Abstract
Meiotic recombination is initiated by DNA double-stranded break (DSB) formation catalyzed by Spo11, a type-II topoisomerase-like transesterificase, presumably via a dimerization-mediated mechanism. We demonstrate the existence of in vivo interactions between Spo11 proteins carrying distinct tags, and the chromatin-binding and DSB activity of tagged Spo11 at innate and targeted DSB sites upon fusion to the Gal4 DNA-binding domain. First we identified the interaction between Spo11-3FLAG and Gal4BD-Spo11 proteins, and established that this interaction specifically occurs at the time of DSB formation. We then observed that presence of the Gal4BD-spo11Y135F (nuclease-deficient) protein allows Spo11-3FLAG recruitment at the GAL2 locus, indicative of the formation of a hetero-complex near the GAL2 UAS sites, but no formation of double- or single-strand breaks. Spo11 self-interaction around the GAL2 DSB site depends on other proteins for DSB formation, in particular Rec102, Rec104 and Rec114. Together, these results suggest that in vivo self-association of Spo11 during meiosis is genetically regulated. The results are discussed in relation to possible roles of Spo11 self-interaction in the control of the cleavage activity.
INTRODUCTION
Meiotic recombination is critical for sexual reproduction, since it is essential for the viability of gametes and their genetic diversity. In meiosis, recombination between homologous chromosomes is initiated by programmed double-stranded DNA breaks (DSBs), which are transiently and meiotically introduced at recombination initiation sites, after the completion of premeiotic DNA replication. In the yeast Saccharomyces cerevisiae, ten genes (SPO11, MEI4, MER2/REC107, REC102, SKI8/REC103, REC104, REC114, MRE11, RAD50 and XRS2) are required for meiotic DSB formation, in addition to two genes for meiosis-specific RNA splicing (MER1 and MRE2) (1). Null mutations of these genes result in defective meiotic recombination and spore inviability.
Spo11, an evolutionarily conserved protein, is the catalytic component of meiotic DNA cleavage, and many studies suggest that Spo11 cleaves DNA via a topoisomerase-like transesterification reaction, forming intermediates by linking to the 5′ ends of the DNA strands (2,3). These intermediates are then further processed via asymmetric endonucleolytic cleavage that is likely mediated by one or more other endonucleases (4). Importantly, Spo11 shares structural similarity with the A subunit of topoisomerase 6 (Top6A) that has been found only in the archaeon Sulfolobus shibatae. Top6A has ATP-dependent DNA relaxation activity in vitro and forms a heterotetramer with Top6B (5). Crystallized Top6A of Methanococcus jannaschii forms a U-shaped dimer with a putative DNA interaction channel (6). To date, whether or not this dimer is similar to the Spo11 complex has not been determined, since it has proved difficult to purify functional Spo11 protein. In S. cerevisiae, the heterozygous combination of various DSB-defective spo11 mutant alleles and SPO11-3HA results in a partial dominant negative phenotype with respect to DSB formation, although the SPO11-3HA/SPO11-3HA homozygous diploid strain has a wild-type level of DSB formation, suggesting that Spo11 functions in dimeric or multimeric form (7). The difficulties experienced in purifying soluble Spo11 led us to investigate the interaction between Spo11 subunits in vivo.
Regulation of Spo11 endonucleolytic activity can be envisaged to occur in at least two ways. In the first model, cleavage activity of Spo11 might be activated or facilitated by other proteins. In S. cerevisiae, at least nine other proteins are known to be required for DSB formation, but their role remains poorly understood; genetic and/or physical studies indicate that they interact with each other (1). Ski8 interacts directly with Spo11, and this interaction, in addition to the interaction of Spo11 with Rec102 and Rec104, is essential for DNA cleavage (8), suggesting that these proteins form a complex with Spo11 to activate the cleavage reaction. Mer2/Rec107, Mei4 and Rec114 interact with each other, and contact Xrs2 of the Mre11/Rad50/Xrs2 complex, which is required for DSB formation and repair, raising the additional question of how DSB formation is coupled to single-strand end processing (8–10). Overexpression of Rec114 in S. cerevisiae is known to inhibit meiotic DSB formation, suggesting that Rec114 is a key regulator of meiotic DSB formation (11), but the molecular basis of this effect has not yet been elucidated. Some additional factors, including histone acetyltransferases and chromatin-remodeling factors, are involved in meiotic alteration of local chromatin structure at DSB sites (12–14), which is a prerequisite for meiotic DSB cleavage. DSB formation is temporally correlated with DNA replication (15), and is also controlled by cell cycle regulators, since inactivation of the S cyclins Clb5-Clb6, CDK and Hsk1 kinasae (S. pombe homolog of CDC7) eliminates DSB formation (15–17).
An alternative but not mutually exclusive model to explain the regulation of Spo11 activity is to invoke chromosomal features (chromatin or higher order chromosome structures) defining permissive or nonpermissive chromosomal domains for DSB formation. Genome-wide studies have shown that DSB regions are not randomly distributed, but are rather distributed nonrandomly in DSB-hot and DSB-cold domains (9,18,19). Whether this reflects the regulation of Spo11 binding or cleavage is not known.
The fusion of Spo11 protein to the Gal4 DNA-binding domain (Gal4BD-Spo11), as accomplished by us in a previous study, produced a novel experimental tool that enabled additional information about molecular mechanism of the DSB cleavage by Spo11 to be determined (20). Expression of Gal4BD-Spo11 in spo11Δ diploids allowed recombinogenic DSB formation at innate DSB sites and wild-type production of viable spores. In addition, Gal4BD-Spo11 expression allowed the targeted stimulation of novel DSB sites, located in the vicinity of Gal4 consensus-binding sites (UAS), such as in the GAL2 locus located within a DSB-cold domain on chromosome XII (20). In that study, we examined the genetic requirements for the formation of these targeted DSBs. Interestingly, we found that DSB formation at the targeted DSB sites required all of the known factors (DSB proteins and Clb5-Clb6) that are indispensable for DSB formation at innate DSB sites. This indicated that Gal4BD-Spo11 catalyzes DSB formation near the Gal4 UAS by locally recruiting the components necessary for DSB formation, whereas they might be absent or improperly localized in DSB-cold domains. In this model, the binding of Spo11 to DSB sites would be the first rate-limiting step for DSB formation. However, the observation that uncleaved DNA intermediates are bound by Spo11 suggests that the activation of Spo11 cleavage is controlled separately from its physical interaction with DSB sites (21). Thus, activation of Spo11 cleavage activity is likely more complex than initially anticipated.
Here, to provide insights into the activation and catalytic processes controlling Spo11 activity, we examined the in vivo interaction between Spo11 and Gal4BD-Spo11 proteins carrying distinct tags, and assayed their chromatin-binding and DSB formation activity at innate (YCR048w) and targeted (GAL2) sites. We demonstrate that Rec102, Rec104 and Rec114 regulate meiotic association between Spo11 subunits on chromatin sites. Such meiotic self-association of Spo11 may be important for the regulation of DNA cleavage activity.
MATERIALS AND METHODS
Yeast strains and media
All yeast strains used in this study were isogenic derivatives of SK1 and are listed in Table 1. The strains were cultured to a concentration of ∼4 × 107 cells per ml in an appropriate medium (SPS: 0.5% yeast extract, 1% peptone, 1% potassium acetate, 0.17% yeast nitrogen base without ammonium sulfate and amino acids, 0.05 M potassium phthalate, 0.5% ammonium sulfate, pH 5.0) with nutritional supplements, and further incubated in a medium with 1% potassium acetate (SPM) with vigorous shaking.
Table 1.
