Abstract
Human telomerase reverse transcriptase (hTERT), the catalytic subunit of human telomerase, contains conserved motifs common to retroviral reverse transcriptases and telomerases. Within the C motif of hTERT is the Leu866-Val867-Asp868-Asp869 tetrapeptide that includes a catalytically essential aspartate dyad. Site-directed mutagenesis of Tyr183 and Met184 residues in HIV-1 RT, residues analogous to Leu866 and Val867, revealed that they are key determinants of nucleotide binding, processivity and fidelity. In this study, we show that substitutions at Val867 lead to significant changes in overall enzyme activity and telomere repeat extension rate, but have little effect on polymerase processivity. All Val867 substitutions examined (Ala, Met, Thr) led to reduced repeat extension rates, ranging from ∼20 to 50% of the wild-type rate. Reconstitution of V867M hTERT and telomerase RNAs (TRs) with mutated template sequences revealed the effect on extension rate was associated with a template copying defect specific to template A residues. Furthermore, the Val867 hTERT mutants also displayed increased nucleotide incorporation fidelity, implicating Val867 as a determinant of telomerase fidelity. These findings suggest that by evolving to have a valine at position 867, the wild-type hTERT protein may have partially compromised polymerase fidelity for optimal and rapid repeat synthesis.
INTRODUCTION
Telomeres are specialized structures at chromosome termini consisting of hundreds to thousands of hexanucleotide repeat sequences complexed with several proteins (1). They prevent chromosome ends from being perceived as DNA damage, effectively protecting chromosomes from degradation and end-to-end joining, thereby rendering chromosomes stable through successive rounds of replication. Telomerase is a specialized cellular reverse transcriptase (RT) employed by eukaryotic cells to restore telomeric DNA that is lost as a consequence of chromosomal replication (2). Structurally, telomerase is a ribonucleoprotein complex, with an RNA-dependent DNA polymerase activity residing in a protein subunit, the telomerase reverse transcriptase (TERT), which utilizes a short, well-defined sequence within an intrinsic RNA subunit, telomerase RNA (TR), to template the synthesis of telomeric repeats.
The initial identification of the TERT protein component of telomerase was facilitated by its homology to other reverse transcriptases (3,4) including human immunodeficiency virus-1 RT (HIV-1 RT). Sequence analysis of TERT proteins reveals the presence of several of the conserved motifs important for RT function. Among these motifs the RT C motif, which contains a common signature sequence ‘YXDD’ (5–7) shared by all retroviral reverse transcriptases, has been shown to be one of the most essential for enzyme activity (8,9). The invariant aspartates in this motif constitute part of an absolutely conserved aspartate triad thought to directly participate in nucleotide addition, coordinated by divalent cations via a two metal ion mechanism (10). TERT proteins also contain a triad of invariant aspartates, two of which are within a ‘hhDD’ (where h = hydrophobic residue) sequence in their C motifs analogous to the YXDD sequence (Figure 1A), that when substituted with alanine result in completely inactive enzymes (4,11,12). The fact that these acidic residues are indispensable underscores their functional importance for telomerase enzymatic activity and suggests that telomerase may also utilize a two metal ion mechanism to catalyze polymerization.
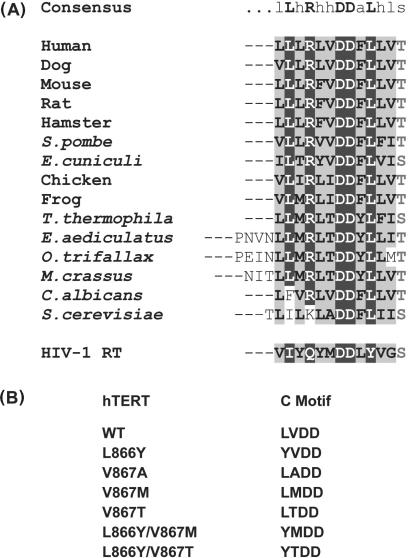
Figure 1.
Sequence alignment of TERT protein C motifs and hTERT C-motif mutants. (A) Alignment of TERT protein C-motif sequences from human (36,37), dog (38), mouse (39), rat (GenBank accession no. AAF62177.1), hamster (40), S. pombe (36), E. cuniculi (41), chicken (42), frog (43), T. Thermophila (44,45), E. aediculatus (4), O. trifallax (44), M. crassus (46), C. albicans (EMBL accession no. CAC37831.1) and S. cerevisiae (4). The HIV-1 RT C-motif sequence (47) is also shown. (B) Sequences of hTERT C-motif mutants studied in this work.
In addition to the catalytic role of the aspartate residues, key roles in other aspects of reverse transcriptase function have been assigned to the tyrosine (Y) and ‘X’ residues of the YXDD sequence. Studies on retroviral RTs (mainly HIV-1 RT) have revealed that these residues influence dNTP binding, polymerase processivity, nucleoside analog resistance and polymerase fidelity (8,13–18). The crystallographic structures of binary (enzyme–nucleic acid) and ternary (enzyme–nucleic acid–nucleotide) complexes of HIV-1 RT place these residues within the active site in close proximity of the incoming dNTP substrate and the extending primer terminus (19,20). These structures show that the ‘X’ amino acid, methionine 184 (Met184) in HIV-1 RT, is positioned against the terminus of the extending primer such that its side chain interacts with the base and deoxyribose ring of the terminal nucleotide. The tyrosine residue, tyrosine 183 (Tyr183) in HIV-1 RT, is positioned such that its side chain is intercalated between the terminal and penultimate primer nucleotides. Thus, the influence of these residues on HIV-1 RT enzymatic properties stems from the central location of these amino acids within the catalytic site, where they can establish multiple contacts with the primer, dNTP and surrounding dNTP-binding pocket residues.
Little is known about the functional significance of the two residues that precede the conserved aspartates in the hhDD sequence of TERTs. In mutagenesis studies on Tetrahymena and yeast TERTs, substitutions at these residues resulted in enzymes with altered processivities (21,22). Curiously, Tetrahymena and yeast TERT variants with substitutions leading to C motifs containing identical YTDD sequences displayed opposite changes in processivity, with the Tetrahymena enzyme becoming more processive and the yeast enzyme less processive. Although the underlying basis for the divergent effects of these mutations is unclear, these observations provide initial indications that these residues, which have major influence on retroviral RT function, may also play important roles in telomerase function. In order to investigate the involvement of the hydrophobic residues of the hhDD sequence in human telomerase activity, we have analyzed the effects of amino acid substitutions in the C motif LVDD sequence in human TERT (hTERT). Our studies reveal that mutation of the leucine residue within the LVDD sequence (leucine 866 (Leu866)) to tyrosine results in only a slight reduction in enzyme activity and no changes in polymerase fidelity or repeat extension rate. Furthermore, unlike Tetrahymena and yeast TERT mutants, no change in processivity was observed for hTERT containing the YTDD sequence. In contrast, substitution at valine 867 (Val867) of the LVDD sequence resulted in significant changes in enzymatic activity, nucleotide insertion rate, polymerase fidelity and telomere repeat extension rate. These findings demonstrate that Val867 plays a central role in nucleotide incorporation by human telomerase and suggest that the residue may provide important interactions with incoming substrate nucleotides.
MATERIALS AND METHODS
Oligonucleotides
Oligonucleotide substrates were purified via denaturing polyacrylamide gel electrophoresis (PAGE) followed by gel extraction and ethanol precipitation.
Site-directed mutagenesis of hTERT
A cassette strategy (23) was used to generate C-motif mutants of hTERT (Figure 1A). An intermediate vector was generated from pNFLAGhTERT (23) by PCR in which the region of hTERT encoding amino acids 864–871 (containing the C motif) was replaced with a pair of Sap I endonuclease sites, resulting in a self-excising sequence. This plasmid, designated pNFLAGhTERT C del, was digested with SapI, to release the self-excising segment and generate a vector backbone with cohesive ends. Pairs of complementary oligonucleotides encoding specific amino acid substitutions (for example, CTCCTGCGTTTGACAGATGATTTCTTG (V867T Top) and CAACAAGAAATCATCTGTCAAACGCAG (V867T Bottom) were used to make the mutant V867T) were phosphorylated, annealed and ligated into the pNFLAGhTERT C del backbone. The resultant constructs were transformed, isolated and sequenced to confirm the presence of the desired changes and the absence of unintended modifications.
