GAA)n runs on transcription are in line with the previous hypotheses implicating unusual secondary structures of FRDA repeats in transcription attenuation (9–12,31). The exact structure responsible for this transcription slowing remains to be elucidated. (GAA)n·(TTC)n runs belong to the class of homopurine–homopyrimidine mirror repeats that are capable of forming intramolecular DNA triplexes, such as H-DNA (32) or ‘sticky’ DNA (33,34). Formation of intramolecular triplexes requires a local unwinding of the DNA, which appears naturally during the course of transcription. If the non-template single-stranded DNA in the transcription bubble contains (GAA)n repeats, it can fold back to form the triplex-like structure, trapping elongating RNA polymerase (11). Another structure that could contribute to transcription stalling is a quasi hairpin built of (GAA)n repeats (35). It can form a roadblock in DNA in front of the elongating RNA polymerase or attenuate the RNA polymerase progression by forming in the newly synthesized RNA. The structural model is also consistent with our observations that chloramphenicol treatment amplifies repeat-caused transcription attenuation in vivo. Chloramphenicol treatment is known to increase plasmid DNA supercoiling in E. coli (36) that in turn facilitates formation of DNA triplexes (37).
A totally unexpected phenomenon observed in the course of our studies is that positioning normal-size (TTC)n runs into the sense strand for transcription results in the degradation of (UUC)n-containing mRNAs in E. coli. This degradation was significantly reduced in the conditionally lethal RNase E mutant at the non-permissive temperature (Figure 6). RNase E is an integral component of the bacterial degradosome, which is crucial for the initial stages of the decay of the majority of mRNAs in E. coli (38). We therefore conclude that (UUC)n stretches serve as targets for the bacterial degradosome. The sequence specificity of RNase E is still unclear. Early efforts to determine the substrate specificity resulted in the following consensus sequence: RAUUU/A (39). Subsequent analysis revealed that RNase E prefers single-stranded AU-rich RNA regions (40). This study points to another motif, (UUC)n: 10 repeats were enough to trigger mRNA decay, and it became highly significant at 20 repeats. These data point to a potential mechanism for gene repression by expandable repeats via accelerated RNA degradation.
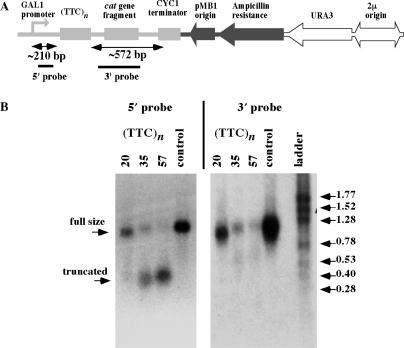
Is this observation relevant for eukaryotic systems? To address this question, we analyzed the effect of (TTC)n repeats on RNA degradation in yeast cells. For this purpose, (GAA)n·(TTC)n repeats of different lengths or a control 188-bp non-repetitive sequence were cloned downstream of the GAL1 promoter such that (TTC)n runs appeared in the sense strand for transcription (Figure 8A). Upon transcription induction by the galactose, total RNA was isolated and analyzed by northern hybridization. Hybridization with the probe, corresponding to the 5′ part of the transcript, revealed the accumulation of short transcription products evidently truncated at the position of the (UUC)n repeat (Figure 8B). The accumulation of these truncations was clearly dependent on the repeat's length; they were non-existent at 20 repeats, evident at 35 repeats and became the main RNA species at 60 repeats. Hybridization with the 3′ probe revealed traces of RNAs, whose lengths corresponded to the distance from the (UUC)n run to the transcription terminator. We therefore believe that RNA truncations are likely caused by the UUC-mediated RNA degradation in yeast, similar to what was observed in bacteria. Since there is no known homolog of RNase E in yeast, it would be of prime interest to identify yeast RNase(s) responsible for this degradation. While UUC-mediated RNA degradation has not yet been observed in cultured mammalian cells (10), our data warrant revisiting this issue. It would also be interesting to study whether other pyrimidine-rich expandable repeats cause RNA degradation in vivo.
Figure 8.
Effects of (TTC)n repeats on RNA expression in yeast S. cerevisiae. (A) The scheme of linearized yeast reporter plasmid showing a yeast transcription cassette (shown in light grey) consisting of GAL1 promoter, 210-bp 5′ part, (TTC)n repeat, and 572-bp 3′ part. 5′ and 3′ probes for northern hybridization are shown. (B) Northern hybridization analysis of the repeat-containing and control RNA transcripts.
ACKNOWLEDGEMENTS
This work was supported by grant GM60987 from NIH to S.M.M. and grants from FARA and MDA foundations to M.M.K. Funding to pay the Open Access publication charge was provided by NIH grant GM60987.
