Abstract
The E. coli protein StpA has RNA annealing and strand displacement activities and it promotes folding of RNAs by loosening their structures. To understand the mode of action of StpA, we analysed the relationship of its RNA chaperone activity to its RNA-binding properties. For acceleration of annealing of two short RNAs, StpA binds both molecules simultaneously, showing that annealing is promoted by crowding. StpA binds weakly to RNA with a preference for unstructured molecules. Binding of StpA to RNA is strongly dependent on the ionic strength, suggesting that the interactions are mainly electrostatic. A mutant variant of the protein, with a glycine to valine change in the nucleic-acid-binding domain, displays weaker RNA binding but higher RNA chaperone activity. This suggests that the RNA chaperone activity of StpA results from weak and transient interactions rather than from tight binding to RNA. We further discuss the role that structural disorder in proteins may play in chaperoning RNA folding, using bioinformatic sequence analysis tools, and provide evidence for the importance of conformational disorder and local structural preformation of chaperone nucleic-acid-binding sites.
INTRODUCTION
Functional RNA molecules have to fold into defined secondary and tertiary structures to reach their active conformation. The RNA folding process can be slow due to stable intermediate folding states, which may represent kinetic traps, or inefficient due to several alternative conformations of comparable structural stability. While many RNAs can reach their active conformation in vitro without the help of proteins, we must assume that in vivo, most RNAs are assisted in their folding and assembly processes by a large variety of proteins (1,2). Such proteins can either recognize specific features of the RNA and thereby stabilize a defined structure or, alternatively, help RNA molecules to fold by resolving kinetically trapped, misfolded conformers. The latter action has been termed RNA chaperone activity (3–5). RNA chaperone activity neither requires a sequence-specific interaction with RNAs nor the recognition of a specific structure (5–7). Another important feature of these proteins is that they do not require ATP or any other energy-providing cofactor for their activity.
A large number of proteins have been identified that display RNA chaperone activity. Some well-characterized examples are the nucleocapsid proteins of HIV-1 (8–10), the E. coli protein StpA (11–14), the cold shock proteins CspA and CspE (15–17), the fragile X mental retardation protein FMRP (18) or hnRNP A1 (19,20). The majority of the proteins with RNA chaperone activity have multiple functions that are often connected with structural transitions of RNAs or the promotion of RNA–RNA interactions. The hnRNP A1 for example, which binds to nascent pre-mRNAs and assists in processing, or the HIV-1 nucleocapsid protein NCp7, which binds to HIV RNA and promotes annealing of the tRNA primer to the primer-binding site, reverse transcription or dimerization of the HIV genome (for reviews see 2,5,21).
The E. coli protein StpA has been identified as a suppressor of a splicing-defective mutant of the phage T4 thymidylate-synthase gene (22). StpA shows high homology to the E. coli protein H-NS, which is involved in nucleoid structure formation and acts as a global transcription regulator (23). StpA shows RNA chaperone activity in a variety of assays. The protein promotes RNA annealing, RNA strand displacement as well as trans- and cis-splicing of the td group I intron (11,24,25). In accordance with the definition of RNA chaperones, StpA can be removed after the folding event is completed without the RNA loosing its native fold (11). The RNA chaperone activity of StpA has also been demonstrated in vivo using a splicing assay that takes advantage of an aberrant exon–intron interaction in the pre-mRNA of the phage T4 thymidylate-synthase gene (26). The presence of StpA leads to an increased accessibility of bases in tertiary structure elements to the methylating agent dimethylsulfate (DMS). By loosening the structure of the RNA, the chaperone enables further conformational searches and refolding of the RNA into a splicing competent structure. This mechanism is significantly different from the tertiary structure stabilization mechanism of the specific group I intron splicing factor Cyt-18 that leads to a compaction of the intron structure by binding specifically to a defined structural element of the intron (13,27,28). The mechanism of action of StpA was further underlined by the observation that intron mutants with a destabilized tertiary structure are sensitive to StpA, suggesting that structural stability of the RNA determines whether StpA is beneficial or detrimental to folding (14). Thus, the RNA chaperone activity of proteins can serve as a quality control for sensing misfolded RNA molecules.
In the work presented here, we address the question how StpA binds to RNA, what type of RNAs StpA prefers, structured or unstructured RNAs, and how binding correlates with RNA chaperone activity. Previous experiments aiming at mapping the StpA-binding site on the td intron RNA failed and no footprint could be detected. It is a classical concept in biochemistry that specific binding of a protein to a given RNA will result in structural stabilisation of the RNA. This leads to the question how a protein that destabilizes RNA structure interacts with the molecule. We anticipated that StpA should not bind tightly to RNA. Indeed, we find that StpA binds weakly to RNA with a preference for unstructured molecules and that a mutation in the nucleic-acid-binding domain that decreases RNA-binding efficiency enhances the RNA chaperone activity. These observations suggest that RNA chaperone activity arises from transient interactions rather than from tight binding.
EXPERIMENTAL PROCEDURES
Plasmids and cloning
All short-exon constructs of the phage T4 thymidylate-synthase pre-mRNA were generated using the vector pTZ18U/tdP6Δ2 as a template for PCR. The plasmid carries the coding sequence of the T4 td gene comprised of exon 1 (549 nt), the td group I intron (lacking the ORF in loop 6) and exon 2 (312 nt) (29). The sequences of the primers used are 5′-GGGGTACCAACGCTCAGTAGATGTTTTC-3′, 5′-GGGGTACCTGAACTTAAATATATGGC-3′, 5′GGGTACCAGTGGCGTGATTTTGG TGG-3′, 5′-GCTCTAGAGCATTATGTTCAGATAAGG-3′, 5′-GCTCTAGAGCTAC AATATGAACTAACG-3′. The different constructs were cloned into the KpnI and XbaI cloning sites of the plasmid pTZ18U. The clones were then used for in vitro transcription to generate RNA.
The plasmid used for cloning and purification of StpA, StpA domains and the mutants was pTWIN1 (#N6951S) from the New England Biolabs IMPACT™-TWIN System (#E6950S). The coding sequences of StpA, NH2–StpA and G126V–StpA were cloned into the vector as a N-terminal fusion to the Mxe GyrA intein into the NdeI and SapI cloning sites. The C-terminal domain of StpA was cloned into the vector as a C-terminal fusion to the Ssp DnaB intein using SapI and PstI as restriction endonucleases. Primers for PCR amplification of the coding sequences were 5′-GGTGGTCATATGTCCGTAATGTTACAAAGTT-3′, 5′-GGTGGTTGCTCTTCCGCACAATAACTCTTCCGGGTTAATT-3′, 5′-GGTGGTCATATG CGCCAGCCGCGTCCGG-3′, 5′-GGTGGTTGCTCTTCCGCAGATCAGGAAATCGTCGA GAG-3′.
In vitro transcription
For the cis-splicing assay, the plasmids containing the different td pre-mRNA constructs were linearized using XbaI. About 10 µg of the linearized plasmid were transcribed under non-splicing conditions to prevent splicing of the precursor RNA. The transcription was performed by T7 RNA polymerase in 40 mM Tris-HCl pH 7.5, 2 mM spermidine, 6 mM MgCl2, 10 mM NaCl, 20 mM DTT, 3 mM ATP, 3 mM GTP, 3 mM CTP, 1 mM UTP and 30 µCi α-35S-UTP. The low Mg2+ concentration and the reaction temperature of 22°C were suboptimal in order to prevent splicing of the precursor RNA during transcription. The reaction was performed overnight and the resulting products were purified on a 5% polyacrylamide gel.
For the filter-binding studies, the different td pre-mRNAs were labelled with 32P, therefore the transcription reaction was performed in 40 mM Tris-HCl pH 7.5, 2 mM spermidine, 6 mM MgCl2, 10 mM NaCl, 20 mM DTT, 3 mM ATP, 3 mM GTP, 3 mM CTP, 0.5 mM UTP, 40 µCi α-32P-UTP and 10 µg of linearized plasmid. The reaction was performed as described above.
