Abstract
The c-myc mRNA coding region determinant-binding protein (CRD-BP) has high affinity for the coding region determinant (CRD) of c-myc mRNA. Such affinity is believed to protect c-myc CRD from endonucleolytic attack. We have recently purified a mammalian endoribonuclease which can cleave within the c-myc CRD in vitro. The availability of this purified endonuclease has made it possible to directly test the interaction between CRD-BP and the endonuclease in regulating c-myc CRD RNA cleavage. In this study, we have identified the coding region of MDR-1 RNA as a new target for CRD-BP. CRD-BP has the same affinity for c-myc CRD nts 1705–1886 and MDR-1 RNA nts 746–962 with Kd of 500 nM. The concentration-dependent affinity of CRD-BP to these transcripts correlated with the concentration-dependent blocking of endonuclease-mediated cleavage by CRD-BP. In contrast, three other recombinant proteins tested which had no affinity for c-myc CRD did not block endonuclease-mediated cleavage. Finally, we have identified RNA sequences required for CRD-BP binding. These results provide the first direct evidence that CRD-BP can indeed protect c-myc CRD cleavage initiated by an endoribonuclease, and the framework for further investigation into the interactions between CRD-BP, c-myc mRNA, MDR-1 mRNA and the endoribonuclease in cells.
INTRODUCTION
The c-Myc protein is an important transcription factor. It influences cell proliferation, differentiation and apoptosis via the activation of a multitude of target genes (1). It has been estimated that c-Myc can influence the expression of ∼10% of all genes in humans (1). c-Myc is implicated in human cancers, and overexpression of c-Myc at the protein and/or mRNA levels has been observed in virtually all types of cancers (2). These observations highlight the significance of understanding c-myc gene regulation.
c-Myc expression is regulated at multiple levels including the level of mRNA stability (3–5). By using a cell-free mRNA decay assay as well as in vivo analysis, it has been shown that c-myc mRNA can be degraded by two different pathways. One pathway involves rapid removal of the poly(A) tail which is then followed by 3′–5′ decay accomplished by 3′–5′ exoribonuclease(s) (6,7). The second decay pathway involves endonucleolytic cleavage within the coding region of c-myc mRNA, a region termed the coding region determinant (CRD) (5). Several studies have subsequently confirmed that the coding region of c-myc mRNA, including the CRD, is involved in the regulation of c-myc mRNA stability in cells (3,8–11).
A cell-free mRNA decay assay involving polysome extracts and a 180 nt RNA sense strand corresponding to the c-myc CRD was found to induce endonucleolytic cleavage within the c-myc CRD and destabilized the transcript 8-fold (5). The effect was highly specific because competitor RNA corresponding to other regions of the c-myc transcript failed to destabilize c-myc mRNA (5). Based on these observations, Ross and co-workers (5) postulated the following model: (i) c-myc mRNA is susceptible to attack by a polysome-associated endoribonuclease; (ii) a protein can interact with and protect the CRD from attack by an endoribonuclease; (iii) the addition of competitor CRD RNAs titrate the protein off c-myc mRNA, leaving the CRD unprotected and susceptible to endonuclease attack. A protein that binds with high affinity in vitro to the c-myc CRD has been purified (12) and cloned (13). This protein termed the c-myc mRNA coding region determinant-binding protein or CRD-BP, was later found to be identical to the IGF-II mRNA-binding protein 1 (IMP-1) (14).
The following observations supported CRD-BP being an oncofetal protein. CRD-BP has been shown to be abundantly expressed in fetal tissues but not in adult tissues (15). CRD-BP-deficient mice exhibited dwarfism, impaired gut development and increased perinatal mortality (16), providing further support that CRD-BP plays a critical role in developmental processes. De novo expression studies on CRD-BP have shown it to be overexpressed in breast cancer (17), colorectal cancer (18), brain tumors (19) and non-small cell lung tumors (19). Further evidence to associate CRD-BP with breast cancer are the observations that ∼30% of human breast cancer cases exhibited amplification of the CRD-BP gene (20), and transgenic mice carrying targeted expression of the protein developed mammary tumors (21).
To date, CRD-BP and its orthologs have been shown to bind to several RNAs, and depending on the RNA in question, multiple functions for these proteins have been proposed. CRD-BP binds to the 5′-untranslated region of IGF-II mRNA and influences its subcellular localization and translation (14,22). CRD-BP also binds to H19 RNA (23) and neuron-specific tau mRNA (24) to influence their localization in cells. The zipcode-binding protein binds to β-actin mRNA and localizes it to the leading edge of fibroblast lamellipodia (25). The IMP binds to the 3′-untranslated region of CD44 mRNA and stabilizes the transcript (26). A recent study has identified CRD-BP as the protein that interacts with the coding region of βTrCP1 mRNA and stabilizes the transcript in response to the β-catenin signaling pathway (27). Finally, as indicated above, CRD-BP has been shown to bind to and is believed to shield the c-myc CRD from endonucleolytic attack thereby prolonging its cytosolic half-life (5).
Three observations support the CRD-BP-c-myc CRD shielding hypothesis. First, overexpression of CRD-BP in colorectal cancer correlates with a modest elevation of c-myc mRNA levels (18). Second, decreased expression of c-myc mRNA subsequent to knockdown of CRD-BP was observed in MCF-7 (28) and colorectal cells (27). Finally, addition of sense RNA corresponding to the CRD resulted in enhanced decay of c-myc mRNA in cells (29). However, the following two recent observations do not support the CRD-BP-c-myc CRD shielding hypothesis: (i) transgenic mice over-expressing CRD-BP in mammary tissues did not exhibit elevated levels of c-myc mRNA (21); and (ii) targeted knockdown of CRD-BP in K562 cells had no effect on c-myc mRNA levels (30). Such unexpected results or discrepancies may in part be due to the role of the third party molecule, namely an endoribonuclease that can cleave the c-myc CRD. The availability of an identified endoribonuclease should permit systematic testing of the CRD-BP-c-myc CRD shielding hypothesis in vitro and in vivo, and may help clarify some of the contradictory reports.
We have recently purified a mammalian endoribonuclease from rat liver based on its ability to cleave within the c-myc CRD (31). Several biochemical properties distinguished the enzyme from previously described vertebrate endonucleases and supported it being a novel mammalian endoribonuclease (31). The enzyme preferentially cleaves between the dinucleotides UA, CA and UG, and it is capable of cleaving other RNAs including MDR-1 and β-globin RNAs (31). With the availability of purified mammalian endoribonuclease and recombinant CRD-BP, we sought to directly test the CRD-BP-c-myc CRD-shielding hypothesis. In this study, we show that CRD-BP is able to dose-dependently bind to c-myc CRD RNA and MDR-1 RNA with Kd of ∼500 nM. This interaction correlates with its ability to dose-dependently inhibit cleavage of c-myc CRD and MDR-1 RNA by the mammalian endoribonuclease.
MATERIALS AND METHODS
Expression and purification of recombinant CRD-BP, Rpp20, Rpp21, Rpp25 and Rpp40
The plasmid pET28b(+)-CRD-BP which contains mouse CRD-BP cDNA was a generous gift from Dr Jeffrey Ross, University of Wisconsin and was prepared as previously described (13). Plasmids pHTT7k-p20, pHTT7k-p21, pHTT7k-p25 and pHTT7k-p40 which contain the Rpp20, Rpp21, Rpp25 and Rpp40 cDNAs were generous gifts from Dr Sidney Altman, Yale University and were prepared as previously described (32–34). All His6-tagged proteins were purified from Escherichia coli BL21(DE3) using the QIAexpress Ni-NTA metal affinity spin kit (QIAGEN) under denaturing conditions. Proteins eluted from the column at either pH 5.4 or 4.5 were subjected to a series of dialysis steps (3 h at each step) pH 5/6 M urea, pH 5.5/4 M urea, pH 6/2 M urea, pH 6.7/1M urea and pH 7.4/0 M urea in a buffer containing 200 mM KCl, 1 mM EDTA, 10% (v/v) glycerol, 1 mM reduced glutathione, 0.1 mM oxidized glutathione, 0.01% (v/v) Triton X-100, 20 mM triethanolamine (13) and EDTA-free protease inhibitor tablets (Roche). Following dialysis, samples were spun at 13,200 rpm for 30 min to remove any precipitated proteins. The purified protein solutions were then quantified and analyzed for purity using Coomasie blue-stained 12% SDS-PAGE.
