Abstract
The transcription factor SOX9 plays a critical role in chondrogenesis as well as in sex determination. Previous work has suggested that SOX9 functions as a DNA-dependent dimer when it activates genes involved in chondrogenesis, but functions as a monomer to activate genes involved in sex determination. We present evidence herein for a third binding configuration through which SOX9 can activate transcription. We have identified four separate SOX consensus sequences in a COL9A1 collagen gene enhancer. The sites are arranged as two pairs, and each pair is similar to previously discovered dimeric SOX9 binding sites. Increasing the spacing between the pairs of sites eliminated enhancer activity in chondrocytic cells, as did the mutation of any one of the four sites. The COL9A1 enhancer is ordinarily inactive in 10T1/2 cells, but cotransfection with a SOX9 expression plasmid was sufficient to activate the enhancer, and mutation of any one of the four sites reduced responsiveness to SOX9 overexpression. These results suggest a novel mechanism for transcriptional activation by SOX9, in which two SOX9 dimers that are bound at the two pairs of sites are required to interact with one another, either directly or indirectly, in order to produce a functional transcriptional activation complex.
INTRODUCTION
The transcription factor SOX9 is critical for normal embryonic development. SOX9 is a member of the high-mobility group (HMG) superfamily and contains an HMG DNA binding domain, as well as a transactivation domain and a DNA-dependent dimerization domain (1). It is related to the testes determining factor gene SRY, and the SOX family of proteins was named for this relationship; SOX stands for SRY-type HMG box (2). SOX9 plays a critical role in chondrogenesis, as demonstrated by knockout mouse experiments in which all cells bearing a homozygous targeted mutation in the SOX9 gene were excluded from the chondrocyte differentiation pathway (3). Evidence from the human disease campomelic dysplasia, which results from mutations in the SOX9 gene, indicates that SOX9 functions in sex determination as well as in chondrogenesis. Campomelic dysplasia is always characterized by severe cartilage abnormalities, but about two-thirds of chromosomally male patients also exhibit XY sex reversal (4–6).
A number of enhancer elements from genes involved in both chondrogenesis and sex determination have been shown to be directly activated by SOX9. We have previously identified three separate chondrocyte-specific enhancer elements in the type XI collagen gene Col11a2 (7,8). Each enhancer binds dimeric SOX9 at paired binding sites arranged in opposite orientation to each other and separated by three or four base pairs (bp). Mutation of either site in a pair inactivated the enhancer, as did changing the spacing between paired sites, indicating that the dimeric SOX9 interaction is essential for enhancer activity. We have also shown that the type XXVII collagen gene COL27A1 contains at least two similar enhancer elements that bind SOX9 at paired sites, and mutation of either site in a pair also inactivated those enhancers (9). Furthermore, enhancers from the cartilage-specific genes Col2a1, Col9a2, and the cartilage-derived retinoic-acid-sensitive protein (CD-RAP) all contain paired binding sites for SOX9 (10–14).
In contrast, enhancers from the anti-Mullerian hormone (AMH) gene and the Steroidogenic Factor 1 (SF1) gene, which both play a critical role in sex determination and are both activated by SOX9, only contain one SOX9 binding site each (12,15). This evidence led to the hypothesis put forth by Bernard et al. in 2003 that dimerization of SOX9 is required for the activation of genes involved in chondrogenesis but not for those involved in sex determination (12). This hypothesis is further supported by a human case of campomelic dysplasia in which a missense mutation (A76E) in the dimerization domain of SOX9 resulted in chondrodysplasia but not sex reversal (12,15). In enhancer/reporter assays, the A76E SOX9 dimerization mutant was found to activate the SF1 and AMH sex determination enhancers normally, but did not activate either the Col11a2 or Col9a2 cartilage enhancers (12,15).
Prior to the publication of the dimerization-for-chondrogenesis hypothesis, work was published identifying an enhancer region in the type IX collagen gene COL9A1 in which only one SOX9 binding site was pinpointed as being essential for enhancer activity in chondrocytic cells (16). We suspected that this enhancer might also be subject to regulation by dimerized SOX9, and upon examining the sequence we identified a second potential SOX9 binding site 4 bp upstream from the first site and oriented appropriately for the binding of dimeric SOX9. Surprisingly, however, the candidate enhancer sequence we initially constructed, which contained the paired sites, had no activity in enhancer/reporter assays. Instead, enhancer activity required the inclusion of 54 additional bp upstream. This upstream sequence included two more previously unidentified SOX consensus sequences. All four sites were found to bind SOX9, and each was essential for the activity of the enhancer element; mutation of any one of the four sites completely abolished enhancer activity in chondrocytic cells.