For filter binding, td mRNA and td ribozyme RNA were transcribed from 2.5 µg of PCR product in a buffer containing 40 mM Tris-HCl pH 7.0, 26 mM MgCl2, 3 mM spermidine, 6.6 mM of ATP, CTP and UTP, 0.25 mM GTP, 20 mM DTT, 75 µCi α-32P-GTP and T7 RNA polymerase. The reaction was carried out at 37°C for 5h.
In vitro cis-splicing assay for RNA chaperone activity
About 0.5 pM 35S-bodylabelled precursor RNA were denatured for 1 min at 95°C and cooled to 37°C. Splicing buffer containing 50 mM Tris-HCl pH 7.3, 0.4 mM spermidine and 5 mM MgCl2 (final concentration) was added followed by the respective protein as indicated in a total volume of 50 µl. Splicing was induced by the addition of 0.5 mM GTP final concentration. About 5 µl aliquots were taken after the following time points (15, 30 and 45 s; 1, 2, 5, 10, 30 and 60 min). The reactions were stopped by the addition of 5 µl RNA loading buffer containing 7 M urea and 1 mM EDTA. The samples were denatured for 1 min at 95°C prior to loading onto denaturing 5% polyacrylamide gels. The gels were quantified using a PhosphoImager and the ImageQuant software. Reaction constants and graphs were calculated using the KaleidaGraph software and the data were fit to a first-order equation with a double exponential: fraction pre-mRNA remaining = A · e(-kA · t) + B · e(-kB · t).
Protein purification
The fusion proteins (StpA-intein, NH2–StpA–intein, G126V–StpA–intein and intein–COOH–StpA) were overexpressed in BL21 (DE3) E.coli cells and the lysate was loaded onto a chitin column for affinity purification. Protein splicing was induced by increasing the DTT concentration or by a pH shift. All the steps of protein purification were performed according to the protocol (New England Biolabs IMPACT™-TWIN System). The proteins were stored in a buffer containing 50 mM Tris-HCl pH 7.5, 500 mM NaCl, 1 mM EDTA, 0.5 mM DTT and 12% glycerol.
Equilibrium filter-binding assay
32P-labelled RNA (2000–4000 cpm) was used for each binding experiment in a final concentration of 100 pM. The reactions were performed in 75 mM Tris-HCl pH 7.5, 0.4 mM spermidine, 5 mM MgCl2, 250 mM NaCl, 0.5 mM EDTA, 0.25 mM DTT and 6% glycerol in a total volume of 200 µl. The RNA was denatured for 1 min at 95°C. After cooling to 37°C the respective protein was added and binding proceeded for 10 min at 37°C. The reaction mix was then applied onto a nitrocellulose filter with a pore size of 0.25 µm (Schleicher & Schuell) and washed three times with 1 ml binding buffer. After air-drying the filter, binding of the RNA was measured by scintillation counting. For binding studies to analyse the influence of Mg2+ or Na+ concentration, the binding buffer contained the indicated concentration of the respective ion.
In vitro selection of StpA-binding RNAs using a genomic E. coli library
A representative genomic library of E. coli was constructed according to the random priming method published by Singer et al. (30). The oligonucleotides cl1 (5′-AGGGGAATTCGGAGCGGGGCAGCNNNNNNNNN-3′) and cl2 (5′-CGGGATCCTCGGGGCTGGGATGNNNNNNNNN-3′) were used for first and second strand synthesis, respectively and library fragments ranging from 50 to 500 nt in length were recovered after preparative denaturing gel electrophoresis. clFOR (5′-CCAAGTAATACGACTCACTATAGGGGAATTCGGAGCGGG-3′), containing the T7-promoter sequence (underlined) and clREV (5′-CGGGATCCTCGGGGCTG-3′) were used for transcription, reverse transcription and amplification during the selection cycles. StpA-binding RNAs were selected via filter binding. The 32P body labelled RNA pool was filtered through the membrane once (nitrocellulose, 0.2 µm pore size) in order to eliminate RNAs that bind non-specifically to the filter. Subsequently 10 µM of the flow through RNA was scintillation counted and incubated together with 1 µM StpA for 30 min at 25°C in a total volume of 100 µl. Two different binding buffers were used, either buffer A (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.8 mM MgCl2 and 0.5 mM DTT) or buffer B (10 mM Tris-HCl pH 8.0, 50 mM NaCl, 50 mM KCl, 2 mM MgCl2 and 0.5 mM DTT). After binding had proceeded, the reaction was filtered through the membrane and four washing steps with 500 µl of the respective binding buffer followed. Bound RNAs were recovered by shaking the filter at maximum rpm in 400 µl FES (7 M urea, 20 mM sodium citrate pH 5.0 and 1mM EDTA) and 500 µl phenol for 10 min at RT, phenol chloroform extraction and ethanol precipitation. The amount of recovered RNA was determined by scintillation counting prior to precipitation and the percentage of recovered RNA in relation to the input RNA was calculated. The protein Hfq served as a positive control and RNAs binding to Hfq were selected under the same conditions using buffer A as a binding buffer. Recovered RNAs were enriched via several rounds of in vitro selection (31, 32 submited for publication). Reverse transcription and amplification were performed using the QIAGEN One Step RT-PCR Kit.
FRET assay for RNA annealing and simultaneous binding of two RNAs
This set-up was modified from an RNA annealing assay published earlier (25). About 21-nt long oligoribonucleotides were obtained from VBC-Genomics (Vienna, Austria) with the 5′-ends labelled with Cy5 and Cy3, respectively. For annealing, the fully complementary oligos 21R+ (5′-Cy5-AUGUGGAAAAUCUCUAGCAGU-3′) and 21R− (5′-Cy3-ACUGCUAGAGAUUUUCCACAU-3′) were used. For dual RNA binding, the non-complementary oligos 21R+ and duplex− (5′-Cy3-CUUUCAUUGGUCGGUCUCUCC-3′) were employed. In a TECAN Genios Pro microplate reader, the two oligoribonucleotides were injected onto either annealing buffer alone (50 mM Tris-HCl pH 7.5, 3 mM gCl2 and 1 mM DTT) or onto annealing buffer with protein. The final concentration of the RNAs was 10 nM in 40 µl annealing buffer. After 2 s of shaking, the Cy3 donor fluorophore was excited at 535 nm once every second and readings were taken at the two emission wavelengths 590 and 680 nm by switching between corresponding glass filters. The reaction proceeded at 37°C for 150 s. The fluoresence resonance energy transfer (FRET) index was calculated as the ratio of acceptor to donor dye fluorescence, and values were normalized at t0. The time-resolved curves were least-square fitted with the second-order reaction equation for equimolar initial reactant concentrations: Pt = P · (1 - 1/(kobs · t + 1)); Pt = fraction annealed, P = maximum EFRET. Note that Pt is only indicative since no absolute FRET can be derived with fluorescence emissions measured with different glass filters. The curve shown is the average of three independent experiments, and kobs was calculated as the average of the three corresponding reaction rates.
Calculation of residue compactness and secondary structure
Originally, the calculation of residue-specific compactness in a protein was used as a (sub)classification tool for protein families. More information on the method and prediction plots for RNA chaperones will be available at: http://www.projects.mfpl.ac.at/rnachaperones/index.html. In brief, the calculation of residue-specific compactness (e.g. inversely related to surface exposure) was based on statistical distributions of 3D atomic coordinates extracted from a subset of the PDB database. From the 3D coordinate files, inter-residue Cα–Cα distances between amino acids A and B were extracted and stored as a function of amino acid types (A, B). Additionally, the primary sequence distance between residues A and B was taken into account (lAB). To describe the spatial neighbourhood of the two amino acids in the 3D structure of the entire protein (e.g. the way the two amino acids are embedded in the 3D fold), the pairwise distance distributions were transformed from cartesian space (distance rAB) to topological space (δAB). The obtained topological parameter δAB reflects the differential structural neighbourhood properties of the two amino acids A and B, respectively. The obtained pairwise distributions ρ(δAB, A, B,lAB) can subsequently be used to predict a topological compactness parameter Ci solely based on the primary sequence. Ci describes the spatial neighbourhood of residue i and is inversely related to residue exposure. Large values of Ci are found for residues located in stable parts of the protein and on average deeply buried in the interior of the structure. In contrast, small values are found for flexible loop regions and/or intrinsically unfolded segments of the polypeptide chain. Secondary structural features were performed using only ρ(δAB, A, B, lAB) distribution functions with small primary sequence differences (lAB < 5).
