Abstract
Over the past few years, small ubiquitin-like modifier (SUMO) modification has emerged as an important regulator of diverse pathways and activities including protein localization and transcriptional regulation. We identified a consensus sumoylation motif (IKEE), located within the N-terminal activation domain of the ATF7 transcription factor and thus investigated the role of this modification. ATF7 is a ubiquitously expressed transcription factor, homologous to ATF2, that binds to CRE elements within specific promoters. This protein is able to heterodimerize with Jun or Fos proteins and its transcriptional activity is mediated by interaction with TAF12, a subunit of the general transcription factor TFIID. In the present article, we demonstrate that ATF7 is sumoylated in vitro (using RanBP2 as a E3-specific ligase) and in vivo. Moreover, we show that ATF7 sumoylation affects its intranuclear localization by delaying its entry into the nucleus. Furthermore, SUMO conjugation inhibits ATF7 transactivation activity by (i) impairing its association with TAF12 and (ii) blocking its binding-to-specific sequences within target promoters.
INTRODUCTION
In eukaryotic cells, post-translational protein modifications are crucial mechanisms to regulate protein function. Polypeptide conjugation to substrate proteins is one of these known mechanisms. Ubiquitin and ubiquitin-like proteins are small polypeptides of about 8–11 kDa that covalently modify various intracellular proteins. Among these, the small ubiquitin-related modifiers (SUMO-1–4) are covalently linked as a 93–97 amino-acid polypeptides to specific target lysine residues (1–4). Sumoylation is catalyzed by an enzymatic machinery analogous to that of ubiquitin-modification (2) and usually takes place at sites matching the sequence ϕKXE (where ϕ stands for I, L or V and X for any amino acid residue). In addition to this core consensus motif, an extended sequence encompassing several acidic residues clustered within the 10-amino-acid region located immediately downstream has been recently described to play an important role in determining the efficiency of substrate sumoylation (5). Sumoylation depends on an activating SAE1/SAE2 E1-enzyme heterodimer, the SUMO conjugating enzyme Ubc9 and recently identified E3 SUMO ligases. In contrast to ubiquitination that targets the conjugated proteins to 26S proteasome-mediated degradation, the biological consequences of SUMO modification include nuclear targeting, the formation of subnuclear structures, the regulation of transcriptional activities and the control of protein stability (1,4,6).
The family of ATF7 (formerly ATFa) transcription factors, structurally and functionally related to ATF2, is composed of at least three members (ATF7-1, 2, 3) (7,8) translated from alternatively spliced messengers issued from a single gene (9). Although the three ATF7 isoforms differ by the presence of small motifs within their N-terminal region, they exhibit indistinguishable activities. Their transcriptional activation domain has been delineated within the N-terminal part of the protein and shown to include a critical zinc-binding element and two conserved threonine residues (T51 and T53, corresponding to the T69 and T71 homologs in ATF2) (10,11). This region, together with sequences located in the C-terminal portion of the ATF7 proteins, contributes to their interaction with the adenovirus E1a oncoprotein (8). ATF7, like ATF2, can also associate with c-Jun or c-Fos proteins through their C-terminal leucine-zipper (b-ZIP) region. While ATF proteins usually bind as homodimers to ATF/CRE promoter elements, they also can bind TRE sequences if heterodimerized with members of the Jun family (10,12). ATF7 proteins also strongly interact with the JNK2 protein kinase (13), although they do not constitute substrates for this kinase but rather serve as a JNK2-docking site for ATF7-associated partners like JunD, (11). Collectively, these data suggest that ATF7 proteins play important functions early in cell signaling.
The transcriptional activity of ATF7 is potentiated by the expression of TAF12, a subunit of TFIID, through direct interactions implicating the N-terminal domains of the two proteins, as well as the TAF12 histone fold domain (14). TAF4, the HFD-containing protein partner of TAF12 within TFIID also interacts with ATF7 and thereby competes for the coactivator effect of TAF12. Interestingly its paralog TAF4b, that plays a role in fertility does not (14,15). In fact, TAF4 has been shown to play a central role in multiple signaling pathways by competing with TAF4b (16) for the presence of various transcription factors, among these CREB, SP1, RAR and ATF7 (17).
In the present study, we investigate the modification of ATF7 protein by SUMO-1. We demonstrate that ATF7 is sumoylated in human cells and that this modification delays its nuclear localization and therefore inhibits ATF7 transcriptional activity by impairing its interaction with TAF12 in TFIID.
MATERIALS AND METHODS
Expression vectors
The recombinant human ATF7 isoform used in the present study is ATF7-1 and will be referred as ATF7, with coordinates corresponding to the amino acid numbering of the largest protein (ATF7-3, 494 residues) (14).
The vectors pG4-ATF7-1 recombinant and corresponding derivatives encode the DNA-binding domain of the yeast Gal4 protein fused to the wild type (wt) or mutated human ATF7-1 polypeptide, as described (10). The cDNAs of ATF7-1 and derivatives, and human ATF2, RanBP2▵FG have also been inserted into the pXJ or pXJ-HA vectors, under the control of the CMV promoter (18), generating the px-ATF7 series and px-hsATF2, px-RanBP2▵FG, respectively.
The (17m5)-TK-Luc reporter (19) contains the luciferase gene, driven by the thymidine kinase promoter and five Gal4 binding sites (see Figure 5A).
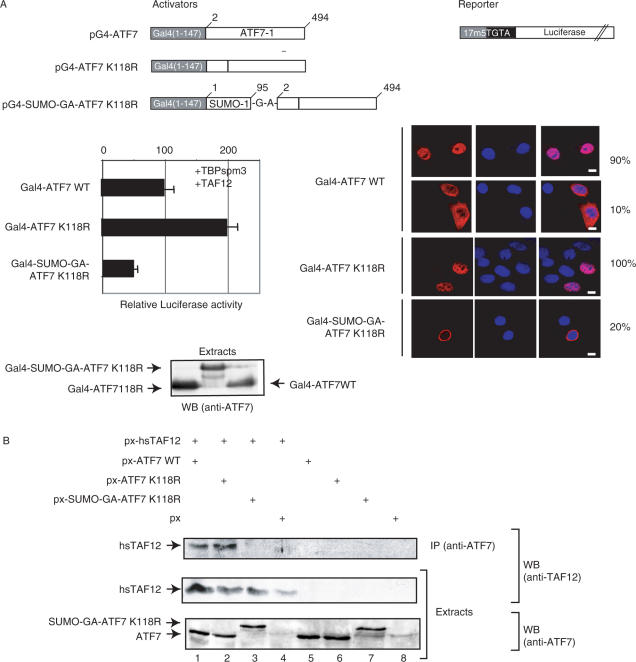
Figure 5.
Sumoylation affects ATF7 transcriptional activity. (A) HeLa-SUMO cells were co-transfected with 0.25 µg of the pG4-ATF7 expression vectors, 2 µg of the luciferase reporter, 0.25 µg of the TBP-spm3 and 0.25 µg of p-hsTAF12 expression vectors. The structures of the pG4-ATF7 activator and the luciferase reporter plasmids are schematized. The results of luciferase assays are presented (arbitrary units). (B) HeLa-SUMO cells were transfected with 0.5 µg px-ATF7 derivatives and hsTAF12. Extracts from transfected cells were immunoprecipitated (IP) with the anti-ATF7 (2F10) monoclonal antibody as described in Materials and Methods. The purified complexes were separated by SDS-PAGE. The presence of hsTAF12 proteins in both IP and extracts, and ATF7 derivatives in the extracts was revealed by western blotting (WB) using specific antibodies.
