Abstract
Raman and Raman optical activity (ROA) spectra were collected for four RNA oligonucleotides based on the EMCV IRES Domain I to assess the contributions of helix, GNRA tetraloop, U·C mismatch base pair and pyrimidine-rich bulge structures to each. Both Raman and ROA spectra show overall similarities for all oligonucleotides, reflecting the presence of the same base paired helical regions and GNRA tetraloop in each. Specific bands are sensitive to the effect of the mismatch and asymmetric bulge on the structure of the RNA. Raman band changes are observed that reflect the structural contexts of adenine residues, disruption of A-form helical structure, and incorporation of pyrimidine bases in non-helical regions. The ROA spectra are also sensitive to conformational mobility of ribose sugars, and verify a decrease in A-type helix content upon introduction of the pyrimidine-rich bulge. Several Raman and ROA bands also clearly show cooperative effects between the mismatch and pyrimidine-rich bulge motifs on the structure of the RNA. The complementary nature of Raman and ROA spectra provides detailed and highly sensitive information about the local environments of bases, and secondary and tertiary structures, and has the potential to yield spectral signatures for a wide range of RNA structural motifs.
INTRODUCTION
Vibrational spectroscopic techniques, such as Raman and Raman optical activity (ROA), have great potential as probes of biomolecular structure (1,2). As the vibrational modes of a molecule are sensitive to the atomic and functional group composition, as well as the environments and interactions of these groups, vibrational spectra contain a great deal of information about molecular structure and behaviour.
Inelastic scattering of electromagnetic radiation, that is scattering resulting in a change in frequency of the incident photons, is known as Raman scattering. Raman spectroscopy measures the intensity of the scattered radiation, with the frequencies of scattered photons generating spectral features referred to as Raman lines or bands. Raman bands that have a lower frequency than the initial radiation are referred to as Stokes bands and those that have a higher frequency are known as anti-Stokes bands (3). As Stokes Raman scattering is more intense than anti-Stokes Raman scattering, it is more widely used. The frequency of each Raman band in the spectrum corresponds to a particular vibrational mode of the molecule, and is sensitive to the atoms and bonds involved in that vibrational mode and their local environment. ROA measures the small difference in the intensity of Raman scattering from chiral molecules in right- and left-circularly polarized light or, equivalently, the small circularly polarized component in the scattered light (4). In other words, ROA measures the chirality associated with molecular vibrational transitions (5). Raman and ROA spectroscopies have attracted great interest as incisive structural probes for proteins and, to a lesser extent, nucleic acids and their components (2,6,7). As these techniques are sensitive to different aspects of molecular structure and can be recorded simultaneously, the data obtained are complementary. To date, however, their combined potential for investigating RNA structure has not been fully explored.
Raman and ROA spectroscopies cannot provide the atomic resolution structural detail of X-ray crystallography or multidimensional NMR but they are more widely applicable as there is no size limit or requirement for diffracting crystals, and they can be used to study RNA under near-physiological conditions. Therefore, Raman and ROA complement these high resolution methods as probes of RNA structure and dynamic behaviour. Raman spectroscopy was first applied to nucleic acids in the 1960s and has subsequently been used to investigate a wide range of natural and synthetic nucleic acids as well as their complexes with proteins, metal ions and drugs (2,8). More recently, Raman spectroscopy has been used to probe some aspects of RNA structure, for example, to identify structural features and conformations such as the GNRA tetraloop (9–11). Early studies on pyrimidine nucleosides (12) and polyribonucleotides (13–15) provided a starting point for the interpretation of ROA spectra of RNA. ROA can provide novel structural information about RNA as exemplified by a study of RNA molecules bound in cowpea mosaic virus (CPMV) capsids (16).
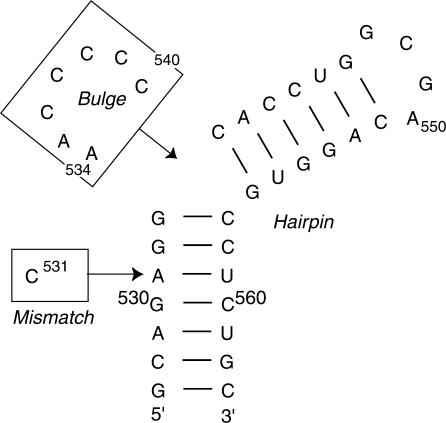
Although Raman and ROA are complementary techniques, to date, they have only been used independently to study RNA structure. Here we demonstrate their combined application to probe RNA structure within a 37-nucleotide RNA fragment from Domain I of the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) RNA (17,18). The IRES RNA guides internal, non-cap dependent, initiation of EMCV protein translation (19,20) and bases critical for IRES function, including a GNRA tetraloop structure, lie within this small fragment (21,22). Significantly, no evidence for protein interaction with this domain has been observed implying that it is the RNA structure of Domain I itself that is vital for function (23). This makes the domain an interesting target for the application of these two spectroscopic techniques as structural probes of a functional RNA and a good model system for establishing rigorous spectral analysis methods. The 37-nucleotide RNA fragment was chosen as a starting point based on several criteria. It adopts a stable independently folded structure with the secondary structure motifs shown in Figure 1, and has been partially characterized structurally by NMR (24). This provides both a basis for developing spectral interpretation and the opportunity to obtain new information about the environment of RNA bases in non-helical elements of the IRES Domain I secondary structure, and the contribution of these regions individually and in combination to the structure of the 37-nucleotide RNA fragment. We show that Raman and ROA spectra are sensitive to both RNA sequence and structure within this RNA domain, and can identify changes in sequence and structure through characteristic spectral changes.