RESULTS
Pre-mRNAs with a two-nucleotide long exon 2 fold inefficiently
In order to study how the E. coli protein StpA aids the td pre-mRNA to fold, we set up a cis-splicing assay, in which StpA was essential for efficient folding. We had previously observed that exon sequences interfere with proper folding of the td intron through the formation of aberrant base pairs (33). We constructed a set of pre-mRNAs with exons of varying lengths in search of a poorly folding RNA. Pre-mRNAs with exon 1 sequences 27 (short), 113 (medium) and 216 (long) nucleotides in length in combination with exon 2 sequences 2 (short) or 48 (long) nucleotides in length were constructed and tested for their splicing efficiency in vitro (Figure 1). Folding efficiencies of these RNAs were monitored via their splicing activity, which is induced by the addition of the guanosine cofactor. While the length of exon 1 has little impact on splicing, the very short exon 2 with only two nucleotides results in inefficiently folding td pre-mRNAs (Figure 1B). Splicing kinetics show that the population of RNA molecules is heterogeneous with fractions of approximately 10, 30 or 50% of the molecules in a fast splicing conformation with a kobs of 0.4 min−1 depending on the construct. About 90, 70 or 50% of the population of molecules are trapped in slow splicing conformations with observed rates of 5 × 10−3 min−1. We chose the construct with short exons 1 and 2, termed sho-sho for further analyses of the effect of StpA on folding of the RNA.
Figure 1.
Splicing behaviour of td pre-mRNA constructs with varying exon lengths. (A) Schematic representation of the different constructs; exon 1 constructs are 216, 113 and 27 nt long; exon 2 constructs are either 2 or 48 nt long. (B) Splicing assays performed with different exon constructs show the influence of exon length on splicing and folding of the td intron. The splicing reaction is induced by the addition of GTP, in the presence of 5 mM Mg2+ at 37°C. The decrease in pre-mRNA versus time is indicated.
StpA enhances folding of the td pre-mRNA in vitro
We studied the effect of different concentrations of StpA on folding and splicing of the sho-sho RNA construct. Splicing experiments were performed in the presence of increasing amounts of StpA. The presence of StpA leads to an increase of RNA molecules in the fast splicing population (Figure 2). The higher the concentration of StpA the more RNA molecules are in a splicing competent conformation and undergo the splicing reaction fast. A maximum of about 64% of the RNAs can be shifted to the correctly folded conformation in the presence of 1.4 µM StpA. This protein concentration seems optimal for chaperone activity. Increasing the protein concentration higher than 1.4 µM leads to a less efficient promotion of folding. StpA does not interfere with the splicing reaction rates. The values for the kobs of the fast-reacting fraction of molecules stay around 0.4 min−1. Values for the second, slower rate remain about two orders of magnitude lower, at 5 × 10−3 min−1 (Table 1).
Figure 2.
StpA-induced splicing of the td pre-mRNA. (A) Splicing assay of the sho-sho RNA in the presence of increasing amounts of StpA. The reactions were performed at 37°C as described in experimental procedures. Increasing the amount of the RNA chaperone leads to an increase of the fast-reacting RNA population with a peak activity at 1.4 µM StpA. Higher StpA concentrations again reduce the activity. (B) Comparison of the splicing behaviour of the sho-sho RNA in the absence and after addition of StpA to the splicing reaction after 10 min of incubation. StpA causes a burst of activity immediately after addition.
Table 1.
Rate constants obtained from cis-splicing assays using td pre-mRNA in the short-exon context (sho-sho/exon 1 = 27 nt, exon 2 = 2 nt) in the presence of increasing amounts of StpA (0.2–2.2 µM).
| Conc. (µM) | Fast reacting | Slow reacting | ||
|---|---|---|---|---|
| % | kobs | % | kobs | |
| [min−1] | [min−1] | |||
| 0 | 34 ± 4 | 0.44 ± 0.14 | 66 ± 4 | 4.7 × 10−3 ± 1.7 × 10−3 |
| 0.2 | 35 ± 3 | 0.50 ± 0.11 | 65 ± 3 | 4.9 × 10−3 ± 1.3 × 10−3 |
| 0.4 | 38 ± 2 | 0.53 ± 0.09 | 62 ± 2 | 4.3 × 10−3 ± 1.1 × 10−3 |
| 0.6 | 43 ± 3 | 0.46 ± 0.10 | 57 ± 3 | 4.7 × 10−3 ± 1.7 × 10−3 |
| 0.8 | 47 ± 3 | 0.51 ± 0.10 | 53 ± 3 | 4.5 × 10−3 ± 1.8 × 10−3 |
| 1.0 | 50 ± 4 | 0.40 ± 0.10 | 50 ± 4 | 5.1 × 10−3 ± 2.6 × 10−3 |
| 1.2 | 54 ± 4 | 0.37 ± 0.08 | 46 ± 4 | 5.7 × 10−3 ± 3.0 × 10−3 |
| 1.4 | 64 ± 3 | 0.41 ± 0.06 | 36 ± 3 | 6.2 × 10−3 ± 2.8 × 10−3 |
| 1.6 | 56 ± 4 | 0.53 ± 0.10 | 44 ± 4 | 5.3 × 10−3 ± 2.5 × 10−3 |
| 1.8 | 51 ± 5 | 0.53 ± 0.15 | 49 ± 5 | 4.6 × 10−3 ± 3.3 × 10−3 |
| 2.0 | 54 ± 4 | 0.56 ± 0.12 | 46 ± 4 | 5.3 × 10−3 ± 2.7 × 10−3 |
| 2.2 | 54 ± 5 | 0.56 ± 0.13 | 46 ± 5 | 5.3 × 10−3 ± 3.0 × 10−3 |
The quantification follows the decrease of the amount of pre-mRNA fit to a first-order equation: pre-mRNA remaining = A · e(-kA · t) + B · e(-kB · t). A and B represent the fraction of fast- and slow-reacting RNA molecules, respectively, whereas kA and kB are the observed rate constants for each fraction.
These results suggest that there is an optimal concentration of StpA for promoting folding, and that StpA increases the proportion of RNA molecules with a fast folding conformation without altering the kobs values. This is consistent with the activities of other proteins like ribosomal protein S12, hnRNP A1 and the HIV nucleocapsid-related proteins (3,4,6,8,34,35). The effect of StpA is very fast, faster than can be monitored by manual pipetting as shown in Figure 2B. As indicated, StpA is added to the splicing reaction after 10 min, leading to a burst of activity with the same reaction rate that is observed in the reactions where StpA is added before starting splicing. These results further suggest that there are at least three types of differently folded RNA molecules. The first population splices fast with an observed reaction rate of ∼0.4 min−1. The rate of the chemical step is still not known, but it is at least 10 min−1 as has been shown earlier (36). The second is a population of slow folders with the kobs of 5 × 10−3 min−1, which are not activated by StpA. The third group of RNA molecules is at least as slow, but is refolded in the presence of StpA into a species with a splicing rate of ∼0.4 min−1.