Purification of the mammalian endoribonuclease
Purification of the mammalian endoribonuclease from rat livers has been previously described (31). One unit of the purified enzyme from the last step of column chromatography, the heparin-sepharose column, was used in all experiments described in this study.
In vitro transcription and labeling of 5′ ends of RNA
To synthesize c-myc CRD RNA, plasmids pUC19-CRDmyc-1705-1792 (31) and pUC19-CRDmyc-1705-1886 were linearized with EcoRI and transcribed with SP6 RNA polymerase using SP6 Megascript kits (Ambion, TX, USA). To make human β-globin RNA corresponding to nts 1–145, the plasmid SPκβc (35) was linearized with FokI and transcribed with SP6 RNA polymerase using SP6 Megascript kit. To make human MDR-1 RNA corresponding to nts 746–962, the plasmid pGEM7Z-MDR-1-V2 was linearized with ApaLI and transcribed with T7 RNA polymerase using T7 Megascript kit (Ambion, TX, USA). For 5′-32P-end labeling of RNA, 5 μg of in-vitro transcribed RNA was first dephosphorylated with 10 units of calf alkaline phosphatase (GE Healthcare) for 30 min at 37°C in a 100-μl reaction according to the manufacturer's instructions. Dephosphorylated RNA was purified by phenol/chloroform extraction and ethanol precipitation. About 2.5 μg of dephosphorylated RNA was incubated in a 25-μl reaction with 30–50 μCi of [γ-32P] ATP at 37°C for 1 h with 40–50 units of T4 polynucleotide kinase (New England Biolabs). The entire sample was then run on a 6% polyacrylamide/7M urea gel and the band containing 5′-labeled 32P-RNA was excised and eluted with elution buffer (0.5 M ammonium acetate, 1 mM EDTA, 0.2% SDS) at 37°C for 6 h. The purified radiolabeled RNA was then concentrated by ammonium acetate/isopropanol precipitation. Specific activity of the RNA was determined by scintillation counting.
Electrophoretic mobility shift assays
Electrophoretic mobility shift assay (EMSA) binding buffers (5 mM Tris-Cl pH 7.4, 2.5 mM EDTA pH 8.0, 2 mM DTT, 5% glycerol, 0.1 mg/ml bovine serum albumin, 0.5 mg/ml yeast tRNA, 5 units RNasin) (13) were prepared on ice prior to each experiment. In order to facilitate RNA denaturation and renaturation, the [32P] RNA sample was heated to 50°C for 5 min and cooled to room temperature before being added to the EMSA-binding buffer. EMSA-binding buffer containing radiolabeled RNA was then incubated with purified recombinant protein in a 20-μl reaction volume at 30°C for 10 min and transferred to ice for 5 min. The reaction was again incubated at 30°C for 10 min and transferred to ice for an additional 5 min. Heparin (Sigma) was then added to a final concentration of 5 mg/ml and the reaction was incubated on ice for a final 5-min period. A total of 2 μl EMSA loading dye (250 mM Tris-Cl pH 7.4, 0.2% bromophenol blue, 0.2% xylene cyanol, 40% sucrose) was added to each reaction and 10 μl of the EMSA reaction was loaded onto an 8% native polyacrylamide gel and resolved at 25 mA for 90 min. Following electrophoresis, the gel was dried and subjected to autoradiography using the Cyclone PhosphorImager and Optiquant Software.
All EMSA saturation-binding experiments were carried out as described above and the dissociation constant (Kd) for CRD-BP against MDR-1 and c-myc CRD was determined by fits to the Hill equation:
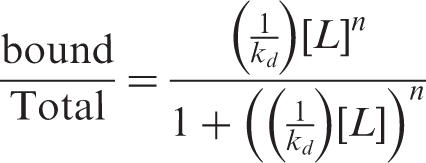 |
where Kd is the dissociation constant and [L] is the concentration of CRD-BP and n is the Hill coefficient. All saturation-binding data was analyzed by densitometry of the autoradiograph using the Cyclone Storage Phosphor-System and Optiquant software. For each reaction the total activity in each lane was determined; this involved summing the total activity in bound complexes with the total activity present in the unbound fraction. It should be noted that in lanes in which both Complex I and Complex II were present, their total activity was added together and therefore represented the total bound fraction. This analysis allowed for the calculation of the percentage of RNA bound to the protein. The percentage of bound RNA and the protein concentration (nM) were inserted into the Hill equation and the results were expressed graphically.
For EMSA competition assays, 5′-[32P] radiolabeled c-myc CRD RNA nts 1705–1886 and unlabeled in vitro transcribed competitor RNA were prepared as described above. EMSA-binding buffers and EMSA reaction mixtures were prepared and 350 nM of CRD-BP was added to the reactions containing one of the following unlabeled competitors: c-myc-CRD-1705-1885, c-myc-CRD 1705-1792, MDR-1 or β-globin RNA. EMSA competition assays involved the pre-incubation between competitor RNA and CRD-BP for 10 min at 30°C. Following the pre-incubation, 40 nM [32P] c-myc CRD RNA was added to the reaction. This was followed by the standard EMSA protocol as described above. The concentrations of unlabeled competitor RNA employed were 1-, 2-, 5-, 10- and 50-fold greater than the 40 nM [32P] c-myc-CRD RNA used.
In vitro endonuclease shielding assay
The first step of this assay involved the binding between one of the [32P] RNA constructs (c-myc CRD-1705-1886, MDR-1 or β-globin) and one of the purified recombinant proteins (CRD-BP, Rpp20, Rpp21, Rpp25 or Rpp40). Binding was facilitated using the standard EMSA protocol. Immediately following the final EMSA step, one unit of the heparin-sepharose purified mammalian endoribonuclease was added to the binding reaction, mixed and incubated at 37°C for 3 min. As a positive control, the dialyzed mammalian endoribonuclease was added to the EMSA buffer in the absence of recombinant protein and incubated at 37°C for 3 min. All reactions were terminated by heating at 100°C for 3 min followed by the addition of 1 µl phenol and 3 µl formamide loading dye (80% formamide, 0.05 mg/ml bromophenol blue, 0.05 mg/ml xylene cyanol, 0.01 M EDTA pH 8.0). Aliquots containing ∼30 000 c.p.m. were loaded onto 8% denaturing polyacrylamide gel and resolved at 30 mA until the bromophenol blue reached the bottom of the gel plate. Gels were fixed in 10% acetic acid, 10% methanol for 15 min and subjected to autoradiography. In some experiments, higher amounts (three units) of the purified endoribonuclease were used. In such cases, similar blocking effects of CRD-BP on endonuclease-mediated cleavage were observed.
RNase T1 digestion and alkaline hydrolysis of radiolabeled c-myc CRD RNA were performed to generate sequencing ladders for cleavage site identification purposes. Alkaline ladders were generated by combining 100 000 c.p.m. [32P] c-myc CRD-1705-1886 RNA and 1× alkaline hydrolysis buffer (50 mM NaHCO3/Na2CO3 pH 9.2, 1 mM EDTA, 3 μg yeast tRNA) in a total reaction volume not exceeding 15 µl. The reaction was incubated at 95°C for 10 min, transferred to ice and combined with 20 µl of formamide loading dye and stored at −20°C until required. RNase T1 digests were generated by incubating 100 000 c.p.m. [32P] c-myc CRD-1705-1886 RNA in EMSA-binding buffer with 1 unit of RNase T1 (Ambion). The RNA/EMSA binding buffer mixture was heated to 55°C for 5 min and cooled to room temperature prior to the addition of RNase T1. The T1 cleavage reaction was incubated at room temperature for 10 min and was terminated by phenol/chloroform extraction. The RNA was extracted using ammonium acetate/isopropanol precipitation and was resuspended in 15 µl of formamide loading dye.
