Abstract
Pyrrolysine (Pyl), the 22nd co-translationally inserted amino acid, is incorporated in response to a UAG amber stop codon. Pyrrolysyl-tRNA synthetase (PylRS) attaches Pyl to its cognate tRNA, the special amber suppressor tRNAPyl. The genes for tRNAPyl (pylT) and PylRS (pylS) are found in all members of the archaeal family Methanosarcinaceae, and in Desulfitobacterium hafniense. The activation and aminoacylation properties of D. hafniense PylRS and the nature of the tRNAPyl identity elements were determined by measuring the ability of 24 mutant tRNAPyl species to be aminoacylated with the pyrrolysine analog N-ε-cyclopentyloxycarbonyl-l-lysine. The discriminator base G73 and the first base pair (G1·C72) in the acceptor stem were found to be major identity elements. Footprinting analysis showed that PylRS binds tRNAPyl predominantly along the phosphate backbone of the T-loop, the D-stem and the acceptor stem. Significant contacts with the anticodon arm were not observed. The tRNAPyl structure contains the highly conserved T-loop contact U54·A58 and position 57 is conserved as a purine, but the canonical T- to D-loop contact between positions 18 and 56 was not present. Unlike most tRNAs, the tRNAPyl anticodon was shown not to be important for recognition by bacterial PylRS.
INTRODUCTION
Every known member of the archaeal family Methanosarcinaceae is able to utilize methylamines as an energy source, a virtually exclusive feature of this family. Crucial to this process are the methylamine methyltransferase enzymes, the successful biosynthesis of which requires suppression of an in-frame UAG codon (1,2). The amino acid incorporated at the UAG position is the 22nd genetically encoded amino acid pyrrolysine (Pyl) (3). Pyl is charged specifically onto the amber suppressor tRNAPyl by the aminoacyl-tRNA synthetase (AARS), pyrrolysyl-tRNA synthetase (PylRS) (4–6). PylRS and SepRS are the only two examples to date of aminoacyl-tRNA synthetases specific for a modified amino acid. The genes for tRNAPyl (pylT) and PylRS (pylS) are found in every known member of the archaeal family Methanosarcinaceae and in the bacterium Desulfitobacterium hafniense (5). As D. hafniense also encodes the machinery for selenocysteine incorporation, it is the only known organism that potentially uses all 22 amino acids in protein translation.
While the archaeal pylT and pylS genes are highly conserved throughout the members of the Methanosarcinaceae family, the bacterial genes differ more markedly from their archaeal counterparts. The archaeal tRNAPyl sequences are highly conserved with only 7 nucleotide changes among the 6 known isoacceptors, while the D. hafniense tRNAPyl has 23 unique residues relative to the archaeal consensus. In addition, in both the archaea and bacteria, the amber suppressor tRNAPyl has several unique structural features when compared to canonical tRNAs. These include (a) an anticodon stem of 6 base pairs instead of 5, (b) a short variable loop of only 3 bases, (c) a single base separating the acceptor and D stems, rather than 2, (d) a small D-loop with only 5 bases, (e) the absence of the almost universally conserved G18G19 sequence in the D-loop and T54Ψ55C56 sequence in the T-Loop and (f) only two modified bases have been proposed in Methanosarcina. barkeri Fusaro tRNAPyl: 4-thiouridine at position 8 and 1-methyl-pseudouridine at position 54 (4).
The archaeal PylRS enzymes have ∼55–75% amino acid identity, while the D. hafniense PylRS shares ∼39% amino acid identity with the archaeal enzymes. The relatively high ∼39% identity between the D. hafniense and Methanosarcinaceae PylRS suggests that the presence of PylRS in D. hafniense may be a product of horizontal gene transfer from an archaeal progenitor (7).
The D. hafniense PylRS also has an approximately 100 amino acid amino-terminal truncation when compared to the archaeal PylRS. The absence of these 100 amino acids brings into question whether D. hafniense PylRS can charge tRNAPyl. In this study we investigate whether the D. hafniense PylRS is able to aminoacylate tRNAPyl in vitro.
Furthermore, as Pyl likely plays an essential role in methylamine methyltransferase activity, the accurate formation of Pyl-tRNAPyl is crucial. Thus, we embarked on determining the molecular recognition of tRNAPyl by PylRS in D. hafniense.
MATERIALS AND METHODS
General
Oligonucleotides synthesis and DNA sequencing were performed by the Keck Foundation Biotechnology Resource Laboratory at Yale University. Uniformly labeled sodium [32P]pyrophosphate [1–60 Ci/mmol (1 Ci = 37 GBq)], and [α-32P]ATP (10 mCi/ml) was from Amersham Biosciences. N-ε-cyclopentyloxycarbonyl-l-lysine (Cyc) was purchased from Sigma.
Purification of D. hafniense PylRS
The D. hafniense pylS gene was cloned into the pET15b vector (Novagen). Escherichia coli BL21-CodonPlus (DE3)-RIL (Stratagene) was transformed with the resulting construct. The overexpression protocol was performed as described (8). Briefly, cells were grown overnight at 37°C until a density of A600 = 6.0 was reached in Luria Broth supplemented with 100 µg/ml ampicillin, 34 µg/ml chloramphenicol, 50 × 52 buffer (0.5% glycerol, 0.05% glucose, 0.2% α-lactose), NPS (50 mM Na2HPO4, 50 mM KH2PO4 and 25 mM (NH4)2SO4) and 1 mM MgSO4. PylRS was purified by Ni-NTA technology according to the manufacturer's protocols (Qiagen).