Strains
| Strain | Genotype |
|---|---|
| ORD5806 | trp1::ADH1-GAL4BD-SPO11-TRP1-KANMX/” spo11Δ::URA3 arg4-bgl/-rV nuc1::LEU2/” |
| YHS113 | SPO11-6HIS-3FLAG-loxP-KANMX-loxP/” arg4/” |
| YHS173 | trp1::ADH1-GAL4BD-SPO11-TRP1-KANMX/” SPO11-6HIS-3FLAG-loxP-KANMX-loxP/” spo11Δ::URA3 arg4/” |
| YHS326 | rad50S::URA3/” nuc1Δ::LEU2/” |
| YHS363 | trp1::ADH1-GAL4BD-SPO11-TRP1-KanMX/” spo11Δ::URA3 rad50S::URA3/” nuc1Δ::LEU2/” |
| YHS395 | aur1::ADH1-SPO11-3F-AUR1-C/” spo11Δ::URA3/” arg4/” |
| YHS397 | aur1::ADH1-SPO11-3F-AUR1-C/” spo11Δ::URA3/” rad50S::URA3/” arg4/” |
| YHS425 | trp1::ADH1-GAL4BD-SPO11-TRP1-KanMX/” aur1::ADH1-SPO11-3F-AUR1-C/” spo11Δ::URA3/” |
| YHS427 | trp1::ADH1-GAL4BD-SPO11-TRP1-KanMX/” aur1::ADH1-SPO11-3F-AUR1-C/” spo11Δ::URA3/” rad50S::URA3/” nuc1Δ::LEU2/” |
| YHS439 | trp1::ADH1-GAL4BD-spo11Y135F-TRP1-KANMX/” rad50S::URA3/” nuc1Δ::LEU2/” |
| YHS517 | trp1::ADH1-GAL4BD-spo11Y135F-TRP1-KanMX/” aur1::ADH1-SPO11-3F-AUR1-C/” spo11Δ::URA3/” rad50S::URA3/” nuc1Δ::LEU2/” |
| YHS518 | trp1::ADH1-GAL4BD-spo11Y135F-TRP1-KanMX/” aur1::ADH1-SPO11-3F-AUR1-C/” spo11Δ::URA3/” |
| YHS550 | trp1::ADH1-GAL4BD-spo11Y135F-TRP1-KanMX/” SPO11-6HIS-3FLAG-loxP-KANMX-loxP/” spo11Δ::URA3/” arg4/” |
| YHS620 | trp1::ADH1-GAL4BD-SPO11-TRP1-KanMX/” aur1::ADH1-SPO11-3F-AUR1-C/” spo11Δ::URA3/” rec102Δ::6HIS-3FLAG- loxP-KANMX-loxP/” |
| YHS639 | trp1::ADH1-GAL4BD-SPO11-TRP1-KanMX/” aur1::ADH1-SPO11-3F-AUR1-C/” spo11Δ::URA3/” rec103Δ::6HIS-3FLAG- loxP-KANMX-loxP/” |
| YHS612 | trp1::ADH1-GAL4BD-SPO11-TRP1-KanMX/” aur1::ADH1-SPO11-3F-AUR1-C/” spo11Δ::URA3/” rec104Δ::6HIS-3FLAG- loxP-KANMX-loxP/” |
| YHS615 | trp1::ADH1-GAL4BD-SPO11-TRP1-KanMX/” aur1::ADH1-SPO11-3F-AUR1-C/” spo11Δ::URA3/” rec107Δ::6HIS-3FLAG- loxP-KANMX-loxP/” |
| YHS616 | trp1::ADH1-GAL4BD-SPO11-TRP1-KanMX/” aur1::ADH1-SPO11-3F-AUR1-C/” spo11Δ::URA3/” rec114Δ::6HIS-3FLAG- loxP-KANMX-loxP/” |
| YHS618 | trp1::ADH1-GAL4BD-SPO11-TRP1-KanMX/” aur1::ADH1-SPO11-3F-AUR1-C/” spo11Δ::URA3/” mei4Δ::6HIS-3FLAG-loxP- KANMX-loxP/” |
| YHS843 | trp1::ADH1-GAL4BD-SPO11-TRP1-KanMX/” aur1::ADH1-spo11Y135F-3F-AUR1-C/” spo11Δ::URA3/” |
| YHS900 | trp1::ADH1-GAL4BD-SPO11-3FLAG-TRP1-KanMX/” spo11Δ::URA3/” arg4/” |
All strains are ho::LYS2/” lys2/” trp/” ura3/” leu2/”.
Plasmid construction
The integration plasmids for expression of Gal4BD–spo11Y135F (pAP1-YF) and Spo11-3FLAG (pAUS) were constructed as follows. The plasmid pAS2-1 (Clontech Laboratories, Inc., USA) was digested with BseRI and NsiI to remove the 2 μm replication origin. The kanMX6 cassette was then inserted into the NruI site. The DNA fragment for the expression of Spo11Y135F was amplified by PCR, followed by insertion into the NdeI/BamHI site (pAP1-YF), which enabled expression of Spo11Y135F under the control of the ADH1 promoter. The plasmid pAUS was constructed as follows. First, a BamHI fragment harboring the ADH1 promoter and terminator derived from pAUR123 (TaKaRa-Bio Co. Ltd, Japan) was inserted into the integrative pAUR101 plasmid digested with SacI and SphI. Then, the SPO11-3FLAG DNA fragment was amplified by PCR and inserted into a KpnI/XbaI site in the multiple cloning site. The linearized pAUS was further integrated into the aur1 locus, followed by selection on YEPD containing 0.5 mg/ml aureobasidin A (TaKaRa-Bio Co. Ltd). Expression was confirmed by immunoblotting using an anti-FLAG peptide (Sigma-Aldrich, Inc., USA) and anti-Gal4 DNA-binding domain (Gal4BD) antibodies (Santa Cruz Biotechnology, Inc., USA).
Immunoprecipitation and chromatin immunoprecipitation
For immunoprecipitation (IP), approximately 5 × 109 cells were harvested from 250 ml of meiotic culture, washed twice with ice-cold water, and suspended in 4 ml of the lysis buffer (50 mM HEPES-KOH at pH 7.5, 300 mM KCl, 0.005% Tween 20, 0.005% NP-40, 10% glycerol, 2 mM NaF, 0.4 mM Na3VO4, 0.5 mM Na-pyrophosphate, 2 mM beta-glycerophosphate, ×2 complete protease inhibitor, Roch) and added 5 μl DNase I. After the cells were disrupted by using a Multibeads-Shocker (Yasui Kikai Co Ltd, Japan), cell lysates were cleaned twice by centrifugation at 15 000 rpm for 15 min. The resulting cell extracts were then incubated for 5 h with 40 μl of protein-G DynaBeads (Invitrogen, Inc., USA) along with the anti-FLAG and anti-Gal4BD antibodies. Immunocomplexes were collected with a magnet and then washed four times with lysis buffer. For the DNase I treatment, the beads were suspended in lysis buffer containing 2 mM MgCl2, incubated for 30 min at 30°C, then washed three times and boiled in ×2 SDS-PAGE sample buffer (4% SDS, 20% glycerol, 10% beta-mercaptoethanol, 0.04% bromophenol blue, 120 mM Tris-HCl pH 6.8). Chromatin immunoprecipitation (ChIP) was carried out as described earlier with some modifications (22). First, 25 ml of the meiotic culture suspensions (2 × 107 cells per ml) were treated with 1% formaldehyde for 10 min at room temperature at the indicated time points. The cells were then pelleted and resuspended in 430 μl of lysis buffer II (50 mM Na-deoxycholate, 1 mM ethylenediaminetetraacetic acid (EDTA), 50 mM HEPES-KOH [pH 7.5], 1% Triton X-100) containing 140 mM NaCl, followed by successive washes in lysis buffer II with 500 mM NaCl.
Multiplex and quantitative PCR of immunoprecipitated DNA
For multiplex PCR analysis, 1-ml aliquots of immunoprecipitate were analyzed using the primer pairs PPGAL2 and PPSMC1 (see Results and discussion section and Figure 2B) at a final concentration of 20 pmol each. The following primers were used for PCR: forward primer for PPGAL2, 5′-CTAGAAAGTTAACTGTGCACATATTC-3′; reverse primer for PPGAL2, 5′-GGCATATTGTTCTCCTCAACTGCC-3′; forward primer for PPSMC1, 5′-AAAGATTTAATCTATAGAGGTGTTC-3′; reverse primer for PPSMC1, 5′-TTATAGGAGACAGTTTTTCCATCAA-3′; forward primer for PP048w, 5′-CGTACGATAACGTGATCCTGCCACAGG-3′; reverse primer for PP048w, 5′-CCGAGACTTGCTCTTCAGGTGTGAAA-3′. The reaction mixture (50 ml) contained 0.025 U ExTaq polymerase (TaKaRa-Bio Co. Ltd), and 5 ml of ×10 PCR buffer supplied by the manufacturer. PCR conditions were as follows: incubation at 95°C for 2 min; 30 cycles of incubations for 10 s at 95°C, 10 s at 55°C and 1 min at 72°C; followed by a final incubation for 7 min at 72°C. The amplified products were separated on 3% Metaphor agarose (Cambrex Bio Science Rockland, Inc.). Real-time PCR was performed using the ABI 7300 system (Applied Biosystems, Inc.). We used SYBR-premixed ExTaq (TaKaRa-Bio Co. Ltd) for the samples and the ROX-dye for the references. After a 2-min incubation at 95°C, reaction cycles (95°C for 10 s, 55°C for 30 s and 72°C for 1 min) were repeated 45 times, followed by a 7-min incubation at 72°C. Then, dissociation curve analysis was conducted to determine if specific products were amplified. The amount of precipitated (IP) DNA and whole-cell extract (WCE) DNA were measured relative to a standard sample of yeast genomic DNA. The results are expressed in ratios of IP versus WCE DNA (INPUT), which are further normalized to those obtained with intragenic region of SMC1 gene, located in a DSB-cold domain.