Mapping of CRD-BP-binding sites using hydroxyl radical foot-printing
The Fe(II)/EDTA cleavage complex (36) was used to map the CRD-BP-binding sites on c-myc CRD-1705-1886. Initially, binding between c-myc CRD (∼45 nM; 100 000 c.p.m./reaction) and CRD-BP (1500 nM) was facilitated using the standard EMSA buffer and reaction conditions as described above. Immediately following the binding reaction, 6 µl of Fe(II)/EDTA cleavage reagent (0.4 mM Fe(II)(NH4)2(SO4)2, 0.8 mM EDTA pH 8.0, 20 mM l-ascorbic acid sodium salt, 0.35% H2O2) was added to the 20 µl binding reaction. The cleavage reaction was incubated at room temperature for 10 min and was terminated by the addition of 5 µl 0.1 M Thiourea (Sigma). The RNA was extracted by ammonium acetate/isopropanol precipitation and the pellet was resuspended in 15 µl formamide loading dye. About 5 µl of the reaction was run on an 8% denaturing polyacrylamide-sequencing gel.
RESULTS
Purified recombinant His6-tagged CRD-BP binds to the full-length c-myc CRD RNA nts 1705–1886 and MDR-1 RNA nts 746–962
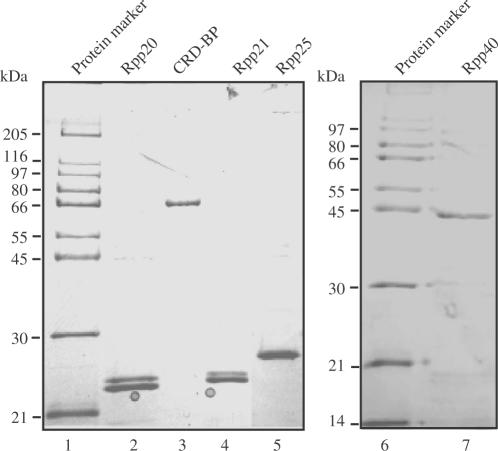
In an effort towards testing the CRD-BP-c-myc CRD shielding hypothesis, it was imperative to demonstrate that purified recombinant His6-tagged CRD-BP could bind avidly to the CRD. The plasmid pET28b(+)-CRD-BP was used to generate the recombinant mouse CRD-BP containing a His6-tag at the C-terminus of the protein. CRD-BP fractions eluted from the Ni-NTA column contained an enriched band at ∼68 kDa when visualized by SDS-PAGE, (refer to lane 3, Figure 1) which is consistent with the predicted and reported molecular weight of recombinant mouse CRD-BP (13). The recombinant His6-tagged CRD-BP of ∼95% purity was consistently obtained and an example of this is shown in lane 3, Figure 1. Additional recombinant proteins, Rpp20, Rpp21, Rpp25 and Rpp40, were used as controls in this investigation. Rpp20, Rpp21, Rpp25 and Rpp40 are protein subunits of human RNase P (32–34) and were chosen on the basis of their ability or inability to bind to nucleic acids. The criteria used to determine if these recombinant proteins possessed this binding potential was based on the presence of putative nucleic-acid-binding domains in their sequence and known RNA–protein interactions. Purity of the recombinant proteins was assessed by visualization using Coomasie blue-stained SDS-PAGE. As shown in Figure 1, the purity of all the recombinant proteins was greater than 95% and their observed molecular weights were consistent with their predicted molecular weights (lanes 2, 4, 5, 7).
Figure 1.
Analysis of purified recombinant His6-tagged proteins by SDS-PAGE. Recombinant His6-tagged mouse CRD-BP was purified using Ni-NTA spin column and dialyzed prior to separation (2 μg) (lane 3) on 12% SDS-PAGE and visualization by Coomasie blue staining. Recombinant His6-tagged human Rpp20 (lane 2), Rpp21 (lane 4), Rpp25 (lane 5) and Rpp40 (lane 7) were also purified and dialyzed in a similar manner before its purity (2 μg) being analyzed on Coomasie blue-stained SDS-PAGE.
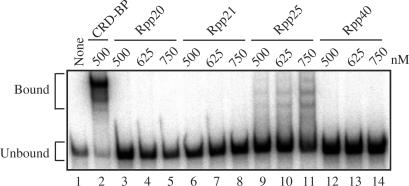
EMSA was performed to determine whether the purified and dialyzed recombinant proteins were capable of binding to c-myc CRD RNA nts 1705–1886. About 500 nM recombinant CRD-BP was incubated with c-myc CRD [32P]RNA and RNA–protein complexes were visualized following non-denaturing gel electrophoresis. As shown in lane 2, Figure 2, two clear bound complexes were observed. In contrast, no bound complexes were observed when 500–750 nM Rpp20 (lanes 3–5), Rpp21 (lanes 6–8) or Rpp40 (lanes 12–14) were incubated with c-myc CRD [32P]RNA. Three low-activity binding complexes were observed when Rpp25 was incubated with c-myc CRD [32P]RNA (lanes 9–11). The ability of Rpp25 to bind to the c-myc CRD was not surprising given the fact that it possesses an Alba RNA-binding motif (37,38) and it is known to bind to several RNA targets including a 71-nt segment of H1 RNA, M1 RNA and E. coli tRNAtyr (34). It should be noted that the intensity of the Rpp25-binding complexes is significantly less than CRD-BP-binding complexes at the same concentration. Further study would be required to determine if Rpp25 has in fact lower affinity than CRD-BP for c-myc CRD or yeast tRNA present in the EMSA reaction actually competed with Rpp25 for binding to the transcript. The inability of Rpp40 to bind with the c-myc CRD was supported by the previous suggestions that this protein in isolation does not play a role in nucleic acid binding (37,38).
Figure 2.
Electromobility shift assay showing binding of recombinant CRD-BP and Rpp25 to c-myc CRD RNA. About 500 nM of purified recombinant CRD-BP (lane 2) and purified recombinant Rpp20 (lanes 3–5), Rpp21 (lanes 6–8), Rpp25 (lanes 9–11) and Rpp40 (lanes 12–14) (at various concentrations as indicated) were incubated with 40 nM [32P] c-myc CRD RNA nts 1705–1886 (∼50 000 c.p.m). None (lane 1) indicates no protein was added. The positions of protein–RNA complexes (Bound) and unbound RNA (Unbound) are indicated on the left.
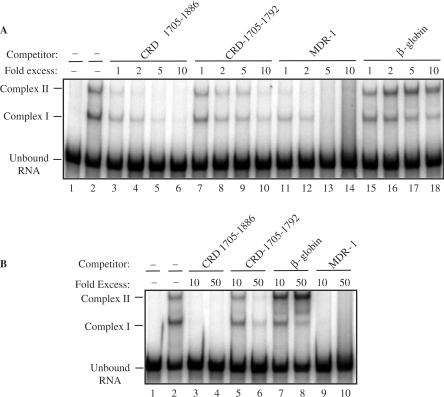
To further determine if the recombinant CRD-BP-c-myc CRD [32P] RNA nts 1705–1886 association was specific, we chalenged the interaction with unlabeled competitor RNA. Our EMSA competition assays revealed that up to 50X molar excess of unlabeled β-globin competitor RNA was unable to dilute the binding between CRD-BP and [32P] c-myc CRD (lanes 15–18, Figure 3A; lanes 7–8, Figure 3B). The inability of unlabeled β-globin competitor to dilute the CRD-BP-c-myc CRD interaction in conjunction with the saturation-binding experiment shown in Figure 4C support the observation that the 145-nt β-globin RNA fragment is not a bona fide CRD-BP target in vitro. The β-globin results from this investigation are also supported by the two earlier reports (12,13).