The pATF-GFP was constructed by cloning the sequence encoding the N-terminal portion of ATF7 (amino acids 1–250) into an engineered XhoI site immediately 3′ to the start codon of GFP in pXRGG (20), a derivative of pGreenLantern (Life Technologies). This plasmid encodes a fusion protein containing the ATF7 activation domain, followed by the hormone-response element of the rat glucocorticoid receptor and by the GFP. pGST-RanBP2·FG was a gift of Anne Dejean.
Point-mutations and deletions were created in the ATF7-1 moiety by oligonucleotide-directed mutagenesis using the Pfu-DNA polymerase (21), PCR-amplification of appropriate DNA fragments, or restriction fragment deletion. All constructions were verified by DNA sequencing.
Cells, transfection and extract preparation
HeLa-SUMO cells, a gift from P. O'Hare (22), were grown as monolayers in Dulbecco medium supplemented with glucose (1 g/l) and 10% fetal calf serum. Huvec-c cells were grown in Ham's F12K medium supplemented with 2 mM l-glutamine, 0.1 mg/ml heparin, 0.05 mg/ml endothelial cell growth supplement (ECGS), and 10% FBS. Cells were transfected with recombinant plasmids, 8 h after plating, using Transfectin reagent (Bio-Rad, USA), with the amounts of recombinant DNA indicated in the figure legends. After 36 h, cells were harvested in phosphate-buffered saline (PBS) and resuspended in RIPA buffer (150 mM NaCl, 10 mM Tris-HCl, pH 7.2, 0.1% SDS, 1% Triton X-100, 1% deoxycholate, 5 mM EDTA, 0.4 mM PMSF, 2.5 ng/ml each of leupeptin, pepstatin, aprotinin, antipain and chymostatin). After 30 min on ice, the resulting crude suspension was cleared by centrifugation for 20 min at 10 000 g.
Raji cells were grown in RPMI 1640 medium containing 10% calf serum. Cos-1 cells, grown as monolayers in Dulbecco medium supplemented with glucose (1 g/l) and 5% fetal calf serum, were transfected with recombinant plasmids, 8 h after plating, using ExGen 500 reagent (Euromedex, France) (23), with the amounts of recombinant DNA indicated in the figure legends. After 48 h, cells were harvested in PBS and resuspended in lysis buffer (0.4 M KCl, 20 mM Tris-HCl, pH 7.5, 20% glycerol, 5 mM dithiothreitol, 0.4 mM PMSF, 2.5 ng/ml each of leupeptin, pepstatin, aprotinin, antipain and chymostatin). After one freeze–thawing cycle in liquid nitrogen, the resulting crude suspension was cleared by centrifugation for 20 min at 10 000 g (24).
Antibodies
Rabbit antisera against ATF7, and monoclonal antibodies specifically recognizing the ATF7 isoforms (2F10, 1A7, 3C12), GFP (2A5) GST (1D10), FLAG (M2) and HA epitopes (12CA5) have been described (13,14). Rabbit antisera against RanBP2 (PA1-082) was purchased from Affinity BioReagen. Rabbit antisera against SUMO-1 was a gift from Dr Jacob Seeler (Institut Pasteur, Paris). The monoclonal antibody against Nuclear Pore Complex proteins (mAb414) was a kind gift from V. Doye (Institut Curie, Paris).
In vitro SUMO-1 conjugation
The SUMO-1 conjugation assay was previously described (25). Briefly, this assay was performed in a system containing an ATP-regenerating system (50 mM Tris-HCl, pH 7.6, 5 mM MgCl2, 10 mM creatine phosphate, 3.5 units/ml creatine kinase, 0.6 units/ml inorganic pyrophosphatase, 2 mM ATP). Twenty-microliter reactions contained 100 ng of purified recombinant hsSAE1/2 (E1), 400 ng of purified recombinant hsUbc9 (E2), 1 µg of purified recombinant human 6His:Myc:SUMO-1(1–97) and either 100 ng purified recombinant protein substrate (ATF7) or 5 µl of in vitro transcription/translation-coupled reactions (ATF2) (10). Reactions were incubated at 37°C for 1–2 h.
NiNTA-agarose affinity chromatography and immunoblotting
Aliquots of cell extracts were incubated (1 h at 4°C) with 50 µl of a NiNTA-agarose bead suspension (Qiagen) in PBS. The beads were washed three times with 1 ml of PBS, 0.5% Nonidet P-40 and NaCl adjusted to 250 mM. The proteins were then dissociated by boiling for 5 min in 20 µl sample buffer, before SDS-10% polyacrylamide gel electrophoresis (SDS-PAGE). Protein analysis by western blotting was carried out as described (13). Briefly, proteins were electrotransferred onto nitrocellulose, reacted with specific primary antibodies (see above) and revealed with peroxidase-linked goat anti-mouse Kappa-light chain or goat anti-rabbit immunoglobulins (SantaCruz) as indicated, using the ECL system (Amersham).
Luciferase assay
Transfected cells were harvested in ice-cold PBS, pelleted, washed once in PBS and resuspended in lysis buffer (100 mM potassium phosphate, pH 7.8). After three freeze–thawing steps in liquid nitrogen, the resulting cell lysate was cleared by centrifugation. Aliquots of the extracts (normalized by protein concentration) were assayed for luciferase activity using a Berthold Centro LB 960 luminometer, as previously described (26,27). In all cases, at least five independent transfections were carried out and the results always agreed within 10%. The results of typical experiments are shown in the figures.
Electrophoretic DNA-binding assay
Gel-retardation experiments were performed as previously described (28). Briefly, about 0.3 ng (5000 cpm) of a 32P-5′-end-labeled double-stranded oligonucleotide probe was incubated with the cell extract in the presence of poly (dI-dC), poly (dI-dC) as non-specific competitor and 1 µg of anti ATF7(3C12) monoclonal antibody when indicated. For competition experiments, the protein fractions were preincubated with 50 ng of unlabeled oligonucleotides before addition of the labeled probe. After 10 min at 25°C, the complexes were separated by electrophoresis on a non-denaturing 4.5% polyacrylamide gel.
RNA expression analysis (RT–PCR)
Total RNA was extracted using TRIzol reagent. One microgram of purified total RNA was used for RT–PCR analysis using the Transcriptor First Strand CDNA Synthesis Kit (Roche Diagnostics GmbH). For PCR reaction, 25 cycles were performed (denaturation: 95°C, 1 min; annealing: 60°C, 2 min; polymerization: 68°C, 2 min). PCR products were separated onto agarose gels and DNA was visualized by ethidium bromide staining. Primers for E-selectin RNA amplification were designed according to the E-selectin mRNA sequence published [NCBI Nucleotide GenBank (accession number: NM-000450) 5′-primer: 5′-TCTCTCAGCTCTCACTTTG-3′, 3′-primer: 5′-TTCTTCTTGCTGCACCTCT-3′]. E-selectin PCR product is 383 bp in length. Primers for β-actin amplification (367 bp PCR product) were as follows: 5′-primer: 5′-CTCACCATGGATGATGATAT-3′ and 3′-primer: 5′-TGGGTCATCTTCTCGCGGTT-3′.
ChIP and quantitative real-time PCR
HeLa-SUMO cells were transfected with either px-ATF7WT px-ATF7K118R or px-SUMO-GA-ATF7K118R vectors. Forty-eight hours after transfection, ChIP assays were performed using a ChIP Assay Kit (Upstate Biotechnology, USA), following the manufacturer recommendations.