Conflict of interest statement. None declared.
REFERENCES
- 1.Romeo G, Menozzi P, Ferlini A, Fadda S, Di Donato S, Uziel G, Lucci B, Capodaglio L, Filla A, Campanella G. Incidence of Friedreich ataxia in Italy estimated from consanguineous marriages. Am. J. Hum. Genet. 1983;35:523–529. [PMC free article] [PubMed] [Google Scholar]
- 2.Cossee M, Schmitt M, Campuzano V, Reutenauer L, Moutou C, Mandel JL, Koenig M. Evolution of the Friedreich's ataxia trinucleotide repeat expansion: founder effect and premutations. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7452–7457. doi: 10.1073/pnas.94.14.7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harding AE. Friedreich's ataxia: a clinical and genetic study of 90 families with an analysis of early diagnostic criteria and intrafamilial clustering of clinical features. Brain. 1981;104:589–620. doi: 10.1093/brain/104.3.589. [DOI] [PubMed] [Google Scholar]
- 4.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 5.Gerber J, Muhlenhoff U, Lill R. An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 2003;4:906–911. doi: 10.1038/sj.embor.embor918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montermini L, Andermann E, Labuda M, Richter A, Pandolfo M, Cavalcanti F, Pianese L, Iodice L, Farina G, et al. The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum. Mol. Genet. 1997;6:1261–1266. doi: 10.1093/hmg/6.8.1261. [DOI] [PubMed] [Google Scholar]
- 7.Pandolfo M. The molecular basis of Friedreich ataxia. Adv. Exp. Med. Biol. 2002;516:99–118. doi: 10.1007/978-1-4615-0117-6_5. [DOI] [PubMed] [Google Scholar]
- 8.Campuzano V, Montermini L, Lutz Y, Cova L, Hindelag C, Jiralerspong S, Trottier Y, Kish SJ, Faucheux B, et al. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Genet. 1997;6:1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- 9.Bidichandani SI, Ashizawa T, Patel PI. The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am. J. Hum. Genet. 1998;62:111–121. doi: 10.1086/301680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohshima K, Montermini L, Wells RD, Pandolfo M. Inhibitory effects of expanded GAA.TTC triplet repeats from intron I of the Freidreich's ataxia gene on transcription and replication in vivo. J. Biol. Chem. 1998;273:14588–14595. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- 11.Grabczyk E, Usdin K. The GAA*TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakamoto N, Ohshima K, Montermini L, Pandolfo M, Wells RD. Sticky DNA, a self-associated complex formed at long GAA*TTC repeats in intron 1 of the frataxin gene, inhibits transcription. J. Biol. Chem. 2001;276:27171–27177. doi: 10.1074/jbc.M101879200. [DOI] [PubMed] [Google Scholar]
- 13.Gacy AM, Goellner GM, Spiro C, Chen X, Gupta G, Bradbury EM, Dyer RB, Mikesell MJ, Yao JZ, et al. GAA instability in Friedreich's ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol. Cell. 1998;1:583–593. doi: 10.1016/s1097-2765(00)80058-1. [DOI] [PubMed] [Google Scholar]
- 14.Potaman VN, Oussatcheva EA, Lyubchenko YL, Shlyakhtenko LS, Bidichandani SI, Ashizawa T, Sinden RR. Length-dependent structure formation in Friedreich ataxia (GAA)n*(TTC)n repeats at neutral pH. Nucleic Acids Res. 2004;32:1224–1231. doi: 10.1093/nar/gkh274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grabczyk E, Fishman MC. A long purine-pyrimidine homopolymer acts as a transcriptional diode. J. Biol. Chem. 1995;270:1791–1797. doi: 10.1074/jbc.270.4.1791. [DOI] [PubMed] [Google Scholar]
- 16.Mèaueler W, Bassili G, Hardt C, Keyl HG, Epplen JT. A complex containing at least one zinc dependent HeLa nuclear protein binds to the intronic (gaa)(n) block of the frataxin gene. Gene. 2001;270:131–143. doi: 10.1016/s0378-1119(01)00487-5. [DOI] [PubMed] [Google Scholar]
- 17.Saveliev A, Everett C, Sharpe T, Webster Z, Festenstein R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–913. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- 18.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat. Chem. Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 19.Krasilnikova MM, Mirkin SM. Replication stalling at Friedreich's ataxia (GAA)n repeats in vivo. Mol. Cell. Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krasilnikov AS, Podtelezhnikov A, Vologodskii A, Mirkin SM. Large-scale effects of transcriptional DNA supercoiling in vivo. J. Mol. Biol. 1999;292:1149–1160. doi: 10.1006/jmbi.1999.3117. [DOI] [PubMed] [Google Scholar]