Figure 3.
Electromobility shift competition assays with c-myc CRD RNA, β-globin RNA or MDR-1 RNA. About 350 nM of purified recombinant His6-tagged CRD-BP was incubated with 40 nM [32P] c-myc CRD RNA nts 1705–1886 (50 000 c.p.m) with or without the indicated molar excess of unlabeled competitor RNA. (A) 1–10 molar excess of unlabeled c-myc CRD RNA nts 1705–1886 (lanes 3–6), c-myc CRD RNA nts 1705–1792 (lanes 7–10), MDR-1 RNA nts 746–962 (lanes 11–14) and β-globin RNA nts 1–145 (lanes 15–18) were used. (B) 10–50 molar excess of unlabeled c-myc CRD RNA nts 1705–1886 (lanes 3 and 4), c-myc CRD RNA nts 1705–1792 (lanes 5 and 6), β-globin RNA nts 1–145 (lanes 7 and 8) and MDR-1 RNA nts 746–962 (lanes 9 and 10) were used.
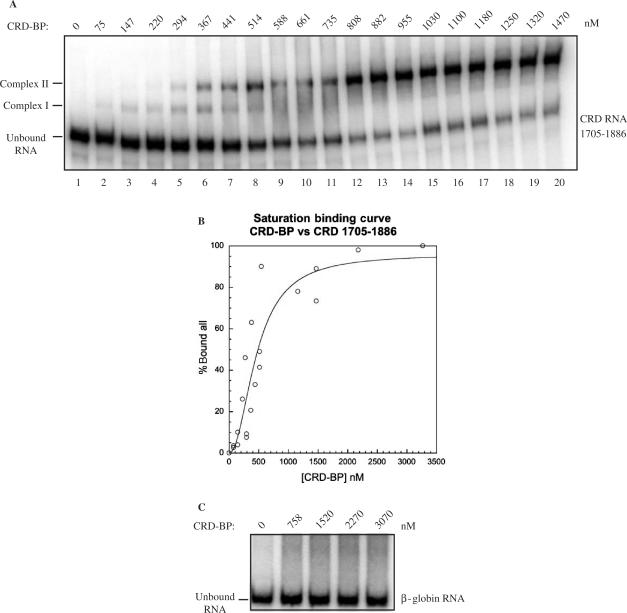
Figure 4.
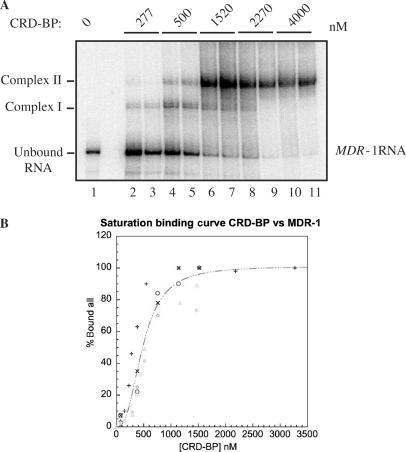
CRD-BP-binding affinity for c-myc CRD RNA and β-globin RNA. (A) An electromobility shift assay showing binding to [32P] c-myc CRD RNA nts 1705–1886 by an increasing amount of purified recombinant His6-tagged CRD-BP (0–1470 nM). (B) Binding activity in (A) was quantified using the PhorphorImager to compare the relative amount of radiolabeled unbound RNA shifted into slower-migrating Complex I and Complex II. Data obtained was then used to plot the saturation binding curve as shown. (C) An electromobility shift assay assessing the ability to bind to [32P] β-globin RNA nts 1–145 by an increasing amount of purified recombinant His6-tagged CRD-BP (0–3070 nM).
Similar to unlabeled c-myc CRD nts 1705–1886 (compare lane 3 to lane 2, Figure 3A), at 1× molar excess, the MDR-1 RNA nts 746–962 was capable of diluting the CRD-BP-[32P] c-myc CRD interaction (compare lane 11 to lane 2, Figure 3A) and at 5×, 10× and 50× molar excess it diluted the interaction completely (compare lanes 13–14 with lane 2, Figure 3A; compare lanes 9–10 with lane 2, Figure 3B). Similar results were observed when the unlabeled competitor c-myc CRD-1705-1886 RNA was used (compare lanes 5–6 with lane 2, Figure 3A; compare lanes 3–4 with lane 2, Figure 3B). The unlabeled competitor c-myc CRD nts 1705–1792 was also observed to dilute the interaction when a minimum of 2× molar excess was added (compare lane 8 to lane 2, Figure 3A) and even at 50× molar excess, the CRD-BP-[32P] c-myc CRD interaction was still visible (compare lanes 5–6 with lane 2, Figure 3B). This result is consistent with previous observation that c-myc CRD nts 1705–1792 has indeed reduced affinity for CRD-BP as compared to the full-length c-myc CRD nts 1705–1886 (13). It is possible that the secondary structure of the c-myc CRD-1705-1886 contains the full complement of recognition and binding elements that facilitate high-affinity-binding interactions with CRD-BP.
Characterizing the binding of CRD-BP to c-myc CRD RNA and MDR1 RNA
Although CRD-BP is known to bind to the c-myc CRD (12,13), the binding characteristics between the two components has not been previously determined. With the availability of purified recombinant CRD-BP as well as MDR-1 RNA, we decided to further characterize and compare the nature of the binding interactions. Saturation-binding experiments were carried out using the standard EMSA conditions and employed a wide range of CRD-BP concentrations. The saturation-binding experiments were conducted three times, using three different purified CRD-BP preparations. A representative result for the binding of CRD-BP to c-myc [32P] CRD RNA nts 1705–1886 is shown in Figure 4A. It was determined experimentally that the minimum concentration of CRD-BP required to generate Complex I (binding of one molecule of CRD-BP, see below for discussion) was about 15 nM (data not shown). At concentrations above 100 nM, the slower migrating Complex II (binding of two molecules of CRD-BP, see below for discussion) was frequently observed (lanes 3–20, Figure 4A). In many instances, at higher concentrations of CRD-BP (e.g. 500–750 nM), Complex I was absent and it would appear that the bound RNA shifted exclusively to Complex II (compare lanes 8–20 to lanes 2–7, Figure 4A). When the saturation-binding data from Figure 4A was fit to the Hill equation and expressed graphically, a sigmoidal curve was generated as shown in Figure 4B (Kd of 500 nM). It is important to point out that the concentration of CRD-BP at which formation of Complexes I and II would take place was highly dependent upon the age of the dialyzed protein and the number of freeze–thaw cycles subjected to the protein. However, the actual dissociation constant may be lower than 500 nM because it cannot be assumed that 100% of the CRD-BP molecules were present in a conformation suitable for binding.
Keeping in mind the limitation of the EMSA, we also performed saturation-binding experiments to determine if CRD-BP could bind with other radiolabeled RNA substrates. When challenged with [32P] β-globin RNA, CRD-BP up to 3070 nM was unable to form an observable RNA–protein complex (Figure 4C). However, when CRD-BP was challenged with [32P] MDR-1, two RNA–protein complexes that exhibited a similar profile to that of the CRD-BP-c-myc CRD interaction were observed; Kd = 500 nM (Figure 5). The analysis of the combined binding data from three separate experiments revealed a level of cooperativity was present between the formation of the first and second binding complexes (Figure 5B). The cooperativity observed is also reflected in the Hill coefficient, which for CRD-BP versus c-myc CRD (Figure 4B) was 2.06, and 2.6 when challenged with MDR-1 RNA (Figure 5B). The Hill coefficients in both cases suggest that two molecules of CRD-BP bind with one molecule of c-myc CRD RNA or with one molecule of MDR-1 RNA at identical or non-identical sites. Evidence to support a two RNA-binding site model is provided by the kinetic analysis of the interactions between IMP-1 and IGF-II RNA (39). In this study, IMP-1 was shown to bind through a cooperative, sequential mechanism via two binding sites within the IGF-II transcript (39). Interestingly, dissociation constant of about 2 nM was reported for the interaction between IMP-1 and IGF-II RNA (39). The significantly higher dissociation constant determined in this study could reflect the inefficiency of renaturation of CRD-BP and c-myc CRD RNA as discussed above, or/and the use of His6-tagged CRD-BP since it has been previously reported that tagged-IMP-1 exhibited poor RNA binding (14).