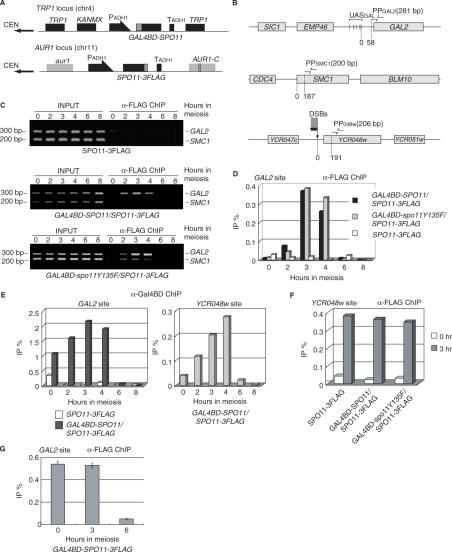
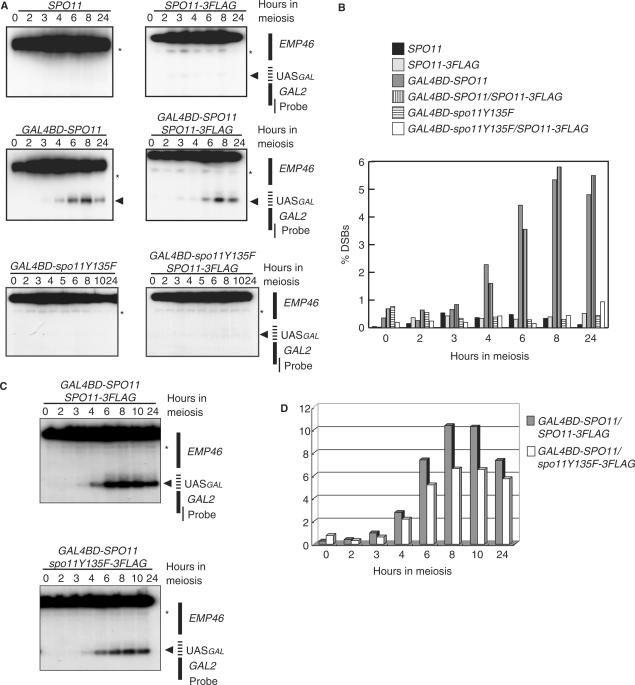
Figure 2.
Gal4BD–Spo11-dependent meiotic association of Spo11-3FLAG with the GAL2 UAS region in vivo. (A) SPO11-3FLAG was integrated into the AUR1 locus so that aureobasidin A could be used as a selection marker. GAL4BD-SPO11 (YHS425) or -spo11Y135F (YHS518) transgenes were integrated into the TRP1 locus. Both transgenes were constitutively expressed under the control of the ADH1 promoter (thick arrows) during the course of meiosis. Gray and black boxes represent the indicated loci. (B) Maps of the regions used for PCR detection in chromatin immunoprecipitation (ChIP). Five UAS sequences (indicated by vertical bars) exist around the GAL2 promoter region. PPGAL2 (indicated by the two arrows in the upper diagram) are located 58 bp away from the GAL2 locus. PPSMC1 (indicated by the two arrows in the middle diagram) amplifies an intragenic region of SMC1 (187 bp away from the BLM10 locus). PP048w (indicated by the two arrows in the lower diagram) is located 191 bp distal to the DSB site of the YCR048w locus. Gray boxes represent the indicated loci. (C) Meiotic binding of Spo11-3FLAG in the GAL2 UAS region in the presence (middle and lower panels) or absence (upper panel) of Gal4BD-fused protein (middle, Gal4BD-Spo11; lower, Gal4BD-spo11Y135F). Cells were crosslinked at 0, 2, 3, 4, 6 or 8 h of meiosis, and harvested to prepare WCEs from the cells expressing Spo11-3FLAG (YHS395), Gal4BD–Spo11/Spo11-3FLAG (YHS425) and Gal4BD–spo11Y135F/Spo11-3FLAG (YHS518). ChIP experiments were conducted using the WCEs and anti-FLAG antibody as described in the Materials and methods section. The immunoprecipitated (IP) DNA and the total genomic DNA from the WCEs (INPUT) were then amplified by 30 cycles of PCR and separated on a 3% agarose gel by electrophoresis. The upper and lower bands corresponding to the DNA fragment amplified by PPGAL2 (the GAL2 UAS region) and PPSMC1 (an internal control) are shown. The numbers at the left and upper sides of the panels indicate the positions of size markers (200 and 300 bp) and hours in SPM (lanes 0, 2, 3, 4, 6 and 8). (D) Quantitative real-time PCR (qPCR) of the immunoprecipitated DNA from samples taken from each time point as described in (C). The graph shows the kinetics of Spo11-3FLAG binding to the GAL2 UAS region during meiosis. The vertical axis indicates IP efficiencies (IP%) for the GAL2 UAS region, normalized with reference to the SMC1 locus. Numbers beneath the graphs are culture time (hours) in SPM. The experiment was performed independently twice. (E) Quantification of immunoprecipitated DNA at the GAL2 (left graph) and innate YCR048w (right graph) DSB sites by using the anti-Gal4BD antibody. The left graph shows the kinetics of Gal4BD–Spo11 binding to the GAL2 UAS site in YHS425 (co-expression of Gal4BD–Spo11 and Spo11-3FLAG) and YHS395 (expression of only Spo11-3FLAG). The right graph shows the ChIP signal at the innate YCR048w DSB site in YHS425. The experiment was performed independently twice. (F) Quantification of the anti-FLAG antibody-immunoprecipitated DNA at the YCR048w DSB sites in cells expressing Spo11-3FLAG alone, Gal4BD–Spo11 plus Spo11-3FLAG or Gal4BD-spo11Y135F plus Spo11-3FLAG. Open and gray bars represent signals at 0 and 3 h of meiosis. The ratios were normalized with reference to the values at the SMC1 locus. (G) Quantification of immunoprecipitated DNA using anti-FLAG antibody at the GAL2 in cells expressing Gal4BD–Spo11-3FLAG (YHS900). Numbers beneath the graphs are culture time (0, 3, 6 h) in SPM. The experiment was performed independently three times.
Detection of DNA breaks during meiosis
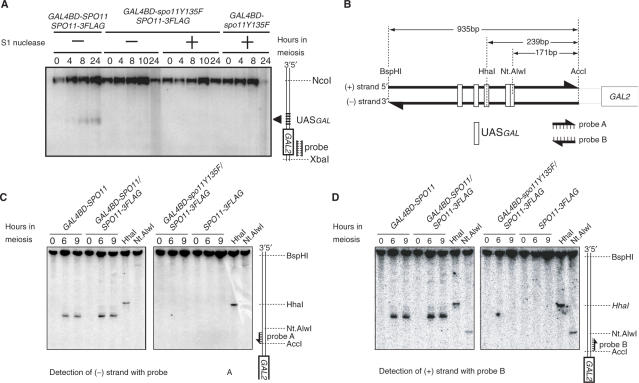
All DNA samples, except those dedicated to SSB detection presented in Figure 6B and C, were prepared in plugs of low-melting-point agarose (23,24). After the plugs were equilibrated with restriction enzyme buffers (New England Biolabs, USA) and melted at 65°C for 10 min, they were digested with AseI (for the YCR048W locus) and NcoI/XbaI (for the GAL2 locus) for 4 h at 37°C. Each digested sample was then separated by electrophoresis in a 1.0% agarose gel (40 cm long) containing TAE. The separated DNA fragments were further alkali-transferred to charged nylon membranes (Biodyne B membrane, PALL, EA). To determine the presence of SSBs, the agarose plugs containing genomic DNA were directly treated with NcoI/XbaI without melting. Thereafter, the plugs were equilibrated extensively with the S1 buffer (30 mM Na-acetate at pH 4.5, 280 mM NaCl, 1 mM ZnSO4) overnight. After the agarose plugs were melted at 65°C for 10 min, S1 nuclease was added and the mixture was incubated for 30 min at 37°C (25,26). To terminate the reaction, 0.5 M EDTA was added to the reaction mixture. To detect SSBs using single-stranded DNA probes, genomic DNA was extracted from cells harvested at 0, 6 and 9 h after transfer to SPM media as described earlier with minor modifications (27). After cells were spheroplasted with Zymolyase-20T (MP Biomedicals, Inc.) in spheroplast buffer (1% 2-mercaptoethanol, 1 M sorbitol, 0.1 M EDTA [pH 8.0]), they were lysed and digested in lysis buffer (50 mM EDTA [pH 8.0], 50 mM Tris [pH 8.0], 0.5% SDS, 200 µg of proteinase K). At the GAL2 locus, the purified genomic DNA was treated with AccI, BspHI, HhaI and Nt.AlwI (New England BioLabs) for several hours. Nt.AlwI was used as a positive control for SSB detection. All samples were purified using a NucleoSpin Extract II kit (Macherey-Nagel) and suspended in TE and loading buffer (95% formamide, 10 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue). After being denatured for 5 min at 100°C, the DNA fragments were separated in 6% polyacrylamide gels containing 8 M urea and ×1 TBE buffer at 240 V for 45 min, and transferred to Nylon membranes (GeneScreen, Perkin-Elmer). The preparation of strand-specific probes was performed as described earlier (28). A 152-bp DNA fragment was amplified using a set of primer (5′-TCCCATCTCAAGATGGGGAG-3′ and 5′-CTACAAAACTTATCCTATCTCCAC-3′), and used as a template for preparation of 32P-labeled strand-specific probes. Probes referred to as probes A and B (see Figure 6B) were prepared by multiple-cycle primer extension using an each primer, respectively.