Quantitative real-time PCR was performed on LightCycler (Roche Diagnostics, Switzerland), as specified by the manufacturer, using LightCycler FastStart DNA Master SYBR Green I reagents with 45 cycles of three-step amplification. The 5′ to 3′ sequences of the primers used in the PCR were: Fwd: GTCATATTAATAAAATTGCATATACGATAT; Rev:TCTCAGGTGGGTATCACTGCTGCCTCTGTC. After PCR, amplified DNA was collected and migrated on 1% agarose gels.
Immunofluorescence
Immunofluorescence staining experiments were carried out as previously described (29). Briefly, 48 h after transfection, HeLa-SUMO cells, grown on coverslips, were fixed with formaldehyde (4% [vol/vol] in PBS) and permeabilized with 0.1% Triton X-100 in PBS. The primary antibodies were diluted in PBS containing 0.1% Triton X-100. Antibodies were used at the following dilutions in PBS containing 0.1% Triton X-100: the anti-ATF7 monoclonal (2F10 or 1A7) and polyclonal antibodies were used both at 1/2000 dilution; the anti-RanBP2 and NCP4 antibody were used at 1/1000 dilution. After incubation at 20°C for 1 h, the coverslips were washed several times in PBS, 0.1% Triton X-100 and then incubated 1 h at 20°C with donkey CY3-conjugated anti-rabbit or anti-mouse IgG and/or donkey Alexa588-labeled anti-rabbit IgG (Sigma), at concentrations recommended by the suppliers. Nuclei were counterstained with Hoechst 33258. After the staining, the coverslips were mounted and analyzed using a confocal laser-scanning microscope (Leica). Image-enhancement software was used to balance signal strength, and 8-fold scanning was used to separate signal from noise.
Time laps experiments
For monitoring the nuclear import of the ATF constructs, HeLa-SUMO cells were seeded on coverslips, grown for 24 h, before mounting in a Life Imaging Services Ludin Chamber kept at 37°C, 5% CO2, placed in a Leica DMIRE2 microscope equipped with a Life Imaging Services temperature control system. The induction of nuclear importation was triggered by addition of dexamethasone (100 µM) to the cell-culture medium. Images were acquired every 25 s during 15 min, using a Leica DC350FX CCD camera piloted by the Leica FW4000 software. Eight fields (corresponding to a minimum of 60 cells) were recorded during each experiment by the use of a Marzhauser automated stage piloted by the FW4000 software. To compare the intensity of the fluorescent signals in the cytoplasm and the nucleus, the sequences were loaded in the Universal Imaging Metamorph software, and average signal intensity were recorded in regions selected in the perinuclear cytoplasm and inside the nucleus. Signal intensity values were transferred in Microsoft Excel where the cytoplasmic and nuclear signals were compared. The equilibration point refers to the time needed for each construct to reach a signal intensity superior in the nucleus than in the cytoplasmic perinuclear region.
RESULTS
ATF7 Sumoylation
To identify polypeptides that are able to interact with the activation domain of ATF7 transcription factor, a two-hybrid screen in yeast was performed to a mouse cDNA fusion library, using the 60% N-terminal portion of ATF7 as a bait. In this screening only the N-terminal portion of ATF7 lacking the leucine-zipper (b-ZIP) was chosen to avoid potential interactions with other b-ZIP-containing molecules. Among the selected clones, one corresponded to mBLM, a DNA helicase implied in the Bloom Syndrome disease (30), another one to mAM (29) a recently described protein shown to facilitate conversion from dimethyl to trimethyl lysine 9 of histone H3 by ESET and cause transcriptional repression (31). Interestingly, a third one corresponded to Ubc9, the SUMO E2-conjugating enzyme (32,33). In addition, protein sequence analysis revealed that ATF7 contains a potential sumoylation target site (I-K-X-E) matching the consensus sequence (Figure 1A) (1,34,35). To investigate whether ATF7 could be sumoylated, we first used an in vitro assay (Figure 1B). Purified bacterially expressed ATF7 proteins were incubated with a minimal SUMO-1 conjugation system composed of purified recombinant human E1 (SAE1/2), E2 (Ubc9) enzymes and purified mature SUMO-1 (aa 1–97) (36,37). Immunoblot analysis revealed an additional band of slower mobility when the ATF7 protein was the substrate (ATF7WT) but not when its sumoylation site was mutated (ATF7K118R). As a negative control, ATF2, a protein closely related to ATF7 that lacks the consensus sumoylation site (Figure 1A), was shown not to be sumoylated in this assay (Figure 1B).
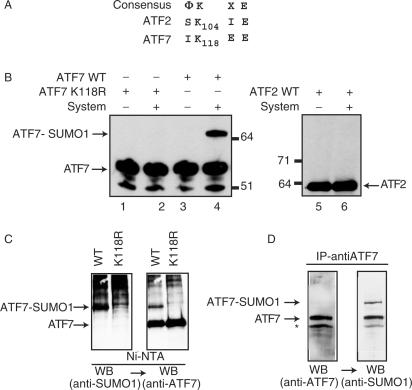
Figure 1.
SUMO-1 modification of recombinant ATF proteins. Peptide sequence alignments between ATF7 and ATF2. (A) Amino acid sequences (one-letter code) of relevant portions of the proteins are given, below the consensus sumoylation target site (Φ, hydrophobic residues; X, any residue). Bold-faced characters represent potential sumoylation sites, with respective coordinates. (B) Purified bacterially expressed ATF7 WT (lanes 3 and 4), ATF7 K118R (lanes 1 and 2) and in vitro synthesized 35S-labeled ATF2 (lanes 5 and 6) were subjected to in vitro SUMO-1 conjugation, as indicated (System refers to the complete assay reaction as described in Materials and Methods—AU refers to arbitrary unit). Reaction products were analyzed by SDS-PAGE, followed by immunoblotting using ATF7 (2F10) monoclonal-antibodies (lanes 1–4) or autoradiography (lanes 5 and 6). (C) HeLa-SUMO cells were transfected with 0.5 µg px-ATF7WT or pXATFK118R. Extracts (500 µg) from transfected cells were passed through Ni-NTA agarose (affinity-purification). Proteins were separated by SDS-PAGE and the presence of sumoylated ATF7, and then ATF7 was revealed by western blotting (WB) using specific antibodies, as indicated. (D) Extracts (500 µg) from Raji cells were immunoprecipitated with the anti-ATF7 (2F10) antibodies. Proteins were separated by SDS-PAGE and the presence of ATF7, and then sumoylated ATF7 was revealed by western blotting (WB) using specific antibodies, as indicated.
Detection of SUMO-1 conjugates in living cells is often difficult because only a small proportion of cellular SUMO-1 target proteins is modified at any given time. For this reason, SUMO conjugation is generally investigated using cells overexpressing SUMO proteins. To verify SUMO-1 modification of ATF7 in a cellular context, we thus transfected a human cell line (HeLa-SUMO) that constitutively expresses elevated levels of 6His:Myc:epitope-tagged human SUMO-1 protein (22) with plasmids expressing either the ATF7WT or the K118R derivative (Figure 1C). Transfected cells were lysed in the presence of SDS and N-ethylmaleimide (NEM) to inhibit SUMO-specific proteases activity. Cellular extracts were passed over nickel-charged agarose resin (Ni-NTA agarose, Qiagen) to purify 6His:Myc:SUMO-1 conjugated proteins. Analysis of Ni-NTA bound fractions was first performed by immunoblotting using SUMO-1 specific antibodies (Figure 1C, left panel). Ladders of many SUMO conjugated proteins retained on the Ni-NTA beads could be observed, but interestingly a major band with an apparent molecular weight compatible with that of a mono-sumoylated form of ATF7 was detected only into the cells transfected with ATF7WT. The same blot was then probed with ATF7-specific antibodies to confirm in both cell-extracts the presence of similar amounts of non-modified ATF proteins that also bind to the resin through their zinc-binding domain (Figure 1C, right panel). Together these results clearly demonstrate that the mutation K118R impairs the symoylation of ATF7.