Figure 5.
CRD-BP binding affinity for MDR-1 RNA. (A) An electromobility shift assay showing binding to [32P] MDR-1 RNA nts 746–962 by an increasing amount of purified recombinant His6-tagged CRD-BP (0–4000 nM). (B) Binding activity in (A) was quantified using the PhorphorImager to compare the relative amount of radiolabeled unbound RNA shifted into slower-migrating Complex I and Complex II. Data obtained from three separate experiments was then used to plot the saturation binding curve as shown.
CRD-BP protects the c-myc CRD and MDR-1 RNA from cleavage by the mammalian endoribonuclease
Having established that recombinant CRD-BP can bind c-myc CRD and MDR-1 RNA, we then sought to determine if the binding of CRD-BP to RNA could directly protect these transcripts from degradation by an endoribonuclease recently discovered in our lab (31).
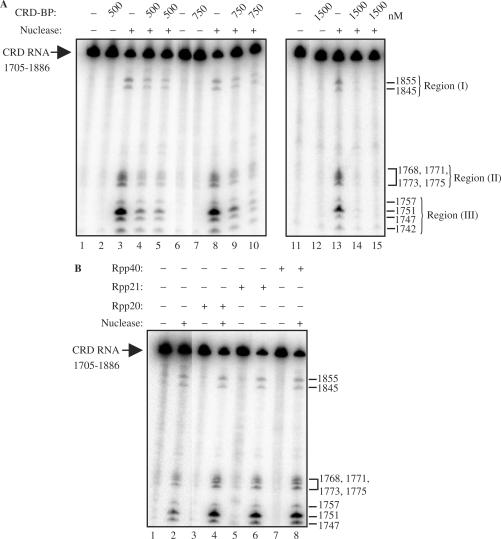
5′-End labeled c-myc CRD nts 1705–1886 was incubated with one unit of the purified endonuclease in the presence or absence of recombinant CRD-BP. As shown in Figure 6A, the typical ten distinct cleavage sites at nts 1742, 1747, 1751, 1757, 1768, 1771, 1773, 1775, 1845 and 1855 were generated upon incubation with the purified endonuclease (lanes 3, 8 and 13) (31). For ease in quantification and categorization, we have named the separate cleavage sites as Region I (1845, 1855), Region II (1768, 1771, 1773, 1775) and Region III (1742, 1747, 1751, 1757) (Figure 6). Incubation of the transcript with recombinant CRD-BP at 500 nM (lane 2), 750 nM (lane 7) and 1500 nM (lane 12) alone had no effect, indicating that CRD-BP lacks RNase activity. It was anticipated that c-myc cleavage would be reduced using CRD-BP concentrations equal to the measured dissociation constant because 50% of c-myc CRD would be bound at 500 nM (Figure 4A and B). This prediction was confirmed by the reduction in cleavage fragments observed in all three regions indicated in Figure 6A (compare lanes 4–5 with lane 3). At 500 nM, CRD-BP was responsible for a 43% reduction in Region I, a 72% reduction in Region II and a 77% reduction in Region III.
Figure 6.
CRD-BP shields the c-myc CRD RNA from endonucleolytic attack by a mammalian endoribonuclease. (A) One unit of the purified mammalian endoribonuclease was incubated with [32P] c-myc CRD RNA nts 1705–1886 as described in the Materials and methods section, with (lanes 4, 5, 9, 10, 14, 15) or without (lanes 3, 8 and 13) the presence of 500, 750 or 1500 nM of purified recombinant His6-tagged CRD-BP as shown. (B) As in (A), the purified endoribonuclease was incubated with the radiolabeled c-myc CRD RNA in the absence (lane 2) or presence of 1500 nM of Rpp20 (lane 4), Rpp21 (lane 6) and Rpp40 (lane 8). Lanes 3, 5 and 7 had the purified recombinant proteins only. Numbers on the right indicate the cleavage sites generated by the mammalian endoribonuclease on the CRD RNA substrates.
It was predicted that cleavage reductions would increase at concentrations above the measured dissociation constant because at 750 and 1500 nM, ∼75 and 90% of c-myc CRD were bound by CRD-BP respectively (Figure 4A and B). At 750 nM, (lanes 9–10, Figure 6A) CRD-BP was responsible for a 45% reduction in Region I, a 75% reduction in Region II and a 80% reduction in Region III. As shown in lanes 14–15 in Figure 6A, at 1500 nM, CRD-BP was responsible for the 95% cleavage reductions observed in all three regions.
To determine if the blocking effect was due to specific binding interactions, we incubated c-myc CRD RNA with Rpp20, Rpp21 and Rpp40 which had been shown to have no affinity for the transcript (Figure 2). Figure 6B shows that at 1500 nM, Rpp20, Rpp21 and Rpp40 were unable to block endonucleolytic cleavage of CRD RNA by the native mammalian endoribonuclease (compare lanes 4, 6 and 8 to lane 2).
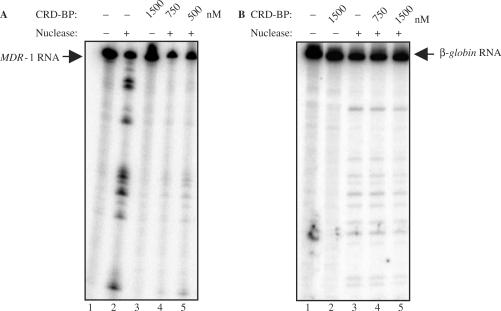
CRD-BP was shown to bind with considerable affinity and specificity to MDR-1 RNA (Figure 5) and we have previously shown that the endonuclease is capable of cleaving MDR-1 RNA (31). It was therefore anticipated that CRD-BP would also be capable of blocking MDR-1 cleavage by the mammalian endoribonuclease. As predicted, CRD-BP was observed to shield MDR-1 RNA from endonucleolytic attack in a concentration-dependent manner (Figure 7A), and the most significant cleavage reductions were observed at 1500 nM (lane 3, Figure 7A). The consistent reduction observed in all regions suggests that the entire MDR-1 RNA was protected via CRD–BP interactions. We have also tested the ability of CRD-BP to block cleavage of β-globin RNA by the endonuclease. As expected, CRD-BP that had no affinity for β-globin RNA (Figure 4C) did not effectively block the endonuclease from cleaving this transcript (Figure 7B).
Figure 7.
CRD-BP is capable of shielding MDR-1 RNA from endonucleolytic attack by the mammalian endoribonuclease. (A) One unit of the purified mammalian endoribonuclease was incubated with [32P] MDR-1 RNA nts 746–962 as described in the Materials and methods section, without (lane 2) or with the presence of 500 nM (lane 5), 750 nM (lane 4) or 1500 nM (lane 3) of purified recombinant His6-tagged CRD-BP. Lane 2 had the input RNA only. (B) As in (A), the purified endoribonuclease was incubated with [32P] β-globin RNA nts 1–145 without (lane 3) or with the presence of 750 nM (lane 4) or 1500 nM (lane 5) of purified recombinant His6-tagged CRD-BP. Lane 1 had the input RNA only.