Cloning and in vitro transcription of the tRNAPyl substrates
Desulfitobacterium hafniense wild type and mutant tRNAPyl were prepared as previously reported for the M. barkeri Fusaro tRNAPyl (9). tRNA genes for in vitro T7 RNA polymerase transcription were cloned by annealing two DNA oligonucleotides and direct ligation into the pUC18 plasmid using BamHI and HindIII restriction sites. The E. coli DH5α was transformed with the pylT containing plasmids. The 3′-end of the transcription template was generated by digestion overnight with BstNI at 60°C. In vitro T7 RNA polymerase run off transcription was conducted according to standard procedures (10). tRNAPyl transcripts were gel purified, then 3′ end-labeled with [α-32P]ATP using the E. coli CCA-adding enzyme (11,12). 3′-labeled tRNAs were gel purified, and the concentration determined by measuring A260.
ATP-[32P]PPi exchange
The assay was used to measure PylRS activation of amino acid substrates, and performed as previously described (13). Reactions (200 µl final volume) were performed at 37°C in 100 mM Na-HEPES, pH 7.2, 10 mM MgCl2, 50 mM KCl, 5 mM DTT, 2 mM KF, 2 mM ATP, 2 mM [32P]PPi (1.6 cpm/pmol), 0.5 µM PylRS enzyme and either 1mM lysine or 10 mM Cyc. [32P]ATP formation was followed by specific absorption on acid-washed Norit [0.2 ml of a 1% suspension (wt/vol) of Norit in a solution of 0.4 M sodium pyrophosphate solution containing 15% (vol/vol) perchloric acid], filtration with Whatman GF/C fiber glass filter disks (Millipore), washed with 15 ml ethanol and 2 times 15 ml water, dried and liquid-scintillation-counted (Beckman Coulter).
Aminoacylation and determination of kinetics parameters
Aminoacylation assays were adapted from a recently described procedure (11). Aminoacylation reactions (10 µl) were carried out at 37°C in 100 mM Na-Hepes, pH 7.2, 25 mM MgCl2, 60 mM NaCl, 5 mM ATP, 1 mM DTT, 10 mM Cyc (14) and 3′-[α-32P]ATP-labeled tRNA ranging in concentration from 0.25- to 5-fold Km. PylRS concentrations ranged from 5 to 90 nM. Reactions were stopped by removing 1 µl reaction mix, adding it to 3 µl of 2.5 units/µl of nuclease P1 (American Bioanalytical) in 0.1 M sodium citrate (pH 4.5) at 25°C for 30 min, thus releasing [α-32P]AMP from uncharged tRNA and Cyc-[α-32P]AMP from charged tRNA. [α-32P]AMP and Cyc-[α-32P]AMP were separated by thin layer chromatography (TLC) on PEI cellulose plates (Baker) in 0.1 M sodium acetate and 5% acetic acid (15). [α-32P]AMP and Cyc-[α-32P]AMP were exposed to phosphoimager plates (FujiFilms), visualized using a Molecular Dynamics Storm 860 scanner (Amersham) and quantified using ImagQuant© software. Kinetic parameters were calculated using nonlinear regression plots of the initial velocity (v) and represent the results of at least three independent experiments.
tRNAPyl footprinting with S1 and T1 nucleases
Partial digestions of D. hafniense tRNAPyl 3′-labeled transcripts with nuclease S1 and T1 were carried out in buffer containing 100 mM NaCl, 50 mM Na-Hepes, pH 7.2, 20 mM MgCl2 and 0.5 µg/µl cold yeast RNA (Ambion) (with 1 mM ZnCl2 for RNase S1 digest only). Each reaction (10 µl) contained 0.25 µM 3’ labeled tRNAPyl (∼1 00 000 Cerenkov counts), and either 50 U of RNase S1 or 1 U of RNase T1 in the absence or presence of 26 µM D. hafniense PylRS. Reactions were preincubated on ice for 10 min, followed by the addition of RNase and 5-min digestion at 25°C. Reactions were stopped by phenol and chloroform extraction and ethanol precipitation. Samples were washed with 70% ethanol, dried, re-suspended in gel loading buffer (95% fomamide, 10 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol), then quantified using a LS 6500 multi-purpose liquid scintillation counter (Beckman Coulter). An equal number of cpms per sample were analyzed by electrophoresis on an 8% denaturing polyacrylamide gel. Gels were exposed to a phosphoimaging plate (FujiFilms) for 12 h, visualized using a Molecular Dynamics Storm 860 scanner (Amersham), and quantified using ImagQuant© software.
tRNA footprinting with phosphorothioate incorporated tRNA
Phosphorothioates were randomly incorporated into the tRNA by inclusion of either 0.05 mM CTP [αS], GTP [αS], ATP [αS] or UTP [αS] (Sp isomers, Amersham & Invitrogen) as described previously (16,17). After transcription, tRNAs were gel purified, and 3’ labeled as described above. tRNAs were co-incubated with PylRS enzyme as described above for the RNase footprinting reactions, only in the absence of any DTT. Digestions were performed by adding 1 µl of 10 mM iodine to the binding reaction for 60 s on ice. Iodine cleavage was stopped by adding 20 µl of stop buffer (100 mM DTT and 1 mg/ml yeast RNA (Ambion), followed by ethanol precipitation. Digests were analyzed by gel electrophoresis as described previously for the RNase footprinting.