To demonstrate that the endogenous ATF7 proteins are covalently modified by SUMO in vivo, we used Raji cells, a human lymphoid cell line in which ATF7 is naturally more abundant than in other cell lines (14). Cells were lysed in the presence of SDS and N-ethylmaleimide (NEM) to inhibit SUMO-specific proteases activity. The cellular extracts were immunoprecipitated using monoclonal anti-ATF7 antibodies and analyzed by western blotting using first ATF7 antibodies (Figure 1D, left panel). A major band corresponding to ATF7 was observed. Interestingly, a minor band with an apparent molecular weight compatible with that of a mono-sumoylated form of ATF7 was also detected. To assess the specificity of this band, the same blot was then probed with anti SUMO-1 polyclonal antibodies. As shown in Figure 1D (right panel), the SUMO-1 antibodies specifically recognized the upper band. Thus the endogenous ATF7 proteins are subjected to modification by SUMO-1 in vivo.
SUMO-1 modification affects ATF7 intracellular localization
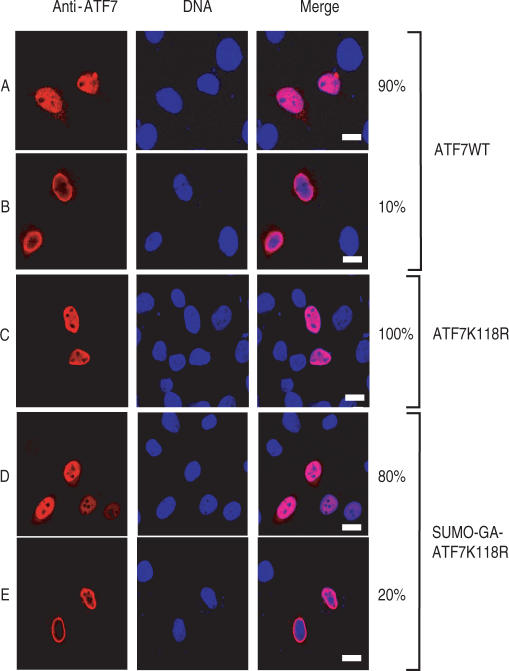
Previous studies had correlated SUMO modification with the recruitment of target proteins to specialized subnuclear domains, (1–3,34,35,38). To address this issue, we transiently expressed ATF7WT and ATF7K118R mutant into HeLa-SUMO cells and followed their intracellular localization by fluorescence microscopy. Immunostaining of transfected cells with anti-ATF7 antibody revealed two patterns of localizations: 90% of the transfected cells mainly showed a nuclear localization for ATF7WT, with a diffuse distribution excluding the nucleoli (Figure 2A) while 10% of the transfected cells showed a distinct pattern restricted to the periphery of the nucleus (Figure 2B). On the other hand, expression of ATF7K118R proteins that cannot be sumoylated showed exclusively the nucleoplasmic localization pattern (Figure 2C). These results suggest that sumoylation may trap the protein at the vinicity of the nuclear envelope. In order to check this hypothesis, we engineered a constitutively sumoylated form of ATF7 by covalently attaching SUMO-1 to ATF7 by gene fusion as previously described for Sp3 or TAF5 (25,39). We therefore generated a ATFK118R derivative composed of the protein SUMO-1 (amino acids 1–96) directly fused N-terminus of ATF7. The C-terminal Gly of full-length SUMO-1 was replaced by an Ala in order to prevent cleavage of the fusion proteins by C-terminal SUMO hydrolases (39). As shown in Figure 2D and E, 20% of the cells transfected with this construct displayed an immunofluorescence staining localized at the periphery of the nucleus. This represents a 2-fold increase compared to the number observed with wild-type ATF7. The observation that not all the cells showed such a perinuclear localization pattern suggests that sumoylation may only transiently block the protein at this level.
Figure 2.
Localization of ATF7 is regulated by SUMO-1. HeLa-SUMO cells were transfected with plasmids encoding wild-type ATF7 (WT, rows A and B) or mutated ATF7 (K118R, row C; SUMO-GA-ATF7 K118R, rows D and E), as indicated. At 36 h post-transfection, cells were processed for immunostaining with monoclonal anti-ATF7 antibodies (2F10) and donkey Cy3-labeled anti-mouse IgG. Nuclei were counterstained with Hoechst 33258. Bar, 10 nm.
RanBP2 functions as a SUMO E3 ligase for ATF7
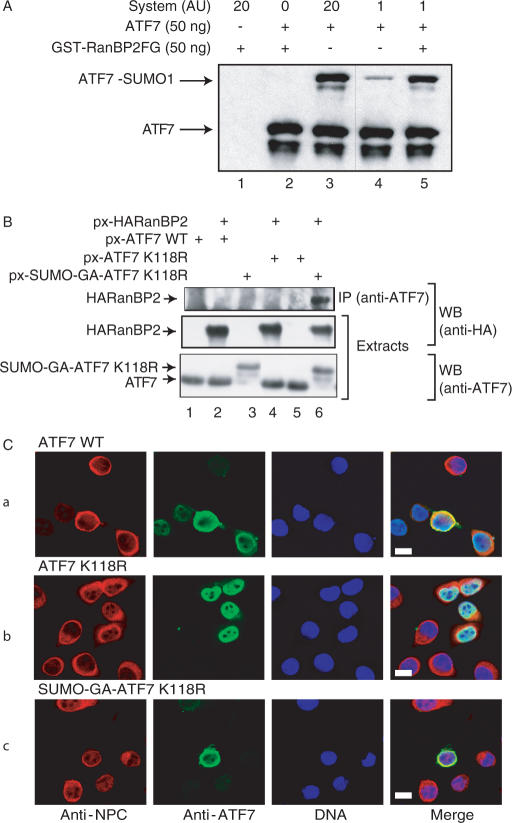
The observed perinuclear localization of the sumoylated form of ATF7 proteins strongly suggests that the corresponding modification process may occur during their entry into the nucleus. Since it has been shown that the nuclear pore complex (NPC) RanBP2 protein strongly enhances SUMO-1 modification of the nuclear body-associated SP100 protein (40) and HDAC4 deacetylase (41), we tested whether this protein could function as a SUMO E3 ligase for ATF7. To verify this hypothesis, our reconstituted in vitro modification assay (see Figure 1) was performed with recombinant ATF7 in the presence or absence of a bacterially produced RanBP2 fusion protein (GST-RanBP2▵FG). This truncated protein was previously described to exhibit an E3 ligase activity (40). As shown in Figure 3A, although the sumoylation of ATF7 occurred to some extent in the absence of SUMO ligase (lane 3), the reaction was strongly stimulated by addition of the RanBP2 derivative (lane 5). To confirm the specificity of the assay, we used GST-PIAS1 instead of RanBP2 in a similar in vitro modification experiment. PIAS1 also exhibits E3 ligase activity but directed towards different transcription factors, like c-jun or p53 (42,43). Whereas sumoylation of ATF7 was greatly enhanced by RanBP2, PIAS1 failed to catalyze SUMO modification of ATF7 in a parallel reaction (data not shown). These observations support the conclusion that RanBP2 functions as a specific SUMO E3 ligase towards ATF7.