Taken together, our overall results suggest that CRD-BP was capable of shielding both c-myc CRD and MDR-1 RNAs in a dose-dependent manner from the cleavage activities of a mammalian endoribonuclease via specific RNA–protein interactions. In addition, the results suggest that CRD-BP binds to a considerable portion of both MDR-1 and c-myc RNAs and at 1500 nM, both RNA transcripts appeared to be entirely protected.
Mapping the CRD-BP-binding sites on c-myc CRD RNA
To map the CRD-BP-binding sites on c-myc CRD, we used the hydroxyl radical foot-printing method (36). The chemical cleavage reagent chosen was the Fe(II)/EDTA complex. The Fe(II)/EDTA complex is a negatively charged species and does not interact electrostatically with the RNA molecule thereby limiting its interference with RNA–protein interactions (36). The complex is known to generate hydroxyl radicals from the reduction of hydrogen peroxide in a reaction known as the Haber–Weiss reaction (36). The hydroxyl radical is thought to cleave the RNA backbone by attacking ribose moieties (36). Regions of the RNA backbone that are occluded by protein are cleaved with decreased efficiency by the radical and the result is a series of cleavage fragments exhibiting reduced intensity (36).
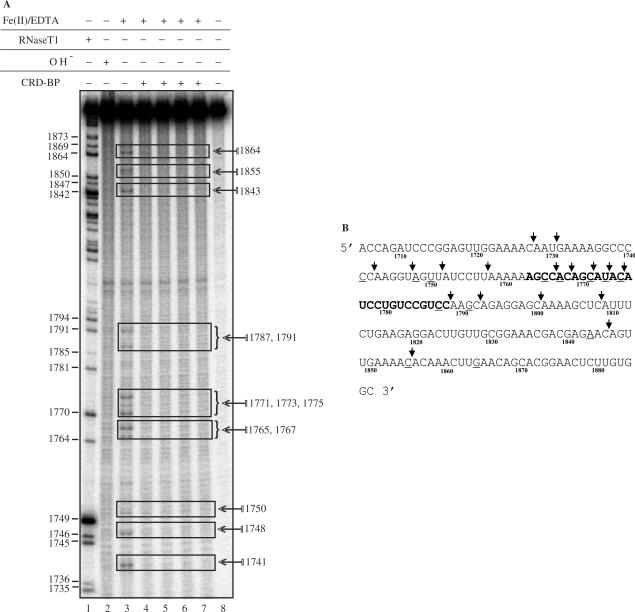
Figure 8A shows a typical CRD-BP-c-myc CRD foot-printing experiment. Lane 3 depicts the Fe(II)/EDTA cleavage profile characteristic of [32P] c-myc CRD-1705–1886 in the absence of CRD-BP. The alkaline hydrolysis ladder (lane 2) exhibited a uniform cleavage profile with the exception of two enriched fragments located between nts 1794 and 1842; which were also observed in lanes 4–9. RNase T1 digest of [32P] c-myc CRD-1705–1886 shown in lane 1 provided mapping for cleavage site identification. The cleavage pattern observed as a result of Fe(II)/EDTA-treatment of c-myc CRD was hallmarked by several enriched fragments which have been mapped and are indicated in Figure 8A. When compared to cleavage reactions containing 1500 nM CRD-BP (lanes 4–7), it is apparent that several regions of cleavage were significantly reduced. The cleavage fragments corresponding to nts 1741 and 1748 (lane 3) were reduced by 75%. An equivalent reduction was also observed at discrete fragments in the nucleotide region spanning 1765–1775 (compare lane 3 with lanes 4–7). At positions 1787 and 1791, a 50% reduction in cleavage intensity was observed and a 75% reduction was also observed at positions 1843, 1855 and 1864 (compare lane 3 with lanes 4–7). Figure 8B shows the identified CRD-BP-binding sites in comparison with the putative CRD-BP-binding sites previously determined using antisense oligonucleotides (29). Six of the binding sites (1765, 1767, 1771, 1773, 1775, 1787) identified in this study were in good agreement with those identified by Coulis et al. (29). Six identified CRD-BP-binding sites (1747, 1767, 1771, 1773, 1855) are within the direct cleavage sites of the mammalian endonuclease and five (1741, 1765, 1787, 1791, 1843) are within one to two nucleotides away from the enzyme cleavage sites (Figure 8B). Such observation is consistent with the notion that CRD-BP is capable of shielding c-myc CRD RNA from attack by the endonuclease.
Figure 8.
Mapping of CRD-BP-binding sites on c-myc CRD RNA using the hydroxyl radical-mediated cleavage reaction. (A) 45 nM [32P] c-myc CRD RNA nts 1705–1886 was subjected to Fe(II)/EDTA cleavage reaction in EMSA-binding buffer in the absence (lane 3) or presence (lanes 4–7) of 1500 nM purified recombinant His6-tagged CRD-BP. Reaction samples were run on a 8% denaturing polyacrylamide gel, fixed, dried and subjected to autoradiography as described in the Materials and methods section. Lane 8 is the input RNA. Lanes 1 and 2 represent RNase T1 digest and alkaline hydrolysis of the radiolabeled c-myc CRD RNA respectively. Regions highlighted by rectangles showed significant reductions in cleavage. Numbers on the left indicate guanosine cleavage sites of RNase T1 digest and numbers on the right indicate the hydroxyl radical-mediated cleavage sites. (B) Identified CRD-BP-binding sites on sequences of c-myc CRD RNA nts 1705–1886. Underlined sequences indicate the CRD-BP-binding sites as determined by the hydroxyl radical foot-printing in (A). Bolded sequences indicate the putative CRD-BP-binding sites as previously determined using antisense oligonucleotides competition method (29). Arrows indicate cleavage sites generated by the purified endoribonuclease (31).
DISCUSSION
CRD-BP belongs to the VICKZ family of RNA-binding proteins that influence various aspects of cellular physiology such as cell growth and differentiation (40). CRD-BP was initially discovered (5) and eventually purified (12) based on its ability to bind to the c-myc CRD. The working model for CRD-BP function is that it binds to and protects the c-myc CRD from endonucleolytic cleavage, resulting in enhanced transcript half-life and expression (5). This hypothesis is supported by evidence from in vitro mRNA decay studies (5,12) as well as studies in cells (3,28). Although this hypothesis has been proposed for a number of years, it has not been directly tested. That is CRD-BP has not been directly shown to block CRD cleavage by an endoribonuclease. This is in part due to the unavailability of a candidate endoribonuclease that can cleave within the c-myc CRD. In this study, we show that recombinant CRD-BP can dose-dependently inhibit cleavage of c-myc CRD and MDR-1 RNA by a mammalian endoribonuclease. This blocking effect correlates with CRD-BP's dose-dependent affinity for c-myc CRD and MDR-1 RNAs.
Specificity of recombinant CRD-BP for the CRD was demonstrated in two ways. First, while three other recombinant proteins, Rpp20, Rpp21 and Rpp40, had no affinity for c-myc CRD, CRD-BP was shown to bind strongly to the transcript (Figure 2). Second, consistent with previous findings (12,13), c-myc CRD competitor RNAs were effective in competing with [32P]c-myc CRD in binding to CRD-BP, with nts 1705–1886 being more effective than nts 1705–1792 (Figure 3). On the contrary, at 50× molar excess, β-globin RNA could not compete with [32P]c-myc CRD in binding to CRD-BP. Unexpectedly, we found that the coding region of MDR-1 RNA nts 746–962 was as effective as c-myc CRD nts 1705–1886 in competing for binding to CRD-BP (Figure 3). This result is consistent with the observed saturating-binding profile upon incubation of excess CRD-BP with [32P]MDR-1 RNA (Figure 5).