In vitro reconstitution of human telomerase
Telomerase activity was reconstituted in vitro by coupled in vitro transcription/translation of hTERT protein in rabbit reticulocyte lysate (Promega) in the presence of purified hTR RNA (23). Reconstitution reactions containing pNFLAGhTERT (or its C-motif mutant derivatives) (0.77 pmol (0.04 µg)/µl) and purified hTR RNA (0.82 pmol (0.12 µg)/µl) were carried out as previously described (23) and stored at −80°C until needed. Aliquots of the reconstitutions were also analyzed via SDS-PAGE to quantitate the in vitro synthesized hTERT protein.
Telomerase activity assay
Telomerase activity was measured via a direct primer extension assay using radiolabeled primer and unlabeled deoxynucleotides. Primer extension reactions contained 2 µl in vitro reconstituted (IVR) telomerase, 1 pmol 5′ 32P end-labeled substrate primer d(TTAGGG)3, 1 mM dATP, 1 mM dGTP and 1 mM TTP in PE reaction buffer (50 mM Tris-Cl pH 8.3, 50 mM KCl, 2 mM DTT, 3 mM MgCl2 and 1 mM Spermidine) in a total volume of 10 µl. Reactions were incubated at 30°C for 30 min and then terminated by addition of an equal volume of stop solution (RNAse A (100 µg/ml) in 10 mM Tris-Cl pH 8.0, 20 mM EDTA) followed by incubation at 37°C for 10 min. Reaction products were then isolated and resolved on 8 M urea −10% polyacrylamide gels (23). Dried gels were then analyzed via phosphoimaging using ImageQuant software (Molecular Dynamics).
Competitive primer challenge assay
A competitive primer challenge (bind and chase) assay was used to assess processive DNA synthesis and measure repeat extension rate (23). IVR hTERT (8 µl) was pre-bound to 5′32P-labeled substrate primer d(TTAGGG)3 (4 pmol) in 1× PE reaction buffer (total volume, 24 µl) for 5 min at 30°C. Primer extension synthesis was then initiated by adding 16 µl of 80 µM unlabeled (cold) competitor (chase) primer plus 2.5 mM dATP, 2.5 mM dGTP and 2.5 mM TTP (in 1× PE salts). The reaction mixture was incubated at 30°C and aliquots (10 µl) were removed at 0.5–30 min and synthesis terminated with an equal volume of stop solution. A pre-chased reaction, where labeled primer (1 pmol), unlabeled primer (160 pmol) and dNTPs (1.25 mM) in 1.25× PE buffer were combined prior to addition of IVR hTERT (2 µl), was also performed to confirm the effectiveness of the unlabeled competitor primer. The products of the primer extension reactions were then processed and analyzed via standard protocol described earlier for telomerase activity assay.
Relative enzyme activities and repeat extension rates were calculated from the quantity and length the modal product band (23) within the population of radiolabeled products associated with actively polymerizing complexes, at a given time point. The population of products associated with actively polymerizing complexes were identified by their ability to be further elongated during the chase period (e.g. see Figure 3, compare wild type 3 and 30-min lanes). In these studies, the modal product was determined from the 3-min reaction time point. This time point allowed for the products of actively polymerizing complexes to be easily distinguished from relatively abundant products resulting from low efficiency translocation characteristically seen over the first few rounds of template copying, as well as for accurate size determination (23).
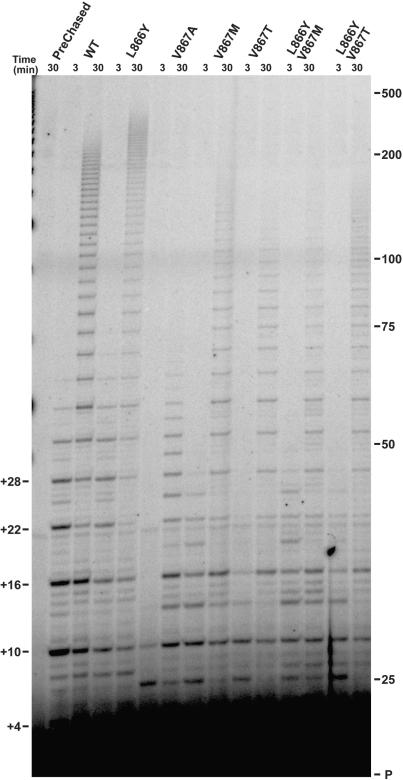
Figure 3.
Competitive primer challenge assay with hTERT C-motif mutants. Primer extension reactions were carried out under competitor challenge conditions as detailed in Materials and Methods section. Following 5-min binding of radiolabeled substrate primer, extension reactions were initiated and chased with excess cold competitor primer. Post-chase aliquots were taken at 3 and 30 min and analyzed via PAGE. Pre-chased lane: 30-min extension reaction where excess competitor primer was added before adding IVR wild-type telomerase. Lysate lane: extension reaction with control IVR containing RRL only. Marker sizes (in nucleotides) are indicated. 85 nt R + 3 product band of V867M telomerase is indicated by an arrowhead.
Repeat extension rate was determined by dividing the length of the modal band (in number of repeats) by time (in this case, 3 min). Relative enzyme activity was calculated from the intensity (corrected for background and normalized against unextended primer) and length of the modal product band using the following equation:
 |
where LMB = modal band length (in nt), IMB = modal band intensity, P = Repeat processivity (see later) and RMB = number of product repeats in the modal band. Since all extension products were equally radiolabeled (i.e. 5′ end-labeled), total extension synthesis can be directly determined from the intensity and length of the modal product band, adjusting for the effects of enzyme dissociation by the factor (P/100)RMB. This adjustment was necessary because the conditions under which enzyme activity was assayed (i.e. in the presence of an unlabeled competitor trap) only allow for the detection of processive synthesis. However, product synthesis by polymerases (such as telomerase) can result from either processive polymerization and/or distributive polymerization. Since only synthesis resulting from the initial primer–enzyme binding would be detected, an adjustment factor was included to account for further primer binding and extension synthesis by dissociated enzyme that would normally have been observed in the absence of competitor. Thus, the observed activity derived from the modal band intensity (LMB * IMB) was adjusted by an enzyme dissociation factor ((P/100)RMB), which was based on the observed processivity (P).
Repeat processivity was determined from the cumulative product attrition observed over multiple successive rounds of extension, as measured by the fraction of total products that have been extended beyond a given repeat. The fraction of products that are extended beyond a given repeat is related to repeat processivity by the following equation:
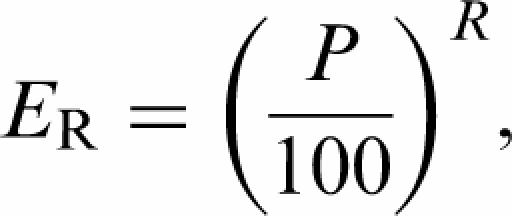 |
where ER = Fraction of total product extension beyond repeat R, P = Repeat processivity, R = repeat number. Repeat processivities were obtained from ER values calculated from the ratio of the amount of products 18 standard repeats or greater to the amount of total products (products < 18 repeats + products > 18 repeats) at the 30-min time point. We chose to obtain ER values from the level of 18th standard repeat to ensure that all products below this level were no longer capable of further extension, based on the repeat extension rate of slowest enzyme being tested (1.39 repeats/min) (Table 2).
Table 2.