Figure 6.
Detection of SSBs in the GAL2 UAS region. (A) Detection of SSBs by S1 nuclease. Plugs containing genomic DNA from wild-type cells were directly equilibrated in restriction digestion buffer and treated with NcoI and XbaI. Genomic DNA recovered from the melted plugs was then incubated in the presence (lanes designated +) or absence (lanes designated −) of S1 nuclease. Numbers above the panels are culture time (hours) in SPM. The diagram is labeled as for Figure 5. (B) Schematic diagram of SSB detection in rad50S strains using denaturing polyacrylamide gel. Nt.AlwI introduces an SSB into specific sequences of DNA (5′-GGATCNNNN/N-3′). The GAL2 promoter has the sequence only on the + strand. Probes A and B were labeled by multiple round primer extension in the presence of radio-labeled dCTP. (C) and (D) Detection of SSBs on the −/+ strand by using strand-specific probes. All strains are homozygous for the rad50S allele. After genomic DNA was prepared from the meiotic cells at the indicated times (hours), it was treated with AccI and BspHI. The right two lanes in each panel contain 5% DNA fragments, which were further digested with HhaI or Nt.AlwI. Strains used were YHS363, YHS397, YHS427 and YHS514.
RESULTS
Gal4BD–Spo11 and Spo11-3FLAG interact at the time of meiotic DSB formation
X-ray crystallography of the Spo11-like core region of the M. jannaschii Top6A subunit revealed that this region can interact stably with itself via a deep groove that we assume acts as a DNA-binding site (6), suggesting that Spo11 has the ability to interact with itself. To examine the interactions between Spo11 subunits, we constructed diploids co-expressing ×3 FLAG-tagged Spo11 (Spo11-3FLAG) and Gal4BD-Spo11 (or Gal4BD-spo11Y135F, a nuclease-deficient mutant of Spo11) proteins under the control of native and ADH1 promoters, respectively. Upon sporulation, tetrad analysis indicated that both co-expressing strains produced a wild-type level of viable spores (247/258 and 235/240, respectively), indicating that meiosis occurred normally in these strains.
Protein expression and the physical interactions between Spo11 subunits were first examined using WCEs and immunoblotting. Cells were harvested at two time points: when cells started sporulation (t = 0 h) and when meiotic DSBs were actively being formed (t = 3.5 h). Western blotting analysis using anti-FLAG or anti-Gal4BD antibodies revealed that all proteins (Spo11-3FLAG, Gal4BD-Spo11 and Gal4BD-spo11Y135F) were present at similar and steady levels in WCEs, when they are expressed under the control of ADH1 promoter, from premeiotic and meiotic cells (Figure 1A–C).
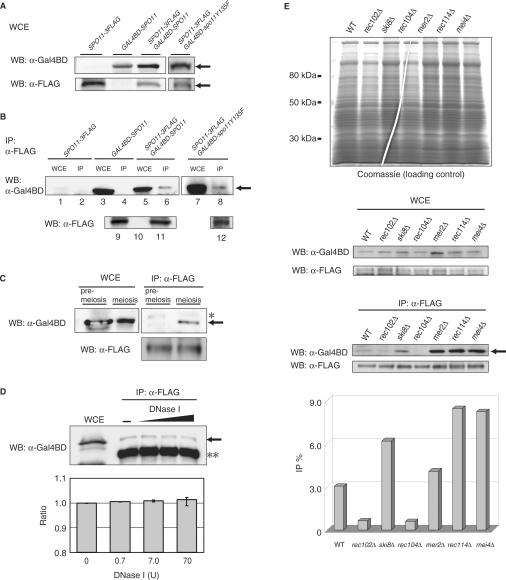
Figure 1.
Physical interactions between Spo11 proteins. (A) Immunoblotting analysis for Gal4BD-fused Spo11 and Spo11-3FLAG expressed in meiotic cells. Whole-cell extracts (WCEs) of meiotic cells were prepared from cells cultured for 3.5 h. Upper panel: anti-Gal4BD; lower panel: anti-FLAG immunoblotting. Arrows indicate Gal4BD–Spo11 (upper panel) or Spo11-3FLAG (lower panel) proteins. ‘WB’ means Western blotting. (B) Immunoprecipitation (IP) of Spo11-3FLAG with Gal4BD–Spo11 (or Gal4BD–spo11Y135F). YHS113 (Spo11-3FLAG), ORD5806 (Gal4BD–Spo11), YHS173 (Gal4BD–Spo11/Spo11-3FLAG) and YHS550 (Gal4BD–spo11Y135F/Spo11-3FLAG) cells were transferred to a sporulation medium (SPM). Meiotic cells were disrupted under non-denaturing conditions. Then, WCEs were immunoprecipitated with anti-FLAG antibody as described in the Materials and methods section. Gal4BD-fused proteins were detected by immunoblotting with anti-Gal4BD antibody. Strains expressing Spo11-3FLAG alone or Gal4BD–Spo11 alone were used as negative controls. In these experiments, all SPO11-derived genes were expressed under the control of the native SPO11 promoter. Lanes marked WCE and IP represent WCEs and immunoprecipitates, respectively. Western blotting of IP fraction with anti-FLAG is shown in lanes 9–12 as a control (lane 9: Spo11-3FLAG, lane 10: Gal4BD–Spo11, lane 11: Gal4BD–Spo11/Spo11-3FLAG, lane 12: Gal4BD–spo11Y135F/Spo11-3FLAG) (C) Comparison of the interactions between Spo11 subunits during premeiosis and meiosis. Two aliquots of the culture were taken after incubation of the YHS425 strain in premeiotic SPS medium. One aliquot was processed to prepare premeiotic WCEs and the rest was further incubated in SPM for 3.5 h to prepare meiotic WCEs. IP experiments were conducted as described in (B). The asterisk indicates a band attributable to a non-specific reaction. The lower panel shows immunoprecipitated Spo11-3FLAG detected with anti-FLAG by immunoblotting analysis. (D) Treatment of the Spo11 complex with DNase I. Immunoprecipitates were treated with DNase I as described in the Materials and methods section. The double asterisk represents the mouse IgG heavy chain and the arrow indicates immunoprecipitated Gal4BD–Spo11 protein. The lower graph shows quantification of the band intensity for Gal4BD–Spo11 protein in upper panel. The data are averages of three independent experiments. YHS425 strain was used in this experiment. (E) Interactions between Spo11 in DSB-defective mutants. WCEs were prepared from wild-type (YHS425), rec102Δ (YHS620), rec103Δ (YHS639), rec104Δ (YHS612), rec107Δ (YHS615), rec114Δ (YHS616) and mei4Δ (YHS618) strains. The upper panel shows the polyacrylamide gel stained with Coomassie brilliant blue as a loading control for each mutant and wild-type WCE. The middle panel shows the results of immunoblotting of each WCE with anti-Gal4BD and anti-FLAG antibodies. The arrow in the lower panel indicates Gal4BD–Spo11 immunoprecipitated by anti-FLAG. This experiment was repeated three times using independent cultures. The bottom graph indicates quantification of the intensity of each band normalized with reference to those of Gal4BD–Spo11 in WCE.
We next examined the interaction between Spo11-3FLAG and Gal4BD-Spo11 by IP using an anti-FLAG antibody. We found that these proteins specifically interact at the time of meiotic DSB formation (Figure 1B, lane 6). In the control strain expressing Gal4BD-Spo11 alone, no anti-FLAG IP signal was detected in WCEs (Figure 1B, lane 4). Furthermore, the meiotic interaction between Spo11-3FLAG and Gal4BD-Spo11 was confirmed in the YHS425 diploid strain, in which both Spo11 constructs were inserted in the AUR1 locus (chromosome 11) and were constitutively expressed under the control of the same ADH1 promoter. At t = 3.5 h but not at t = 0 h, an interaction between Spo11-3FLAG and Gal4BD-Spo11 was detected in meiotic WCEs (Figure 1C). Quantitatively, the efficiency of Gal4BD-Spo11 IP with Spo11-3FLAG in meiosis was estimated to be ∼3% of the total Gal4BD-Spo11 protein in the WCEs (normalized with reference to the IP efficiency of Spo11-3FLAG; Figure 1E). Similarly, as shown in Figure 1B, this interaction was also detected in meiotic cells co-expressing Spo11-3FLAG and Gal4BD-spo11Y135F, indicating that the tyrosine residue of the Spo11 catalytic site is dispensable for the interaction between Spo11 subunits. We conclude that these Spo11 subunits interact in a meiosis-specific manner at the time of DSB formation, as suggested by studies on Spo11-oligonucleotide complex (4).
Since Spo11 forms a cleavage intermediate by covalently linking to the DSB ends (3), it is possible that the above interaction between Spo11 subunits is mediated by protein–DNA interactions. To test this hypothesis, we examined Spo11 interactions after treating the IP complex with various concentrations of DNase I in the presence of Mg2+. Irrespective of DNase I concentration, and thus after extensive digestion of the IP material, the Spo11-3FLAG and Gal4BD-Spo11 interaction was still detected (Figure 1D), indicating that their interaction is not mediated by protein–DNA interactions.