RESULTS
D. hafniense PylRS aminoacylates tRNAPyl
Pyrrolysine is not a commercially available substrate, and its synthesis has proven both difficult and expensive (4,14,18). A pyrrolysine analog, N-ε-cyclopentyloxycarbonyl-l-lysine (Cyc), was recently shown to be a reasonable substrate for M. barkeri PylRS; Cyc can be aminoacylated onto tRNAPyl both in vitro and in vivo (14).
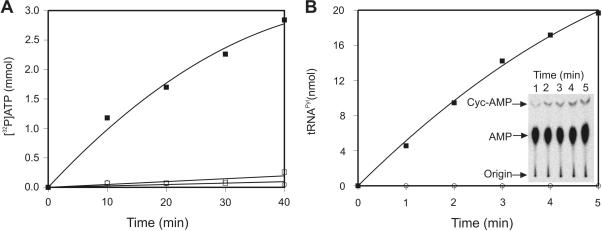
The ability of Cyc to promote the D. hafniense PylRS catalyzed ATP-[32P]PPi exchange reaction was assayed. PylRS was able to activate Cyc but not lysine (Figure 1A). Subsequently Cyc was used to assay PylRS ability to aminoacylate tRNAPyl in vitro (Figure 1B). The tRNAPyl transcripts was labeled at its 3′-terminal adenosine with [α-32P]ATP by the E. coli CCA-adding enzyme (11) and used in the aminoacylation reaction. At designated time points, aliquots of the aminoacylation reaction were digested with nuclease P1 at acidic pH in order to prevent deacylation of any Cyc-tRNAPyl formed. Nuclease P1 treatment released [32P]AMP from uncharged tRNAs, and Cyc-[32P]AMP from charged tRNAs. These two species were then separated by thin layer chromatography (TLC), [32P]AMP was visualized using a phosphoimager plate (Figure 1B, insert), and the fraction of charged versus uncharged tRNA was quantified as described in the Materials and Methods section. No charging was observed in the no amino acid control (Figure 1B). In the presence of excess PylRS enzyme, 80–90% of the tRNAPyl transcript could be aminoacylated with Cyc (data not shown).
Figure 1.
Activation and ligation onto D. hafniense tRNAPyl transcript of the pyrrolysine analogues N-ε-cyclopentyloxycarbonyl-L-lysine (Cyc) by D. hafniense PylRS. (A) Time course for the activation of pyrrolysine analogues: Cyc (▪), Lysine (□), no amino acid control (○). (B) Time course for in vitro aminoacylation of tRNAPyl transcript with Cyc (▪) or with no amino acid (○) as visualized by thin layer chromatography. The inset shows a TLC plate demonstrating the separation of the substrate (AMP; center of TLC plate) and product (Cyc-AMP; top of TLC plate). The reactions were spotted at the bottom of the plate (Origin). ATP-[32P]PPi exchange reaction and tRNAPyl transcript aminoacylation with Cyc were conducted as described in the Material and Methods section.
Identity elements of D. hafniense tRNAPyl
Extensive studies of tRNA identity elements have revealed that for most tRNAs the anticodon, the first base pair of the acceptor stem and the discriminator base N73 are important for tRNA aminoacylation by the cognate synthetase (19). To investigate the role of these standard identity elements in recognition of tRNAPyl by PylRS, a series of D. hafniense tRNAPyl variants with mutations at these positions were cloned and transcribed in vitro. The D. hafniense tRNAPyl mutants were assayed for their ability to be charged by D. hafniense PylRS with Cyc and kcat and Km were measured. The loss of aminoacylation efficiency relative to wild type tRNAPyl (L) was determined (L = [(kcat/Km)wild type]/[(kcat/Km)mutant)]) (Table 1).
Table 1.
Kinetic parameters of pyrrolyslation of wild-type and mutant D. hafniense tRNAPyl by D. hafniense PylRS
| tRNA | Km (µM) | kcat (s−1) × 10−2 | kcat/Km × 10−3 | La |
|---|---|---|---|---|
| WT | 2.9 ± 1.5 | 1.9 ± 0.45 | 6.4 | 1 |
| Anticodon CUA → UUU | 5.3 ± 2.3 | 1.8 ± 0.61 | 3.5 | 1.8 |
| Anticodon CUA → CUU | 2.1 ± 0.96 | 0.39 ± 0.098 | 1.9 | 3.4 |
| Anticodon CUA → AAA | 3.7 ± 0.68 | 0.15 ± 0.023 | 3.9 | 1.6 |
| G8U | 8.0 ± 1.1 | 3.3 ± 0.11 | 0.41 | 4.6 |
| G8C | 4.1 ± 2.0 | 0.32 ± 0.013 | 0.80 | 8.0 |
| G8A | 11.4 ± 1.3 | 0.57 ± 0.02 | 0.50 | 12.8 |
| G10A & C25U | 6.4 ± 0.69 | 0.11 ± 0.019 | 0.17 | 38.2 |
| A11G & U24C | 8.8 ± 2.3 | 0.21 ± 0.069 | 0.24 | 26.8 |
| G73A | 8.0 ± 3.7 | 0.052 ± 0.015 | 0.064 | 99.6 |
| G73U | 10.3 ± 2.1 | 0.22 ± 0.079 | 0.21 | 30.1 |
| G73C | 6.93 ± 2.9 | 0.097 ± 0.023 | 0.14 | 45.7 |
| G1A, C72U | 13.8 ± 1.6 | 0.14 ± 0.025 | 0.10 | 62.7 |
| G2A, C71U | 1.84 ± 0.66 | 0.24 ± 0.082 | 1.3 | 4.8 |
| G1A,C72U & G2A,C71U | 14.6 ± 3.2 | 0.088 ± 0.013 | 0.060 | 106.4 |
| A58G | NA | NA | NA | >1000 |
| A58U | NA | NA | NA | >1000 |
| A58C | NA | NA | NA | >1000 |
| A57G | 1.5 ± 0.42 | 0.18 ± 0.047 | 1.2 | 5.3 |
| A57U | 6.4 ± 2.4 | 0.095 ± 0.028 | 0.15 | 43.3 |
| A57C | 5.6 ± 1.2 | 0.11 ± 0.016 | 0.20 | 31.6 |
| A56G | 1.6 ± 0.55 | 0.19 ± 0.078 | 1.2 | 5.3 |
| A56U | 1.9 ± 0.66 | 0.18 ± 0.024 | 0.97 | 6.6 |
| A56C | 3.1 ± 0.88 | 0.21 ± 0.028 | 1.4 | 4.6 |
aL refers to the loss in aminoacylation efficiency compared to the wildtype; L is calculated as the ratio [kcat(wild type)/Km(wild type)]/[kcat(mutant)/Km(mutant)]. NA; not determined because below detection.