StpA preferentially binds to unstructured RNA
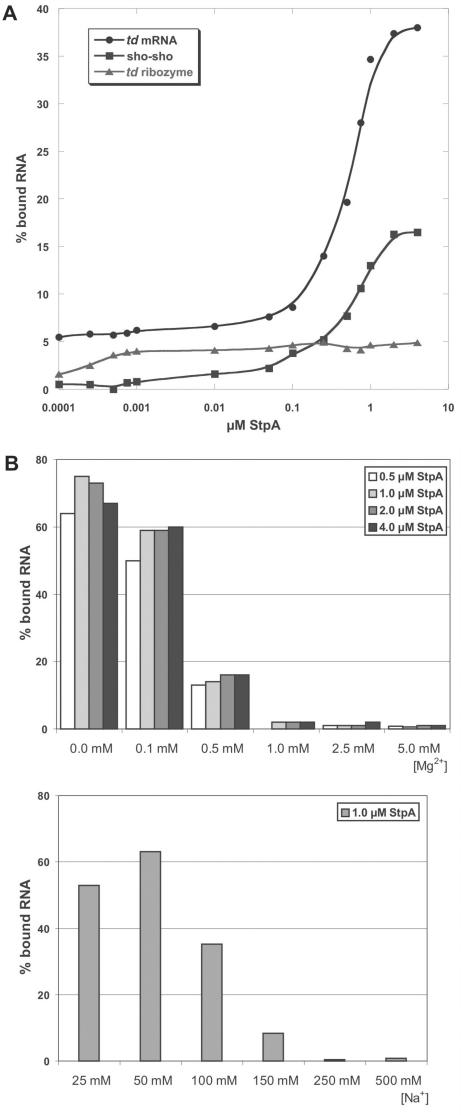
To test how well StpA binds to differently structured RNAs, we prepared three types of the td RNA: (1) the mature mRNA without the intron sequences as an example of a mostly unstructured RNA, (2) the sho-sho-type pre-mRNA with short exon sequences as an example of a mixed RNA with a structured core but with unstructured flanking sequences and (3) the ribozyme core as an example of a compactly folded RNA. Folding of this construct is well characterized and it folds into a homogenous population of molecules (37,38). As can be seen in Figure 3A, none of these RNAs bind efficiently to 4 µM StpA. Under these conditions, 38% of the mRNA, 16% of the pre-mRNA and only trace amounts of the ribozyme are bound to StpA. This clearly demonstrates that StpA preferentially binds to unstructured RNAs. The dissociation constants are 0.58 μM for the intronless mRNA and 0.73 μM for the sho-sho pre-mRNA construct. The K1/2 for the ribozyme construct could not be unambiguously determined due to inefficient binding. At 1.4 µM, the previously observed optimal protein concentration for RNA chaperone activity, binding is significant although only a small fraction of the RNA molecules is bound to the protein.
Figure 3.
RNA-binding assays (A) Different RNA constructs were incubated with increasing amounts of protein (1 pM to 4 µM). The td ribozyme construct lacks exon sequences and misses 7 nt of the 5′ end of the intron and 5 nt at the 3′ end; (B) Influence of mono- and divalent metal ions on the binding behaviour of the RNA chaperone to td ribozyme RNA. Increasing amounts of mono- and divalent ions lead to a drastic drop in the binding efficiency of StpA.
We further tested binding of StpA to a short single-stranded 21-mer RNA (21R+) and to a double-stranded 21-mer RNA (21 R+ and 21R− duplex) and to a short hairpin loop closed by a tetraloop as an example for a structured small RNA. In the filter-binding assay, the single-stranded 21-mer bound to StpA with a similar affinity as the intronless mRNA construct, the RNA duplex and the RNA hairpin loop did not bind to StpA (data not shown). This further confirms that StpA binds to unstructured or single-stranded RNA molecules.
To test the binding behaviour of StpA to td RNA under different salt conditions, equilibrium filter-binding studies with various ion concentrations were performed (Figure 3B). In these experiments, we used the td ribozyme RNA that shows only very weak binding to the protein under the conditions used in the chaperone assay. At low Mg2+ concentrations (0.1 mM), binding to StpA is efficient and 70% of the RNA is bound to the protein. By increasing the MgCl2 concentration to 0.5 mM a significant amount of the RNA is already displaced from the protein. The same effect can be seen in the presence of increasing Na+ concentrations. At the lowest concentrations, the protein binds to a high extent to the RNA (between 50 and 60%) while at 150 mM Na+ the RNA is displaced.
These effects can be caused either by the fact that at very low ion concentrations the RNA is not able to fold into its compact conformation and therefore StpA interacts with its unstructured regions. As shown before, the protein prefers binding to unstructured stretches of RNA (Figure 3A). On the other hand, the positive charges of the metal ions may simply displace the protein from the RNA. This would indicate that binding of the RNA chaperone to RNA is based mostly on electrostatic interactions to the negatively charged backbone of the RNA molecule. This is in good agreement with the fact that no footprint of StpA could be monitored in DMS protection assays (13).
Genomic selection of StpA-binding RNAs
We further asked the question if there are any endogenous StpA-binding RNAs in E. coli. In order to identify potential candidates, we performed a genomic selection. A genomic library of E. coli that contains overlapping fragments of approximately 50–500 nt in length was constructed. This library was used for several rounds of selection to enrich RNAs that bind to StpA. We used two different binding buffers for parallel selections. Buffer A was chosen to resemble physiological conditions and buffer B was adopted from Brescia et al. (39) as the authors studied the binding of rpoS mRNA and the non-coding RNA DsrA to the StpA homologue H-NS under these conditions. For the binding reaction, we used RNA (10 µM) in a tenfold molar excess over protein (1 µM). The protein Hfq, which is known to bind several RNAs of E. coli, served as a positive control for the selection procedure, and RNAs binding to Hfq were selected using buffer A. Results of the selections are shown in Figure 4. As expected, Hfq-binding RNAs were enriched over five consecutive selection cycles and up to 2.7% of the input RNA could be recovered. It should be noted that due to the tenfold molar excess of RNA over protein the maximal recovery rate is 10% assuming a single binding site. However, no StpA-binding RNAs could be enriched, neither for buffer A (data not shown) nor for buffer B and the recovery of bound RNAs remained at background levels throughout the selection procedure. We therefore assume that StpA has no specific RNA target in E. coli, at least under the tested conditions. This result stresses the non-specificity of StpA–RNA interactions and suggests that StpA is a general RNA chaperone.
Figure 4.
In vitro selection of StpA- and Hfq-binding RNAs from a genomic E. coli library. The percentage of recovered RNA in relation to the input RNA is shown for both proteins for eight consecutive selection cycles. For the Hfq, used here as a positive control, RNAs are enriched and increasing amounts of the input RNA were recovered. No StpA-binding RNAs could be selected and recovery remained at background levels throughout the procedure.
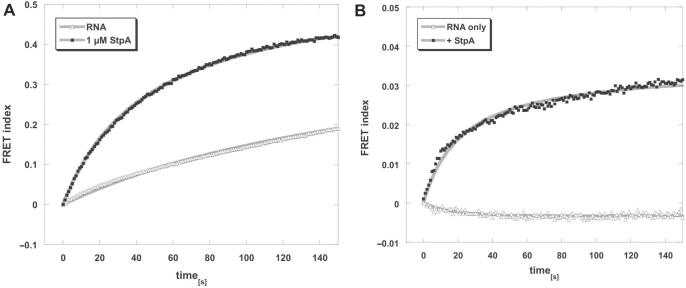
StpA binds two RNAs simultaneously to promote annealing
To further investigate the binding properties of StpA, we modified a fluorescence assay, which we used previously to show that StpA promotes annealing of short RNAs (25). In this assay, two fully complementary 21-mer RNAs labelled with Cy3 or Cy5, respectively, are co-incubated at 37°C. Upon annealing, the two fluorophores come into close vicinity and give rise to a time-resolved FRET signal, from which the reaction rate can be derived (Figure 5A). Since the two RNAs used are unstructured, they can anneal by themselves with a kobs of 0.005 s−1. The presence of 1 µM StpA accelerates this reaction 4-fold to a kobs of 0.021 s−1.