Figure 1.
The secondary structure of the 37-nucleotide domain from the EMCV IRES. The Mismatch and Bulge RNAs are generated by the addition of the single structural motif of interest (boxed sequences), the U·C mismatched base pair and pyrimidine-rich asymmetric bulge respectively, to the Hairpin RNA. The EMCV RNA contains both of these motifs and all four RNAs contain the GNRA tetraloop (GCGA, nucleotides 547–550).
MATERIALS AND METHODS
Sample preparation and data collection
Three oligonucleotides were designed to allow the contribution of each secondary structure motif to the Raman and ROA spectra of the 37-nucleotide RNA (Figure 1) to be determined:
The fully base paired helix with a GNRA (GCGA) tetraloop (Hairpin RNA).
Hairpin RNA incorporating a U·C mismatch (A531 → C change) (Mismatch RNA).
Hairpin RNA incorporating a pyrimidine-rich (AACCCCC) asymmetric bulge (Bulge RNA).
The fragment with the authentic IRES sequence incorporating the tetraloop, mismatch and bulge structures (EMCV RNA) was synthesized by in vitro transcription and purified by denaturing PAGE as described previously (24,25). This RNA contains an additional 5'-G nucleotide (not shown in Figure 1) to provide a strong promoter for transcription. Chemically synthesized HPLC-purified oligonucleotides were purchased from CureVac/VH Bio as lyophyilized powders. These were dissolved directly in 110 μl of buffer (50 mM MOPS, pH 6.5 with 150 mM KCl) to give final RNA concentrations in the range 40–50 mg/ml (4.2–4.3 mM), based upon absorbance measurements at 260 nm and using calculated extinction coefficients. Initial Raman spectra collected for these samples were found to have two major problems: (1) a high level of background fluorescence and (2) a considerably more complex band structure relative to the in vitro transcribed PAGE-purified EMCV RNA sample. Each chemically synthesized RNA was therefore subjected to PAGE purification followed by dialysis back into the original buffer and concentration using Centricon and Microcon YM-3 devices (Amicon) to 120–150 μl. We estimate based on sample loading (up to 5 μg RNA) and the detection limit that RNA sample purity is better than 99%. The final concentration for each sample used in Raman and ROA spectra collection was in the range 19–35 mg/ml (1.6–2.8 mM).
Each sample was centrifuged at maximum speed in a microcentrifuge before loading into a quartz microfluorescence cell and centrifuged again at low speed for 5 min in the cell to remove dust particles prior to data collection. Raman and ROA spectra were measured using a ChiralRaman Spectrometer (BioTools Inc.) (26) at a wavelength of 533 nm, laser power at the sample of 0.65 W and total data accumulation times of 6–24 h. Raman and ROA spectra were acquired simultaneously for each sample. Although high quality Raman spectra could be measured for these samples in <1 s, each Raman spectrum reported here was accumulated for the same time period as the corresponding ROA spectrum to ensure consistency. The sample integrity was assessed before and after data collection by denaturing acrylamide gel electrophoresis and ethidium bromide staining. No deterioration of the sample was observed for any RNA.
Analysis of Raman and ROA spectra
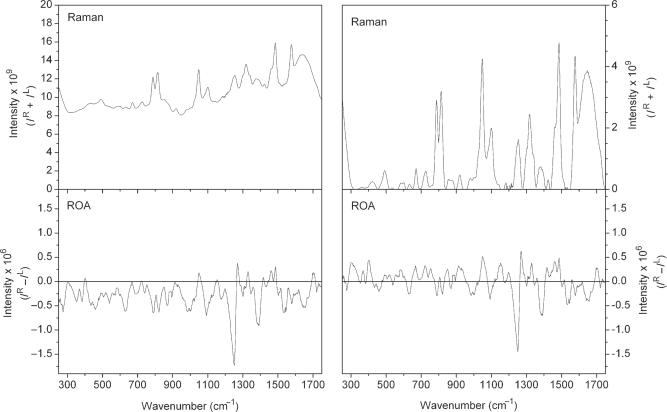
Despite the presence of residual fluorescent impurities in the RNA samples, high quality Raman and ROA spectra were recorded. Examples of the raw Raman and ROA spectra, without any modification, are presented for Hairpin RNA in Figure 2. Only minimal distortion of the baseline is evident in the ROA spectrum and this originates in changes to the background fluorescence during data accumulation. This fluorescence gives rise to the elevated background in the Raman spectrum at high wavenumbers. Such distortions are common in Raman and ROA studies of nucleic acids and other biological molecules but do not interfere with qualitative analysis of the spectra. However, quantitative analysis of the spectra requires careful control of such distortions, typically through electronic processing of the raw spectra, and all spectra were background subtracted before further analysis. Those regions known to be of zero intensity in spectra of nucleic acids were anchored to the baseline and between these regions, a new baseline was interpolated. Any residual errors in the spectral baseline are unlikely to have a significant effect on the data analyses or interpretation presented here.
Figure 2.
Raman and ROA spectra for the Hairpin RNA. Raw Raman (top panel) and ROA (bottom panel) spectra are shown in the left column, and the corresponding baseline corrected and normalized spectra shown in the right column. Raman spectra are presented as intensity sums (IR + IL) and ROA spectra as intensity differences (IR − IL).