The results presented herein suggest a novel mechanism by which gene expression can be regulated by the transcription factor SOX9. It appears that, in the COL9A1 enhancer, two interdependent SOX9 dimers are required for transcriptional activation. Mutations that inactivated either dimer by preventing binding of one of the SOX9 molecules left the other dimeric binding site incapable of transcriptional activation. Likewise, insertion mutations that increased the spacing between the two dimerization sites eliminated enhancer activity, demonstrating the necessity of a dimer–dimer interaction for transcriptional activation.
EXPERIMENTAL PROCEDURES
Plasmid construction
COL9A1 D1/D2 4x enhancer/reporter plasmid
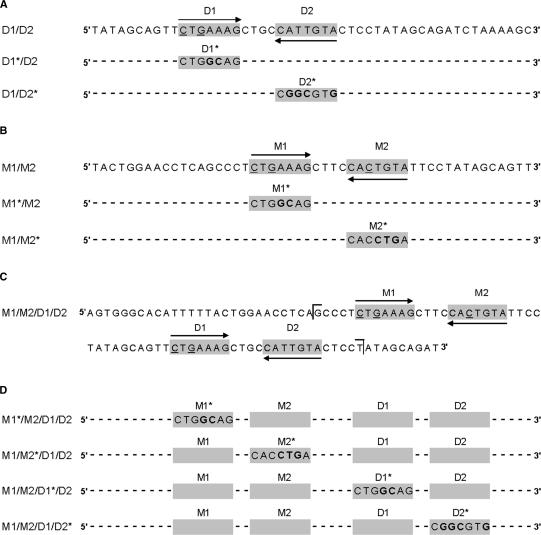
Complementary oligonucleotides were designed for a 50-bp portion of the COL9A1 D enhancer region described by Zhang et al. (16), containing either wild-type paired SOX9 binding sites (D1/D2) or substitution mutations in either the upstream or downstream site within each pair to generate D1*/D2 and D1/D2*, respectively (Figure 1A). Mutant sites contained a total of four mismatches with the consensus SOX binding sequence, A/TA/TCAAA/TG. Complementary single-strand DNA oligonucleotides were purified by denaturing polyacrylamide gel electrophoresis, annealed and cloned into the BamHI-BglII site of the p89Col2a1Bs plasmid. The enhancer elements were multimerized to four tandem copies as previously described and transferred with the minimal promoter to the luciferase reporter plasmid p95Luc, resulting in four repeats of the enhancer upstream of the Col2a1 minimal promoter and luciferase reporter gene (7). Constructs were verified by sequencing in the BYU DNA Sequencing Center (BYU DNASC).
Figure 1.
Sequences from the COL9A1 enhancer region. SOX9 binding consensus sequences are marked with grey background, and arrows indicate site orientation. Mismatches with the SOX9 consensus sequence are underlined, and introduced mutations are bold. (A) The D1/D2 element contains the SOX9 binding site (D2) that was previously shown by Zhang et al. (16) to be critical for COL9A1 enhancer activity. Mutations were introduced into the D1 site (D1*/D2) or the D2 site (D1/D2*) to produce a total of four mismatches with the SOX binding consensus sequence A/TA/TCAAA/TG. (B) The M1/M2 element includes the newly identified binding sites upstream of the D1/D2 element. M1*/M2 and M1/M2* mutants contain four mismatches with the SOX binding consensus sequence. (C) Sequence of the full-length M1/M2/D1/D2 enhancer. Brackets indicate the boundaries of the probes used for EMSA. (D) Mutants of the M1/M2/D1/D2 enhancer.
COL9A1 M1/M2/D1/D2 1x enhancer/reporter plasmid
To create the COL9A1 M1/M2/D1/D2 enhancer plasmid, PCR was used to amplify the 96-bp COL9A1 D enhancer region described by Zhang et al. (16) (Forward TTAAGGATCCAGTGGGCACATTTTTAC, Reverse GGCGAGATCTATCTGCTATAGGAGTAC) (Figure 1C). DNA template was extracted from human white blood cells using the DNeasy Tissue Kit (Qiagen). The amplified enhancer region was cloned into the BamHI-BglII site of the p89Col2a1Bs plasmid. The enhancer and minimal promoter were then transferred to the luciferase reporter plasmid p95Luc, and correct construction was verified by DNA sequencing in the BYU DNASC.