Figure 5.
StpA promotes RNA annealing by simultaneoulsly binding two RNAs. In a microplate reader, 10 nM each of two fluorophore-labelled oligoribonucleotides were injected, mixed and then incubated at 37°C in the absence or presence of 1 µM StpA. With the Cy3 donor dye excited, fluorescence emissions of Cy3 and Cy5 were measured every second, and the FRET index was calculated as a ratio of acceptor to donor fluorescence. (A) To assay RNA annealing, fully complementary RNA 21 mers (21R+, 21R−) were used that comprise no significant secondary structures and therefore can anneal by themselves. The presence of 1 µM StpA accelerates this reaction 4-fold. (B) Using a non-complementary RNA pair (21R+, duplex−) the two 21 mers alone cannot hybridize. However, with StpA present, fluorescence resonance energy transfer (FRET) takes place, indicating simultaneous binding of both RNAs to the protein. Graphs were fitted with the second-order reaction equation for equimolar initial reactant concentrations.
When one of the RNAs is changed to a non-complementary one (duplex− instead of 21R−) the RNAs can no longer anneal (Figure 5B). However, in the presence of StpA, FRET can be monitored (with a kobs of 0.028 s−1). In the absence of protein, no FRET signal can be detected when the two non-complementary RNAs are co-incubated. Although in this set-up, absolute FRET values cannot be derived, the 10 times lower FRET index of the binding reaction compared to the annealing reaction indicates either that in the binding reaction the two fluorophores are further apart than in an RNA duplex, or that only a fraction of the Cy3 and Cy5-labelled RNA population is bound at the correct ratio to yield a high FRET signal. As both RNAs bind similarly to StpA, we think that the FRET signal is derived from simultaneous binding of both RNAs, but that the two fluorophores are further apart than when they form a duplex. This strongly points to a simultaneous binding of at least two differently labelled 21-mer RNAs by StpA. Other proteins such as E. coli ribosomal protein S1, which cannot promote RNA annealing, do not show this effect (data not shown).
This observation supports the ‘RNA crowding’ mechanism for unspecific RNA annealing activity of StpA: the simultaneous binding of two RNAs will lead to an increase of their local concentration, and thereby, the annealing of complementary RNA strands is promoted.
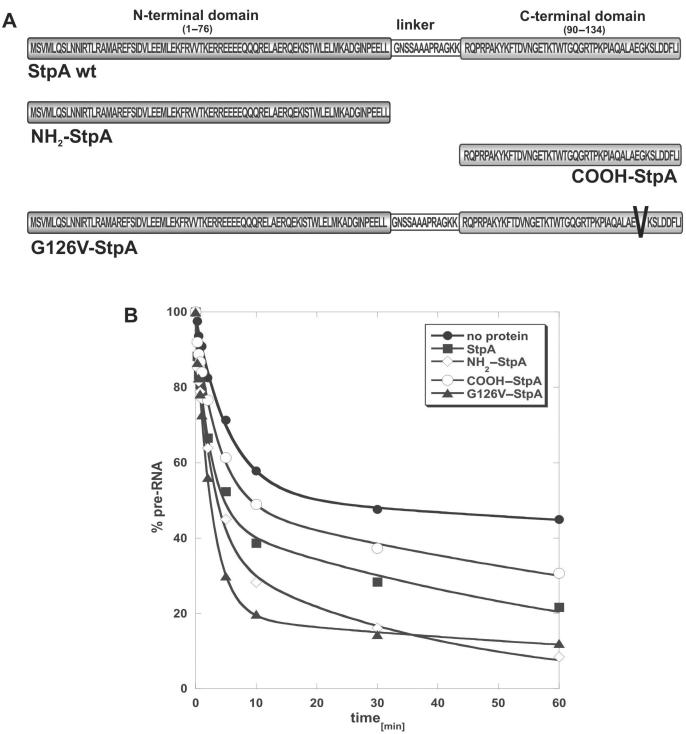
A StpA mutant with an altered nucleic-acid-binding domain has decreased RNA-binding efficiency but increased RNA chaperone activity
Like its paralogue H-NS, StpA has a two-domain structure (40). The negatively charged N-terminal region is responsible for dimerization, whereas the positively charged C-terminus is needed for nucleic acid binding. The two domains are interconnected by a flexible linker region (Figure 6A) (42). The C-terminal domain had previously been reported to exert RNA annealing activity and also to promote trans-splicing in vitro (40). To map the RNA chaperone activity of StpA, we purified both domains separately (Figure 6A). We performed cis-splicing assays with the N-terminal and C-terminal domains separately and compared the activities to that of the full-length protein. We were able to confirm that the C-terminal domain shows RNA chaperone activity although to a lesser extent than the full-length protein. The N-terminal domain also showed RNA chaperone activity in the cis-splicing assay (Figure 6B and Table 2). The kobs value of the second phase is raised fourfold in the presence of NH2–StpA, suggesting that it can activate the population of molecules that is not induced by the full-length protein. Furthermore, from the X-ray crystal structure of the N-terminal domain of an H-NS homologue, it has recently been proposed that the N-terminal domain contributes to DNA recognition (42).
Figure 6.
StpA mutant and domains. (A) Schematic representation of the different protein constructs used to specify the regions responsible for the RNA chaperone activity of StpA. The N-terminal (NH2–StpA) and the C-terminal domains (COOH–StpA) as well as a mutant with a glycine to valine change at position 126 (G126V–StpA) in the DNA-binding domain were prepared. (B) In splicing assays, 1.4 µM of the different protein constructs were tested for RNA chaperone activity with 0.5 pM of the sho-sho pre-mRNA. The graphs show the decrease of pre-mRNA during splicing.
Table 2.
Influence of various forms of StpA (1.4 μM protein) on splicing and folding of the sho-sho RNA construct.
| Fast reacting | Slow reacting | |||
|---|---|---|---|---|
| % | kobs | % | kobs | |
| [min−1] | [min−1] | |||
| no protein | 35 ± 3 | 0.42 ± 0.12 | 65 ± 3 | 6.7 × 10−3 ± 1.5 × 10−3 |
| StpA | 63 ± 4 | 0.40 ± 0.04 | 37 ± 4 | 7.2 × 10−3 ± 4.5 × 10−3 |
| NH2–StpA | 57 ± 7 | 0.35 ± 0.10 | 43 ± 7 | 2.6 × 10−2 ± 8.8 × 10−3 |
| COOH–StpA | 47 ± 4 | 0.26 ± 0.04 | 53 ± 4 | 7.3 × 10−3 ± 2.0 × 10−3 |
| G126V–StpA | 78 ± 3 | 0.38 ± 0.03 | 22 ± 3 | 5.2 × 10−3 ± 4.2 × 10−3 |
Values and reaction rates were calculated as described in experimental procedures and in Table 1.
A mutant of StpA with a single amino acid alteration in the C-terminal nucleic-acid-binding domain, a glycine to valine change at position 126, was further analysed for RNA-binding and RNA chaperone activity (Figure 6A). Glycine 126 of E. coli StpA is a highly conserved amino acid in various H-NS and StpA homologues of different organisms and is part of a consensus region (GKSLDDFLI) in the very C-terminal part of the protein (39,42). We performed equilibrium filter-binding experiments with wt StpA and the G126V–StpA mutant using the construct with short exons, which is also used in the cis-splicing assay. While 17% of the RNA was bound to wild-type StpA, no binding to the RNA was detected for the mutant protein. Even raising the protein concentration of G126V–StpA 10 times over the concentration where RNA chaperone activity is detected did not result in binding (Figure 7A). Binding of StpA wild-type and the G126V–StpA mutant was also tested with an intron mutant variant, tdC865U, which has a disturbed base triple interaction in the intron core resulting in a molecule with destabilized tertiary structure (14). In Figure 7B, the results of the filter-binding experiments show that both wild type and G126V–StpA bind more efficiently to the mutant intron than to the wild-type intron. Approximately 27% of the tdC865U pre-mRNA are bound to wild-type StpA and 7% to G126V–StpA. This indicates that G126V–StpA retains RNA-binding affinity to some extent, preferentially binding to less well-folded RNA. These results further confirm that StpA prefers to interact with more flexible RNA molecules. The better an RNA is folded, the less efficient is its interaction with StpA. Furthermore, amino acid Q126 in H-NS, which is immediately upstream of the G127 (corresponding to G126 in StpA), showed a strong chemical shift in NMR spectra in the presence of DNA, stressing its involvement in nucleic acid interaction (44).