Surprisingly, the tRNAPyl anticodon, one of the most common identity elements in tRNAs, is not recognized by the PylRS. Changing the tRNAPyl anticodon from CUA to UUU, CUU or AAA did not decrease the aminoacylation efficiency (Table 1).
The discriminator base proved to be an important identity element, as changing G73 to U and C decreased aminoacylation efficiency by 30- and 45-fold, respectively, and G73A resulted in nearly 100-fold loss. Discriminator base changes showed only 2–3-fold increases in Km, and the majority of the loss in aminoacylation was dominated by a decrease in kcat. The first and second base pairs of the acceptor stem were changed from G·C to A·U independently and together. While G1A·C72U reduced aminoacylation efficiency by almost 63-fold, the G2A·C71U) alone had very little effect (L = 4.8). The simultaneous changes of both base pairs further reduced aminoacylation efficiency beyond G1A·C72U alone to 106-fold. As the change to G2A·C71U only impacted aminoacylation when present in conjunction with the first base pair G1A·C72U mutation, it is likely that G2·C71 is not recognized specifically. G1·C72 and G2·C71 are conserved in all tRNAPyl isoacceptors, while the remainder of the acceptor stem sequence is distinct in D. hafniense and archaea. The requirement for G·C pairs at the top of the acceptor stem may be essential for a specific conformational change the tRNA must undergo upon PylRS binding. Alternatively, the effect these pairs have on the structure of the acceptor stem helix may be important. In either case, it demonstrates that the top of the acceptor stem is an important part of the RNA–protein interface.
Footprinting of D. hafniense tRNAPyl with D. hafniense PylRS
In order to search for other possible identity elements, and to look more generally at the PylRS–tRNAPyl complex, the D. hafniense tRNAPyl was probed in the presence or absence of PylRS using two methods of analysis: RNase digestion and iodine cleavage of phosphorothioate-containing transcripts.
RNase footprinting of tRNAPyl
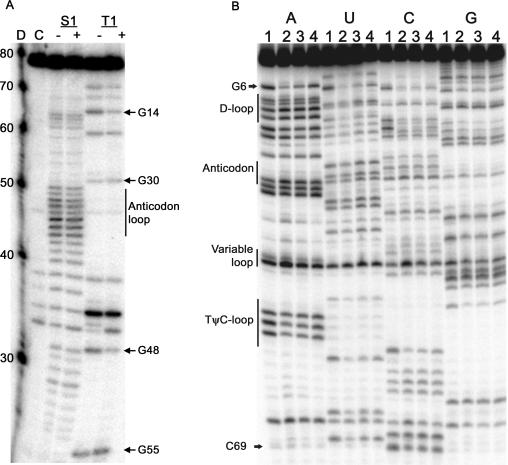
The structure of tRNAPyl was probed with RNase S1 (digests at single-stranded RNA) and RNase T1 (digests at single-stranded G) in the presence and absence of D. hafniense PylRS (Figure 2A). Under the experimental conditions for the RNase footprinting, approximately 90% of the tRNA was confirmed to be in the bound state by a nitrocellulose filter binding assay (data not shown). Due to intrinsic RNA instability, there is non-specific degradation in the anticodon loop, and in the variable loop region (Figure 2A). In the absence of PylRS, RNase T1 digested tRNAPyl at G5, G8, G14, G22, G30, G42, G44, G48 and G55. Other than G5, which is wobble paired to U68, no cleavage is observed in the acceptor stem as G1 to G4 and G6 are G·C paired and not accessible to RNase T1. The G30·U40 wobble pair in the anticodon stem is also disrupted, while G29·C41 is not. Cleavage at G22 indicates the G22·C13 pair at the end of the D-stem is subject to opening. Relative to the other positions, there is enhanced cleavage at G44, but no change in intensity in the presence or absence of PylRS. In the presence of PylRS, tRNAPyl was protected from RNase T1 digestion at nucleotides G14 in the D-loop and G48 in the variable loop. In the PylRS–tRNAPyl complex the anticodon loop was not protected from RNase S1 digestion as expected since the tRNAPyl anticodon sequence is not important for aminoacylation by PylRS. RNase S1 and T1 probing of the bacterial tRNAPyl agreed with the proposed secondary structure of the M. barkeri tRNAPyl (20).
Figure 2.