In order to allow direct comparisons between the spectra measured for different oligonucleotides the spectra were normalized for differences in experimental parameters. Figure 3 shows Raman and ROA spectra for the Hairpin, Mismatch and Bulge RNAs after normalization to the same sample concentration and accumulation time. Differences in spectrometer performance after instrument realignment prevented use of this normalization procedure for the EMCV RNA spectra. However, the Raman band at ∼1098 cm−1 is invariant of base composition (27), allowing the EMCV RNA Raman spectrum to be normalized to the intensity of this band in the Raman spectrum of Bulge RNA, after adjusting for the one additional nucleotide in EMCV RNA. As all ROA spectra were collected simultaneously with their corresponding Raman spectra, the same normalization ratios were applied to directly compare ROA spectra. We have adopted a rigorous set of procedures consistently applied to the analysis of spectra and conservatively estimate that the relative Raman and ROA band intensities presented here are accurate to ±5% for strong and medium bands.
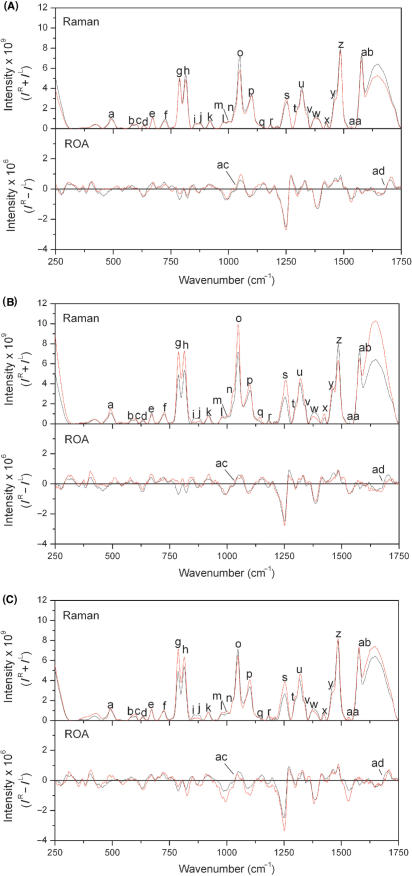
Figure 3.
Raman and ROA spectra for all oligonucleotides. Pairwise comparisons of baseline corrected and normalized Raman (top panel) and ROA (bottom panel) spectra for Hairpin RNA (black) and (A) Mismatch RNA, (B) Bulge RNA and (C) EMCV RNA (red). The letters identifying each peak in the spectra correspond to those given in Table 1.
There are differences between all three oligonucleotide spectra for the intensity of the broad Raman band from ∼1600 to 1650 cm−1, that originates from the O–H bending mode of solvent water molecules. These differences are a product of the intensity normalization procedure as the spectra were normalized with respect to oligonucleotide concentration, while water concentration is constant. Therefore, the absolute Raman intensity of the O–H bending mode from solvent water will vary between the spectra presented in Figure 3 but this will not affect analysis of the spectra outside of this narrow region. As water is achiral and does not generate ROA bands, the ROA spectra are unaffected.
RESULTS AND DISCUSSION
Raman and ROA spectra for all RNAs, following baseline subtraction and normalization procedures, are presented in Figure 3. The bands observed in these spectra, assignments and relative intensities are summarized in Table 1. We discuss in detail later the Raman and ROA bands measured for these oligonucleotides, specifically in the context of the U·C mismatch and asymmetric pyrimidine-rich bulge motifs. Several of these bands are sensitive to the environment of bases while others reflect ribose–phosphate backbone conformation.
Table 1.
Raman and ROA bands for the Hairpin, Mismatch, Bulge and EMCV RNAs
| Raman band (cm−1)a | Assignment | Intensity relative to Hairpin (%)b | |||
|---|---|---|---|---|---|
| Mismatch | Bulge | EMCV | |||
| Bands arising from ribose/phosphate/backbone | |||||
| c | 601 wk | Ribose (32) | +6 | +30 | +55 |
| h | 813 str | O–P–O symmetric stretching, marker for A-form helix (2,9–11,28,32,34,36) | −9 | +36 | +18 |
| i | 856 wk | Ribose–phosphate (32) | −19 | −68 | −56 |
| k | 919 wk | Ribose–phosphate, –C–O– stretching (32,34) | −15 | −5 | 0 |
| l | 977 wk | Ribose–phosphate, –C–O– stretching (32,34) | 0 | +21 | +24 |
| o | 1047 str | P–O stretch, sugar phosphate –C–O– stretching (30,34) | −18 | +38 | −10 |
| p | 1098 med |
 symmetric stretching (2,9–11,28,32,34,36) symmetric stretching (2,9–11,28,32,34,36) |
−2 | +18 | +21 |
| q | 1136 wk | Stretching at 2’ position of ribose (32) | +5 | +71 | +22 |
| Bands arising from bases | |||||
| a | 489 wk | G out of plane ring deformations (34) | −15 | +35 | +13 |
| b | 580 wk | A/U/G/C out of plane ring deformations (34) | +16 | +34 | +13 |
| d | 631 wk | A/U/G/C (10,28,34) | +5 | +49 | +46 |
| e | 670 wk | G out of plane ring deformations (9–11,32,34) | +4 | −13 | −1 |
| f | 726 wk | A ring stretching (9,11,27,32,34) | −21 | +23 | +4 |
| g | 787 str | C/U breathing/stretching (27,28,32–34,36) | +3 | +47 | +45 |
| j | 876 wk | A/U/G/C out of plane ring deformations, backbone (10,28,34) | −16 | −50 | −56 |
| r | 1182 wk | A/U/G/C ring; external C–N stretching (9,10,28,34) | +9 | −24 | +32 |
| s | 1253 med | U/C ring stretching (2,9,11,27,32–34) | +9 | +62 | +44 |
| t | 1300 sh | C/A ring stretching (10,11,32–34) | −10 | −20 | +12 |