Figure 7.
Equilibrium filter-binding assays with wt and mutant StpA. (A) Increasing amounts of wt StpA or G126V–StpA mutant (50 pM to 15 µM) were bound to 100 pM of sho-sho pre-mRNA construct of wild-type intron sequence or (B) to a sho-sho pre-mRNA construct with a mutation (C865U) in the joining region of J3/4 of the intron RNA. This mutant has a destabilized tertiary structure (14).
The G126V–StpA mutant has a higher RNA chaperone activity than wild-type StpA. In the cis-splicing assay, G126V–StpA promotes folding of about 80% of the RNA molecules (Table 2). Compared to wt StpA and the single-protein domains, G126V–StpA is the most active RNA chaperone (Figure 6B). The change from a glycine to valine results in a shift from a small and neutral amino acid in respect to its function to a bulky and hydrophobic one. The position 126 in StpA lies next to the region responsible for DNA binding (44). The insertion of a hydrophobic and bulky moiety at this position might disturb the nucleic-acid-binding domain of the protein.
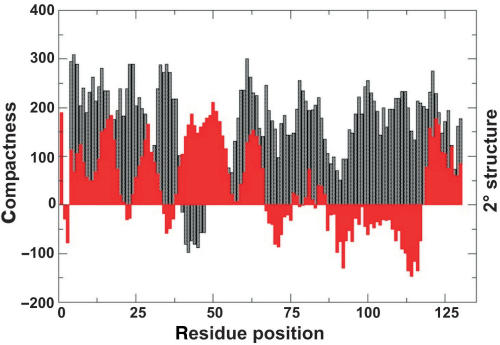
In silico analysis suggests that StpA is a highly disordered protein
Recently, Tompa and Csermely (45) reported that proteins with RNA chaperone activity belong to the class of highly unstructured proteins. We further addressed this question, because structural disorder would be a good candidate property for explaining the mode of interaction of StpA with RNA. Figure 8 shows the in silico prediction for StpA of residue compactness and local secondary structure as a function of residue position. The average compactness value of StpA (or average residue exposure (ARE)) is about 160, which is considerably smaller than average ARE values of stably folded proteins. For example, the average residue exposure of the proteins stored in the PDB database was found to be about 300 (R.K., unpublished results). The considerably smaller ARE value found for StpA is consistent with the notion that conformational disorder plays a significant role in RNA chaperoning interactions (45). The conformational disorder for StpA was found to be 73.13% using the PONDR algorithm. In contrast to existing disorder predictors, however, our algorithm provides residue-specific information about residue compactness (indirectly related to solvent exposure) and local secondary structural features. It thus allows for a per residue analysis of local structural preformation and the availability of individual residues for putative intermolecular interactions with nucleic acids. Interestingly, the location of secondary structure elements convincingly agrees with structural features recently observed by heteronuclear NMR spectroscopy for the StpA homologue H-NS (44). Specifically, the extended or β-strand conformation between residues Y97 and W109 as well as the C-terminal α-helical segments was reproduced by the prediction algorithm. Additionally, consistently smaller compactness values were observed for residues 80–115 for which experimental NMR chemical shift changes indicated an involvement in DNA binding (44). The N-terminal part of StpA comprises three α-helices between residues α1:1–19 (with a slight bend), α2:26–33 and α3:41–53. In the crystal structure of H-NS, the α-helices α1 and α2 comprise the dimerization motif (42). Interestingly, residues within helix α3 also display significantly smaller compactness values that is indicative of an additional preformed interaction site.
Figure 8.
Residue compactness and secondary structure plot of StpA. Predicted compactness and local secondary structural features of StpA are shown as a function of residue position. Large compactness values (black) indicate residue positions typically buried in the interior of the 3D structure, whereas small values are found for residues exposed to the solvent. Corresponding local secondary structure elements (red) are overlayed. Positive values are indicative of α-helical segments. In contrast, continuous negative values are typical for extended or β-strand regions.
DISCUSSION
DMS modification of the td pre-RNA in the presence of StpA revealed an increased accessibility of several bases involved in the formation of tertiary structure elements but no StpA-specific footprint or protection of the bases from DMS modification could be detected. This suggested the lack of a high-affinity binding site on the td pre-mRNA (13). Nevertheless, it was clearly demonstrated that StpA accelerates folding of the td pre-mRNA both in vivo and in vitro by loosening the tertiary structure of the intron (13,24). Specific and high-affinity binding of a protein to an RNA molecule is expected to lead to an increased structural stability of the RNA. It is part of the definition of an RNA chaperone that once the RNA is correctly folded, the chaperone is no longer required for RNA function and is then displaced from the RNA (3,4). For example, Cyt-18, a protein which specifically recognizes and binds group I introns with high affinity, is required to stabilize the intron RNA throughout the splicing process (46–48). In contrast, for ribosomal protein S12 (3) and for StpA (11) it was shown that they can be removed after folding of the RNA without the RNA loosing its structure. Therefore, it is conceivable that proteins with RNA chaperone activity should not bind tightly to the RNA, especially not to natively folded RNAs. We are interested in understanding how these proteins interact with RNA leading to the promotion of folding.
Inverse correlation between RNA-binding and RNA chaperone activities of StpA
For the E. coli protein StpA, we show that a mutation in the DNA-binding domain leads to a decrease in RNA binding compared to wt StpA. On the other hand, due to this change from a neutral to a hydrophobic (glycine 126 change to valine) amino acid next to the nucleic-acid-binding region, the protein acquires a higher RNA chaperone activity. We further show that binding of StpA to RNA is inefficient, that StpA has a clear preference for unstructured RNAs and that binding is of electrostatic nature. From all these observations, we propose that there is an inverse correlation between the RNA-binding efficiency of the protein and its RNA chaperone activity. The stronger a protein interacts with the nucleic acid, the less efficient might be its ability to promote folding by structure destabilization. The low binding efficiency and affinity of StpA to RNA as well as the fact that low amounts of mono- and divalent metal ions can easily impede protein binding suggest that the interaction between StpA and RNA is weak and most probably highly transient. Furthermore, the rates at which StpA accelerates folding, which is faster than can be monitored by manual pipetting, also suggests that the interactions are rapid and transient, rather than tight and persistent.
Members of the E. coli cold shock protein family, like CspA or CspE, have been found to exert RNA chaperone activity both in vitro and in vivo (49). For both of them it has been demonstrated that mutations in the RNA-binding domains affect binding and chaperone activity. By replacing aromatic amino acids with arginine the binding affinity of CspE increases by one order of magnitude. At the same time the in vitro nucleic acid melting activity drops significantly. In vivo, these mutations lead to the loss of the ability to cause transcription antitermination (15–17,49).
The bacterial FinO protein, which represses F-plasmid conjugative transfer, facilitates sense–antisense RNA interactions by accelerating RNA strand exchange. Like StpA, FinO binds poorly to RNA with an affinity of 0.2–1 µM and mutant variants of FinO that have partially lost their RNA strand exchange acceleration ability bind with higher affinity to RNA than the wild type. Thus, like for StpA, an inverse correlation between RNA chaperone and RNA-binding activity has been observed for FinO (50).