Processivities and repeat extension rates of hTERT C-motif mutants
| Processivitya | Extension ratea | |||||
|---|---|---|---|---|---|---|
| hTERT | % | SDb | Relativec | Repeats/min | SDb | Relativec |
| WT | 95.79 | 0.13 | 1.00 | 7.33 | 0.69 | 1.00 |
| 866Y | 96.12 | 2.22 | 1.00 | 8.06 | 0.24 | 1.10 |
| 867A | 95.05 | 1.12 | 0.99 | 1.39 | 0.00 | 0.19 |
| 867M | 95.99 | 1.14 | 1.00 | 3.69 | 0.39 | 0.50 |
| 867T | 98.90 | 0.05 | 1.03 | 3.52 | 0.22 | 0.48 |
| YM | 92.06 | 1.00 | 0.96 | 2.91 | 0.20 | 0.40 |
| YT | 96.01 | 0.65 | 1.00 | 3.25 | 0.43 | 0.44 |
aProcessivities and extension rates were determined as described in Materials and Methods section. Processivity values represent averages of two independent experiments while extension rates represent averages of three independent experiments.
bSD = Standard deviation.
cRelative to wild type.
Nucleotide exclusion assay
Nucleotide misincorporation by telomerase was measured via primer extension in the absence of one of the three substrate nucleotides. Telomerase activity assays were carried out under standard conditions (as detailed earlier) except that only two of the three substrate nucleotides (dATP, dGTP and TTP) were present (at 1 mM) in the extension reaction. Under these conditions, <10% of starting substrate primer is consumed, ensuring single hit kinetics. In addition, control reactions were performed where all three substrate nucleotides necessary for telomere synthesis were present. Following primer extension, the DNA products of these reactions were processed as described earlier for the standard telomerase activity assay and background-corrected product band intensities obtained via phosphoimaging.
Nucleotide incorporation fidelity was calculated from the product band intensities as the ratio of the total band intensity of products longer than the barrier product, i.e. the sum of IB+1 + IB+2 + ·, etc. to the band intensity of the barrier product, IB, (i.e. the product corresponding to copying up to the barrier site).
RESULTS
Enzymatic activity of C-motif mutants of human TERT
To assess the contribution of the hTERT C-motif residues Leu866 and Val867 to telomerase enzymatic activity and function, a panel of hTERT protein mutants was generated (Figure 1B). These mutants included L866Y, V867A, M, or T, and, the double substitution mutants L866Y/V867M and L866Y/V867T. Among these variants were mutants with substitutions that resulted in hTERT proteins whose C-motif sequences were converted to ciliate (LTDD) and yeast (LADD) TERT sequences, as well as to the HIV-1 RT C-motif (YMDD) sequence. Reconstituted telomerase complexes were assembled in rabbit reticulocyte lysates from hTERT protein synthesized in vitro in the presence of purified, in vitro transcribed human TR (hTR) RNA. The reconstituted enzymes were then assayed for telomerase activity in a direct primer extension assay using radiolabeled telomeric (d(TTAGGG)3) substrate primer. All of the mutants were active to some extent (Figure 2, Table 1), although in every case overall activity was lower than with wild-type hTERT. While some substitutions resulted in modest reductions in activity (L866Y) others had severe effects (V867T) (Table 1). Interestingly, the double substitutions L866Y/V867M and L866Y/V867T yielded enzymes with activities at least as active as the single 867M and 867T substitutions.
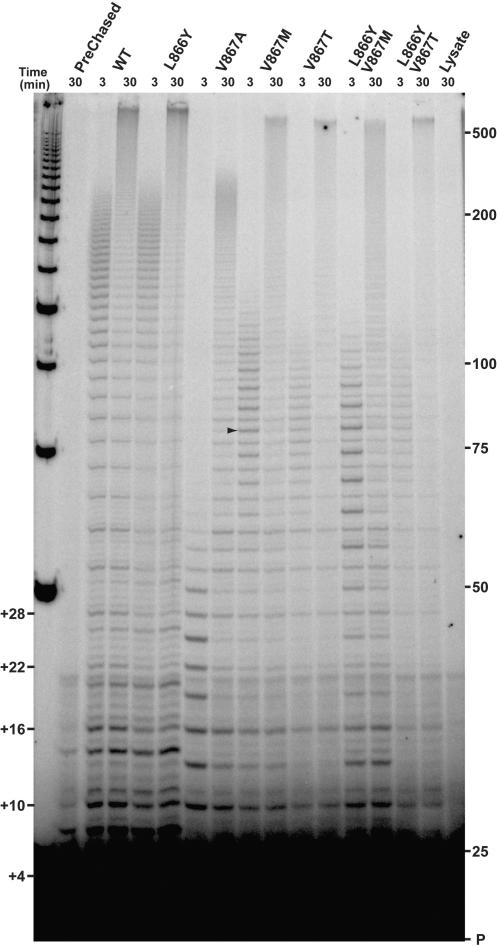
Figure 2.
Primer extension by hTERT C-motif mutants. (A) In vitro reconstituted telomerase mutants were assayed for telomerase activity via direct primer extension as described in Materials and Methods section. Lysate lane: extension reaction with control IVR containing RRL only. Unextended d(TTAGGG)3 substrate primer (P) within each reaction served as loading control (as it represented >95% of the recovered material). Numbers on left (+4, +10, etc.) indicate the positions of products corresponding to the end of each round of template copying (expressed as number of nucleotides added to the primer). Marker sizes (in nucleotides) are indicated. A representative repeat + 3 (R + 3) product is indicated by arrowhead. (B) Schematic diagram of major synthesis products from primer extension reactions. Potential product alignments of R + 3 and repeat + 6 (R + 6) products with the template RNA are shown. Nucleotides added during each round of primer elongation are shown in lower case. hTR nucleotide positions are indicated next to template sequence.
Table 1.
Activity of hTERT C-motif mutants
| hTERT | Relative activitya | SDb |
|---|---|---|
| WT | 1.00 | – |
| 866Y | 0.66 | 0.13 |
| 867A | 0.28 | 0.07 |
| 867M | 0.30 | 0.03 |
| 867T | 0.15 | 0.01 |
| YM | 0.48 | 0.05 |
| YT | 0.22 | 0.06 |
aRelative telomerase activities, expressed as a fraction of wild type, were determined from competitive primer challenge assays as described in Materials and Methods section. Activities represent averages of two independent experiments.
bSD = Standard deviation.
Val867 mutants display altered product synthesis
In addition to differences in overall activity, most of the C-motif mutants displayed two distinct qualitative differences in product synthesis. First, the length of the longest products synthesized by all of the mutants containing Val867 substitutions was considerably shorter than those produced by the wild-type enzyme (Figure 2). This was particularly evident in the telomeric repeat products of the V867A mutant. Second, the band pattern of the products synthesized by all of the Val867 variants was significantly different from that of wild-type hTERT. Specifically, rather than the six-nucleotide ladder characteristic of wild-type synthesis, the ladder of primer extension produced by the Val867 mutants displayed a three nucleotide periodicity (Figure 2). A second major product species presumably representing copying of the first three nucleotides of the template sequence (nucleotides 51CCA49) is seen in addition to the usual major product which accumulates at the end of a cycle of template copying. The accumulation of this repeat + 3 (R + 3) product indicated that reverse transcription had either slowed down, stalled or the telomerase had dissociated from the primer substrate completely, after copying nucleotide A49. Taken together, the appearance of the R + 3 products and the observed decrease in the length of the longest extension products suggested that the effect of mutation of Val867 on template copying may result in changes in enzyme repeat extension rate and/or processivity.
Substitutions at Val867 affect repeat extension rate but not processivity
In order to establish whether hTERT C-motif mutations could influence the rate of telomere repeat synthesis or enzyme processivity, primer extension reactions were performed under competitor challenge (bind and chase) conditions (Figure 3). When assayed under these conditions both repeat extension rate and processivity can be readily determined (23). The results of this assay revealed that all of the mutant enzymes were quite capable of processive synthesis (compare 3 and 30-min time points). In fact, the processivities of the mutants, ranging from ∼92% (L866Y/V867M) to 99% (V867T) were essentially equivalent to wild-type hTERT (∼96%) (Table 2). Competitor-challenged assays performed at 20-fold lower (50 µM) nucleotide concentration revealed only slight differences in processivities amongst the enzymes tested (Figure 4 and Table 3).
Figure 4.