Mutations were made to each of the four putative SOX9 binding sites located within the COL9A1 M1/M2/D1/D2 enhancer using QuikChange II Site-Directed Mutagenesis (Stratagene) resulting in the mutants M1*/M2/D1/D2, M1/M2*/D1/D2, M1/M2/D1*/D2 and M1/M2/D1/D2* (Figure 1D) where each mutated SOX9 binding site contained a total of four mismatches to the SOX binding consensus sequence. To construct mutants M1/M2/A*/D1/D2 and M1/M2/B*/D1/D2, 4 bp (A, CTAT; B, GTCA) were inserted between the two pairs of SOX9 binding sites using QuikChange II Site-Directed Mutagenesis (Stratagene).
SOX9 Δ66-75 expression plasmid
Mutations were introduced into the wild-type SOX9-pcDNA-5′-UT expression plasmid using QuikChange II Site-Directed Mutagenesis (Stratagene). Mutant SOX9 Δ66–75 is a de novo human mutation described by Sock et al. (15), which results in the loss of amino acids 66–75 in the conserved DNA-dependent dimerization domain.
Tissue culture and transfections
Rat chondrosarcoma (RCS) cells and 10T1/2 cells were a gift from Dr. Benoit de Crombrugghe of the University of Texas M.D. Anderson Cancer Center. Cells were cultured at 37°C with 5% CO2 in supplemented Dulbecco's modified Eagle medium (D-MEM) (Gibco). Supplemented medium contained 50 U/ml penicillin (Cellgro), 50 µg/ml streptomycin (Cellgro), 2 mM l-glutamine (Cellgro) and 10% fetal bovine serum (HyClone). RCS and 10T1/2 cells were transiently transfected using Lipofectamine Reagent with Plus Reagent (Invitrogen), according to the manufacturer's instructions. The plasmid pSV-β-galactosidase (Promega) was included in every reaction as an internal control for transfection efficiency, and a total of 2.0 µg of DNA for RCS transfections and 1.4 µg of DNA for 10T1/2 transfections were used in each 10-cm2 well.
Following transfection, cellular extracts were prepared and enzyme activity assays performed. Luciferase activity was measured using the Luciferase Assay system (Promega), and β-galactosidase activity was measured using the Galacto-Light Plus system (Tropix), both according to manufacturer's instructions. Results were measured in a TD 20/20 luminometer (Turner Designs) and are presented as luciferase units per β-galactosidase unit ± standard error. Results were analyzed for statistical significance using Student's t-test.
Electrophoretic mobility shift assay (EMSA)
EMSAs were performed using wild-type and mutant versions of the COL9A1 D1/D2 element, the M1/M2 element and the M1/M2/D1/D2 element as DNA probes (see Figure 1 for sequences), which were 32P radiolabeled by end-filling with Klenow fragment. SOX9 was synthesized by in vitro transcription/translation of SOX9-pcDNA-5′-UT expression plasmid using the TNT Coupled Wheat Germ Extract System (Promega).
DNA-protein binding reactions were performed with radiolabeled probes and SOX9, either wild type or the Δ66–75 mutant, with or without anti-SOX9 antibody. Reactions were carried out at room temperature for 45 min in a DNA-binding buffer containing 20 mM Hepes (pH 7.9), 10% glycerol, 50 mM KCl, 0.05% Nonidet NP-40, 0.5 mM EDTA, 0.5 mM DTT, and 1 mM PMSF. Poly(dG-dC)·poly(dG-dC) (0.5–2 µg) was added as a nonspecific competitor. When anti-SOX9 antibody was included, it was pre-incubated with SOX9 for 20–30 min before the addition of radiolabeled probes. Binding reactions were separated by electrophoresis for 3–4 h at 150 V.