Recently, single-molecule FRET has been used to monitor conformational dynamics during folding of the bI5 group I intron in the presence of its specific splicing factor CBP2 (51). Two different types of interaction could clearly be detected. Before CBP2 binds with high affinity to the RNA, a non-specific mode of interaction is observed, which causes conformational fluctuations in the intron RNA corresponding to various conformations. This non-specific mode of interaction is a mechanism, which could be also used by proteins with RNA chaperone activity. It is conceivable, that many proteins, which interact specifically with an RNA molecule, first undergo this type of non-specific interaction promoting conformational searching before they achieve successful specific and high-affinity binding. This might explain, why so many specific RNA-binding proteins have RNA chaperone activity in addition to specific RNA-binding properties.
RNA annealing is promoted by simultaneous binding of both RNAs
The term RNA chaperone activity is being used for a wide and not very well-defined set of activities. Using different assays to monitor RNA chaperone activity, different activities can be detected. For example, StpA can accelerate RNA annealing and strand displacement, while ribosomal protein S1 can promote strand displacement but not RNA annealing and in contrast, the E. coli protein Hfq can accelerate annealing but not strand displacement (O.M. and L.R. unpublished results). Thus it becomes evident that RNA annealing and strand displacement are different activities that require different mechanisms. While strand displacement requires RNA unfolding activity, this is not the case for RNA annealing, providing that the RNAs are not highly structured. We show here, that the mechanism of RNA annealing acceleration acts via simultaneous binding of both RNAs. Low levels of FRET demonstrated this after incubation of non-complementary fluorophore-labelled RNAs with StpA.
Protein disorder and RNA chaperone activity
A recent review by Jane Dyson and Peter Wright discusses the role of structural disorder for protein function (52). Intrinsically unstructured proteins or unstructured regions in proteins do, in contrast to the classical view, exert function. In favour of this view is also a recent model proposed by Tompa and Csermely for the mode of action of proteins with RNA chaperone activity. They observed that RNA chaperones belong to the class of proteins with the highest degree of structurally disordered regions (45). Structural flexibility and dynamics facilitate intermolecular interactions by promoting conformational fluctuation in search for tight binding. This model would fit well with the behaviour of several well-studied RNA chaperones (53). For example, the N-terminal region of the FinO protein, which is essential for strand exchange activity, was also found to be unstructured, at least in the free protein (52,54). Very recently, an algorithm for disorder prediction (DisProt VL3-14) was used to search for the RNA chaperone domain in the Drosophila retrovirus Gypsy Gag gene, and the region could be correctly predicted (55). Here, we analysed structural features of StpA using a novel tool for prediction of residue compactness and local secondary structure elements based on the primary sequence. The results obtained revealed that StpA exists in solution as a largely unfolded polypeptide chain lacking considerable stabilizing tertiary interactions. However, despite its loosely packed structure it comprises local secondary structure elements, which also display significantly smaller calculated compactness values than the average of proteins. StpA is thus prone to intermolecular interactions with possible binding partners. Most importantly, the location of these exposed preformed regions coincide with experimentally validated DNA-recognition sites found for its homologue H-NS (44). StpA thus constitutes an additional member of the growing class of intrinsically unfolded proteins with preformed molecular recognition elements.
ACKNOWLEDGEMENTS
We thank Rupert Grossberger for providing the G126V mutant of StpA and Paul Watson for his efforts to improve the language of the manuscript. Many thanks also due to Marlene Belfort for providing plasmids coding for StpA and Andrew Feig for the gift of Hfq. We are grateful to all the members of the Schroeder group for constant discussions, input and for comments on the manuscript. This project was funded by the Austrian Science Fund FWF grant F1703 to R.S. and F1719 to R.K. Funding to pay the Open Access publication charge was provided by Austrian Science Fund “FWF”.
Conflict of interest statement. None declared.
REFERENCES
- 1.Weeks KM. Protein-facilitated RNA folding. Curr. Opin. Struct. Biol. 1997;7:336–342. doi: 10.1016/s0959-440x(97)80048-6. [DOI] [PubMed] [Google Scholar]
- 2.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell. Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 3.Coetzee T, Herschlag D, Belfort M. Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes Dev. 1994;8:1575–1588. doi: 10.1101/gad.8.13.1575. [DOI] [PubMed] [Google Scholar]
- 4.Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 5.Cristofari G, Darlix JL. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- 6.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 7.Portman DS, Dreyfuss G. RNA annealing activities in HeLa nuclei. EMBO J. 1994;13:213–221. doi: 10.1002/j.1460-2075.1994.tb06251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsuchihashi Z, Khosla M, Herschlag D. Protein enhancement of hammerhead ribozyme catalysis. Science. 1993;262:99–102. doi: 10.1126/science.7692597. [DOI] [PubMed] [Google Scholar]
- 9.Urbaneja MA, Wu M, Casas-Finet JR, Karpel RL. HIV-1 nucleocapsid protein as a nucleic acid chaperone: spectroscopic study of its helix-destabilizing properties, structural binding specificity, and annealing activity. J. Mol. Biol. 2002;318:749–764. doi: 10.1016/S0022-2836(02)00043-8. [DOI] [PubMed] [Google Scholar]
- 10.Windbichler N, Werner M, Schroeder R. Kissing complex-mediated dimerisation of HIV-1 RNA: coupling extended duplex formation to ribozyme cleavage. Nucleic Acids Res. 2003;31:6419–6427. doi: 10.1093/nar/gkg873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang A, Derbyshire V, Salvo JL, Belfort M. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA. 1995;1:783–793. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang A, Rimsky S, Reaban ME, Buc H, Belfort M. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]
- 13.Waldsich C, Grossberger R, Schroeder R. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo. Genes Dev. 2002;16:2300–2312. doi: 10.1101/gad.231302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossberger R, Mayer O, Waldsich C, Semrad K, Schroeder R. Influence of RNA structural stability on the RNA chaperone activity of the E. coli portein StpA. Nucleic Acids Res. 2005;33:2280–2289. doi: 10.1093/nar/gki515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 16.Phadtare S, Inouye M, Severinov K. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 2002;277:7239–7245. doi: 10.1074/jbc.M111496200. [DOI] [PubMed] [Google Scholar]
- 17.Phadtare S, Tyagi S, Inouye M, Severinov K. Three amino acids in Escherichia coli CspE surface-exposed aromatic patch are critical for nucleic acid melting activity leading to transcription antitermination and cold acclimation of cells. J. Biol. Chem. 2002;277:46706–46711. doi: 10.1074/jbc.M208118200. [DOI] [PubMed] [Google Scholar]
- 18.Gabus C, Mazroui R, Tremblay S, Khandjian EW, Darlix JL. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Res. 2004;32:2129–2137. doi: 10.1093/nar/gkh535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pontius BW, Berg P. Rapid assembly and disassembly of complementary DNA strands through an equilibrium intermediate state mediated by A1 hnRNP protein. J. Biol. Chem. 1992;267:13815–13818. [PubMed] [Google Scholar]