Competitive primer challenge assay with hTERT C-motif mutants under reduced nucleotide concentration. Primer extension reactions were carried out under competitor challenge conditions as detailed in Materials and Methods section, except that extension reactions were performed at a dNTP concentration of 50 µM rather than the standard 1 mM. Following 5-min binding of radiolabeled substrate primer, extension reactions were initiated and chased with excess cold competitor primer. Post-chase aliquots were taken at 3 and 30 min and analyzed via PAGE. Pre-chased lane: 30-min extension reaction where excess competitor primer was added before adding IVR wild-type telomerase. Marker sizes (in nucleotides) are indicated.
Table 3.
Processivities of hTERT C-motif mutants under reduced nucleotide concentration
| Processivitya | ||
|---|---|---|
| hTERT | % | Relativeb |
| WT | 84.19 | 1.00 |
| 866Y | 86.72 | 1.03 |
| 867A | NDc | NDc |
| 867M | 81.63 | 0.97 |
| 867T | 77.33 | 0.92 |
| YM | 67.24 | 0.80 |
| YT | 80.45 | 0.96 |
aProcessivities were determined as described in Materials and Methods section, except that ER values were calculated from the 4th standard repeat rather than the 18th standard repeat. Processivity values were obtained from reactions shown in Figure 4.
bRelative to wild type.
cNot determined.
In contrast, substitutions within the C-motif had quite substantial effects on repeat extension rate (Table 2). In fact, among the mutants tested, only the L866Y mutant displayed unaltered repeat extension rate. All hTERT substitutions involving replacement of Val867 led to decreases in repeat extension rate, which ranged from ∼50% (V867M) to 80% (V867A) reduction in extension rate compared to wild-type Val867 hTERT. Thus, these data indicate that the difference between the length of the longest products of wild-type hTERT and those of the Val867 variants reflects reduced repeat extension rate and not lower processivity.
The competitor-challenge assays also revealed that the majority of the R + 3 products associated with primer extension by the Val867 mutants represent paused synthesis rather than stalled or terminated polymerization. A comparison of extension products from the 3 and 30-min reaction time points shows a decrease in the band intensity of shorter R + 3 products as the competitor chase period proceeds (e.g. compare the 85 nt R + 3 band of V867M, indicated by an arrowhead in Figure 3, 3 and 30-min lanes). The concomitant increase in longer extension products indicates that most of the R + 3 products have continued to be elongated, demonstrating that R + 3 products are predominantly paused species.
Template copying by V867M hTERT is sensitive to dTTP concentration
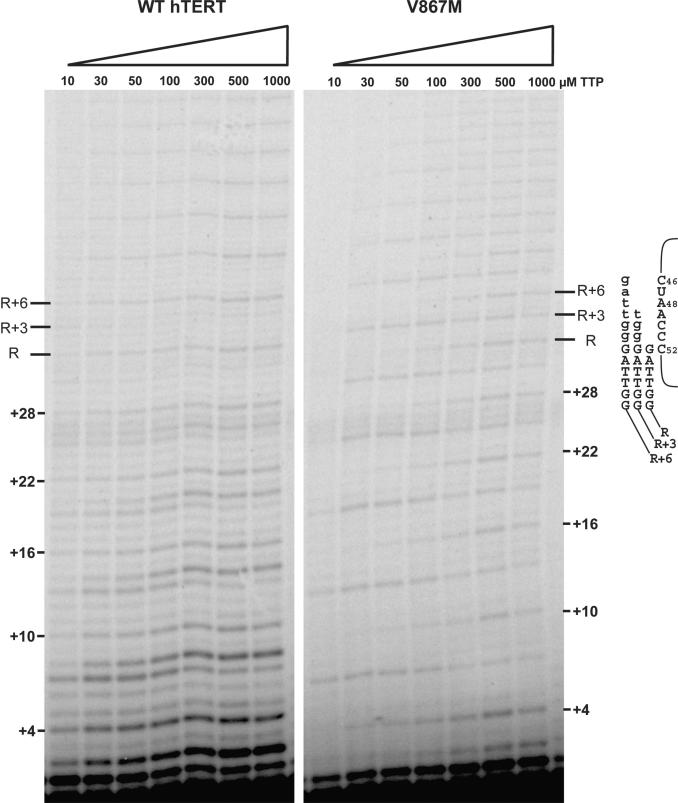
The characteristic six-nucleotide ladder of in vitro synthesis products observed for wild-type human telomerase results as a consequence of repeat translocation being slower and less efficient than template copying. To better understand the nature of the slowed polymerization that gave rise to the R + 3 products, we asked whether R + 3 accumulation was related to changes in nucleotide insertion efficiency. Specifically, since template copying appeared to proceed normally up to residue A49, this implied that the Val867 mutants were having difficulty inserting a dT residue opposite A48. Therefore, we examined the effect of dTTP concentration on R + 3 product formation by Val867 mutants. We chose the V867M hTERT as a representative Val867 mutant for these experiments since it was the most active of the single mutants.
Primer extension assays were performed in the presence of varied dTTP concentrations and the results are depicted in Figure 5. As seen from the results of these assays, repeat synthesis by V867M mutant telomerase was quite sensitive to dTTP concentration. At 10 µM dTTP (the lowest concentration tested) the mutant enzyme was only capable of adding a few repeats onto the substrate primer. In contrast, the wild-type telomerase, while exhibiting some effects on template copying (see later), was still able to extend primers with many repeats at the same dTTP concentration. Furthermore, the proportion of R + 3 products to total products synthesized by the V867M mutant was also quite sensitive to dTTP concentration. As TTP concentration was decreased, R + 3 products constituted a higher percentage of the total products, indicating that the accumulation of R + 3 products was the result of impaired dT insertion opposite A48. In fact, at dTTP concentrations lower than 50 µM the R + 3 band was the sole major product observed (Figure 5). This was of particular significance as it indicated that at reduced dTTP concentrations, the rate (and/or efficiency) of copying past A48 is even lower than that of repeat translocation for the V867M mutant. While the effect of dTTP concentration on template copying by wild-type hTERT was not as dramatic as with the V867M mutant, reduced dTTP concentrations did lead to the appearance of R + 3 product. However, unlike the mutant, the R + 3 product was never more abundant than the full repeat product.
Figure 5.
Effect of dTTP concentration on primer extension activity. Primer extension synthesis by wild type and V867M telomerases was measured in primer extension reactions in the presence of increasing concentrations of TTP. Extension reactions containing 1 mM dATP, 1 mM dGTP and TTP at the concentration indicated, were performed under standard conditions as detailed in Materials and Methods section. Representative repeat, R, as well as R + 3 and R + 6 products are indicated, along with potential product alignments with template RNA at right. Nucleotides added during each round of primer elongation are shown in lower case. hTR nucleotide positions are shown next to template sequence. Both wild type (left) and V867M (right) panels are from the same gel.
V867M hTERT exhibits a template copying defect specific to template A residues
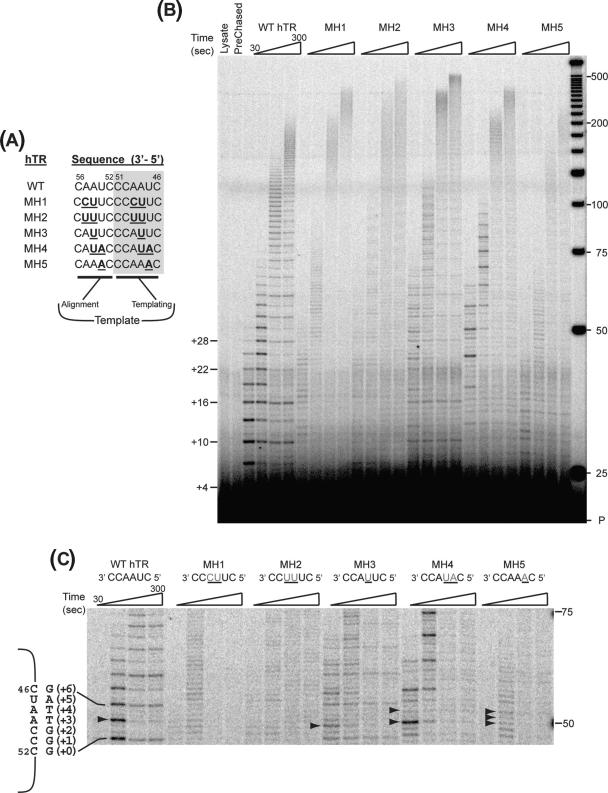
Based on the activity assays it appeared that the reduction in repeat extension rate was correlated to the difficulty Val867 mutants had copying past residue A48. This raised the question of whether the copying defect is specific to residue A48 (or hTR nucleotide 48) or a general defect copying template A residues. In order to answer this question, template copying by V867M hTERT was examined using a panel of hTR mutants with modified template sequences described previously (23). The template sequences of these mutant hTRs contained different substitutions within the template sequence 49AAU47, resulting in sequences with zero to three template A residues (Figure 6A). Four out of the five mutants that were tested contained A48U substitutions. In addition, as these hTR template mutants were designed to allow for efficient processive repeat synthesis, the alignment sequences (nucleotides 55AAU53) in these mutants were also modified to reflect the changes to the templating nucleotides (49AAU47) (Figure 6A).