Arora et al. and other groups revealed that Spo11 directly or indirectly interact with other DSB proteins such as Rec102, Ski8/Rec103, Red104, Mer2/Rec107, Rec114 and Mei4 (8,29,30). Thus, we next investigated the interaction between Spo11 subunits in diploid deletion mutants for those proteins, that co-expressed Spo11-3FLAG and Gal4BD-Spo11 (Table 1). Western blotting analysis for the wild-type and DSB-deficient mutant strains revealed that Spo11-3FLAG and Gal4BD-Spo11 were present at similar levels in the WCEs (Figure 1E). However, in the anti-FLAG immunoprecipitated sample, the amount of Gal4BD-Spo11 significantly varied: it was reduced to background level in rec102Δ and rec104Δ mutants (Figure 1E), but increased 2–3-fold in ski8/rec103Δ, mer2/rec107Δ, rec114Δ and mei4Δ mutants. These results indicate that the meiotic interaction between the Spo11 subunits (assayed in WCEs) does not depend upon meiotic DSB formation, but is genetically controlled by the Spo11-interacting Rec102 and Rec104, which are essential regulator of meiotic DSB formation on meiotic chromosomes (29–31). The increased Spo11 interaction observed in the other DSB-deficient mutants can be explained by the formation of non-functional Spo11 aggregates, which form in the absence of the DSB proteins in question, and likely accumulate in the cytoplasm (21).
Gal4BD-Spo11 and Gal4BD-spo11Y135F recruit Spo11-3FLAG to the GAL2 locus
To investigate the interaction of Spo11 subunits on the chromosomal substrate, we examined the interaction between Spo11-3FLAG and Gal4BD-Spo11 or Gal4BD-spo11Y135F by using ChIP assays, using anti-FLAG and anti-Gal4BD antibodies (Figure 2B). The binding of these proteins was examined by multiplex and quantitative real-time PCR using three pairs of primers, allowing DSB-proficient and -deficient regions to be monitored. The PP048w primer pair was used to detect Spo11 binding to the YCR048w region, which contains the strongest innate DSB hotspot on chromosome III; and the GAL2 region was monitored using a pair of primers located 58 bp downstream of the Gal2 UAS sites, away from the DSB cleavage sites (Murakami, unpublished dots). As a control, the third primer pair located within the SMC1 DSB-cold domain was used (9,19). ChIP experiments using high-resolution genome tiling arrays have revealed that little Spo11 binds to this region (within ∼7 kb 5′ and 3′ of the region amplified with PPSMC1; K. Kugou et al., unpublished results).
As expected, in the strain expressing Spo11-3FLAG (YHS395), 3–4 h after entry into meiosis, we observed Spo11-3FLAG binding to the YCR048w region (0.3–0.4% of ChIP efficiency; Figure 2F), but not to the GAL2 region (less than 0.03% of ChIP efficiency; Figure 2C (upper panel) and D). In contrast, in strains co-expressing Spo11-3FLAG and Gal4BD-Spoll (YHS425) or Gal4BD-spo11Y135F (YHS518) proteins, Spo11-3FLAG accumulated efficiently at both the YCR048w and GAL2 regions (Figure 2C and D). Importantly, we also noted that the recruitment of Spo11-3FLAG to the GAL2 DSB region peaked at 3 h and 4 h (an IP: WCE DNA ratio is about 0.3%, an average of 3 and 4 h time points, Figure 2D), corresponding to the time of DSB formation, although the Gal4BD fusion proteins were constitutively expressed in premeiotic and meiotic cells. This indicates that Spo11 interactions occurring on the chromatin of the DSB sites are meiotically and temporally regulated.
In addition, we also examined the behavior of Gal4BD-Spo11 during meiosis by ChIP using an anti-Gal4BD antibody. Although all strains used here carried the wild-type GAL4 gene, only a small IP signal was detected in the GAL2 region in the absence of Gal4BD-SPO11 under the experimental condition used (Figure 2E). In contrast, in cell constitutively expressing Gal4BD-Spo11, the Gal4BD ChIP signal at the GAL2 region stays at constant levels from premeiosis through to the period of DSB formation (an IP:WCE DNA ratio is about 2.0%, an average of 3 and 4 h time points, Figure 2E, left), whereas it accumulated over time at the innate YCR048w DSB region (Figure 2E, right) followed by a reduction after 6 h of meiosis at both sites. These control experiments suggest that the Gal4BD ChIP signal in the presence of overexpressed Gal4BD-Spo11 was mainly due to the binding of Gal4BD-Spo11, but little endogenous Gal4 protein, to the GAL2 UAS site.
The ChIP signal of Gal4BD-Spo11-3FLAG at GAL2 UAS site by anti-FLAG antibody was 0.54% (Figure 2G), while it was 2.0% by anti-Gal4BD antibody. Therefore, ChIP efficiency by anti-Gal4BD antibody is about 3.7-fold higher than that by anti-FLAG antibody. Considering such difference in IP efficiency by distinct antibodies, we estimated that the molecular ratios of Gal4BD-Spo11 (or Gal4BD-spo11Y135F) versus Spo11-3FLAG at GAL2 UAS and YCR048w regions (1:0.6 and 1:5.8, respectively). The ratio at GAL2 UAS regions seems reasonable, given that free Gal4BD-Spo11/Gal4BD-spo11Y135F and Spo11-3FLAG can equally access to preloaded Gal4BD-Spo11/Gal4BD-spo11Y135F on GAL2 UAS site. Less binding of Gal4BD-Spo11 at YCR048w region may be due to the difference in the number of Gal4 UAS consensus sequences within GAL2 UAS and YCR048w regions (five and one UAS sequences, respectively). In contrast with the case of GAL2 UAS region, the binding of Spo11-FLAG in the YCR048w region is generally unaffected by the presence of Gal4BD-Spo11/Gal4BD-spo11Y135F. From these, we conclude that the Spo11-3FLAG and Gal4BD-Spo11/Gal4BD-spo11Y135F proteins form a chromatin-associated heterocomplex in the GAL2 UAS region during meiosis.
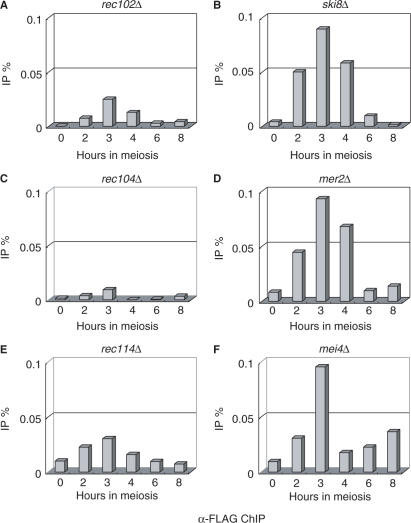
To address the genetic control of the meiotic association of Spo11-3FLAG with the GAL2 region in the presence of Gal4BD-Spo11, we examined the effects of deleting other DSB genes (Figure 3). A ChIP assay indicated that the absence of Rec102, Ski8/Rec103, Rec104, Mer2/Rec107, Rec114 or Mei4 severely impaired the recruitment of Spo11-3FLAG by Gal4BD-Spo11 to chromatin of the GAL2 UAS region. Quantitatively, the ChIP ratio was reduced to 0.01–0.1%, compared with 0.3–0.4% in the wild-type strain (compare Figures 2D and 3). Interestingly, two mutant categories were distinguishable: the first class, which included the rec102Δ, rec104Δ and rec114Δ mutants, exhibited almost complete loss of the Spo11-3FLAG ChIP signal, whereas the second class, which included the ski8/rec103Δ, mer2/rec107Δ and mei4Δ mutants, were associated with less severe defects (approximately 25% of the wild-type levels) (Figure 3). The severe deficiencies of the rec102Δ and rec104Δ mutants observed in the Spo11-3FLAG ChIP assay correlate with the lack of stable interaction between the Spo11 subunits observed by IP in WCEs. However, this was not the case for Rec114 (see Figure 1E), suggesting that Rec114 more specifically controls the chromatin-associated interaction of Spo11-3FLAG and Gal4BD-Spo11 proteins. We also noted that all members of the second class of mutants exhibited increased levels of Spo11 interaction in WCE IP experiments (Figure 1E), indicating that the Spo11 heterocomplexes form cytoplasmic and/or nuclear aggregates that cannot be recruited to the DSB sites (21).
Figure 3.
Quantification of ChIP at the GAL2 UAS site in DSB-defective mutants. The ChIP method was performed as described in Figure 2C, and two independent replicates of this experiment were conducted. The strains used in this experiment were the same as those in Figure 1E. ChIP experiments were performed as described in the Materials and methods section.