The Kd for the CRD-BP-c-myc-1705–1886 interaction was significantly lower than that of CRD-BP-c-myc-1705–1792 interaction. It is apparent that one of the proposed CRD-BP-binding sites (nts 1843–1864) is absent from the 1705–1792 CRD region, and may attributed to the differences in specificity and stability observed in the RNA–protein interactions. The binding profiles exhibited by the CRD-BP-c-myc CRD and CRD-BP–MDR-1 interactions were hallmarked by the presence of two binding complexes. These RNA–protein interactions are believed to occur via a sequential, cooperative binding mechanism. The formation of the faster migrating Complex I at low concentrations of CRD-BP, the emergence of the slower migrating Complex II at intermediate concentrations of CRD-BP and the predominance of Complex II at concentrations greater than (or) equal to the calculated Kd are suggestive of a sequential binding mechanism. It is possible that the formation of Complex I may represent an intermediate state in which one molecule of CRD-BP is bound to one RNA molecule. The emergence of Complex II suggests that a second molecule of CRD-BP engages in interactions with a Complex I species in a concentration-dependent manner. We propose that Complex II may function to increase the stability and specificity of the RNA–protein interaction. Such interactions have also been observed and proposed for IMP-1-IGF-II RNA (39) and Vg1RBP-VLE 3′-UTR RNA (41).
Our data demonstrated that the blocking of endonuclease-mediated c-myc CRD cleavage by CRD-BP is concentration dependent and this correlated with concentration-dependent binding of CRD-BP to the transcript. We propose that at 500 nM CRD-BP, most c-myc CRD RNA molecules are bound by only one molecule of CRD-BP. At 1500 nM CRD-BP, c-myc CRD RNA is likely to be bound by two molecules of CRD-BP as observed by the formation of Complex II. In this scenario, it is highly likely that the entire transcript is bound and protected by CRD-BP as evidenced by the hydroxyl radical foot-printing experiments (Figure 8). Our observations that CRD-BP binds to larger stretches of nucleotides rather than to specific positions is supported by evidence that members of the VICKZ family bind to larger elements rather than to individual nucleotides (40). Further evidence to support this came from a study using complementary oligonucleotides to map the CRD-BP-binding sites within the c-myc CRD (29). Our findings of the presence of CRD-BP-binding sites within the regions targeted by the mammalian endoribonuclease support the hypothesis that CRD-BP serves as a stabilizing factor for c-myc in vitro. The evidence provided in this study confirms that CRD-BP is in fact capable of blocking a mammalian endoribonuclease from cleaving c-myc CRD via specific interactions with RNA sequences.
To our knowledge, this is the first study to demonstrate the ability of CRD-BP to directly protect c-myc CRD cleavage by an endoribonuclease. This is a critical first step towards directly testing the interaction between CRD-BP and endoribonuclease in regulating c-myc CRD cleavage in cells. As indicated in the Introduction, four studies indirectly supported this hypothesis (18,27,28,29). However, two recent observations on the lack of changes in c-myc mRNA and protein levels upon changes in CRD-BP expression (21,30) are inconsistent with this working model. As previously suggested (21,30), the known multilevel regulation of c-myc gene expression could contribute to these observations. Alternatively, the absence of the responsible endoribonuclease may in part explain these unexpected findings. The latter hypothesis can be investigated upon availability of identified endoribonucleases that can cleave within the c-myc CRD.
Our finding that CRD-BP has high affinity for MDR-1 RNA was totally unexpected and has important implications. MDR-1 is a plasma membrane drug-transporter protein and is associated with multidrug resistance phenotypes in cultured cells as well as in human cancers (42). Enhanced mRNA stability for MDR-1 and c-myc mRNA has been reported in liver cancer (43) and MDR-1 mRNA decay intermediates corresponding to nts 746–962 of MDR-1 have been detected in the normal liver but not liver cancer (44). Although CRD-BP expression has not been studied in liver cancer, results to date are consistent with the model that c-myc and MDR-1 mRNAs are short-lived in normal adult tissues (43) because CRD-BP is absent or present at low levels. However, in cancerous liver cells, enhanced MDR-1 and c-myc mRNAs (43) are observed because CRD-BP may be overexpressed.
In conclusion, our data provides direct evidence that CRD-BP can indeed protect c-myc mRNA from cleavage by an endoribonuclease, and supports its proposed role in regulating mRNA stability in cells. We have also identified MDR-1 RNA as a new target for CRD-BP, and the transcript can also be protected from endonucleolytic cleavage by CRD-BP. These findings provided further support for the significant and global roles that CRD-BP play in regulating mRNA stability in cancer.
ACKNOWLEDGEMENTS
This work was supported by a grant from Natural Science & Engineering Research Council and Canadian Liver Foundation. C.H.L. is a Research Scientist of the National Cancer Institute of Canada supported with funds provided by the Canadian Cancer Society. We thank Andrea Gorrell for her assistance in the preparation of saturation-binding curves. Funding to pay the Open Access publication charge was provided by the NSERC Discovery Grant 227158.
Conflict of interest statement. None declared.
REFERENCES
- 1.Levens D. Reconstructing MYC. Genes Dev. 2003;17:1071–1077. doi: 10.1101/gad.1095203. [DOI] [PubMed] [Google Scholar]
- 2.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 3.Herrick DJ, Ross J. The half-life of c-myc mRNA in growing and serum-stimulated cells: influence of the coding and 3′ untranslated regions and role of ribosome translocation. Mol. Cell. Biol. 1994;14:2119–2128. doi: 10.1128/mcb.14.3.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiu RPC, Watson PH, Dubik D. C-myc oncogene expression in estrogen-dependent and -independent breast cancer. Clin. Chem. 1993;39:353–355. [PubMed] [Google Scholar]
- 5.Bernstein PL, Herrick DJ, Prokipcak RD, Ross J. Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region determinant. Genes Dev. 1992;6:642–654. doi: 10.1101/gad.6.4.642. [DOI] [PubMed] [Google Scholar]
- 6.Brewer G. Evidence for a 3′-5′ decay pathway for c-myc mRNA in mammalian cells. J. Biol. Chem. 1999;274:16174–16179. doi: 10.1074/jbc.274.23.16174. [DOI] [PubMed] [Google Scholar]
- 7.Brewer G. Regulation of c-myc mRNA decay in vitro by a phorbol ester-inducible, ribosome-associated component in differentiating megakaryoblasts. J. Biol. Chem. 2000;275:33336–33345. doi: 10.1074/jbc.M006145200. [DOI] [PubMed] [Google Scholar]
- 8.Lemm I, Ross J. Regulation of c-myc mRNA decay by translational pausing in a coding region instability determinant. Mol. Cell. Biol. 2002;22:3959–3969. doi: 10.1128/MCB.22.12.3959-3969.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morello D, Lavenu A, Pournin S, Babinet C. The 5′ and 3′ non-coding sequences of the c-myc gene, required in vitro for its post-transcriptional regulation, are dispensable in vivo. Oncogene. 1993;8:1921–1929. [PubMed] [Google Scholar]
- 10.Wisdom R, Lee W. The protein coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitor. Genes Dev. 1991;5:232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- 11.Yielding NM, Lee WMF. Coding elements in exons 2 and 3 target c-myc mRNA downregulation during myogenic differentiation. Mol. Cell. Biol. 1997;17:2698–2707. doi: 10.1128/mcb.17.5.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prokipcak RD, Herrick DJ, Ross J. Purification and properties of a protein that binds to the C-terminal coding region of human c-myc mRNA. J. Biol. Chem. 1994;269:9261–9269. [PubMed] [Google Scholar]