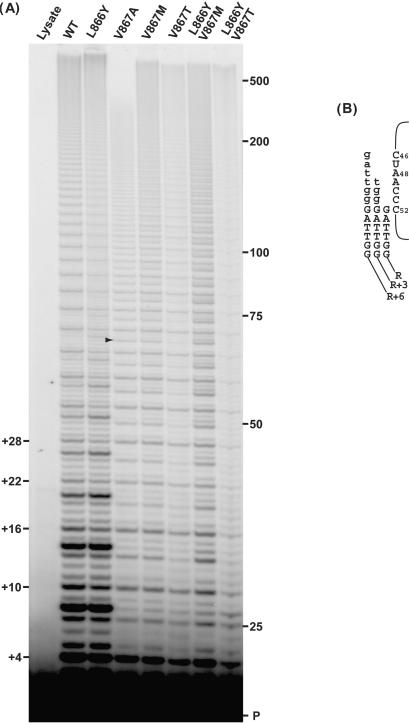
Figure 6.
Effect of hTR template sequence on primer extension by 867M hTERT. (A) Wild type and V867M hTERT proteins were reconstituted with wild type and template mutant (MH) hTR RNAs (23) shown. (B) Primer extension reactions were carried out under competitor challenge conditions as detailed in Materials and Methods section. Post-chase aliquots were taken at 3 and 30 min and analyzed via PAGE. Pre-chased lane: 30-min extension reaction where excess competitor primer was added before adding IVR wild-type telomerase. Lysate lane: extension reaction with control IVR containing RRL only. (C) Enlarged view of region between 48 and 75 nt. Sequence of the WT hTR template (nucleotides 46–51) is shown along with the sequence of the wild-type product. The number of nucleotides added onto a previously copied full repeat is indicated in parentheses. Arrowheads indicate product accumulation prior to copying a template A residue.
Telomerase complexes were assembled in vitro with V867M hTERT and each individual mutant hTR and the reconstituted telomerases assayed for activity. Primer extension reactions were performed under competitor challenge conditions to allow for measurement of repeat extension rate, as well as for discrimination of slowed synthesis from stalled/terminated primer elongation. These assays revealed that V867M hTERT was capable of efficient repeat synthesis using each of the mutant templates (Figure 6B). Similar to previous observations with wild-type hTERT (23), repeat synthesis with the mutant template RNAs was processive, as evidenced by the high proportion of actively engaged primer substrate being further elongated during the chase period (Figure 6B). Moreover, extension rate measurements (Table 4) indicated a clear influence of template sequence on V867M telomerase extension rate. Interestingly, each of the mutant hTRs reconstituted enzymes with increased extension rates compared to V867M hTERT reconstituted with wild-type hTR. This was not entirely unexpected as we have previously shown with wild-type hTERT that template sequence can affect the repeat extension rate (23). However, it was surprising that in the majority (four out of five) of the cases, the relative changes in extension rate seen with the V867M hTERT/mutant hTR enzymes did not parallel the changes observed with wild-type hTERT/mutant hTRs. Specifically, reconstitution of V867M hTERT with MH1, MH2, MH3 and MH5 hTRs led to extension rates that were 2.14×, 2.23×, 3.94× and 1.41× faster, respectively, than wild-type hTR (Table 4). In contrast, reconstitution of wild-type hTERT with MH1, MH2, MH3 and MH5 hTRs yielded enzymes whose extension rates were 0.85×, 1.17×, 1.97× and 1.76× as fast as the rate of wild-type hTERT with wild-type hTR (Table 4). If it is assumed that the relative differences in telomerase extension rate between enzymes assembled with wild-type or mutant hTRs are inherent to the template sequence, then these differences should be constant regardless of the hTERT in the complex. Yet, a distinct hTERT-specific modulation of template sequence-governed extension rate is evident, which can be expressed as the ratio of 867M extension rate/wild-type extension rate (867MEXT/867VEXT) (Table 4). When expressed as 867MEXT/867VEXT values, one finds the mutant TERT has no effect on the inherent extension rates of the MH2 and MH3 hTRs, (ratios ≈ 1), slight increase of MH1 rate (ratio = 1.28) and substantial decrease of extension rates of MH4, MH5 and wild-type hTRs (ratios = 0.54, 0.4 and 0.50, respectively). Conversely, considered from the perspective of template utilization, the V867M and wild-type enzymes performed repeat synthesis at the same rates with the MH2 and MH3 hTRs, nearly the same with MH1 but substantially different with MH4, MH5 and wild-type hTRs. Furthermore, 867MEXT/867VEXT values appear to correlate with template A residue content. The MH1, MH2 and MH3 hTRs which contain the fewest (0–1) template A bases had the highest 867MEXT/867VEXT values, the MH5 hTR, with three A residues, had the lowest ratio while the WT and MH4 hTRs, with two A residues, had intermediate ratios (Table 4). These data indicate increasing template A content results in reduced repeat extension rate for V867M telomerase.
Table 4.
Extension rate of V867M hTERT reconstituted with hTR template mutants
| V867M | Wild typea | |||||||
|---|---|---|---|---|---|---|---|---|
| HTR | Repeats/min | SDb | Relative extension ratec | Repeats/min | SDb | Relative extension ratec | 867MER/867VERd | Template A residues |
| WT | 3.69 | 0.39 | 1.00 | 7.33 | 0.69 | 1.00 | 0.50 | 2 |
| MH1 | 7.97 | 0.12 | 2.14 | 6.22 | 0.22 | 0.85 | 1.28 | 0 |
| MH2 | 8.56 | 0.47 | 2.23 | 8.56 | 1.17 | 1.17 | 1.00 | 0 |
| MH3 | 14.06 | 0.71 | 3.94 | 14.46 | 1.75 | 1.97 | 0.97 | 1 |
| MH4 | 8.06 | 1.18 | 2.41 | 15.06 | 1.90 | 2.05 | 0.53 | 2 |
| MH5 | 5.14 | 0.12 | 1.41 | 12.93 | 0.80 | 1.76 | 0.40 | 3 |
aData for the wild type is taken from Drosopoulos et al. (23).
bSD = Standard deviation.
cExtension rates are expressed relative to wild-type hTR for the given hTERT.
dRatio of extension rate of 867M hTERT to extension rate of 867V hTERT. Rates represent averages of two independent experiments.
While it is possible that differences in extension rates seen with the mutant templates may reflect substrate alignment or affinity differences, it is likely that altered substrate affinity or alignment would only affect initial binding of substrate primer and not extension synthesis. This is because once the mutant template is copied, the nascent product is completely complementary to both the alignment and templating (copied) nucleotides of the mutant template (as indicated in Figure 6A). Thus, repeat extension rates would more probably be influenced by product synthesis (template copying) and translocation rates rather than substrate affinity or initial alignment.