Figure 3.
Nuclear-membrane markers co-localize with ATF7. (A) Purified bacterially expressed ATF7WT was subjected to in vitro SUMO-1 conjugation, in the presence or in the absence of purified bacterially expressed GST-RanBP2. Reactions in lanes 4–5 were performed with 20-fold less in vitro sumoylation reaction mix (System) than in lanes 1–3. Reaction products were analyzed by SDS-PAGE and by immunoblotting using ATF7 antibodies. (B) HeLa-SUMO cells were transfected with 0.5 µg px-ATF7 derivatives and px-HA-RanBP2▵FG as indicated. Extracts from transfected cells were immunoprecipitated (IP) with the anti-ATF7 (2F10) monoclonal antibodies as described in Materials and Methods. The purified complexes were separated by SDS-PAGE. The presence of HA-RanBP2▵FG proteins in both IP and extracts, and ATF7 derivatives in the extracts was revealed by western blotting (WB) using specific antibodies. (C) HeLa-SUMO cells were transfected with plasmids encoding mutated ATF7WT (a), ATF7 K118R (b) or SUMO-GA-ATF7 K118R (c), as indicated. At 36 h post-transfection, cells were processed for immunostaining with monoclonal anti-ATF7 antibodies (2F10) and donkey Alexa488-labeled anti-mouse IgG or rabbit polyclonal anti-NPC and donkey Cy3-labeled anti-rabbit IgG. Nuclei were counterstained with Hoechst 33258. Bar: 10 nm.
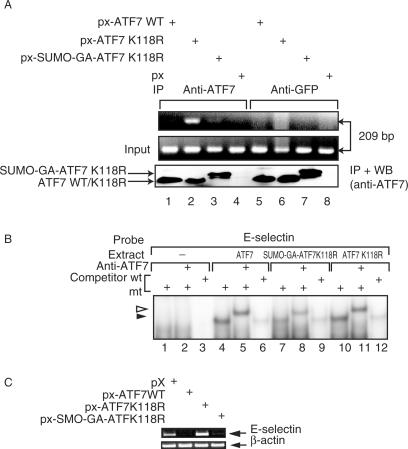
We next analyzed the ability of the fusion SUMO-GA-ATF7 and RanBP2 proteins to interact within a cellular context. To that end, HeLa-SUMO cells were transfected with plasmids expressing either px-ATF7WT, px-ATF7K118R, px-SUMO-GA-ATF7K118R and px-HA-RanBP2·FG (Figure 3B). Whole cell extracts were then submitted to immunoprecipitation (IP) with antibodies against ATF7 and the precipitated proteins were analyzed by western blotting using anti-HA antibodies. As shown in Figure 3B, RanBP2 was detected in the immunoprecipitates only when coexpressed with the constitutively sumoylated ATF7 construct, but not in the presence of ATF7WT or ATF7K118R. Therefore, these results strongly suggest that sumoylation promotes the interaction between ATF7 and RanBP2, supporting the idea that RanBP2 is the ATF7 specific E3 ligase. The fact that no interaction could be detected between ATF7WT and RanBP2 can be explained by a very low amount of the sumoylated ATF7WT protein in HeLa-SUMO-transfected cells.
In order to refine the location of sumoylated ATF7 within the nucleus of transfected cells, we performed co-localization experiments using antibodies against nuclear pore complex proteins (anti-NPC) and ATF7 (Figure 3C).
As expected, the anti-NPC antibodies primarily stained the nuclear membrane. While the non-sumoylatable ATF7 derivative (ATF7K118R) was clearly nucleoplasmic (row b), the ATF7 WT (row a) and its constitutively sumoylated derivative (SUMO-GA-ATF7K118R; row c) were confined to the nuclear periphery, within up to 20% of the transfected cells, in agreement with the results shown in Figure 2. Furthermore, as revealed by the double immunostaining, these ATF7 proteins clearly accumulate in close proximity to the nuclear membrane, in a region documented to be transcriptionally inactive (4).
Sumoylation delays the entry into the nucleus
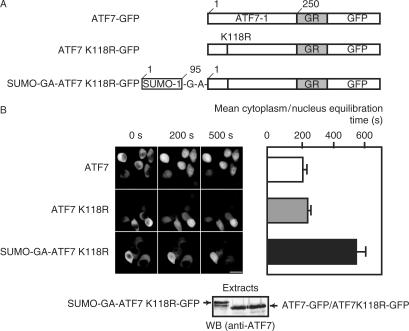
The results of our immunofluorescence experiments suggest that the entry of ATF7 into the nuclei is a dynamic phenomenon which may be modulated by sumoylation. In order to visualize ATF7 diffusion into the nucleoplasm more precisely, time laps microscopy experiments using GFP-tagged constructs were performed. We used hormone-inducible GFP-tagged chimeric ATF7 protein (ATF7-GR-GFP) (Figure 4A) to study the ATF7 nuclear import in living cells. The first 250 amino acid residues of ATF7 (to delete the ATF7 endogenous NLS sequence) were fused to the hormone-responsive element from the glucocorticoid receptor–GFP (GFP-GR) chimera.
Figure 4.
Sumoylation delays protein entry into the nucleus. (A) The structures of the different ATF7 constructs are schematized. (B) Time laps microscopy monitoring of cells expressing GFP fused constructs ATF7WT, ATF7K118R or SUMO-GA-ATF7. HeLa-SUMO cells were transfected with 0.25 µg of the different plasmids as indicated. Twenty-four hours later 10 nM Dexamethasone was added to the culture medium (t = 0). The level of fluorescence in the nucleus was monitored using time laps microscopy. Bar: 20 nm. Quantification of the mean time required to reach a nuclear signal of same intensity as the one measured in the cytoplasm perinuclear region (cytoplasm/nuclear equilibration). Constitutively sumoylated ATF requires twice as much time as the other constructs to equilibrate the nuclear signal to the level of the cytoplasm.
To assess the effect of sumoylation, the import of ATF7WT, ATFK118R and SUMO-GA-ATF7K118R GR-GFP derivatives was examined in HeLa-SUMO cells (Figure 4B). In the absence of steroid, the chimeric proteins were all localized in the cytoplasm and translocation of the three variants into the nucleus was observed only after the addition of dexamethasone (Figure 4B). For each construct, we measured the mean time required to reach a nuclear signal of same intensity as the one observed in the cytoplasmic perinuclear region. Clearly, constitutively sumoylated ATF requires twice as much time as the other proteins to equilibrate the nuclear signal to the level of the cytoplasm. This indicates that sumoylation specifically delays the import of the ATF7 protein into the nucleus.
Sumoylation of ATF7 interferes with its transcriptional activity
To get an insight of the effect of sumoylation on the transcriptional activation mediated by ATF7, we analyzed the effect of K118R mutation specifically impairing the sumoylation of ATF7 on its transcriptional activity. We investigated the interplay of this modification using the Gal4 protein fusion system (44), in which the ATF7 protein is linked to the yeast Gal4 DNA binding domain (1–147). The activity of the chimeric products was assayed in cotransfection experiments with a luciferase reporter gene whose promoter contained five Gal4 binding sites (44). Cos-1 cells were transfected with the pG4-ATF7 vector or derivatives and the luciferase reporter. The results of a typical experiment are shown in Figure 5A. The K118R mutant stimulated transcription of the reporter gene 2-fold more than the WT protein. In contrast, the Gal4-SUMO-GA-ATF7 K118R construction was about 4-fold less active than the WT protein while equal levels of Gal-ATF7 fusions were expressed and showed identical cellular distribution as the non-fused proteins. These results indicate that a large part of the WT protein activity is due to its non-sumoylated form.