- 20.Herschlag D, Khosla M, Tsuchihashi Z, Karpel RL. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer O, Windbichler N, Wank H, Schroeder R. Protein-Induced RNA Switches in Nature. In: Silvermann SK, editor. Nucleic Acid Switches and Sensors. Landes Bioscience, Georgetown, Texas, USA; 2005. [Google Scholar]
- 22.Zhang A, Belfort M. Nucleotide sequence of a newly-identified Escherichia coli gene, stpA, encoding an H-NS-like protein. Nucleic Acids Res. 1992;20:6735. doi: 10.1093/nar/20.24.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hommais F, Krin E, Laurent-Winter C, Soutourina O, Malpertuy A, Le Caer JP, Danchin A, Bertin P. Large scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleiod-associated protein, H-NS. Mol. Microbiol. 2001;40:20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 24.Mayer O, Waldsich C, Grossberger R, Schroeder R. Folding of the td pre-RNA with the help of the RNA chaperone StpA. Biochem. Soc. Trans. 2002;30:1175–1180. doi: 10.1042/bst0301175. [DOI] [PubMed] [Google Scholar]
- 25.Rajkowitsch L, Semrad K, Mayer O, Schroeder R. Assays for the RNA chaperone activity of proteins. Biochem. Soc. Trans. 2005;33:450–455. doi: 10.1042/BST0330450. [DOI] [PubMed] [Google Scholar]
- 26.Clodi E, Semrad K, Schroeder R. Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J. 1999;18:3776–3782. doi: 10.1093/emboj/18.13.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohr G, Zhang A, Gianelos JA, Belfort M, Lambowitz AM. The neurospora CYT-18 protein suppresses defects in the phage T4 td intron by stabilizing the catalytically active structure of the intron core. Cell. 1992;69:483–494. doi: 10.1016/0092-8674(92)90449-m. [DOI] [PubMed] [Google Scholar]
- 28.Guo Q, Lambowitz AM. A tyrosyl-tRNA synthetase binds specifically to the group I intron catalytic core. Genes Dev. 1992;6:1357–1372. doi: 10.1101/gad.6.8.1357. [DOI] [PubMed] [Google Scholar]
- 29.Salvo JL, Coetzee T, Belfort M. Deletion-tolerance and trans-splicing of the bacteriophage T4 td intron. Analysis of the P6-L6a region. J. Mol. Biol. 1990;211:537–549. doi: 10.1016/0022-2836(90)90264-m. [DOI] [PubMed] [Google Scholar]
- 30.Singer B, Shtatland T, Brown D, Gold L. Libraries for genomic SELEX. Nucleic Acids Res. 1997;25:781–786. doi: 10.1093/nar/25.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 32.Lorenz C, von Pelchrzim F, Schroeder R. Genomic SELEX for the identification of functional RNAs independent of their expression levels. Nature Protocols. 2006;1:2204–2212. doi: 10.1038/nprot.2006.372. [DOI] [PubMed] [Google Scholar]
- 33.Semrad K, Schroeder R. A ribosomal function is necessary for efficient splicing of the T4 phage thymidylate synthase intron in vivo. Genes Dev. 1998;12:1327–1337. doi: 10.1101/gad.12.9.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cristofari G, Ivanyi-Nagy R, Gabus C, Boulant S, Lavergne JP, Penin F, Darlix JL. The hepatitis C virus Core protein is a potent nucleic acid chaperone that directs dimerization of the viral (+) strand RNA in vitro. Nucleic Acids Res. 2004;32:2623–2631. doi: 10.1093/nar/gkh579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vo MN, Barany G, Rouzina I, Musier-Forsyth K. Mechanistic studies of mini TAR RNA/DNA annealing in the absence and presence of HIV-I nucleocapsid protein. J. Mol. Biol. 2006;363:244–261. doi: 10.1016/j.jmb.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 36.Pichler A, Schroeder R. Folding problems of the 5′ splice-site containing P1 stem of the group I td intron: substrate binding inhibition in vitro and mis-splicing in vivo. J. Biol. Chem. 2002;26:26. doi: 10.1074/jbc.M111798200. [DOI] [PubMed] [Google Scholar]
- 37.Brion P, Schroeder R, Michel F, Westhof E. Influence of specific mutations on the thermal stability of the td group I intron in vitro and on its splicing efficiency in vivo: a comparative study. RNA. 1999;5:947–958. doi: 10.1017/s1355838299990477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brion P, Michel F, Schroeder R, Westhof E. Analysis of the cooperative thermal unfolding of the td intron of bacteriophage T4. Nucleic Acids Res. 1999;27:2494–2502. doi: 10.1093/nar/27.12.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cusick ME, Belfort M. Domain structure and RNA annealing activity of the Escherichia coli regulatory protein StpA. Mol. Microbiol. 1998;28:847–857. doi: 10.1046/j.1365-2958.1998.00848.x. [DOI] [PubMed] [Google Scholar]
- 41.Rimsky S. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 2004;7:109–114. doi: 10.1016/j.mib.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Bloch V, Yang Y, Margeat E, Chavanieu A, Auge MT, Robert B, Arold S, Rimsky S, Kochoyan M. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat. Struct. Biol. 2003;10:212–218. doi: 10.1038/nsb904. [DOI] [PubMed] [Google Scholar]
- 43.Bertin P, Benhabiles N, Krin E, Laurent-Winter C, Tendeng C, Turlin E, Thomas A, Danchin A, Brasseur R. The structural and functional organization of H-NS like proteins is evolutionarily conserved in gram-negative bacteria. Mol. Microbiol. 1999;31:319–329. doi: 10.1046/j.1365-2958.1999.01176.x. [DOI] [PubMed] [Google Scholar]
- 44.Shindo H, Ohnuki A, Ginba H, Katoh E, Ueguchi C, Mizuno C, Yamazaki T. Identification of the DNA binding surface of H-NS protein from Escherichia coli by heteronuclear NMR spectroscopy. FEBS Lett. 1999;455:63–69. doi: 10.1016/s0014-5793(99)00862-5. [DOI] [PubMed] [Google Scholar]
- 45.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 46.Caprara MG, Mohr G, Lambowitz AM. A Tyrosyl-tRNA snthetase protein induces tertiary folding of the group I intron catalytic core. J. Mol. Biol. 1996;257:512–531. doi: 10.1006/jmbi.1996.0182. [DOI] [PubMed] [Google Scholar]
- 47.Caprara MG, Myers CA, Lambowitz AM. Interaction of the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase (CYT-18 protein) with the group I intron P4-P6 domain. Thermodynamic analysis and the role of metal ions. J. Mol. Biol. 2001;308:165–190. doi: 10.1006/jmbi.2001.4581. [DOI] [PubMed] [Google Scholar]
- 48.Chen X, Mohr G, Lambowitz AM. Interaction of the Neurospora crassa CYT-18 protein C-terminal RNA binding domain helps stabilize interdomain tertiary interactions in group I introns. RNA. 2001;10:634–644. doi: 10.1261/rna.5212604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bae W, Phadtare S, Severinov K, Inouye M. Characterization of Escherichia coli CspE, whose product negatively regulates transcription of CspA, the gen for the major cold shock protein. Mol. Microbiol. 1999;31:1429–1441. doi: 10.1046/j.1365-2958.1999.01284.x. [DOI] [PubMed] [Google Scholar]
- 50.Arthur DC, Ghetu AF, Gubbins MJ, Edwards RA, Frost LS, Glover JN. FinO is an RNA chaperone that facilitates sense-antisense RNA interactions. EMBO J. 2003;22:6346–6355. doi: 10.1093/emboj/cdg607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bokinsky G, Nivon LG, Liu SL, Chai G, Hong M, Weeks KW, Zhuang X. Two distinct binding modes of a protein cofactor with its target RNA. J. Mol. Biol. 2006;361:771–784. doi: 10.1016/j.jmb.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell. Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 53.Ivanyi-Nagy R, Davidovic L, Khandjian EW, Darlix JL. Disordered RNA chaperone proteins:from functions to disease. Cell. Mol. Life Sci. 2005;62:1409–1417. doi: 10.1007/s00018-005-5100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghetu AF, Arthur DC, Kerppola TK, Glover JN. Probing FinO-FinP RNA interactions by site-directed protein-RNA crosslinking and gelFRET. RNA. 2002;8:816–823. doi: 10.1017/s1355838202026730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gabus C, Ivanyi-Nagy R, Depollier J, Bucheton A, Pelisson A, Darlix JL. Characterization of a nucleocapsid-like region and of two distinct primer tRNALys,2 binding sites in the endogenous retrovirus Gypsy. Nucleic Acids Res. 2006;34:5764–5777. doi: 10.1093/nar/gkl722. [DOI] [PMC free article] [PubMed] [Google Scholar]