(A) Autoradiography of the 3’-end labeled D. hafniense tRNAPyl transcripts after enzymatic cleavage with RNase S1 (S1) and RNase T1 (T1) in the presence (+) and absence (−) of D. hafniense PylRS. D is Decade Marker (Ambion) that contains 32P-labeled RNA oligos whose nucleotide length is indicated on the left. C is untreated tRNA. RNase S1 (S1) cuts in single stranded regions, and no protection was observed in the anticodon stem. RNase T1 (T1) cuts at single stranded G and protection was observed at G14 and G48. (B) Autoradiography of the 3'-end labeled, phosphorothioate-containing D. hafniense tRNAPyl transcripts after cleavage by iodine in the presence and absence of D. hafniense PylRS. The footprinting experiments were performed using transcripts statistically substituted with ATP[αS] (A lanes), UTP[αS] (U lanes), CTP[αS] (C lanes), and GTP[αS] (G lanes). Lanes are numbered to indicate the PylRS concentration: (1) no PylRS, (2) 28 μM PylRS, (3) 14 μM PylRS, and (4) 7 μM PylRS.
Iodine probing of phosphorothioate-containing tRNAPyl
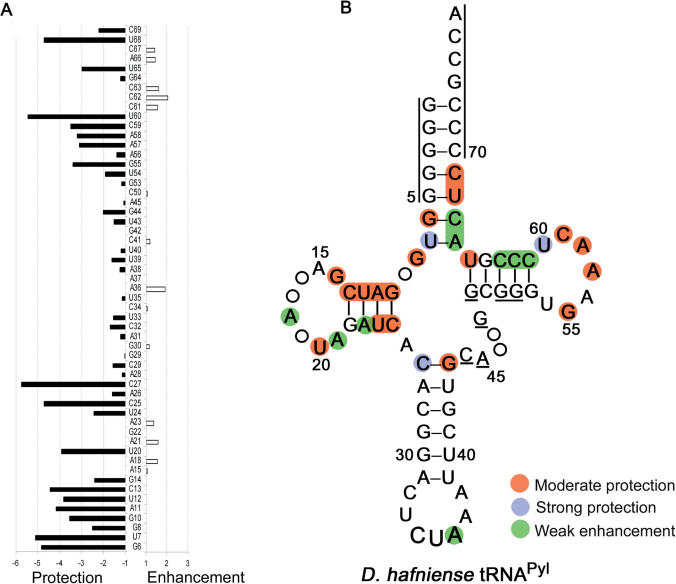
Transcripts of tRNAPyl with approximately 5% of the nucleotides statistically substituted with phosphorothioates were probed with iodine in the presence and absence of D. hafniense PylRS (Figure 3A). Phosphorothioate-containing transcripts were digested with iodine in the presence of 7, 14 and 28 µM PylRS, and band intensities were quantified and compared to look for changes in cleavage intensity that were dependent on the PylRS concentration (Figure 2B). The cleaved phosphates are located 5′ to the nucleotides listed. Changes in cleavage intensities in the presence of PylRS relative to the no enzyme control were labeled weak if they were less than 2-fold, moderate if they were 2- to 5-fold, and strong if they were greater than 5-fold (17).
Figure 3.
(A) Quantification of protection or enhancement of iodine cleavage of the phosphodiester bonds of D. hafniense tRNAPyl transcripts in the absence and presence of 28 μM PylRS. Protection or enhancement factors are represented by filled bars (for example, a protection of 2 means 50% reduction in band intensity in the presence of PylRS). Nucleotides not listed could not be visualized by the applied technique. (B) Summary of the probing of phosphorothioate-containing D. hafniense tRNAPyl transcript with iodine. Areas marked with lines indicate untested regions. Weak enhancement was less than 2 fold, moderate protection was 2 to 5 fold, and strong protection was greater than 5 fold.
Strong and moderate protection from iodine cleavage was observed in the D-stem (G10, A11, U12, C13, U24, C25), D-Loop (G14, U20), T-loop (G55, A57, A58, C59, U60), one base in the T-stem (U65) and the acceptor stem (G6, U7, U68, C69) (Figure 3). Moderate protection was also observed at residue G8, the single base separating the acceptor stem and the D-stem. Other than protection at the first base pair C27·G44, the anticodon stem digestion pattern is the same regardless of the presence or absence of PylRS. As with the RNase probes, no protection is observed in the anticodon loop. Since enhancement of cleavage was weak, significant opening of the tRNA structure to solvent is not occurring upon PylRS binding (Figure 3B).
In tRNAs, the DT-region is where the T-loop and D-loop make contacts with each other that maintain the L-shape structure. The conserved D- to T- loop contacts are between bases, and the phosphate backbone is left exposed in some areas. The backbone of the tRNAPyl T-Loop is occluded from solvent when bound to the synthetase, as expected if the synthetase uses the DT-region in the outer corner of the L-shape structure for recognition.
Aminoacylation efficiency of D. hafniense tRNAPyl variants
Based on the contacts observed in the footprinting experiments, a series of D. hafniense tRNAPyl variants were made and assayed for their ability to be aminoacylated with Cyc by PylRS.
Within the tertiary structure of a tRNA molecule the D-arm is stacked on the anticodon stem and is favorably positioned to play a role in interacting with the cognate synthetase. Moderate losses in aminoacylation efficiency resulting from the changes G10A·C25U (L = 38.2) and A11G·U24C (L = 26.8) in the D-arm suggest that this area, which is conserved throughout the tRNAPyl species, is important for forming a productive RNA–protein interface.