| u | 1316 med | A/G ring stretching (2,9–11,28,32,34) | −7 | +12 | +13 |
| v | 1336 med/sh | A/G/U (2,9–11,28,32,34) | −22 | +22 | +12 |
| w | 1380 wk | A/G/U (10,11,28,32,34,36) | −11 | −39 | −22 |
| x | 1425 wk | A/G ring stretching, CH deformations (28,32,34) | −14 | +66 | +17 |
| y | 1460 med | U/C ring stretching, CH deformations (28,32,34) | −17 | +8 | −19 |
| z | 1485 str | A/G ring stretching/planar vibrations (9,10,28,32,34) | −3 | −20 | +3 |
| aa | 1532 wk | A/G/C ring stretching (10,27,28,32,34) | +78 | −100 | +110 |
| ab | 1578 str | A/G ring stretching (9,10,27,28,32,34) | −6 | −10 | +1 |
| Bands with no previous assignment | |||||
| m | 1002 wk | +3 | +16 | +26 | |
| n | 1025 med/sh | −16 | +63 | 0 | |
| ROA band (cm−1) | Assignment | Intensity relative to Hairpin (%) | |||
|---|---|---|---|---|---|
| Mismatch | Bulge | EMCV | |||
| ac | 994-1093 | Triplet; A-form RNA (13) | +26 | −16 | +46 |
| ad | 1650-1750 | Couplet; right-handed helix (13) | +14 | −19 | +2 |
aLetters identifying each peak correspond to those given in Figure 3. Band positions given are those observed in the spectrum of the Hairpin RNA, and may differ slightly for the other three oligonucleotides. Intensity key: wk: weak, med: medium, str: strong, sh: shoulder.
bThe intensities for each of the other three oligonucleotides are given as percentages of the Hairpin RNA intensity for each band.
Bands arising from ribose/phosphate/backbone
A number of weak Raman bands observed between ∼400 and 1140 m−1 have been previously assigned to ribose and phosphate linkages (2,8–11,27–36). As expected, the majority of these bands do not show significant differences in intensity between the Hairpin and Mismatch RNA spectra as these oligonucleotides are the same length and, consequently, have the same ribose and phosphate content. The same bands do show an increase in intensity for the Bulge and EMCV RNA spectra, relative to the Hairpin RNA spectrum, due to the addition of seven and eight residues, respectively, in these oligonucleotides.
The Raman band observed at ∼813 cm−1 originates from O–P–O stretching, therefore reflecting the length of the oligonucleotide, and has been noted to be an indicator of A-form helical structure (2,9–11,28,32,34,36). The introduction of a mismatched base pair is expected to disrupt such a structure. This can be observed as a decrease in intensity of this Raman band for the Mismatch RNA compared to that of the Hairpin RNA, and for the EMCV RNA compared to the Bulge RNA. A similar decrease in intensity is observed for the Raman band recorded at ∼1047 cm−1, originating from P–O stretching (30), although the decrease in intensity for the EMCV RNA relative to the Bulge RNA is more marked than for that observed at ∼813 cm−1.
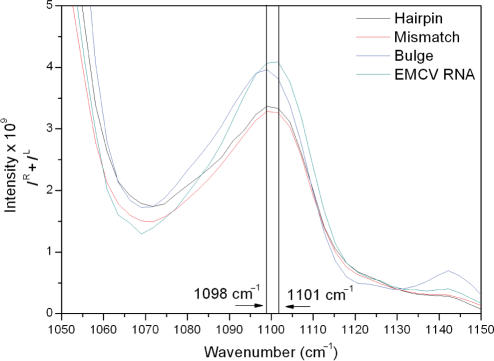
The Raman band recorded at ∼1098 cm−1, previously assigned to  stretching (30), shows a greater intensity in the spectrum of the Bulge RNA compared to that of the Hairpin and Mismatch RNA, with this increase due to the addition of the seven extra residues in the pyrimidine-rich bulge. Close examination of the Raman spectra presented in Figure 3 shows that there is a small shift in frequency of this band, from 1098 to 1101 cm−1, for the EMCV RNA (Figure 4). The position of this band therefore appears to be sensitive to contributions from unusual backbone conformations of nucleotides due to the interaction of the U·C mismatch base pair with the pyrimidine-rich asymmetric bulge.
stretching (30), shows a greater intensity in the spectrum of the Bulge RNA compared to that of the Hairpin and Mismatch RNA, with this increase due to the addition of the seven extra residues in the pyrimidine-rich bulge. Close examination of the Raman spectra presented in Figure 3 shows that there is a small shift in frequency of this band, from 1098 to 1101 cm−1, for the EMCV RNA (Figure 4). The position of this band therefore appears to be sensitive to contributions from unusual backbone conformations of nucleotides due to the interaction of the U·C mismatch base pair with the pyrimidine-rich asymmetric bulge.
Figure 4.
Close-up view of Raman spectra for all oligonucleotides in the region of 1050–1150 cm−1. Comparison of Raman spectra for Hairpin RNA (black), Mismatch RNA (red), Bulge RNA (blue) and EMCV RNA (green). The peak maximum for the  symmetric stretching mode for EMCV RNA is shifted to 1101 cm−1.
symmetric stretching mode for EMCV RNA is shifted to 1101 cm−1.