Co-expression of the Spo11-3FLAG and Gal4BD-spo11Y135F proteins allows DSB formation in the YCR048w region but not at GAL2
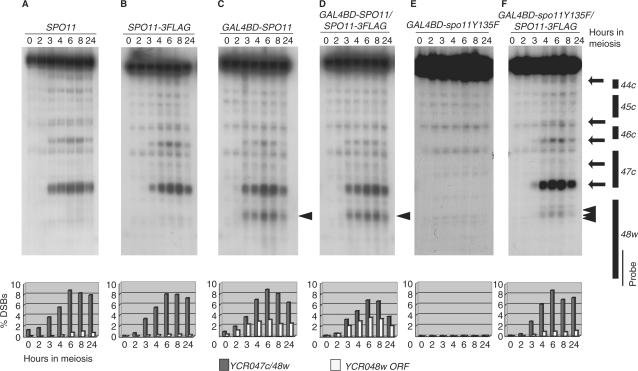
Next, we analyzed DSB formation in the YCR048w and GAL2 regions in cells co-expressing the Spo11-3FLAG and Gal4BD-Spo11 or Gal4BD-spo11Y135F proteins (Figure 4). These DSB analyses were performed using a rad50S mutant background that allowed the accumulation of unresected meiotic DSB fragments (32). In both SPO11 and SPO11-3FLAG control diploids, DSBs became detectable in the YCR048w promoter region 2 h after entry into meiosis, and had accumulated by 6 h to reach an absolute frequency of ∼8% per DNA molecule (Figure 4A and B). In the GAL4BD-SPO11 cells, as reported earlier (20), DSBs were formed not only in the YCR048w promoter region, but also in the vicinity of the fortuitous Gal4 UAS sites located in the YCR048w coding region (Figure 4C, arrowhead). The distribution and timing of DSB formation was similar in cells co-expressing Spo11-3FLAG and Gal4BD-Spo11 (Figure 4C, and D).
Figure 4.
Meiotic DSB formation around the YCR048w DSB hotspot. YHS326 (A, wild type); YHS363 (B, SPO11-3FLAG); YHS397 (C, GAL48D-SPO11); YHS427 (D, GAL4BD–SPO11/SPO11-3FLAG); YHS439 (E, GAL4BD–spo11Y135F); and YHS517 (F, GAL4BD–spo11Y135F/SPO11-3FLAG) cells were taken at the indicated times (hours) after meiotic induction. Genomic DNA was prepared in agarose plugs and digested with AseI. Southern blot analysis was performed with a probe for the YCR048w region (indicated by a thin bar in the right diagram). All strains were homozygous for the rad50S allele, which prevents DNA end processing and causes accumulation of meiotic DSBs. The graphs (panel below the DSB data) show quantified data for the most prominent DSB at the YCR048w DSB hotspot (gray vertical bars; locations are indicated by the lowest arrow in the right diagram) and UAS-dependent DSB at the YCR048w open reading frame (open vertical bars; locations are indicated by arrowhead in the right diagram). Arrows in the right diagram indicate the positions of DSB sites. Numbers beneath the graphs are culture time (hours) in SPM.
The results of DSB analysis in cells expressing the nuclease-deficient spo11Y135F mutation are shown in Figures 4 and 5. We first examined the YCR048w region (Figure 4). As expected, no DSBs were detected in the homozygous GAL4BD-spo11Y135F strain (Figure 4E), and diploid cells co-expressing Spo11-3FLAG and Gal4BD-spo11Y135F exhibited a DSB pattern that was very similar to those observed in diploids expressing Spo11 or Spo11-3FLAG alone (2,20) (compare Figure 4F and A–C, respectively). These results indicate that expression of the Gal4BD-spo11Y135F cleavage-deficient protein hardly interferes with the DSB activity of wild-type and Spo11-3FLAG proteins at innate DSB regions (see discussion).
Figure 5.
Absence of DSB formation in the GAL2 UAS region in cells co-expressing Gal4BD–spo11Y135F and Spo11-3FLAG. (A) DSB detection in the GAL2 UAS region (dashed bar in the right diagram). Strains and plugged DNA used in this experiment were the same as those for Figure 4. The plugged DNA was treated with NcoI and XbaI and probed for a GAL2 internal region (thin vertical bar in the right diagram). An asterisk and a filled triangle represent non-specific bands (EMP46 loci) and meiotic DSB bands introduced in the GAL2 UAS region, respectively. Note that no DSB band was detected in cells that co-express Gal4BD–spo11Y135F and Spo11-3FLAG. (B) Quantification of DSB in (A). DSB intensities (expressed as percentages relative to the total band intensity) are absolute and have not been corrected for background intensity. (C, D) DSB detection and quantification in cells co-expressing Gal4BD–Spo11 and spo11Y135F-3FLAG (YHS843). This strain is rad50S homozygous background. An asterisk in EMP46 loci presents non-specific bands. Quantification was performed as described in (B).
In contrast, as in the GAL4BD-spo11Y135F strain (Figure 5A), no DSBs were formed at the GAL2 locus in cells co-expressing the Gal4BD-spo11Y135F and Spo11-3FLAG proteins (Figure 5A), although the nuclease-active Spo11-3FLAG was meiotically recruited in the presence of the Gal4BD-spo11Y135F protein (see Figure 2D). We also examined whether or not spo11Y135F-3FLAG protein exhibits a dominant negative effects on DSB formation at GAL2 UAS region, when it is co-expressed with Gal4BD-Spo11. In this case, we detected impaired levels of DSB formation (60–70% of the wild-type levels) at GAL2 UAS region (Figure 5C and D).
In the gel image for analysis of DSBs at the YCR048w locus in the GAL4BD-spo11Y135F/SPO11-3FLAG strain, we noted the presence of a faint and diffuse DSB signal within the YCR048w coding region, close to the fortuitous Gal4 UAS consensus-binding sequence (Figure 4F, arrowheads). This weak DSB signal was ∼10-fold weaker than that for cells expressing Gal4BD-Spo11 alone or Gal4BD-Spo11 plus Spo11-3FLAG (compare Figure 4F with C and D). We do not know whether this reflects residual activity of the heterocomplex not detected in the GAL2 region, or results from an indirect effect of the binding of Gal4BD-spo11Y135F in this region, sufficient to locally open the chromatin structure, and thus facilitate access by Spo11-3FLAG alone.
Finally, to further address the potential cleavage activity of the Gal4BD-spo11Y135F/Spo11-3FLAG heterocomplex associated with the GAL2 UAS region, we examined the presence or absence of single-stranded breaks (SSBs). To visualize potential nicks, we used two experimental methods. First, we attempted to detect the presence of SSBs by extensively treating the genomic DNA preparation with the single-strand DNA-specific endonuclease S1, and visualizing the DNA fragments by Southern blot hybridization (see Materials and methods section). As shown in Figure 6A, no band indicating the formation of SSBs at the GAL2 locus was detected upon S1 treatment of the GAL4BD-spo11Y135F/SPO11-3FLAG or control GAL4BD-spo11Y135F DNA samples.
In the second experimental method, which has been used earlier to map DSB sites to nucleotide-level resolution (33–36), native genomic DNA preparations were run on denaturing polyacrylamide gels and hybridized with strand-specific probes. As a positive control for the detection of nicked fragments, DNA preparations were digested with the Nt.AlwI restriction enzyme, which introduces SSBs at the specific 5′-GGATCNNNN/N-3′ sequence. Conveniently, a Nt.AlwI site is located in the GAL2 region, near the Gal4 UAS sites (Figure 6B). Figure 6C and D illustrate the results obtained upon hybridization of the meiotic DNA extracted from the GAL4BD-spo11Y135F/SPO11-3FLAG strain and various positive and negative control strains, using GAL2 Watson and Crick strand-specific probes, respectively. Clearly, no band indicating the formation of meiotic SSBs was detected in the GAL4BD-spo11Y135F/SPO11-3FLAG strain, nor in the SPO11-3FLAG strain. Based on quantification of minor bands, we estimated that the sensitivity of detection of this method is in the range of 0.1–0.2% of total DNA, which is much lower than the intensity of major SSB signals (∼17%) appeared in the wild-type strain. Therefore, we conclude that the wild-type Spo11-3FLAG protein remains inactive for single- and double-strand DNA break formation at GAL2 UAS site, although it is recruited to the chromatin of a potentially cleavable region by association with the nuclease-deficient Gal4BD-spo11Y135F protein.
DISCUSSION
In the present study, we examined the in vivo interaction between Spo11 proteins carrying distinct tags, and assayed chromatin binding and DSB formation at innate (YCR048w) and targeted (GAL2) sites upon fusion of Spo11 to the Gal4 DNA-binding domain. We established that: (1) At the time of DSB formation, but not at premeiotic stages, the Spo11-3FLAG and Gal4BD-Spo11 proteins can form a heterocomplex; (2) In the presence of the Gal4BD-Spo11 protein, Spo11-3FLAG meiotically interacts with the preloaded Gal4BD-Spo11 protein and bound to the chromatin of the GAL2 region; (3) This interaction depends on other DSB proteins, since it is reduced in all DSB protein mutants, but more severely in rec102Δ, rec104Δ and rec114Δ mutants; and (4) Spo11-3FLAG is recruited to the GAL2 region in the presence of the nuclease-deficient Gal4BD-spo11Y135F protein, but remains inactive for DSB as well as potential SSB cleavage activity.