In the canonical tRNA structure, there are two residues (U8 and N9) separating the acceptor stem and D-arm. The highly conserved U8 interacts with A14 in the D-loop. While this A–U pair is preserved in the archaeal tRNAPyl, in D. hafniense positions 8 and 14 are both G. The changes G8C (L = 8.0), and G8A (L = 12.8) had only small affects on aminoacylation. The impact of G8A was dominated by an increase in Km. Interestingly, G8U (L = 4.6) increased Km, but also increased kcat slightly, resulting in a very slight overall loss in aminoacylation efficiency. While the base at position 8 appears not to have a large impact on aminoacylation efficiency, the 2′-OH of the ribose sugar of residue 8 may be involved in stabilizing the overall tRNA secondary structure, as it has been shown in tRNAPro where the 2’-OH interacts with G46 in the variable loop (21). Thus, it is possible the base changes are not important because the phosphate backbone is engaged in relevant contacts which are not affected by the mutations.
In the T-loop of tRNAPyl, protection was observed at A57 and A58. Changes of A58 to U, C or G were all deleterious to the aminoacylation efficiency of tRNAPyl (L > 1000). A58 is almost universally conserved in tRNAs due to the interaction it makes with U54, both of which are conserved in all tRNAPyl isoacceptors. Thus, the U54·A58 contact is likely to be present in tRNAPyl, and essential for maintaining the tRNA tertiary structure. The formation of this bond would expose the phosphate backbone to solvent, which the footprinting results indicate is protected by the PylRS during binding.
At A57, changes to U and C had moderate affects on aminoacylation efficiency of L = 43 and 31, respectively, while the purine-to-purine change of A57G had only L = 5.6. Position 57 is generally conserved as a purine in canonical tRNAs because it is involved in a stacking interaction important for stabilization of the T-loop structure. This explains the larger loss in aminoacylation of the U and C mutations relative to G, and the conservative change G57 in the archaeal tRNAs.
A unique feature of the archaeal and bacterial tRNAPyl is the absence of the almost universally conserved sequences of T54Ψ55C56 in the T-loop, and G18G19 in the D-loop. When canonical tRNA molecules adopt their characteristic L-shape tertiary structure these two motifs are brought into contact and interactions between them help stabilize the overall tertiary structure. Specifically, the interactions occur between G18·Ψ55 and G19· C56. This also leaves other nucleotides exposed to solvent and available for interactions with a synthetase. As both the T54Ψ55C56 and G18G19 sequences are absent in tRNAPyl, understanding how the D- to T- loop contacts are established is of interest.
We found that changing A56 in D. hafniense tRNAPyl to any of the other three nucleotides resulted in only small changes in aminoacylation relative to wild-type (L < 10). There is evidence that the highly conserved G18·Ψ55 and G19·C56 sequences are not essential in certain tRNAs, such as E. coli tRNAAlaCUA (22). These contacts in tRNAPyl may be compensated for by creating other stabilizing interactions.
DISCUSSION
Recognition of a tRNA by its cognate aminoacyl-tRNA synthetase is dependent on the interaction of several factors. These include specific nucleotide–protein contacts, the overall tRNA structure, and the ability of the tRNA to undergo conformational changes upon binding by the cognate synthetase. The interplay of these factors ensures correct and productive interactions between a synthetase and tRNA by creating an exclusive RNA-protein interface upon which binding occurs. In the case of D. hafniense tRNAPyl, the discriminator base and the first base pair were identified as important identity elements for PylRS. The discriminator base G73 could not be changed to any of the other three nucleotides without significantly reducing aminoacylation efficiency. Although changing the first base pair of the acceptor stem G1·C72 to A1·U72 decreases aminoacylation efficiency (L = 62.7), A2·U71 decreases aminoacylation levels only in the presence of the A2·U72 change (L = 106.4), and not by itself. The increased loss in aminoacylation efficiency upon changing both the first and second base pairs in the acceptor stem suggests that the A2·U71 does not make direct contact with the PylRS. The discriminator base, G1·C72 and G2·C71 are conserved throughout the archaeal tRNAPyl as well, suggesting these residues may play an important role in tRNAPyl aminoacylation across species.
The anticodon was shown not to be recognized by the D. hafniense PylRS. Most AARSs recognize the tRNA anticodon through a specialized RNA-binding domain appended to the catalytic core at the amino or carboxyl terminus of the synthetase. For example, the amino terminal of the class II synthetases AspRS and LysRS function as anticodon-binding domains (23,24). PylRS from D. hafniense has a natural 100 amino acid amino-terminal truncation when compared to the archaeal counterparts. Thus, it is still possible that the PylRS in Methanosarcinaceae uses this amino-terminal domain for anticodon recognition. Consequently, these results may differ in the Methanosarcinaceae context.
Although the anticodon stem and loop are relatively conserved between the archaeal and bacterial tRNAPyl isoacceptors, no contact was observed between PylRS and tRNAPyl in this region other than the first base pair of the anticodon stem. Instead, contacts were observed along the TΨC and D-arms and acceptor stem, which is also where the majority of the nucleotide changes between the bacterial and archaeal tRNAPyl are found. Of the 23 nucleotides unique to D. hafniense tRNAPyl, 5 are in the anticodon stem, while 18 are in the D- or T-arms, or acceptor stem. This leaves open the possibility that the archaeal PylRS core relies on contacts different from those found here in the bacterial context.
In addition, the mutations in tRNAPyl may be in part rooted in the higher GC content of the D. hafniense genome (47%) relative to the archaeal genomes (39–42%). Of the 23 residue changes in the D. hafniense tRNAPyl, 15 are changes to G or C, and only 8 are to A or T. Similarly, the D. hafniense pylS has 51.3% GC content, while the M. barkeri pylS has 42.7% GC.