Identical results were obtained with the three ATF7 isomers (9), namely ATF7-1, ATF7-2 and ATF7-3 (not shown). Thus sumoylation plays a negative role in the ATF7-induced transactivation process under these experimental conditions, possibly by sequestering the proteins in the periphery of the nucleus.
We previously showed that the transcriptional activity of ATF7 is potentiated by expression of TAF12 and that the two proteins are interacting in vivo (14). We investigated the ability of the fusion SUMO-GA-ATF7 and TAF12 proteins to interact within a cellular context. To that end, HeLa-SUMO cells were transfected with plasmids expressing p-hsTAF12 together with px-ATF7, px-ATF7K118R and px-SUMO-GA-ATF7K118R (Figure 5B). Whole cell extracts were then submitted to immunoprecipitation (IP) with antibodies against ATF7, and the precipitated proteins were analyzed by western blotting using anti-TAF12 antibodies. As shown in Figure 5B (lanes 1–4), TAF12 was detected in the immunoprecipitates only when coexpressed with ATF7WT or ATF7K118R, but not when coexpressed with the constitutively sumoylated ATF7 construct. In HeLa-SUMO transfected cells, the majority of the overexpressed ATF7WT protein is not sumoylated. Therefore, these results strongly suggest that sumoylation may prevent the interaction between ATF7 and TAF12, and this loss of interaction would thus account for the transcriptional inhibitory effect of ATF7 sumoylation. To gain some insight into the molecular mechanism of this differential action on transcription, we first performed a chromatin immunoprecipitation (ChIP) experiment on the E-selectin gene. ATF7 has previously been shown to specifically bind to the NF-ELAM1 promoter element of the E-selectin gene and to interact with TAF12 (14). HeLa-SUMO cells were transfected with vectors expressing either ATF7WT, K118R ATF7, or SUMO-GA-ATF7K118R under conditions where equal levels of proteins were expressed, as verified by immunoblotting with specific antibodies (Figure 6A). We also checked by gel-shift assays that the three proteins were able to bind the NF-ELAM1 promoter element with very similar efficiencies (Figure 6B). As shown in Figure 6A, the E-selectin promoter was immunoprecipitated more efficently in ATF7K118R expressing cells than in ATF7WT or SUMO-GA-ATF7K118R transfected cells. These results clearly indicate that, in the context of a natural promoter (and in a steady-state level), non-sumoylated ATF7 form is more strongly associated with the chromatin. Therefore sumoylation prevents the association of ATF7 with the target promoter or facilitates its removal in vivo. In addition, a direct measurement of transcription was performed to analyze the changes in an ATF7 target gene expression upon the changes in ATF7 sumoylation. RT-PCR analysis clearly shows that transcription of the E-selectin gene is enhanced in the presence of ATF7K118R (Figure 6C) whereas the expression level of the control gene (β-actin) is unaffected. Altogether, these observations clearly confirm the inhibitory effect of sumoylation of ATF7 on the transactivation process: sumoylation not only impairs the interaction between ATF7 and TAF12 but blocks the access of ATF7 to promoters.
Figure 6.
Sumoylation affects ATF7 activity. (A) ChIP analysis of genomic E-selectin promoter: HeLa-SUMO cells were transfected with px-ATF7WT(lanes 1,3) or px- ATF7K118R (lanes 2,4). Two days later, cells were subjected to ChIP assay with antibodies against ATF7 (lanes 1 and 2) or Flag (lanes 3 and 4). Endogenous E-selectin promoter DNA, coprecipitated with the indicated antibodies, was detected by PCR. (B) Recombinant pSG5-based vectors (0.5 µg) directing the expression of the various chimeras were transfected into HeLa-SUMO cells, and cell lysates were analyzed by EMSA: 20 µg of extracts were preincubated with either 50 ng of non-specific (mt) or specific (wt) competitor before addition of E-selectin CRE probe. Where indicated, reactions were further incubated (30 min, 0°C) with the anti-ATF7 antibody before loading on the gel. The black arrowhead points to the specific complex and the white one to the supershifted one. (C) Recombinant pX-based vectors (0.5 µg) directing the expression of the various chimeras were transfected into HUVEC-C-cells. Two days later, RNA was isolated and E-selectin mRNA expression was analyzed by semi-quantitative RT–PCR method using β-actin as internal control. Ethidium bromide-stained gel is shown in the figure.
DISCUSSION
Small ubiquitin-like modifier (SUMO) modification has emerged as an important regulator of diverse pathways and activities including protein targeting, transcriptional regulation, modulation of enzyme activity and protein turnover (for reviews, see References (1,3,35)). In the present article, we demonstrate that the ATF7 transcription factor is sumoylated in vitro and in vivo, and that sumoylation affects its intranuclear localization and its transcriptional activity by impairing the interaction between ATF7 and TAF12, thereby blocking the access of ATF7 to its specific promoters.
ATF7, but not ATF2, is sumoylated
Since their discovery, ATF7 and ATF2 although encoded by distinct genes (9,45) have always been described as two closely related proteins, with very similar properties (7,46). For example, the two proteins mediate transactivation by the adenovirus oncoprotein E1a (8,47) and heterodimerize with Jun or Fos proteins (10). They are also phoshorylated by p38 and JNK MAP-kinases (11,48,49) and their transcriptional activity is mediated by TAF12 (14). By contrast, a survey of the protein sequences derived from all sequenced eukaryotic genomes clearly reveals a consensus sumoylation site (ϕKXE) within all known ATF7 protein sequences, whereas it is absent in all ATF2 proteins. We clearly show here that ATF7, but not ATF2, is sumoylated in vivo at the lysine residue (K118) belonging to this element. Though this sequence is located in the previously described transactivating domain, it is the first time that such a striking difference is observed between the activity of the two proteins. This indicates that, notwithstanding their high functional and structural homologies, these two ATF proteins likely play distinct roles in the cell.
It has been suggested that SUMO modification could affect the subcellular localization of proteins (3). As shown for many nuclear proteins, sumoylation can direct some transcription factors to the so-called PML nuclear bodies (50), thus sequestering them away from the DNA and taking them out of action (50–52). Furthermore, sumoylation can also be coupled with the transport of proteins between the cytoplasm and the nucleus (53,54).
As revealed by our localization studies, the wild-type ATF7 (ATF7WT) is dispersed throughout the nucleoplasm in the majority of the cells (about 90%), while it is restricted to a perinuclear distribution in approximately 10% of the cells. The fact that a non-sumoylatable mutant protein (ATF7K118R) never accumulates at the nuclear periphery, strongly suggests that the nucleoplasmic ATF7 corresponds to the non-sumoylated fraction of the protein. On the other hand, the in vivo perinuclear localization is in good agreement with the fact that the ATF7 sumoylation process requires (i) Ubc9, a conjugating enzyme which is associated to both cytoplasmic and nucleoplasmic fibrils of the nuclear pore complex (NPC) in vivo (55), and (ii) the E3 ligase RanBP2, a protein also belonging to the NPC (40). Thus, it appears that the ATF7 localization is affected by sumoylation, as previously reported for other transcription factors, like Sp3 (39), CtBP (56), Elk-1 (57) or SATB2 (58). In the latter case, a very similar correlation between sumoylation and subnuclear localization has also been observed: sumoylated SATB2 was predominantly localized at the nuclear periphery, whereas a sumoylation-deficient SATB2 protein had a more diffuse nuclear localization (58).