Spo11 interactions are under genetic control
Our time-course analysis of the Spo11-FLAG and Gal4BD-Spo11 interaction in diploid cells expressing both proteins under the control of the ADH1 promoter (Figure 1C) shows that the Gal4BD-Spo11-mediated recruitment of Spo11-3FLAG is enhanced during the period of DSB formation, although both proteins are present at generally constant levels during premeiosis and meiosis. In addition, they are co-immunoprecipitated only in the meiotic cell extracts. All these data demonstrate that the self-interaction of Spo11 is controlled in a meiosis-dependent manner.
Our analysis of mutants indicates that the self-association of Spo11 is genetically controlled by the activity of other known meiotic DSB proteins (Figures 1E and 3). In cells lacking Rec102 and Rec104, which physically interact with each other and with Spo11, the meiotic self-interaction of Spo11 decreases dramatically (Figure 1E), suggesting that Rec102 and Rec104 may mediate the interaction between Spo11 subunits during meiosis via a physical interaction with Spo11 (Figure 7A) (29–31).
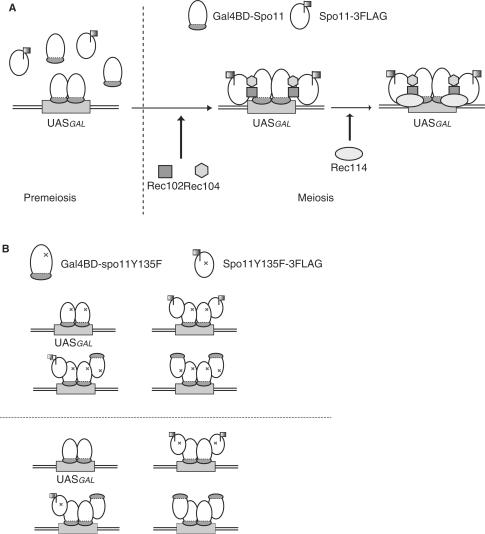
Figure 7.
Schematic diagram of the complexes that are thought to assemble at the GAL2 UAS region at the time of meiotic DSB formation. (A) Spo11 does not self-interact during premeiosis, but Spo11-3FLAG is meiotically recruited to the GAL2 UAS site, at which Gal4BD–Spo11 already exists, as shown in Figure 2D. This association was not detected in rec102Δ, rec104Δ or rec114Δ cells by ChIP assay. (B) In the cells co-expressing Gal4BD-fused protein and Spo11-3FLAG, four types of Spo11 complex may assemble at the GAL2 UAS regions: homocomplexes consisting of either Gal4BD-fused protein alone or heterocomplexes consisting of both Gal4BD-fused protein and Spo11-3FLAG. The homocomplexes of Gal4BD–spo11Y135F cannot introduce DSBs in the GAL2 UAS regions, but the heterocomplex of Gal4BD–spo11Y135F/Spo11-3FLAG is expected to catalyze DSB formation at least partly. However, neither DSBs nor SSBs were detected in cells harboring such heterocomplexes (upper diagram). In cells expressing both Gal4BD–Spo11 and spo11Y135F-3FLAG, the heterocomplex of Gal4BD–Spo11/spo11Y135F-3FLAG formed in the GAL2 UAS regions is inactive (lower diagram).
Interestingly, the genetic requirements for the self-interaction of Spo11 as determined by the IP and ChIP experiments are somewhat different: Rec102 and Rec104 were absolutely required for the Spo11 self-interaction in the cell extracts, whereas Rec114 in addition to Rec102 and Rec104 was essential for the Gal4BD-Spo11-mediated recruitment of Spo11-3FLAG to the chromatin at the GAL2 UAS site (Figures 4 and 5). It is most likely that Rec114 is not involved in the Spo11 self-interaction, but has other roles in facilitating the initial chromatin binding of Spo11 (Figure 7A). This idea is consistent with the previous finding that the chromatin binding of Spo11 to DSB sites is not observed in rec114Δ strains (21). In addition, the idea seems reasonable, since overproduction of Rec114 perturbs global DSB formation, presumably by an abortive chromatin loading of Spo11 proteins in the presence of an excess amount of Rec114 (11).
We also found that other meiotic DSB proteins, Mei4, Mer2 and Ski8, are partially required for Spo11-3FLAG recruitment to the GAL2 UAS site, although they are dispensable for the Spo11 self-interaction in meiotic cell extracts. However, mei4Δ, mer2Δ and rec114Δ mutants had higher levels of Spo11 self-interaction (Figure 1E). Prieler et al. have shown that Spo11-Myc forms aggregates in spread nuclei of rec114Δ mutants in addition to normal Spo11 foci, whereas no Spo11-Myc foci are detected at all in rec102Δ mutants (21). Thus, it is likely that polycomplex aggregates of Spo11 are formed in these three mutants.
It should be noted that Ski8 is known to interact with Spo11, and assists in the nuclear import of Spo11 during meiosis (8). Presumably Ski8 is not involved in controlling the Spo11 self-interaction, but has other roles as a direct partner of Spo11, for example acting to carry Spo11 into meiotic nuclei, converting Spo11 to a nuclease-active state, or stabilizing the binding of pre-DSB complexes to DSB sites.
Spo11 self-interaction and its cleavage activity
We found that the Gal4BD-spo11Y135F-assisted recruitment of the functional Spo11-FLAG subunit to the GAL2 region is not active with respect to DSB or SSB activity. This may be most simply explained by an excess occupancy of Gal4BD-spo11Y135F at GAL2 UAS site over the binding of Spo11-FLAG. However, we think this may not be the case for the following reason. Dimerization of Gal4BD-fused Spo11 may be facilitated via the dimerization between Gal4BD (37). Therefore, it is very likely that a homo-dimer Gal4BD-spo11Y135F preoccupies at the GAL2 UAS site. The molecular ratio of Gal4BD-spo11Y135F versus Spo11-3FLAG at GAL2 UAS site is expected to be 1:0.6 (see above). These results suggest that on average at least one wild-type Spo11 molecule is present at GAL2 UAS region forming a heterocomplex with Gal4BD-spo11Y135F, but cannot catalyze DNA cleavage reaction. Therefore, it is suggested that multimeric assembly of active Spo11 monomers at DSB sites may be critical in DNA cleavage reaction.
In this notion, one can predict that co-expression of Gal4BD-Spo11 and spo11 Y135F may exhibit a dominant negative effect on DSB formation at GAL2 UAS region. In fact, we observed 30–40% reduction of DSB formation at GAL2 UAS region in this situation (Figure 5C and D). The partial (but not total) reduction of DSB formation at GAL2 UAS region can be explained as follows. At GAL2 UAS region in meiotic cells co-expressing Gal4BD–Spo11 and Spo11Y135F, there should be four types of Spo11 complexes (illustrated in Figure 7B, upper panel). Spo11-3FLAG homocomplex is present in nuclei, but cannot bind to GAL2 UAS regions without guidance by Gal4BD-spo11Y135F. Therefore, the GAL2 UAS is likely to be bound either by homocomplexes consisting of only Gal4BD-spo11Y135F, or by heterocomplexes consisting of Gal4BD-spo11Y135F and Spo11-3FLAG, which are all supposed to be inactive in the hypothesis. The lower diagram of Figure 7B illustrates four potential situations at GAL2 UAS region in cells co-expressing Gal4BD-Spo11 and spo11Y135F. In this case, three types (one type is half active) out of four possible combinations are assumed to catalyze DSB formation, thereby dominant negative effects may be only partial.
Since purification of active Spo11 proteins has not yet been achieved, it is difficult to estimate the exact number of Spo11 subunits present in an individual complex. However, biochemical and structural analyses of the archaeal Top6A protein complex suggest the participation of an anti-parallel dimer of Spo11 subunits in DSB cleavage (5,6). In this idea, the location of the catalytic site of each protomer is closer to the magnesium-binding pocket in the Toprim domain of the other protomer than it is to its own (6). The notion of anti-parallel interaction between Spo11 subunits is consistent with the partial dominant negative defects of DSB formation in heterozygous combinations of some DSB-defective alleles and Spo11-3HA (7).
In vitro biochemical analysis of yeast topoisomerase II has revealed that the catalytic tyrosine residue of one protomer causes SSBs in association with the metal-binding pocket of the other protomer (38,39). Therefore, if Spo11 forms a dimer that functions via a topoisomerase-II-like mechanism, the heterocomplex of Gal4BD-spo11Y135F and Spo11-3FLAG should introduce an SSB at the GAL2 UAS. However, this is not the case (Figure 6). It seems likely that the mechanism by which Spo11 cleaves DNA is slightly different from the way that topoisomerase II functions. Presumably, two active catalytic tyrosine residues in a dimeric configuration are essential to function in concert to generate DSBs.