Identity elements in tRNAs can confer global specificity through different mechanisms. One perspective is that each nucleotide makes essential contributions, and changes at these positions each result in large losses in aminoacylation efficiency (generally over 100-fold). This is the case for tRNAs such as yeast tRNAPhe, where the anticodon, and the bases A73 and G20 all act independently (25) and were shown to be sufficient in a transplantation experiment (26). Alternatively, the additive affect of many moderate identity elements can be the dominant mode of recognition. The functional relationship between identity elements of tRNAAsp were investigated and shown that combined effects of multiple identity elements could be additive (27). For D. hafniense tRNAPyl only moderate identity elements were found; the D-stem base pairs G10·C25 and A11·U24, the base G8, the first base pair G1·C72 and discriminator base G73. These may have a combined effect that would be important for PylRS recognition. While we were unable to find discrete residues with larger (over 100-fold) contributions to aminoacylation efficiency, this may still be sufficient to ensure specific aminoacylation of tRNAPyl by PylRS as the major identity elements G73 and G1·C72 are only found together in two other tRNAs in D. hafniense, tRNAAsp and tRNASer. In the in vivo competitive environment, the mischarging of these tRNAs by PylRS is unlikely as they are lacking both the tRNAPyl secondary identity elements and unique secondary structure. The synthetases responsible for tRNAAsp and tRNASer aminoacylation depend on additional identity elements absent in tRNAPyl (19) which presumably prevent misacylation of tRNAPyl with aspartate or serine. Therefore, specific aminoacylation of tRNAPyl by PylRS is likely to result from a combination of direct interactions with the identity set revealed here, and recognition of the overall non-canonical tertiary structure of tRNAPyl. Future studies will determine whether the same is true for the archaeal PylRS and tRNAPyl.
Interestingly, tRNAPyl shares many unique structural features with the Bos taurus mitochondrial tRNASer with the UGA anticodon sequence (tRNASerUGA) including the elongated 6 base pair anticodon stem, a short 5 nucleotide D-loop, the presence of a single residue separating the acceptor and D-stems, and a 3 residue variable loop (20). Although non-canonical structural similarities are present, there is little sequence homology between tRNASerUGA and tRNAPyl. A solution NMR study indicated that the packing of the of tRNASerUGA core is relatively compact due to the missing nucleotides (28). As the compact core was determined to be a result of the structural feature mentioned above, and not specific residues, it is likely that these conclusions hold true for tRNAPyl. The presence of a compact tRNA core has precedent as being a stabilizing factor in the tRNA structure and subsequently leads to enhanced protein binding (12) by reducing the entropic contributions to the binding free energy (29). Furthermore, the structure of B. taurus mitochondrial SerRS indicates the enzyme does not recognize the tRNASer anticodon, as is the case for tRNAPyl (30). The results presented here, taken together with these structural studies, suggest a global identity of D. hafniense tRNAPyl that is likely dependent on the cumulative effect of several minor identity elements, along with the overall compact core structure.
This report also establishes the functionality of D. hafniense PylRS, despite the absence of approximately 100 amino acids from the amino-terminus of the bacterial enzyme relative to those from archaea. Further downstream of the pyl genes, the open reading frame pylSn encodes a putative protein with ∼36% sequence identity to the archaeal PylRS amino-terminus, which initially raised the possibility of a split PylRS in bacteria. However, the PylSn protein is not required for D. hafniense PylRS activity in vitro. The high level of conservation between PylSn and the archaeal PylRS amino-terminus, however, suggests that PylSn may have another role in vivo. Whether the amino-terminus from the Methanosarcinacea PylRS enzymes is important for enzyme activity or amino acid incorporation during translation in vivo will be determined in future studies.
ACKNOWLEDGEMENTS
We thank Sotiria Palioura for critical reading of this manuscript. This work was supported by NIH grant GM22854. Funding to pay the Open Access publication charge was provided by NIH grant GM22854.
Conflict of interest statement. None declared.