Our results clearly indicate that sumoylation does not block the nuclear entry of ATF7 since a constitutively sumoylated ATF7 derivative (SUMO-GA-ATF7K118R) exhibits a distribution pattern similar to the WT protein, as revealed by classical immunofluorescence microscopy experiments based on snapshot pictures of fixed cells. Time laps microscopy further extended our views. Indeed, the stably sumoylated protein takes twice as long as the non-sumoylated form (ATF7K118R) to enter the nucleus and evenly to redistribute over the nucleoplasm. These kinetic experiments strongly suggest that ATF7 sumoylation is a dynamic process that affects its in vivo localization behavior. Thus an equilibrium between the relative amounts of sumoylated and non-sumoylated forms of the protein might exist in the cell.
Sumoylation affects ATF7 transcriptional activity
Besides its contribution to the subcellular localization of ATF7, a functional role for the sumoylation was inferred from the analysis of the SUMO-deficient mutant protein (ATF7K118R). First, chromatin immunoprecipitation experiments revealed that the SUMO-deficient mutant ATF7 derivative associates more efficiently than ATF7WT with the CRE promoter element of the endogenous E-selectin gene. Second, SUMO-deficient ATF7 stimulated expression from a cotransfected reporter plasmid more efficiently than ATF7WT. By contrast, fusion of SUMO to the K118R mutant of ATF7 (as in SUMO-GA-ATF7K118R) resulted in a decrease of its transcriptional activation potential, an observation most likely related to the negative effect of SUMO on the ability of ATF7 to interact with its TAF12 partner. As previously shown, TAF12 potentiates ATF7-induced transcriptional activation through direct interaction with the N-terminal ATF7 transactivation domain (14). It appears therefore that the SUMO modification clearly contributes to the regulation of the ATF7 transcriptional activity by impairing the interaction between ATF7 and TAF12. Interestingly, both TAF5 and TAF12—TFIID subunits—have been shown to be sumoylated (25). However, only SUMO conjugation of TAF5 interferes with binding of TFIID to promoter DNA, whereas modification of TAF12 has no detectable effect in this respect. Furthermore, SUMO conjugation has also no detectable effect on nuclear import or intranuclear distribution of these two TAFs. Clearly sumoylation may differentially affect proteins involved in a same function, i.e. transactivation of target genes. It is difficult to directly correlate the functional differences observed between native ATF7WT and its non-sumoylatable counterpart (ATF7K118R), in terms of transcriptional activity or occupation of its target promoters, with their actual respective sumoylation levels. Indeed, the action of endogenous SUMO proteases (Senp1 and/or 2), released into the cell extracts, likely alters the apparent proportion of sumoylated proteins. This is best illustrated in Figure 5, where ATF7WT and ATF7K118R exhibit significantly different transcriptional and promoter binding activities (panels A and D), while showing equally low levels of sumoylation, compared to the Senp-resistant SUMO-GA-ATF7K118R derivative (panel B). A model has recently been proposed (4), in which the pool of sumoylated transcription factors constitutes a built-in portion of more general transcriptional repression complexes. Within these structures, factors could be desumoylated while remaining associated with the complex of repression, independent of SUMO. Thus, SUMO would be required to initiate but not to maintain repression (4). However, upon sustained desumoylation, the overall number of repression complexes might ultimately decrease. Such a scenario has been described in the case of the Elk-1 transcription factor whose phosphorylation by MAP-kinases is concomitant with its desumoylation and the activation of transcription (59). The fact that transcriptional activity of ATF7 is enhanced when the p38 MAP-kinase pathway is stimulated supports this model. Consequently, it would be advisable to check the relationships between phosphorylation and sumo/desumoylation of ATF7.
ACKNOWLEDGEMENTS
We thank P. O'Hare, D. Bailey, V. Doye, J.L. Bocco, B. Wasylyk, A. Werner, J. Seeler, M.Donzeau and J. De Mey for gifts and helpful discussions.
This work was supported by funds and/or fellowships from the Centre National de la Recherche Scientifique, the Université Louis Pasteur de Strasbourg, the French Ministry of Research, the Institut National de la Santé et de la Recherche Médicale, the Association pour la Recherche sur le Cancer (contract 3712) and the Ligue Nationale contre le Cancer—Comités Alsace (Haut-Rhin et Bas-Rhin) et Vosges. Funding to pay the Open Access publication charge was provided by the Association pour la Recherche sur le Cancer.
Conflict of interest statement. None declared.
REFERENCES
- 1.Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 3.Gill G. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Hay RT. SUMO a history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Yang SH, Galanis A, Witty J, Sharrocks AD. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006;25:5083–5093. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson ES. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 7.Gaire M, Chatton B, Kedinger C. Isolation and characterization of two novel, closely related ATF cDNA clones from HeLa cells. Nucleic Acids Res. 1990;18:3467–3473. doi: 10.1093/nar/18.12.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatton B, Bocco JL, Gaire M, Hauss C, Reimund B, Goetz J, Kedinger C. Transcriptional activation by the adenovirus larger E1a product is mediated by members of the cellular transcription factor ATF family which can directly associate with E1a. Mol. Cell Biol. 1993;13:561–570. doi: 10.1128/mcb.13.1.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goetz J, Chatton B, Mattei MG, Kedinger C. Structure and expression of the ATFa gene. J. Biol. Chem. 1996;271:29589–29598. doi: 10.1074/jbc.271.47.29589. [DOI] [PubMed] [Google Scholar]
- 10.Chatton B, Bocco JL, Goetz J, Gaire M, Lutz Y, Kedinger C. Jun and Fos heterodimerize with ATFa, a member of the ATF/CREB family and modulate its transcriptional activity. Oncogene. 1994;9:375–385. [PubMed] [Google Scholar]
- 11.De Graeve F, Bahr A, Sabapathy KT, Hauss C, Wagner EF, Kedinger C, Chatton B. Role of the ATFa/JNK2 complex in Jun activation. Oncogene. 1999;18:3491–3500. doi: 10.1038/sj.onc.1202723. [DOI] [PubMed] [Google Scholar]
- 12.Chatton B, Bahr A, Acker J, Kedinger C. Eukaryotic GST fusion vector for the study of protein- protein associations in vivo: application to interaction of ATFa with Jun and Fos. Biotechniques. 1995;18:142–145. [PubMed] [Google Scholar]
- 13.Bocco JL, Bahr A, Goetz J, Hauss C, Kallunki T, Kedinger C, Chatton B. In vivo association of ATFa with JNK/SAP kinase activities. Oncogene. 1996;12:1971–1980. [PubMed] [Google Scholar]
- 14.Hamard PJ, Dalbies-Tran R, Hauss C, Davidson I, Kedinger C, Chatton B. A functional interaction between ATF7 and TAF12 that is modulated by TAF4. Oncogene. 2005;24:3472–3483. doi: 10.1038/sj.onc.1208565. [DOI] [PubMed] [Google Scholar]
- 15.Freiman RN, Albright SR, Zheng S, Sha WC, Hammer RE, Tjian R. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science. 2001;293:2084–2087. doi: 10.1126/science.1061935. [DOI] [PubMed] [Google Scholar]