- 21.Krasilnikova MM, Samadashwily GM, Krasilnikov AS, Mirkin SM. Transcription through a simple DNA repeat blocks replication elongation. EMBO J. 1998;17:5095–5102. doi: 10.1093/emboj/17.17.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Komissarova N, Kireeva ML, Becker J, Sidorenkov I, Kashlev M. Engineering of elongation complexes of bacterial and yeast RNA polymerases. Methods Enzymol. 2003;371:233–251. doi: 10.1016/S0076-6879(03)71017-9. [DOI] [PubMed] [Google Scholar]
- 24.Amann E, Ochs B, Abel KJ. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 25.Napierala M, Bacolla A, Wells RD. Increased negative superhelical density in vivo enhances the genetic instability of triplet repeat sequences. J. Biol. Chem. 2005;280:37366–37376. doi: 10.1074/jbc.M508065200. [DOI] [PubMed] [Google Scholar]
- 26.Pollard LM, Sharma R, Gâomez M, Shah S, Delatycki MB, Pianese L, Monticelli A, Keats BJ, Bidichandani SI. Replication-mediated instability of the GAA triplet repeat mutation in Friedreich ataxia. Nucleic Acids Res. 2004;32:5962–5971. doi: 10.1093/nar/gkh933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Mahdawi S, Pinto RM, Ruddle P, Carroll C, Webster Z, Pook M. GAA repeat instability in Friedreich ataxia YAC transgenic mice. Genomics. 2004;84:301–310. doi: 10.1016/j.ygeno.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, Hagerman PJ. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raca G, Siyanova EY, McMurray CT, Mirkin SM. Expansion of the (CTG)n repeat at a reporter's gene 5′-UTR impedes translation. Nucleic Acids Res. 2000;28:3943–3949. doi: 10.1093/nar/28.20.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenneson A, Zhang F, Hagedorn CH, Warren ST. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum. Mol. Genet. 2001;10:1449–1454. doi: 10.1093/hmg/10.14.1449. [DOI] [PubMed] [Google Scholar]
- 31.Grabczyk E, Usdin K. The GAA*TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirkin SM, Frank-Kamenetskii MD. H-DNA and related structures. Annu. Rev. Biophys. Biomol. Struct. 1994;23:541–576. doi: 10.1146/annurev.bb.23.060194.002545. [DOI] [PubMed] [Google Scholar]
- 33.Vetcher AA, Napierala M, Iyer RR, Chastain PD, Griffith JD, Wells RD. Sticky DNA, a long GAA center dot GAA center dot TTC triplex that is formed intramolecularly, in the sequence of intron 1 of the frataxin gene. J. Biol. Chem. 2002;277:39217–39227. doi: 10.1074/jbc.M205209200. [DOI] [PubMed] [Google Scholar]
- 34.Sakamoto N, Chastain PD, Parniewski P, Oshima K, Pandolfo M, Griffith JD, Wells RD. Sticky DNA: self association properties of long GAA.TTC repeats in R.R.Y triplex structures from Freidreich's ataxia. Mol. Cell. 1999;3:465–475. doi: 10.1016/s1097-2765(00)80474-8. [DOI] [PubMed] [Google Scholar]
- 35.Heidenfelder BL, Makhov AM, Topal MD. Hairpin formation in Friedreich's ataxia triplet repeat expansion. J. Biol. Chem. 2003;278:2425–2431. doi: 10.1074/jbc.M210643200. [DOI] [PubMed] [Google Scholar]
- 36.Dayn A, Malkhosyan S, Duzhy D, Lyamichev V, Panchenko Y, Mirkin S. Formation of (dA-dT)n cruciforms in Escherichia coli cells under different environmental conditions. J. Bacteriol. 1991;173:2658–2664. doi: 10.1128/jb.173.8.2658-2664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohwi Y, Malkhosyan SR, Kohwi-Shigematsu T. Intramolecular dG.dCdC triplex detected in Escherichia coli cells. J. Mol. Biol. 1992;223:817–822. doi: 10.1016/0022-2836(92)90242-c. [DOI] [PubMed] [Google Scholar]
- 38.Kushner SR. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 2002;184:4658–4665. doi: 10.1128/JB.184.17.4658-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ehretsmann CP, Carpousis AJ, Krisch HM. Specificity of Escherichia coli endoribonuclease RNase E: in vivo and in vitro analysis of mutants in a bacteriophage T4 mRNA processing site. Genes Dev. 1992;6:149–159. doi: 10.1101/gad.6.1.149. [DOI] [PubMed] [Google Scholar]
- 40.Rauhut R, Klug G. mRNA degradation in bacteria. FEMS Microbiol. Rev. 1999;23:353–370. doi: 10.1111/j.1574-6976.1999.tb00404.x. [DOI] [PubMed] [Google Scholar]