Fewer band assignments have been made for ROA than for Raman spectra of RNA, but most bands observed from ∼950 to 1150 cm−1 are expected to be associated with the sugar–phosphate backbone. A negative–positive–negative triplet at ∼994, 1049 and 1086 cm−1 has been observed for several nucleic acids and is associated with the C3′-endo ribose sugar pucker present in A-form double helices (13,16). Overall, the relative intensities of this triplet suggest that the introduction of a mismatch into the sequence actually increases the intensity of this ROA triplet. It is unlikely that this signifies an increase in A-type helix content, rather that one or more of the ribose sugars is more constrained in the presence of the mismatch in terms of conformational mobility. ROA spectra are sensitive to conformational mobility, with more rigid chiral structures generating more intense ROA spectral features (6). Conversely, the introduction of the pyrimidine bulge decreases the intensity of this triplet, again monitoring a difference in sugar-pucker conformations, but it is also likely that this corresponds to a decrease in A-form helical content for the Bulge RNA. The greatest intensity is observed for the EMCV RNA indicating that the presence of both motifs in a sequence has a cooperative effect, resulting in a structure with more constrained C3′-endo sugar-pucker conformation than found for sequences only containing one of these two motifs. Cooperative effects on local RNA structure such as these are possible as the mismatch pair in EMCV RNA is located only three residues away from the pyrimidine-rich bulge (Figure 1).
Bands arising from bases
In addition to bands arising from ribose, phosphate and backbone vibrations, the ∼400 to 1140 cm−1 region also contains a number of bands arising from the bases. For a number of these, the absolute intensity changes are small and can originate from any or all of the bases. Therefore, the origins of these bands are not discussed in detail.
The Raman band observed at ∼489 cm−1 shows a decrease in intensity for the Mismatch RNA compared to the Hairpin RNA. Although this band has been previously assigned to C=O deformations in guanine (34) it is possible that it may also reflect the same deformations in adenine residues. If this is the case then this decrease directly reflects the substitution of an adenine residue with a cytosine residue in the Mismatch RNA sequence. Similarly, the intensity of this band is greatest for the Bulge RNA due to the two additional adenine residues in its sequence. The intensity of this band for the EMCV RNA shows a small increase as this oligonucleotide has the two additional adenine residues included in the pyrimidine bulge, but also reflects the loss of one adenine residue due to the presence of the mismatched base pair. The Raman band observed at ∼726 cm−1, present for all four oligonucleotides, has been previously assigned to adenine residues (9,11,27,28,32–34). This band also shows a decrease in intensity for the Mismatch RNA compared to the Hairpin RNA spectrum, and for the EMCV RNA compared to the Bulge RNA. Together, these observations show that this band does indeed originate from adenine and that the decrease in Raman intensity at ∼726 cm−1 is due to the adenine to cytosine base substitution.
The Raman band observed at ∼787 cm−1 is assigned to the breathing or stretching vibrational modes of cytosine and uracil bases (27,28,32–34,36). The differences in intensity of the ∼787 cm−1 band between the four oligonucleotides indicate that this band may provide direct information on the environment of these bases. Incorporation of the single cytidine nucleotide into a region of A-form duplex to form the U·C mismatched base pair is only reflected by a small increase of 3% in intensity, within the estimated experimental error. In contrast, a significant increase in intensity is observed when the five cytidine bases of the pyrimidine-rich bulge loop are present, corresponding to an increase of ∼10% per residue. The increase in intensity for this band is significantly greater than the proportional increase in the cytosine/uracil content for both the Bulge RNA and EMCV RNA. This shows that the Raman band at ∼787 cm−1 is sensitive to the environment of the pyrimidine bases and particularly those in non-helical regions. While high resolution structural information is not currently available for this specific RNA sequence, the NMR structure of the Bacillus subtilis T-box antiterminator RNA containing a similar 7-nucleotide asymmetric bulge between two helical regions has been determined (37). The bulge exhibits conformational flexibility and significant deviation from A-form helical geometry including predominantly C2′-endo sugar pucker for some nucleotides. The structure of the loop produces a sharp bend in the RNA fragment, and while the majority of the bases stack locally, they are not involved in stacking with the flanking helical regions. The bases in the pyrimidine-rich bulge of the EMCV are similarly likely to be less constrained by extended stacking interactions and may therefore contribute more strongly to this Raman band.
Similarly, at ∼1253 cm−1 is a Raman band that has previously been assigned to cytosine, adenine and uracil (2,9,11,27,32–34). The intensity of this band is lowest for the Hairpin RNA and highest for the Bulge RNA, suggesting that changes in this band originate from cytosine vibrations. The Raman band observed at ∼1300 cm−1 is thought to originate from stretching modes of adenine and cytosine residues (10,11,32–34). Comparisons between the spectra suggest that in this case, changes in band intensity are due to the addition of adenine: the band intensity for EMCV RNA is highest, followed by the Hairpin and the Mismatch RNAs, reflecting the order of decrease in adenine content. The changes in the Raman bands observed at ∼1316 and 1336 cm−1, assigned to adenine and guanine (2,9–11,28,32,34), also appear to reflect the number of adenine bases in each RNA. These Raman bands have lowest intensity in the case of the Mismatch RNA spectrum and highest intensity for the Bulge and EMCV RNAs.