Self-interaction of Spo11 and regional control of DSB cleavage
In cells co-expressing Gal4BD-spo11Y135F and Spo11-3FLAG, we detected only faint levels of DSB formation at the UAS within the YCR048w coding region, suggesting that DSB formation is impaired, but not absent at this locus, whereas no DSB was detected at the GAL2 UAS site. The former locus is in a chromosomal domain that is very active in meiotic DSB formation, but the latter site is in a DSB-cold domain. Differences in the effects on DSB formation at the two UAS-containing sites may reflect regional conditions that limit or favor DSB formation. In DSB-cold domains, it is possible that Spo11 and its activating factors may be absent or present only in small amounts, so that DSB formation is more severely affected in comparison with the situation at DSB-hot domains, at which Spo11 and its activating factors may be more readily available. In fact, the molecular ratios of Gal4BD-Spo11 (or Gal4BD-spo11Y135F) versus Spo11-3FLAG at GAL2 UAS and YCR048w regions are 1:0.6 and 1:5.8, respectively. Therefore, Spo11 may be able to form larger multimers in DSB-hot domains than in DSB-cold domains. In such larger Spo11 multimers, a small portion of active complexes can exist and sufficiently catalyze DSB formation; thereby the presence of Gal4BD-spo11Y135F hardly interferes with the DSB activity of active Spo11 proteins at innate DSB sites in DSB-hot domains. It will be interesting to test the possibility of regional regulation of DSB formation in terms of the interaction between Spo11 subunits.
Activation of Spo11 is controlled separately from the control of its physical interaction with DSB sites, since there are uncleaved DNA intermediates bound by Spo11, without introducing any nicks (21). We also found that Spo11 can bind to DSB-cold regions such as centromeric regions (K. Kugou et al., unpublished results), and that Gal4BD-Spo11 binds to GAL2 UAS sites without cleaving DNA (9). It is therefore possible that Spo11 complexes at DSB-cold regions do not participate in the DNA cleavage reaction unless they are converted to active complexes with the assistance of other DSB factors. Since Rec102 and Rec104 are involved in the meiotic self-interaction of Spo11, these meiotic DSB factors are assumed to play important roles in the regulation of Spo11 activity by controlling the self-interaction between Spo11 monomers, thereby facilitating the formation of the pre-DSB complex.
The present analyses reveal that Spo11 molecules can interact with each other, and are genetically controlled by meiotic DSB proteins such as Rec102, Rec104 and Rec114. This finding permits new insights into the molecular mechanisms of DSB cleavage and regulation of cleavage activity by Spo11.
ACKNOWLEDGEMENTS
We thank K. Hirota and K. Kugou for their help, advice and comments; and H. Seo for critical discussion regarding the Gal4BD-spo11Y135F/Spo11-3FLAG in vivo assay. We are also grateful to other laboratory members for helpful discussions. A.N. thanks RIKEN and T. Shibata for support from the RIKEN eminent scientist program. This work was supported by grants for basic research from the Bio-oriented Technology Research Advancement Institution (to K.O. and T.S.) and grants-in-aid for scientific research on priority areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. Funding to pay the Open Access publication charge was provided by the grants listed above.
REFERENCES
- 1.Keeney S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 2.Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 3.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 4.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gadelle D, Filee J, Buhler C, Forterre P. Phylogenomics of type II DNA topoisomerases. Bioessays. 2003;25:232–242. doi: 10.1002/bies.10245. [DOI] [PubMed] [Google Scholar]
- 6.Nichols MD, DeAngelis K, Keck JL, Berger JM. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 1999;18:6177–6188. doi: 10.1093/emboj/18.21.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz RL, Alcid AD, Berger JM, Keeney S. Identification of residues in yeast Spo11p critical for meiotic DNA double-strand break formation. Mol. Cell Biol. 2002;22:1106–1115. doi: 10.1128/MCB.22.4.1106-1115.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arora C, Kee K, Maleki S, Keeney S. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell. 2004;13:549–559. doi: 10.1016/s1097-2765(04)00063-2. [DOI] [PubMed] [Google Scholar]
- 9.Borde V, Lin W, Novikov E, Petrini JH, Lichten M, Nicolas A. Association of Mre11p with double-strand break sites during yeast meiosis. Mol. Cell. 2004;13:389–401. doi: 10.1016/s1097-2765(04)00034-6. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Hooker GW, Roeder GS. Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics. 2006;173:1969–1981. doi: 10.1534/genetics.106.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bishop DK, Nikolski Y, Oshiro J, Chon J, Shinohara M, Chen X. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells. 1999;4:425–444. doi: 10.1046/j.1365-2443.1999.00273.x. [DOI] [PubMed] [Google Scholar]
- 12.Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohta K, Nicolas A, Furuse M, Nabetani A, Ogawa H, Shibata T. Mutations in the MRE11, RAD50, XRS2, and MRE2 genes alter chromatin configuration at meiotic DNA double-stranded break sites in premeiotic and meiotic cells. Proc. Natl. Acad. Sci. U. S. A. 1998;95:646–651. doi: 10.1073/pnas.95.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamada T, Mizuno K, Hirota K, Kon N, Wahls WP, Hartsuiker E, Murofushi H, Shibata T, Ohta K. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 2004;23:1792–1803. doi: 10.1038/sj.emboj.7600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith KN, Penkner A, Ohta K, Klein F, Nicolas A. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Curr. Biol. 2001;11:88–97. doi: 10.1016/s0960-9822(01)00026-4. [DOI] [PubMed] [Google Scholar]
- 16.Henderson KA, Kee K, Maleki S, Santini PA, Keeney S. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell. 2006;125:1321–1332. doi: 10.1016/j.cell.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogino K, Hirota K, Matsumoto S, Takeda T, Ohta K, Arai K, Masai H. Hsk1 kinase is required for induction of meiotic dsDNA breaks without involving checkpoint kinases in fission yeast. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8131–8136. doi: 10.1073/pnas.0602498103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baudat F, Nicolas A. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl. Acad. Sci. U. S. A. 1997;94:5213–5218. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pecina A, Smith KN, Mezard C, Murakami H, Ohta K, Nicolas A. Targeted stimulation of meiotic recombination. Cell. 2002;111:173–184. doi: 10.1016/s0092-8674(02)01002-4. [DOI] [PubMed] [Google Scholar]
- 21.Prieler S, Penkner A, Borde V, Klein F. The control of Spo11's interaction with meiotic recombination hotspots. Genes Dev. 2005;19:255–269. doi: 10.1101/gad.321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 23.Borde V, Goldman AS, Lichten M. Direct coupling between meiotic DNA replication and recombination initiation. Science. 2000;290:806–809. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- 24.Borde V, Wu TC, Lichten M. Use of a recombination reporter insert to define meiotic recombination domains on chromosome III of Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:4832–4842. doi: 10.1128/mcb.19.7.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiegand RC, Godson GN, Radding CM. Specificity of the S1 nuclease from Aspergillus oryzae. J. Biol. Chem. 1975;250:8848–8855. [PubMed] [Google Scholar]
- 26.Sambrook J, Fritcsh EF, Maniats T. Molecular Cloning, A Laboratory Manual. 2nd edn 1989. [Google Scholar]
- 27.Vedel M, Nicolas A. CYS3, a hotspot of meiotic recombination in Saccharomyces cerevisiae. Effects of heterozygosity and mismatch repair functions on gene conversion and recombination intermediates. Genetics. 1999;151:1245–1259. doi: 10.1093/genetics/151.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Wu TC, Lichten M. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. EMBO J. 1995;14:4599–4608. doi: 10.1002/j.1460-2075.1995.tb00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kee K, Keeney S. Functional interactions between SPO11 and REC102 during initiation of meiotic recombination in Saccharomyces cerevisiae. Genetics. 2002;160:111–122. doi: 10.1093/genetics/160.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiao K, Salem L, Malone R. Support for a meiotic recombination initiation complex: interactions among Rec102p, Rec104p, and Spo11p. Mol. Cell Biol. 2003;23:5928–5938. doi: 10.1128/MCB.23.16.5928-5938.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kee K, Protacio RU, Arora C, Keeney S. Spatial organization and dynamics of the association of Rec102 and Rec104 with meiotic chromosomes. EMBO J. 2004;23:1815–1824. doi: 10.1038/sj.emboj.7600184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 33.de Massy B, Rocco V, Nicolas A. The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in Saccharomyces cerevisiae. EMBO J. 1995;14:4589–4598. doi: 10.1002/j.1460-2075.1995.tb00138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleckner N. Meiosis: how could it work? Proc. Natl. Acad. Sci. U. S. A. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lichten M, Goldman AS. Meiotic recombination hotspots. Annu. Rev. Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 36.Petes TD, Pukkila PJ. Meiotic sister chromatid recombination. Adv. Genet. 1995;33:41–62. doi: 10.1016/s0065-2660(08)60330-2. [DOI] [PubMed] [Google Scholar]
- 37.Marmorstein R, Carey M, Ptashne M, Harrison SC. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 38.Liu Q, Wang JC. Identification of active site residues in the “GyrA” half of yeast DNA topoisomerase II. J. Biol. Chem. 1998;273:20252–20260. doi: 10.1074/jbc.273.32.20252. [DOI] [PubMed] [Google Scholar]
- 39.Liu Q, Wang JC. Similarity in the catalysis of DNA breakage and rejoining by type IA and IIA DNA topoisomerases. Proc. Natl. Acad. Sci. U. S. A. 1999;96:881–886. doi: 10.1073/pnas.96.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]