- 16.Mengus G, Fadloun A, Kobi D, Thibault C, Perletti L, Michel I, Davidson I. TAF4 inactivation in embryonic fibroblasts activates TGF beta signalling and autocrine growth. EMBO J. 2005;24:2753–2767. doi: 10.1038/sj.emboj.7600748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davidson I, Kobi D, Fadloun A, Mengus G. New insights into TAFs as regulators of cell cycle and signaling pathways. Cell Cycle. 2005;4 doi: 10.4161/cc.4.11.2120. [DOI] [PubMed] [Google Scholar]
- 18.Xiao JH, Davidson I, Matthes H, Garnier JM, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 19.Lavigne AC, Gangloff YG, Carre L, Mengus G, Birck C, Poch O, Romier C, Moras D, Davidson I. Synergistic transcriptional activation by TATA-binding protein and hTAFII28 requires specific amino acids of the hTAFII28 histone fold. Mol. Cell Biol. 1999;19:5050–5060. doi: 10.1128/mcb.19.7.5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love DC, Sweitzer TD, Hanover JA. Reconstitution of HIV-1 rev nuclear export: independent requirements for nuclear import and export. Proc. Natl. Acad. Sci. U.S.A. 1998;95:10608–10613. doi: 10.1073/pnas.95.18.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cline J, Braman JC, Hogrefe HH. PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey D, O'Hare P. Herpes simplex virus 1 ICP0 co-localizes with a SUMO-specific protease. J. Gen. Virol. 2002;83:2951–2964. doi: 10.1099/0022-1317-83-12-2951. [DOI] [PubMed] [Google Scholar]
- 23.Pollard H, Remy JS, Loussouarn G, Demolombe S, Behr JP, Escande D. Polyethylenimine but not cationic lipids promotes transgene delivery to the nucleus in mammalian cells. J. Biol. Chem. 1998;273:7507–7511. doi: 10.1074/jbc.273.13.7507. [DOI] [PubMed] [Google Scholar]
- 24.Kumar V, Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988;55:145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- 25.Boyer-Guittaut M, Birsoy K, Potel C, Elliott G, Jaffray E, Desterro JM, Hay RT, Oelgeschlager T. Sumo-1 modification of human TFIID complex subunits: Inhibition of TFIID promoter binding activity through sumo-1 modification of hsTAF5. J. Biol. Chem. 2005;280:9937–45. doi: 10.1074/jbc.M414149200. [DOI] [PubMed] [Google Scholar]
- 26.de Wet JR, Wood KV, DeLuca M, Helinski DR, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steghens JP, Min KL, Bernengo JC. Firefly luciferase has two nucleotide binding sites: effect of nucleoside monophosphate and CoA on the light-emission spectra. Biochem. J. 1998;336(Pt 1):109–113. doi: 10.1042/bj3360109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scholer HR, Ciesiolka T, Gruss P. A nexus between Oct-4 and E1A: implications for gene regulation in embryonic stem cells. Cell. 1991;66:291–304. doi: 10.1016/0092-8674(91)90619-a. [DOI] [PubMed] [Google Scholar]
- 29.De Graeve F, Bahr A, Chatton B, Kedinger C. A murine ATFa-associated factor with transcriptional repressing activity. Oncogene. 2000;19:1807–1819. doi: 10.1038/sj.onc.1203492. [DOI] [PubMed] [Google Scholar]
- 30.Bahr A, De Graeve F, Kedinger C, Chatton B. Point mutations causing Bloom's syndrome abolish ATPase and DNA helicase activities of the BLM protein. Oncogene. 1998;17:2565–2571. doi: 10.1038/sj.onc.1202389. [DOI] [PubMed] [Google Scholar]
- 31.Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, Tempst P, Roeder RG, Zhang Y. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell. 2003;12:475–487. doi: 10.1016/j.molcel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 32.Gong L, Kamitani T, Fujise K, Caskey LS, Yeh ET. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 1997;272:28198–28201. doi: 10.1074/jbc.272.45.28198. [DOI] [PubMed] [Google Scholar]
- 33.Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 34.Seeler JS, Dejean A. SUMO: of branched proteins and nuclear bodies. Oncogene. 2001;20:7243–7249. doi: 10.1038/sj.onc.1204758. [DOI] [PubMed] [Google Scholar]
- 35.Gill G. Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr. Opin. Genet. Dev. 2003;13:108–113. doi: 10.1016/s0959-437x(03)00021-2. [DOI] [PubMed] [Google Scholar]
- 36.Desterro JM, Rodriguez MS, Kemp GD, Hay RT. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 1999;274:10618–10624. doi: 10.1074/jbc.274.15.10618. [DOI] [PubMed] [Google Scholar]
- 37.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 38.Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 39.Ross S, Best JL, Zon LI, Gill G. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell. 2002;10:831–842. doi: 10.1016/s1097-2765(02)00682-2. [DOI] [PubMed] [Google Scholar]
- 40.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 41.Kirsh O, Seeler JS, Pichler A, Gast A, Muller S, Miska E, Mathieu M, Harel-Bellan A, Kouzarides T, et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 2002;21:2682–2691. doi: 10.1093/emboj/21.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt D, Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2872–2877. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu F, Green MR. A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1a protein. Cell. 1990;61:1217–1224. doi: 10.1016/0092-8674(90)90686-9. [DOI] [PubMed] [Google Scholar]
- 45.Nagase T, Sudo T, Maekawa T, Yoshimura T, Fujisawa J, Yoshida M, Ishii S. Promoter region of the human CRE-BP1 gene encoding the transcriptional regulator binding to the cyclic AMP response element. J. Biol. Chem. 1990;265:17300–17306. [PubMed] [Google Scholar]
- 46.Maekawa T, Matsuda S, Fujisawa J, Yoshida M, Ishii S. Cyclic AMP response element-binding protein, CRE-BP1, mediates the E1A-induced but not the tax-induced trans-activation. Oncogene. 1991;6:627–632. [PubMed] [Google Scholar]
- 47.Zu YL, Maekawa T, Matsuda S, Ishii S. Complete putative metal finger and leucine zipper structures of CRE-BP1 are required for the E1A-induced trans-activation. J. Biol. Chem. 1991;266:24134–24139. [PubMed] [Google Scholar]
- 48.Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong S, Muller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
- 51.Sternsdorf T, Jensen K, Reich B, Will H. The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem. 1999;274:12555–12566. doi: 10.1074/jbc.274.18.12555. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi Y, Lallemand-Breitenbach V, Zhu J, de The H. PML nuclear bodies and apoptosis. Oncogene. 2004;23:2819–2824. doi: 10.1038/sj.onc.1207533. [DOI] [PubMed] [Google Scholar]
- 53.Rangasamy D, Woytek K, Khan SA, Wilson VG. SUMO-1 modification of bovine papillomavirus E1 protein is required for intranuclear accumulation. J. Biol. Chem. 2000;275:37999–38004. doi: 10.1074/jbc.M007777200. [DOI] [PubMed] [Google Scholar]
- 54.Comerford KM, Leonard MO, Karhausen J, Carey R, Colgan SP, Taylor CT. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc. Natl. Acad. Sci. U.S.A. 2003;100:986–991. doi: 10.1073/pnas.0337412100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang H, Saitoh H, Matunis MJ. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell Biol. 2002;22:6498–6508. doi: 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 57.Salinas S, Briancon-Marjollet A, Bossis G, Lopez MA, Piechaczyk M, Jariel-Encontre I, Debant A, Hipskind RA. SUMOylation regulates nucleo-cytoplasmic shuttling of Elk-1. J. Cell Biol. 2004;165:767–773. doi: 10.1083/jcb.200310136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dobreva G, Dambacher J, Grosschedl R. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 2003;17:3048–3061. doi: 10.1101/gad.1153003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang SH, Jaffray E, Hay RT, Sharrocks AD. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol. Cell. 2003;12:63–74. doi: 10.1016/s1097-2765(03)00265-x. [DOI] [PubMed] [Google Scholar]