The Raman band observed at ∼1380 cm−1 has been previously assigned to adenine, guanine or cytidine bases (10,11,28,32,34,36) but here appears to directly reflect changes in the adenine content of the RNAs. The ∼1380 cm−1 band intensity for the Mismatch and Bulge RNAs is less than that for the Hairpin and EMCV RNAs, respectively, by a similar amount (11 and 17%). In both cases, the difference between these RNAs is the addition of one adenine (U·C mismatch versus U–A base pair in each case), suggesting that the change in band intensity is the same for this alteration in structure regardless of the overall context. However, the context in which adenine residues are added or removed does appear to influence the actual contribution the change makes to the band intensity. An increase in band intensity is observed on addition of the adenine residue within the helical region. In contrast, the addition of two adenine residues in the pyrimidine-rich bulge causes a large decrease in band intensity. Finally, the differences in band intensity between the Hairpin RNA and the Mismatch (−11%) and Bulge (−39%) RNAs are not fully reflected in the EMCV RNA (−22%). This suggests, as might be anticipated, that there are cooperative changes in the RNA structure upon combining multiple structural motifs that is reflected in different alterations in band intensity. This type of behaviour is observed for a number of other Raman bands (e.g. those at ∼580, 1047, 1253 and 1300 cm−1) including intense bands indicating that this observation is not due to experimental noise or errors in baseline subtractions.
The Raman band observed at ∼1425 cm−1 has also been assigned to adenine and guanine residues in the literature (28,32,34). This band is more intense in the Raman spectrum for the Bulge RNA compared to that of the EMCV RNA, suggesting that changes in this band originate from adenine residues in these oligonucleotides. The differences in intensity for the Raman band observed at ∼1460 cm−1 between our spectra are not fully explained by the current assignment of this band for uracil, cytosine or ribose (28,32,34). However, for these oligonucleotides it is possible that the band at ∼1460 cm−1 is an indicator of the number of uracil residues involved in base pair interactions. The intensity of this band is greater in the Raman spectra for the Hairpin and Bulge RNAs, where there are four adenine–uracil base pairs, and lower for the Mismatch and EMCV RNA spectra, where there are only three.
Raman bands that arise from the rings of nucleic acids can exhibit significant changes in intensity and/or frequency under different experimental conditions (32). One cause of this intensity or frequency change is the phenomenon known as Raman hyperchromism, where Raman band intensity increases with increased base stacking interactions (27). A hyperchromic band can also show a decrease in intensity as the base stacking of the RNA decreases. The opposite effect, Raman hypochromism, results in a decrease in Raman band intensity with increased face-to-face stacking of the bases (32,38). Such changes in band intensity can be affected or induced by temperature, due to its influence on base stacking interactions (32,38).
A number of Raman bands that appear in these oligonucleotide spectra have been assigned as being hypochromic. However, several of the assignments regarding Raman hypo- and hyperchromism in published literature are contradictory and care must be exercised in their analysis. The Raman bands measured at ∼1380, 1485 and 1578 cm−1 are all assigned to adenine and guanine residues (9–11,27,28,32,35). Two of these bands have been noted to be hyperchromic, those observed at ∼1380 and 1485 cm−1 (27,34), whereas other authors have suggested that those observed at ∼1485 and 1578 cm−1 are hypochromic (9,27). The data presented here are consistent with the assignment that these bands are hyperchromic, especially as the introduction of the pyrimidine bulge is likely to reduce the face-to-face base stacking of the RNA, and the observed decrease in band intensity.
In ROA spectra, the broad negative–positive couplet between ∼1600 and 1650 cm−1 is thought to be indicative of A-type helix (13). Although this couplet is present in all the ROA spectra shown in Figure 3, there is a reduction in intensity of this couplet in the Bulge RNA spectrum compared to the other spectra. This indicates that the introduction of the pyrimidine bulge significantly disrupts the A-form helical structure of the RNA. The introduction of a large asymmetrical bulge loop is likely to produce a sharp turn in the RNA helix, separating and potentially unstacking the two helical regions on either side (Figure 1). In this case, the changes observed in intensity of this ROA couplet going from Hairpin to Bulge RNA appear to reflect the expected changes in RNA structure. However, in comparing the Hairpin and Mismatch RNAs, the introduction of a U·C mismatch produces an apparent increase in A-form helical content. This is also reflected in the intensity of this band for the EMCV RNA which also contains the pyrimidine-rich bulge but maintains an apparent helical content, derived from the band intensity, much greater than the Hairpin RNA. It is possible, as discussed earlier for Raman bands that the presence of both motifs in a sequence may lead to some degree of interaction between these two motifs, such that the presence of the pyrimidine-rich bulge has less of an effect on the structure of the RNA in this case. As discussed earlier for the ROA triplet from ∼994 to 1093 cm−1, it is possible that the intensity of this couplet may depend not only on the A-type helix content in complex RNA structures but also on other factors that require further detailed investigation.
CONCLUSION
Raman and ROA spectroscopies are able to reveal detailed information about RNA structure. A number of bands in the spectra presented here have previously been assigned to specific vibrational modes of the RNA backbone or bases, with the literature on Raman spectroscopy at present being more extensive than for ROA spectroscopy. In this work we have used known Raman and ROA band assignments to investigate the effects of a mismatched base pair and pyrimidine-rich asymmetric bulge on the structure of a small domain from the EMCV IRES RNA. Analysis of both Raman and ROA spectra of model RNA structural motifs provides not only qualitative but also, potentially, quantitative information on RNA secondary and tertiary structure and stability, and the subtle interactions between different structural motifs. We have shown that the combination of band patterns in Raman and ROA spectra of RNAs provides a set of features that are uniquely sensitive to conformation of the phosphodiester backbone and interactions of bases. Changes in these Raman and ROA band patterns generate signatures for the U·C mismatch and pyrimidine-rich asymmetric bulge secondary structure motifs. Fewer band assignments were made for the ROA spectra presented here, reflecting the current much lower understanding of the origin of ROA bands compared to those of Raman spectra. Though significant changes are observed in our ROA spectra, indicating that they contain much potentially useful information, the corresponding Raman spectra appear to display greater variation. This may in part be a consequence of the common stem and GNRA tetraloop structures in each of the oligonucleotides. Whereas Raman is highly sensitive to the environments of bases and backbone conformations, changes in ROA spectra may reflect more global RNA structural features. Further studies will show if this observation holds for more structurally diverse RNAs, where the greater sensitivity of ROA to differences in secondary and tertiary structure is expected to be more evident.