- 13.Doyle GAR, Betz NA, Leeds PL, Fleisig AJ, Prokipcak RD, Ross J. The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic Acid Res. 1998;26:5036–5044. doi: 10.1093/nar/26.22.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol. Cell. Biol. 1999;19:1262–1270. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leeds P, Kren BT, Boylan JM, Betz NA, Steer CJ, Gruppuso PA, Ross J. Developmental regulation of CRD-BP, an RNA-binding protein that stabilizes c-myc mRNA in vitro. Oncogene. 1997;14:1279–1286. doi: 10.1038/sj.onc.1201093. [DOI] [PubMed] [Google Scholar]
- 16.Hansen TVO, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, Christiansen J. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol. Cell. Biol. 2004;24:4448–4464. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ioannidis P, Mahaira L, Papaioannou D, Teixera MR, Heim S, Andersen JA, Evangelou E, Dafni U, Pandis N, et al. 8q24 Copy number gains and expression of the c-myc mRNA stabilizing protein CRD-BP in primary breast carcinomas. Int. J. Cancer. 2003;104:54–59. doi: 10.1002/ijc.10794. [DOI] [PubMed] [Google Scholar]
- 18.Ross J, Lemm I, Berberet B. Overexpression of an mRNA-binding protein in human colorectal cancer. Oncogene. 2001;20:6544–6550. doi: 10.1038/sj.onc.1204838. [DOI] [PubMed] [Google Scholar]
- 19.Ioannidis P, Kottaridi C, Dimitriadis E, Courtis N, Mahaira L, Talieri M, Giannopoulos A, Iliadis K, Papaioannou D, et al. Expression of the RNA-binding protein CRD-BP in brain and non-small cell lung tumors. Cancer Lett. 2004;209:245–250. doi: 10.1016/j.canlet.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 20.Doyle GA, Bourdeau-Heller JM, Coulthard S, Meisner LF, Ross J. Amplification in human breast cancer of a gene encoding a c-myc mRNA-binding protein. Cancer Res. 2000;60:2756–2759. [PubMed] [Google Scholar]
- 21.Tessier CR, Doyle GA, Clark BA, Pitot HC, Ross J. Mammary tumor induction in transgenic mice expressing an RNA-binding protein. Cancer Res. 2004;64:209–214. doi: 10.1158/0008-5472.can-03-2927. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen J, Nielsen FC, Kristenses MA, Koch G, Christiansen J. Cytoplasmic trafficking of IGF-II mRNA-binding protein by conserved KH domains. J. Cell Sci. 2002;115:2087–2097. doi: 10.1242/jcs.115.10.2087. [DOI] [PubMed] [Google Scholar]
- 23.Runge S, Nielsen FC, Nielsen J, Lykke-Andersen J, Wewer UM, Christiansen J. H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. J. Biol. Chem. 2000;275:29562–29569. doi: 10.1074/jbc.M001156200. [DOI] [PubMed] [Google Scholar]
- 24.Atlas R, Behar L, Elliot E, Ginzburg I. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J. Neurochem. 2004;89:613–626. doi: 10.1111/j.1471-4159.2004.02371.x. [DOI] [PubMed] [Google Scholar]
- 25.Ross AF, Oleynikov Y, Kislaukis EH, Taneja KL, Singer RH. Characterization of a β-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vikesaa J, Hansen TVO, Jonson L, Borup R, Wewer UM, Christiansen J, Nielsen FC. RNA-binding IMPs promote cell adhesion and invadopodia formation. EMBO J. 2006;25:1456–1468. doi: 10.1038/sj.emboj.7601039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noubissi FK, Elcheva I, Bhatia N, Shakoori A, Ougolkov A, Liu J, Minamoto T, Ross J, Fuchs SY, et al. CRD-BP mediates stabilization of βTrCP1 and c-myc mRNA in response to β-catenin signaling. Nature. 2006;441:898–901. doi: 10.1038/nature04839. [DOI] [PubMed] [Google Scholar]
- 28.Ioannidis P, Mahaira LG, Perez SA, Gritzapis AD, Sotiropoulou PA, Kavalakis GJ, Antsaklis AI, Baxevanis CN, Papamichail M. CRD-BP/IMP1 expression characterizes cord blood CD34+ stem cells and affects c-myc and IGF-II expression in MCF-7 cancer cells. J. Biol. Chem. 2005;280:20086–20093. doi: 10.1074/jbc.M410036200. [DOI] [PubMed] [Google Scholar]
- 29.Coulis CM, Lee C, Nardone V, Prokipcak RD. Inhibition of c-myc expression in cells by targeting an RNA-protein interaction using antisense oligonucleotides. Mol. Pharmacol. 2000;57:485–494. doi: 10.1124/mol.57.3.485. [DOI] [PubMed] [Google Scholar]
- 30.Liao B, Patel M, Hu Y, Charles S, Herrick DJ, Brewer G. Targeted knockdown of the RNA-binding protein CRD-BP promotes cell proliferation via insulin-like growth factor II-dependent pathway in human K562 leukemia cells. J. Biol. Chem. 2004;279:48716–48724. doi: 10.1074/jbc.M405853200. [DOI] [PubMed] [Google Scholar]
- 31.Bergstrom K, Urquhart JC, Tafech A, Doyle E, Lee CH. Purification and characterization of a novel mammalian endoribonuclease. J. Cell. Biochem. 2006;98:519–537. doi: 10.1002/jcb.20726. [DOI] [PubMed] [Google Scholar]
- 32.Jarrous N, Reiner R, Wesolowski D, Mann H, Guerrier-Takada C, Altman S. Function and subnuclear distribution of Rpp21, a protein subunit of the human ribonucleoprotein ribonuclease P. RNA. 2001;7:1153–1164. doi: 10.1017/s1355838201010469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarrous N, Eder PS, Guerrier-Takada C, Hoog C, Altman S. Autoantigenic properties of some protein subunits of catalytically active complexes of human ribonuclease P. RNA. 1998;4:407–417. [PMC free article] [PubMed] [Google Scholar]
- 34.Guerrier-Takada C, Eder PS, Gopalan V, Altman S. Purification and characterization of Rpp25, an RNA-binding protein subunit of human ribonuclease P. RNA. 2002;8:290–295. doi: 10.1017/s1355838202027954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawn RM, Efstratiadis A, O'Connell C, Maniatis T. The nucleotide sequence of the human β-globin gene. Cell. 1980;21:647–651. doi: 10.1016/0092-8674(80)90428-6. [DOI] [PubMed] [Google Scholar]
- 36.Tullius T, Dombroski BA. Hydroxyl radical footprinting: high resolution information about DNA-protein contacts and applications to lambda repressor and Cro protein. Proc. Natl. Acad. Sci. U.S.A. 1986;83:5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang T, Guerrier-Takada C, Altman S. Protein-RNA interactions in the subunits of human nuclear RNase P. RNA. 2001;7:937–941. doi: 10.1017/s1355838201010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen J, Kristensen MA, Willemoes M, Nielsen FC, Christiansen J. Sequential dimerization of human zipcode-binding protein IMP1 on RNA: a cooperative mechanism providing RNP stability. Nucleic Acids Res. 2004;32:4368–4376. doi: 10.1093/nar/gkh754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yisraeli JK. VICKZ proteins: a multi-talented family or regulatory RNA-binding proteins. Biol. Cell. 2005;97:87–96. doi: 10.1042/BC20040151. [DOI] [PubMed] [Google Scholar]
- 41.Git A, Standart N. The KH domains of Xenopus Vg1RBP mediate RNA binding and self-association. RNA. 2002;8:1319–1333. doi: 10.1017/s135583820202705x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ambudkar SV, Kimchi-Sarfaty C, Sauna ZE, Gottesman MM. P-glycoprotein: from genomics to mechanism. Oncogene. 2003;22:7468–7485. doi: 10.1038/sj.onc.1206948. [DOI] [PubMed] [Google Scholar]
- 43.Lee CH, Bradley G, Ling V. Increased P-glycoprotein messenger RNA stability in rat liver tumors in vivo. J. Cell. Physiol. 1998;177:1–12. doi: 10.1002/(SICI)1097-4652(199810)177:1<1::AID-JCP1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 44.Lee CH, Rehaume VE, Shandro J. Identification of in vivo P-glycoprotein mRNA decay intermediates in normal liver but not in liver tumors. J. Cell. Physiol. 2005;204:638–645. doi: 10.1002/jcp.20313. [DOI] [PubMed] [Google Scholar]