The patterns of extension products further corroborated the conclusion that in general, polymerization slows down when template A bases are encountered by V867M telomerase. For example, significant product accumulation is observed prior to copying past A49 of the MH3 template, A49 (and to some extent A47) of the MH4 template and A49, A48 and A47 of the MH5 template (Figure 6C, arrows) in addition to products seen when the modified templates are copied by wild-type hTERT (23). In fact, the R + 2 products (which result from impeded copying past A49) predominate in the MH4 pattern, indicating that with the MH4 template the rate of polymerization past A49 is even slower than the rate of repeat translocation. In contrast, copying of the MH1 and MH2 templates, which lack template A residues, did not reveal any specific new sites of substantial product accumulation when compared to copying by wild-type hTERT (these mutants produce weak band patterns with wild-type hTERT (23)). These results indicate that the template copying defect displayed by V867M hTERT appears to be primarily a general defect in copying past template A residues. However, additional factors appear to modulate this defect. Although both wild type and MH4 templates possess two A residues, polymerization past only one (A48 in wild type and A49 in MH4) seemed particularly impaired (Figure 6C). Thus, these observations suggest that there is at least a partial effect of template position and/or sequence context on the copying defect.
Val867 hTERT mutants display increased nucleotide incorporation fidelity
Since it has been well established that the HIV-1 RT counterparts of Leu866 and Val867 (Tyr183 and Met184) are key fidelity determinants, it seemed logical that hTERT C-motif mutants might display altered fidelities. We chose to measure the fidelity of the mutants via a nucleotide exclusion assay. In this assay, primer extension reactions are carried out where either all three substrate nucleotides (dATP, dGTP and TTP) are present or one of the three is omitted (excluded) from the reaction. Under conditions of single nucleotide exclusion, faithful template copying proceeds until the first template position requiring the incorporation of the absent dNTP is encountered. Any polymerization opposite or beyond this ‘barrier’ site results from misincorporation of incorrect nucleotides. Thus, measurement of synthesis beyond a barrier site provides an index of polymerase fidelity.
Nucleotide exclusion assays were performed with the Leu866 and Val867 mutant telomerases. In reactions where TTP was excluded, little difference could be discerned between the wild type and mutant enzymes. Synthesis beyond the barrier site in these reactions appeared as a single nucleotide ladder extending approximately 12 nt (data not shown). However, it is likely that this ladder represents strand slippage synthesis rather than incorrect nucleotide incorporation. Studies on Tetrahymena telomerase have demonstrated that similar ladders, composed of poly-(dG), are synthesized in the absence of TTP (24,25), and they are presumed to be the products of strand slippage copying within the template homopolymeric C run.
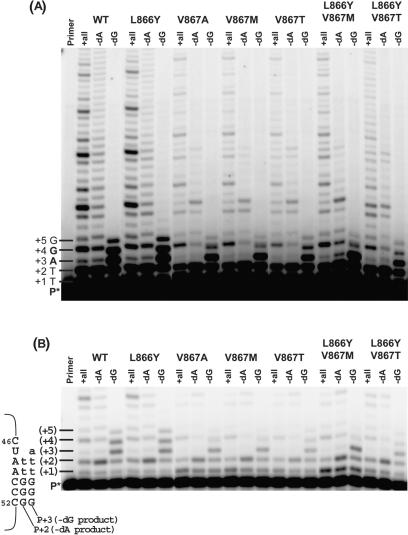
In contrast, reactions lacking dATP or dGTP, revealed major differences in post-barrier site synthesis. With the exception of the L866Y mutant, all of the mutants exhibited significantly lower primer extension past both the ‘U’ barrier site (U47) and the ‘C’ barrier site (C46) compared to wild-type hTERT (Figure 7). In the absence of dATP both wild type and L866Y telomerase could repeatedly extend beyond the U47 barrier position (Figure 7A, WT and L866Y −dA lanes) leading to extension products of over 150 nt (data not shown). Under identical conditions, template copying of U47 by all mutants containing Val867 substitutions was much less efficient, with essentially no extension proceeding beyond the third encounter of the barrier (Figure 7A, −dA lanes). The Val867 mutants were also less efficient than wild type or the L866Y mutant at primer extension past U47 in the absence of dGTP (Figure 7B and C, compare −dG lanes). Careful examination of the post-barrier synthesis products of both dATP and dGTP-excluded reactions (Figure 7A and B) reveals a shift in migration relative to the corresponding products of reactions with all nucleotides present, indicative of true misincorporation.
Figure 7.
Nucleotide exclusion assay with hTERT C-motif mutants. Primer extension reactions were performed under standard conditions with all three substrate nucleotides (dATP, dGTP and TTP) present (+all), only dGTP and TTP present (−dA), or only dATP and TTP present (−dG) in the extension reaction. The sequence of correctly copied product along with the number of nucleotides added to the radiolabeled primer (P*) is indicated. (B) Lower exposure of (A). Expected full-length products of accurate extension synthesis in the absence of dATP (−dA product) and dGTP (−dG product) are shown, with newly copied nucleotides in lower case. The number of nucleotides added to the radiolabeled primer resulting from extension by telomerase is indicated in parentheses.
A quantitative measure of fidelity, calculated from the ratio of the amount (band intensity) of barrier product (i.e. product corresponding to copying up to the barrier site) to the amount of products longer than the barrier product, is presented in Table 5. We chose to quantify fidelity from the dGTP-excluded assays since copying opposite or beyond the C46 barrier site in these reactions was limited to only two main products, P + 4 and P + 5 (the two template positions copied following C46 (i.e. C51 and C50) are also C residues), facilitating accurate measurement. Based on the fidelity value obtained for the wild-type telomerase (Table 5), almost 45% of primer extended to the barrier site by the enzyme was further elongated via nucleotide misincorporation. All of the Val867 substituted enzymes, in contrast, exhibited significantly lower misincorporation synthesis which was reflected in increased fidelities, that ranged from 2.2-fold (V867T) to 13.5-fold (L866Y/V867M) higher than wild type (Table 5). These findings demonstrate that the Val867 residue in hTERT is a determinant of telomerase polymerase fidelity.
Table 5.
Fidelity of hTERT C-motif mutants
| hTERT | Fidelitya | Relative fidelity |
|---|---|---|
| WT | 0.82 | 1.0 |
| 866Y | 0.85 | 1.0 |
| 867A | 0.21 | 3.9 |
| 867M | 0.16 | 5.2 |
| 867T | 0.38 | 2.2 |
| 867YM | 0.06 | 13.5 |
| 867YT | 0.29 | 2.8 |
aValues were calculated from the −dG reaction products shown in Figure 7B.
DISCUSSION
Although telomerase is a quite specialized reverse transcriptase, with regard to catalysis, it is likely that polymerization by telomerase is mechanistically similar to other RTs. Like other RTs, absolutely conserved aspartate residues within conserved RT motifs are essential for telomerase's RNA-dependent DNA polymerase activity. Furthermore, mutation of conserved residues in RT motifs 1, 2 and A of Tetrahymena TERT lead to activity changes that can be rationalized from the roles played by the equivalent retroviral RT amino acids in the active site dNTP-binding pocket of RT (26). The implication from such observations is one of common structure/function relationships for homologous residues in TERT and other RTs. Consistent with this view, we identify here an amino acid in hTERT that functions as a determinant of polymerase fidelity, a role it shares with its counterpart in HIV-1 RT.
In HIV-1 RT Tyr183 and Met184, the cognate amino acids to hTERT Leu866 and Val867, play direct roles in enzymatic activity, processivity, fidelity, dNTP utilization and nucleoside analog inhibitor resistance (8,13,14,17,18). We find that only Val867 has significant influence on several of these properties in human telomerase, specifically, enzymatic activity, dNTP utilization and fidelity. Mechanistically, these properties can all be related through nucleotide incorporation efficiency. Overall enzymatic activity is dependent on efficient dNTP incorporation while the fidelity of DNA synthesis is related to the efficiency that an incorrect nucleotide can be incorporated. Thus, our findings indicate that Val867 plays a key role in nucleotide incorporation and suggest that this residue may provide important interactions with incoming substrate nucleotides.