This work forms a foundation for more extensive studies on the interactions between RNA secondary structure motifs, their effects on tertiary structure stability and the role of ions or proteins in folding and stabilizing RNA structures. The level of information obtained from these studies will increase as the number of RNA structures investigated increases, generating new and more detailed band assignments. The wide applicability of Raman and ROA spectroscopies makes them particularly suitable for investigating conformational transitions associated with RNA folding and protein–RNA interactions, indicating the great potential these optical spectroscopies have for RNA structural biology.
ACKNOWLEDGEMENTS
The authors would like to thank Lorna Ashton for helpful comments on the manuscript. This work was funded by The Wellcome Trust (grant ref. 076060). Funding to pay the Open Access publication charge was provided by The Wellcome Trust.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Barron LD, Hecht L, Blanch EW, Bell AF. Solution structure and dynamics of biomolecules from Raman optical activity. Prog. Biophys. Mol. Biol. 2000;73:1–49. doi: 10.1016/s0079-6107(99)00017-6. [DOI] [PubMed] [Google Scholar]
- 2.Benevides JM, Overman SA, Thomas GJ. Raman, polarized Raman and ultraviolet resonance Raman spectroscopy of nucleic acids and their complexes. J. Raman Spectrosc. 2005;36:279–299. [Google Scholar]
- 3.Aroca R. Surface-enhanced Vibrational Spectroscopy. Chichester, UK: John Wiley & Sons Ltd; 2006. [Google Scholar]
- 4.Nafie LA. Infrared and Raman vibrational optical activity: theoretical and experimental aspects. Annu. Rev. Phys. Chem. 1997;48:357–386. doi: 10.1146/annurev.physchem.48.1.357. [DOI] [PubMed] [Google Scholar]
- 5.Blanch EW, Hecht L, Barron LD. Vibrational Raman optical activity of proteins, nucleic acids, and viruses. Methods. 2003;29:196–209. doi: 10.1016/s1046-2023(02)00310-9. [DOI] [PubMed] [Google Scholar]
- 6.Barron LD, Hecht L, McColl IH, Blanch EW. Raman optical activity comes of age. Mol. Phys. 2004;102:731–744. [Google Scholar]
- 7.Blanch EW, McColl IH, Hecht L, Nielsen K, Barron LD. Structural characterization of proteins and viruses using Raman optical activity. Vib. Spectrosc. 2004;35:87–92. [Google Scholar]
- 8.Thomas GJ. Raman spectroscopy of protein and nucleic acid assemblies. Annu. Rev. Biophys. Biomol. Struct. 1999;28:1–27. doi: 10.1146/annurev.biophys.28.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez B, Baumruk V, Leulliot N, Gouyette C, Huynh-Dinh T, Ghomi M. Thermodynamic and structural features of ultrastable DNA and RNA hairpins. J. Mol. Struct. 2003;651:67–74. [Google Scholar]
- 10.Leulliot N, Baumruk V, Abdelkafi M, Turpin PY, Namane A, Gouyette C, Huynh-Dinh T, Ghomi M. Unusual nucleotide conformations in GNRA and UNCG type tetraloop hairpins: evidence from Raman markers assignments. Nucleic Acids Res. 1999;27:1398–1404. doi: 10.1093/nar/27.5.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leulliot N, Baumruk V, Gouyette C, Huynh-Dinh T, Turpin PY, Ghomi M. Aqueous phase structural features of GNRA tetraloops formed in short hairpins as evidenced by UV absorption and Raman spectroscopy. Vib. Spectrosc. 1999;19:335–340. [Google Scholar]
- 12.Bell AF, Hecht L, Barron LD. Vibrational Raman optical activity of pyrimidine nucleosides. J. Chem. Soc. Faraday Trans. 1997;93:553–562. [Google Scholar]
- 13.Bell AF, Hecht L, Barron LD. Vibrational Raman optical activity as a probe of polyribonucleotide solution stereochemistry. JACS. 1997;119:6006–6013. [Google Scholar]
- 14.Bell AF, Hecht L, Barron LD. Evidence for global mobility in the premelting of a polynucleotide from temperature-dependent Raman optical activity. Biospectroscopy. 1998;4:107–111. doi: 10.1002/(sici)1520-6343(1998)4:2<107::aid-bspy3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 15.Bell AF, Hecht L, Barron LD. Evidence for a new transition in polyribonucleotides from Raman optical activity. J. Raman Spectrosc. 1999;30:651–656. [Google Scholar]
- 16.Blanch EW, Hecht L, Syme CD, Volpetti V, Lomonossoff GP, Nielsen K, Barron LD. Molecular structures of viruses from Raman optical activity. J. Gen. Virol. 2002;83:2593–2600. doi: 10.1099/0022-1317-83-10-2593. [DOI] [PubMed] [Google Scholar]
- 17.Kolupaeva VG, Pestova TV, Hellen CUT, Shatsky IN. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 1998;273:18599–18604. doi: 10.1074/jbc.273.29.18599. [DOI] [PubMed] [Google Scholar]
- 18.Vandervelden A, Kaminski A, Jackson RJ, Belsham GJ. Defective point mutants of the encephalomyocarditis virus internal ribosome entry site can be complemented in trans. Virology. 1995;214:82–90. doi: 10.1006/viro.1995.9952. [DOI] [PubMed] [Google Scholar]