REFERENCES
- 1.Burke SA, Lo SL, Krzycki JA. Clustered genes encoding the methyltransferases of methanogenesis from monomethylamine. J. Bacteriol. 1998;180:3432–3440. doi: 10.1128/jb.180.13.3432-3440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paul L, Ferguson DJ, Jr, Krzycki JA. The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. J. Bacteriol. 2000;182:2520–2529. doi: 10.1128/jb.182.9.2520-2529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, Chan MK. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 4.Polycarpo C, Ambrogelly A, Bérubé A, Winbush SM, McCloskey JA, Crain PF, Wood JL, Söll D. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc. Natl. Acad. Sci. USA. 2004;101:12450–12454. doi: 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 6.Blight SK, Larue RC, Mahapatra A, Longstaff DG, Chang E, Zhao G, Kang PT, Green-Church KB, Chan MK, et al. Direct charging of tRNACUA with pyrrolysine in vitro and in vivo. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 7.Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 9.Polycarpo C, Ambrogelly A, Ruan B, Tumbula-Hansen D, Ataide SF, Ishitani R, Yokoyama S, Nureki O, Ibba M, et al. Activation of the pyrrolysine suppressor tRNA requires formation of a ternary complex with class I and class II lysyl-tRNA synthetases. Mol. Cell. 2003;12:287–294. doi: 10.1016/s1097-2765(03)00280-6. [DOI] [PubMed] [Google Scholar]
- 10.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfson AD, Uhlenbeck OC. Modulation of tRNAAla identity by inorganic pyrophosphatase. Proc. Natl. Acad. Sci. USA. 2002;99:5965–5970. doi: 10.1073/pnas.092152799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bullock TL, Sherlin LD, Perona JJ. Tertiary core rearrangements in a tight binding transfer RNA aptamer. Nat. Struct. Biol. 2000;7:497–504. doi: 10.1038/75910. [DOI] [PubMed] [Google Scholar]
- 13.Ambrogelly A, Korencic D, Ibba M. Functional annotation of class I lysyl-tRNA synthetase phylogeny indicates a limited role for gene transfer. J. Bacteriol. 2002;184:4594–4600. doi: 10.1128/JB.184.16.4594-4600.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polycarpo C, Herring S, Bérubé A, Wood JL, Söll D, Ambrogelly A. Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett. 2006;580:6695–6700. doi: 10.1016/j.febslet.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bullock TL, Uter N, Nissan TA, Perona JJ. Amino acid discrimination by a class I aminoacyl-tRNA synthetase specified by negative determinants. J. Mol. Biol. 2003;328:395–408. doi: 10.1016/s0022-2836(03)00305-x. [DOI] [PubMed] [Google Scholar]
- 16.Rudinger J, Hillenbrandt R, Sprinzl M, Giegé R. Antideterminants present in minihelixSec hinder its recognition by prokaryotic elongation factor Tu. EMBO J. 1996;15:650–657. [PMC free article] [PubMed] [Google Scholar]
- 17.Kreutzer R, Kern D, Giegé R, Rudinger J. Footprinting of tRNAPhe transcripts from Thermus thermophilus HB8 with the homologous phenylalanyl-tRNA synthetase reveals a novel mode of interaction. Nucleic Acids Res. 1995;23:4598–4602. doi: 10.1093/nar/23.22.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao B, Zhao G, Kang PT, Soares JA, Ferguson TK, Gallucci J, Krzycki JA, Chan MK. Reactivity and chemical synthesis of L-pyrrolysine- the 22nd genetically encoded amino acid. Chem. Biol. 2004;11:1317–1324. doi: 10.1016/j.chembiol.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theobald-Dietrich A, Frugier M, Giegé R, Rudinger-Thirion J. Atypical archaeal tRNA pyrrolysine transcript behaves towards EF-Tu as a typical elongator tRNA. Nucleic Acids Res. 2004;32:1091–1096. doi: 10.1093/nar/gkh266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yap LP, Musier-Forsyth K. Transfer RNA aminoacylation: identification of a critical ribose 2′-hydroxyl-base interaction. RNA. 1995;1:418–424. [PMC free article] [PubMed] [Google Scholar]
- 22.Zagryadskaya EI, Kotlova N, Steinberg SV. Key elements in maintenance of the tRNA L-shape. J. Mol. Biol. 2004;340:435–444. doi: 10.1016/j.jmb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Commans S, Plateau P, Blanquet S, Dardel F. Solution structure of the anticodon-binding domain of Escherichia coli lysyl-tRNA synthetase and studies of its interaction with tRNA(Lys) J. Mol. Biol. 1995;253:100–113. doi: 10.1006/jmbi.1995.0539. [DOI] [PubMed] [Google Scholar]
- 24.Berthet-Colominas C, Seignovert L, Hartlein M, Grotli M, Cusack S, Leberman R. The crystal structure of asparaginyl-tRNA synthetase from Thermus thermophilus and its complexes with ATP and asparaginyl-adenylate: the mechanism of discrimination between asparagine and aspartic acid. EMBO J. 1998;17:2947–2960. doi: 10.1093/emboj/17.10.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sampson JR, Behlen LS, DiRenzo AB, Uhlenbeck OC. Recognition of yeast tRNAPhe by its cognate yeast phenylalanyl-tRNA synthetase: an analysis of specificity. Biochemistry. 1992;31:4161–4167. doi: 10.1021/bi00132a002. [DOI] [PubMed] [Google Scholar]
- 26.Frugier M, Florentz C, Schimmel P, Giegé R. Triple aminoacylation specificity of a chimerized transfer RNA. Biochemistry. 1993;32:14053–14061. doi: 10.1021/bi00213a039. [DOI] [PubMed] [Google Scholar]
- 27.Putz J, Puglisi JD, Florentz C, Giegé R. Additive, cooperative and anti-cooperative effects between identity nucleotides of a tRNA. EMBO J. 1993;12:2949–2957. doi: 10.1002/j.1460-2075.1993.tb05957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashi I, Kawai G, Watanabe K. Higher-order structure and thermal instability of bovine mitochondrial tRNASerUGA investigated by proton NMR spectroscopy. J. Mol. Biol. 1998;284:57–69. doi: 10.1006/jmbi.1998.2151. [DOI] [PubMed] [Google Scholar]
- 29.Yamasaki S, Nakamura S, Terada T, Shimizu K. Mechanism of the difference in the binding affinity of E.coli tRNAGln to glutaminyl-tRNA synthetase caused by non-interface nucleotides in variable loop. Biophys. J. 2006 doi: 10.1529/biophysj.106.093351. biophysj.106.093351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chimnaronk S, Gravers Jeppesen M, Suzuki T, Nyborg J, Watanabe K. Dual-mode recognition of noncanonical tRNAsSer by seryl-tRNA synthetase in mammalian mitochondria. EMBO J. 2005;24:3369–3379. doi: 10.1038/sj.emboj.7600811. [DOI] [PMC free article] [PubMed] [Google Scholar]