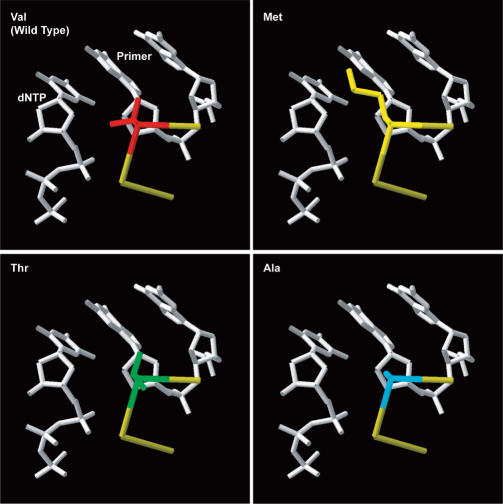
Structural models of wild type and nucleoside analog-resistant Met184Ile and Met184Val mutant HIV-1 RTs may offer some insight into possible nature of Val867 interactions in hTERT. In wild-type HIV-1 RT, the side chain of Met184 projects into the active site between the primer terminal nucleotide and the incoming dNTP. However, the side chain only contacts the primer 3′ terminus (19,20). Crystallographic and computer-generated structures of isoleucine- and valine-substituted β-l-nucleoside analog-resistant mutants indicate that the methyl group of these β-branched side chain residues protrudes into the dNTP-binding pocket and creates an additional contact with the dNTP sugar ring (20,27,28). Consequently, a more sterically constrained dNTP-binding pocket would result that is unable to accommodate the unnatural l-conformation nucleoside rings of β-l-nucleoside inhibitors, but still able to efficiently incorporate normal β-d-dNTPs. If the side chain of Val867 were positioned similarly in the active site of hTERT then it would be making contact with the sugar ring of the incoming dNTP. A requirement for such a contact could furnish a possible explanation for the effects of Val867 mutations on hTERT function. In wild-type hTERT the Val184 side chain–dNTP interaction may aid in proper dNTP positioning and/or add stability to the dNTP–enzyme complex, resulting in efficient nucleotide utilization. Substitution with alanine or methionine removes this interaction and while threonine substitution maintains the side chain β-methyl branch, interactions of its polar hydroxyl with the primer terminus may prevent proper positioning of the methyl group (Figure 8). The absence of this contact would make dNTP binding and incorporation more dependent on the remaining dNTP–enzyme interactions such as Watson–Crick hydrogen bonding and base stacking. Increased dependence on proper Watson–Crick bonding would result in increased nucleotide insertion fidelity, as was observed for all Val867 substitutions. It could also partially account for the specific reduction in TTP utilization efficiency seen in these mutants. Furthermore, the lack of residue 867–dNTP sugar interactions may account for the similar reduction in overall activity exhibited by the methionine mutant, which has significant potential for interactions by virtue of its long side chain, and the alanine mutant, whose short side chain affords it much less interactive capacity.
Figure 8.
Schematic representation of hTERT residue 867 substitutions. Potential orientations of the side chains of the Val (wild type), Met, Thr, Ala 867 residues within the active site of hTERT are depicted. These orientations were generated by mutating the analogous HIV-1 RT residue Met184 in the HIV-1 RT ternary complex structure using the published coordinates (PDB1RTD) of Huang et al. (20). Individual substitutions (M184V, A, or T) were created in the published HIV-1 RT structure using the DeepView/Swiss-pdbViewer program. The ‘best’ fit rotamers are presented.
Our studies reveal that substitution of Val867 with other amino acids, particularly methionine, increases the fidelity of DNA synthesis by hTERT (Table 5). Curiously, we and others (13,14) have found the reverse substitution in HIV-1 RT, M184V, also increases its nucleotide insertion fidelity. Furthermore, while alanine substitution of Val867 led to increased fidelity in hTERT, the M184A HIV-1 RT variant displayed markedly reduced fidelity (14). The opposite effects of these comparable substitutions, along with the similar effects of the reverse Val/Met mutations, point to distinct differences in the nucleotide-binding pockets of hTERT and HIV-1 RT. Structural data and modeling indicate M184V substitution leads to a more closed dNTP-binding site (see above). Because of the presence of the side chain β-methyl group, greater steric demand is imposed on the incoming dNTP for proper base-pair geometry and this in turn may result in the observed increase in fidelity. This would imply that the Val867 β-methyl branch in hTERT may not generate as much of a steric constraint on the dNTP-binding pocket and therefore less steric selection pressure on the incoming dNTP. Moreover, this would suggest that hTERT has a more open nucleotide-binding site than HIV-1 RT, with Val867 positioned further from the dNTP substrate than Val184. A relatively more open active-site geometry would support our proposed explanation of the effects of hTERT Val867 substitutions. Alternatively, the differences in the effects of Val867 and Met184 mutations on fidelity may be the result of differences in primer/template repositioning along with smaller alterations in the dNTP-binding pocket.
One of the consequences of Val867 mutation was reduced repeat extension rate (Table 2). The underlying basis for this appears to involve a general template copying defect specific to template A residues linked to impaired TTP utilization. Interestingly, our studies with varied hTR templates (Figure 6C) revealed that pausing in templates with multiple A residues (such as wild type) did not always occur equivalently at all A residues. Furthermore, pausing did not always occur at common A residue sites (e.g. A49) (Figure 6C, compare WT to MH3, MH4 and MH5 templates) suggesting effects of template position and/or sequence context. If template position is modulating the copying defect then this would suggest that hTERT active site geometry changes as the template traverses the active site. This appears to be consistent with studies on nucleoside analog utilization by Tetrahymena telomerase (29), where a similar explanation was used to account for template position-specific differences in the efficiency of analog incorporation.
Another curious finding was that mutation of either Leu866 or Val867 had no or very limited effect on hTERT processivity. Based on previous reports that substitution at homologous sites in other RTs result in significant changes in processivity, one could have expected that the YTDD substituted hTERT would display altered processivity, similar to the equivalent yeast and in vitro reconstituted Tetrahymena mutants. One explanation for the absence of such effects may lie in the inherently high processivity of human telomerase. In contrast to yeast and in vitro reconstituted Tetrahymena telomerases, in vitro reconstituted (and native) wild-type human telomerase is almost completely processive in in vitro assays ((30); this work, Table 2). This likely reflects multiple important contributions by several regions in the enzyme including the anchor region (31–33) and the TR pseudoknot and template elements (34) to human telomerase processivity. While these features are not unique to human telomerase, it may not be merely their presence but the strength of their contribution to product–enzyme complex stability that is particularly substantial. For example, some of the highly conserved TR elements, e.g. the pseudoknot (CR2–CR3) and CR4–CR5 (Helix IV) domains, are considerably larger in hTR as compared to Tetrahymena TR. This affords these elements more potential sites for interaction in human versus Tetrahymena telomerase. Thus, it is possible that in human telomerase the contributions of these (and/or other) elements to processivity are particularly significant and can effectively compensate for the loss of primer–Leu866/Val 867 interactions.
Mutation of Val867 leads to reductions in overall enzyme activity and efficiency of TTP utilization, resulting in reduced repeat extension rate, yet increased polymerase fidelity. Based on these observations, it appears that the hTERT protein has evolved to have valine at 867 because it is optimal for activity and rapid repeat synthesis, even at the cost of polymerase fidelity. The in vitro error rate of human telomerase has been estimated to be 2 × 10−3 per nucleotide or one non-canonical repeat per hundred normal repeats (35). Presumably, the level of fidelity afforded by Val867 is sufficient to maintain the minimal telomere sequence integrity necessary for specific recognition by telomere-binding proteins.
ACKNOWLEDGEMENTS
The authors wish to thank the AECOM DNA Sequencing Facility. This work was supported by Public Health Service grants K01 CA87542 (Howard Temin Award) to W.C.D. and R01 AI030861 to V.R.P. Funding to pay the Open Access publication charge was provided by Public Health Service grant R01 AI030861.
Conflict of interest statement. None declared.