- 19.Jackson RJ, Howell MT, Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem. Sci. 1990;15:477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- 20.Jackson RJ, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts LO, Belsham GJ. Complementation of defective picornavirus internal ribosome entry site (IRES) elements by the coexpression of fragments of the IRES. Virology. 1997;227:53–62. doi: 10.1006/viro.1996.8312. [DOI] [PubMed] [Google Scholar]
- 22.Robertson MEM, Seamons RA, Belsham GJ. A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA. 1999;5:1167–1179. doi: 10.1017/s1355838299990301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark AT, Robertson MEM, Conn GL, Belsham GJ. Conserved nucleotides within the J domain of the encephalomyocarditis virus internal ribosome entry site are required for activity and for interaction with eIF4G. J. Virol. 2003;77:12441–12449. doi: 10.1128/JVI.77.23.12441-12449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phelan M, Banks RJ, Conn G, Ramesh V. NMR studies of the structure and Mg2+ binding properties of a conserved RNA motif of EMCV picornavirus IRES element. Nucleic Acids Res. 2004;32:4715–4724. doi: 10.1093/nar/gkh805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker SC, Avis JM, Conn GL. General plasmids for producing RNA in vitro transcripts with homogeneous ends. NAR Methods. 2003;31:e82. doi: 10.1093/nar/gng082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hug W, Hangartner G. A novel high-throughput Raman spectrometer for polarization difference measurements. J. Raman Spectrosc. 1999;30:841–852. [Google Scholar]
- 27.Fish SR, Hartman KA, Stubbs GJ, Thomas GJ. Studies of virus structure by laser Raman spectroscopy. 9. Structural studies of Tobacco Mosaic Virus and its components by laser Raman spectroscopy. Biochemistry. 1981;20:7449–7457. doi: 10.1021/bi00529a019. [DOI] [PubMed] [Google Scholar]
- 28.Baumruk V, Gouyette C, Huynh-Dinh T, Sun JS, Ghomi M. Comparison between CUUG and UUCG tetraloops: thermodynamic stability and structural features analyzed by UV absorption and vibrational spectroscopy. Nucleic Acids Res. 2001;29:4089–4096. doi: 10.1093/nar/29.19.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erfurth SC, Kiser EJ, Peticolas WL. Determination of backbone structure of nucleic acids and nucleic acid oligomers by laser Raman scattering. PNAS. 1972;69:938–941. doi: 10.1073/pnas.69.4.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan Y, Thomas GJ. Vibrational analysis of nucleic acids. III. Conformation-dependent Raman markers of the phosphodiester backbone modeled by dimethyl phosphate. J. Mol. Struct. 1996;379:31–41. doi: 10.1016/S0006-3495(96)79474-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lafleur L, Rice J, Thomas GJ. Raman studies of nucleic acids. VII. PolyA · polyU and polyG · poly-C. Biopolymers. 1972;11:2423–2437. doi: 10.1002/bip.1972.360111205. [DOI] [PubMed] [Google Scholar]
- 32.Small EW, Peticolas WL. Conformational dependence of Raman scattering intensities from polynucleotides. III. Order-disorder changes in helical structures. Biopolymers. 1971;10:1377–1416. doi: 10.1002/bip.360100811. [DOI] [PubMed] [Google Scholar]
- 33.Small EW, Peticolas WL. Conformational dependence of Raman scattering intensities from polynucleotides. Biopolymers. 1971;10:69–88. doi: 10.1002/bip.360100107. [DOI] [PubMed] [Google Scholar]
- 34.Thomas GJ. Raman spectral studies of nucleic acids. III. Laser-excited spectra of ribosomal RNA. Biochim. Biophys. Acta. 1970;213:417–429. doi: 10.1016/0005-2787(70)90049-3. [DOI] [PubMed] [Google Scholar]
- 35.Thomas GJ, Hartman KA. Raman studies of nucleic acids. 8. Estimation of RNA secondary structure from Raman scattering by phosphate group vibrations. Biochim. Biophys. Acta. 1973;312:311–322. doi: 10.1016/0005-2787(73)90376-6. [DOI] [PubMed] [Google Scholar]
- 36.Thomas GJ, Medeiros GC, Hartman KA. Dependence of Raman scattering on conformation of Ribosomal RNA. Biochem. Biophys. Res. Commun. 1971;44:587. doi: 10.1016/s0006-291x(71)80123-7. [DOI] [PubMed] [Google Scholar]
- 37.Gerdeman MS, Henkin TM, Hines JV. Solution structure of the Bacillus subtilis T-box antiterminator RNA: seven nucleotide bulge characterized by stacking and flexibility. J. Mol. Biol. 2003;326:189–201. doi: 10.1016/s0022-2836(02)01339-6. [DOI] [PubMed] [Google Scholar]
- 38.Tomlinson BL, Peticolas WL. Conformational dependence of Raman scattering intensities in polyadenylic acid. J. Chem. Phys. 1970;52:2154–2156. doi: 10.1063/1.1673270. [DOI] [PubMed] [Google Scholar]