REFERENCES
- 1.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Chan SR, Blackburn EH. Telomeres and telomerase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:109–121. doi: 10.1098/rstb.2003.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 5.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doolittle RF, Feng DF, Johnson MS, McClure MA. Origins and evolutionary relationships of retroviruses. Q. Rev. Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 7.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larder BA, Purifoy DJ, Powell KL, Darby G. Site-specific mutagenesis of AIDS virus reverse transcriptase. Nature. 1987;327:716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- 9.Kaushik N, Rege N, Yadav PN, Sarafianos SG, Modak MJ, Pandey VN. Biochemical analysis of catalytically crucial aspartate mutants of human immunodeficiency virus type 1 reverse transcriptase. Biochemistry. 1996;35:11536–11546. doi: 10.1021/bi960364x. [DOI] [PubMed] [Google Scholar]
- 10.Steitz TA, Smerdon SJ, Jager J, Joyce CM. A unified polymerase mechanism for nonhomologous DNA and RNA polymerases. Science. 1994;266:2022–2025. doi: 10.1126/science.7528445. [DOI] [PubMed] [Google Scholar]
- 11.Harrington L, Zhou W, McPhail T, Oulton R, Yeung DS, Mar V, Bass MB, Robinson MO. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 13.Wainberg MA, Drosopoulos WC, Salomon H, Hsu M, Borkow G, Parniak M, Gu Z, Song Q, Manne J, et al. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science. 1996;271:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]
- 14.Pandey VN, Kaushik N, Rege N, Sarafianos SG, Yadav PN, Modak MJ. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996;35:2168–2179. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 15.Kaushik N, Singh K, Alluru I, Modak MJ. Tyrosine 222, a member of the YXDD motif of MuLV RT, is catalytically essential and is a major component of the fidelity center. Biochemistry. 1999;38:2617–2627. doi: 10.1021/bi9824285. [DOI] [PubMed] [Google Scholar]
- 16.Kaushik N, Chowdhury K, Pandey VN, Modak MJ. Valine of the YVDD motif of moloney murine leukemia virus reverse transcriptase: role in the fidelity of DNA synthesis. Biochemistry. 2000;39:5155–5165. doi: 10.1021/bi992223b. [DOI] [PubMed] [Google Scholar]
- 17.Harris D, Kaushik N, Pandey PK, Yadav PN, Pandey VN. Functional analysis of amino acid residues constituting the dNTP binding pocket of HIV-1 reverse transcriptase. J. Biol. Chem. 1998;273:33624–33634. doi: 10.1074/jbc.273.50.33624. [DOI] [PubMed] [Google Scholar]
- 18.Harris D, Yadav PN, Pandey VN. Loss of polymerase activity due to Tyr to Phe substitution in the YMDD motif of human immunodeficiency virus type-1 reverse transcriptase is compensated by Met to Val substitution within the same motif. Biochemistry. 1998;37:9630–9640. doi: 10.1021/bi980549z. [DOI] [PubMed] [Google Scholar]
- 19.Ding J, Das K, Hsiou Y, Sarafianos SG, Clark AD, Jr, Jacobo-Molina A, Tantillo C, Hughes SH, Arnold E. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 A resolution. J. Mol. Biol. 1998;284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- 20.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 21.Bryan TM, Goodrich KJ, Cech TR. A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem. 2000;275:24199–24207. doi: 10.1074/jbc.M003246200. [DOI] [PubMed] [Google Scholar]
- 22.Peng Y, Mian IS, Lue NF. Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell. 2001;7:1201–1211. doi: 10.1016/s1097-2765(01)00268-4. [DOI] [PubMed] [Google Scholar]
- 23.Drosopoulos WC, Direnzo R, Prasad VR. Human telomerase RNA template sequence is a determinant of telomere repeat extension rate. J. Biol. Chem. 2005;280:32801–32810. doi: 10.1074/jbc.M506319200. [DOI] [PubMed] [Google Scholar]
- 24.Collins K, Greider CW. Utilization of ribonucleotides and RNA primers by Tetrahymena telomerase. EMBO J. 1995;14:5422–5432. doi: 10.1002/j.1460-2075.1995.tb00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilley D, Lee MS, Blackburn EH. Altering specific telomerase RNA template residues affects active site function. Genes Dev. 1995;9:2214–2226. doi: 10.1101/gad.9.18.2214. [DOI] [PubMed] [Google Scholar]
- 26.Miller MC, Liu JK, Collins K. Template definition by Tetrahymena telomerase reverse transcriptase. EMBO J. 2000;19:4412–4422. doi: 10.1093/emboj/19.16.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarafianos SG, Das K, Clark AD, Jr, Ding J, Boyer PL, Hughes SH, Arnold E. Lamivudine (3TC) resistance in HIV–1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10027–10032. doi: 10.1073/pnas.96.18.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chong Y, Chu CK. Molecular mechanism of dioxolane nucleosides against 3TC resistant M184V mutant HIV. Antiviral Res. 2004;63:7–13. doi: 10.1016/j.antiviral.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Strahl C, Blackburn EH. The effects of nucleoside analogs on telomerase and telomeres in Tetrahymena. Nucleic Acids Res. 1994;22:893–900. doi: 10.1093/nar/22.6.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maine IP, Chen SF, Windle B. Effect of dGTP concentration on human and CHO telomerase. Biochemistry. 1999;38:15325–15332. doi: 10.1021/bi991596+. [DOI] [PubMed] [Google Scholar]
- 31.Harrington LA, Greider CW. Telomerase primer specificity and chromosome healing. Nature. 1991;353:451–454. doi: 10.1038/353451a0. [DOI] [PubMed] [Google Scholar]
- 32.Collins K, Greider CW. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 1993;7:1364–1376. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- 33.Morin GB. Recognition of a chromosome truncation site associated with alpha-thalassaemia by human telomerase. Nature. 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- 34.Chen JL, Greider CW. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 2003;22:304–314. doi: 10.1093/emboj/cdg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreiter M, Irion V, Ward J, Morin G. The fidelity of human telomerase. Nucleic Acids Symp. Ser. 1995;33:137–139. [PubMed] [Google Scholar]
- 36.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 37.Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 38.Nasir L, Gault E, Campbell S, Veeramalai M, Gilbert D, McFarlane R, Munro A, Argyle DJ. Isolation and expression of the reverse transcriptase component of the Canis familiaris telomerase ribonucleoprotein (dogTERT) Gene. 2004;336:105–113. doi: 10.1016/j.gene.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 39.Greenberg RA, Allsopp RC, Chin L, Morin GB, DePinho RA. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- 40.Guo W, Okamoto M, Lee YM, Baluda MA, Park NH. Enhanced activity of cloned hamster TERT gene promoter in transformed cells. Biochim. Biophys. Acta. 2001;1517:398–409. doi: 10.1016/s0167-4781(00)00306-7. [DOI] [PubMed] [Google Scholar]
- 41.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 42.Delany ME, Daniels LM. The chicken telomerase reverse transcriptase (chTERT): molecular and cytogenetic characterization with a comparative analysis. Gene. 2004;339:61–69. doi: 10.1016/j.gene.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 43.Kuramoto M, Ohsumi K, Kishimoto T, Ishikawa F. Identification and analyses of the Xenopus TERT gene that encodes the catalytic subunit of telomerase. Gene. 2001;277:101–110. doi: 10.1016/s0378-1119(01)00684-9. [DOI] [PubMed] [Google Scholar]
- 44.Bryan TM, Sperger JM, Chapman KB, Cech TR. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karamysheva Z, Wang L, Shrode T, Bednenko J, Hurley LA, Shippen DE. Developmentally programmed gene elimination in Euplotes crassus facilitates a switch in the telomerase catalytic subunit. Cell. 2003;113:565–576. doi: 10.1016/s0092-8674(03)00363-5. [DOI] [PubMed] [Google Scholar]
- 47.Wain-Hobson S, Sonigo P, Danos O, Cole S, Alizon M. Nucleotide sequence of the AIDS virus, LAV. Cell. 1985;40:9–17. doi: 10.1016/0092-8674(85)90303-4. [DOI] [PubMed] [Google Scholar]