RESULTS
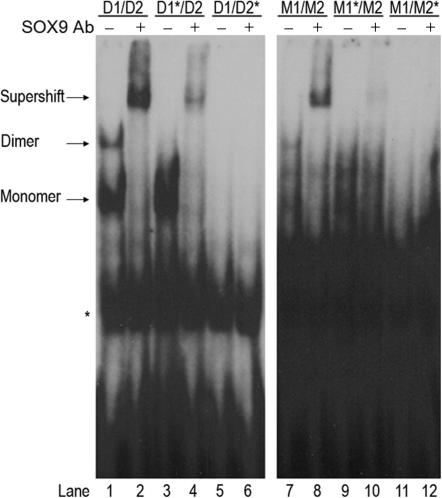
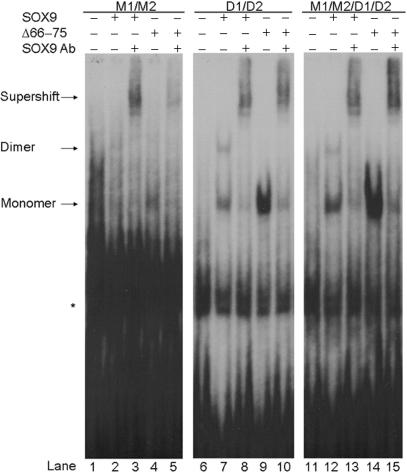
After the putative SOX9 binding site D1 was identified adjacent to the known site, D2, EMSAs were performed to determine whether SOX9 binds to D1 in vitro. Probes used included wild-type D1/D2, as well as mutants D1*/D2 and D1/D2* (Figure 1A). Mutant binding sites contained four mismatches with the SOX binding consensus sequence A/TA/TCAAA/TG. The EMSAs showed that SOX9 bound as a dimer and a monomer to the wild-type COL9A1 D1/D2 element (Figure 2, lane 1). Inclusions of anti-SOX9 antibody supershifted both complexes, confirming that they both contain SOX9 (Figure 2, lane 2). Mutation of the D1 site (D1*/D2) allowed for monomeric binding at the D2 site, but prevented dimeric SOX9 binding (Figure 2, lanes 3 and 4). Mutation of the D2 site (D1/D2*), however, prevented both dimeric and monomeric binding, even though the D1 site was still intact (Figure 2, lanes 5 and 6). This result suggested that binding of SOX9 at the D1/D2 pair of sites occurs cooperatively and sequentially, with binding at the D2 site required before binding at the D1 site can occur. The D1 site is not capable of binding a SOX9 monomer.
Figure 2.
Dimeric and monomeric SOX9 bind to the COL9A1 D1/D2 and M1/M2 elements. EMSA was performed using SOX9 made in vitro and wild-type or mutant probes as indicated. Anti-SOX9 antibody supershifted both monomeric and dimeric complexes of SOX9 with the wild-type D1/D2 probe (compare lanes 1 and 2). Mutation of the D1 site (D1*/D2) allowed only monomeric SOX9 binding (lanes 3 and 4). Mutation of the D2 site (D1/D2*) completely prevented all SOX9 binding (lanes 5 and 6). Anti-SOX9 antibody also supershifted both monomeric and dimeric complexes of SOX9 with the wild-type M1/M2 probe (compare lanes 7 and 8). Mutation of the M1 site (M1*/M2) prevented dimeric SOX9 binding (lanes 9 and 10). Mutation of the M2 site (M1/M2*) completely prevented all SOX9 binding (lanes 11 and 12). The right and left panels contain lanes from the same EMSA experiment, but the panel on the right was exposed longer because SOX9 binding to the M1/M2 element was very weak compared to D1/D2 binding. * denotes a nonspecific band.
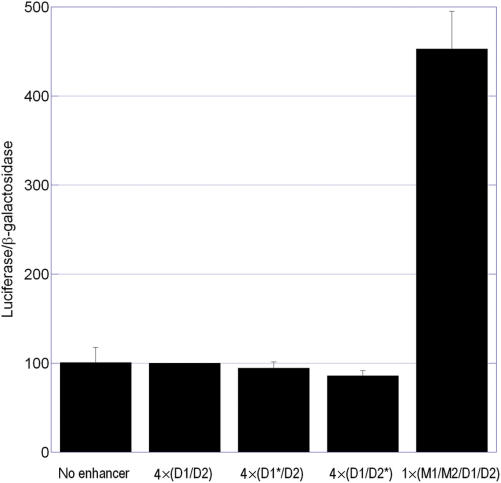
To determine whether the binding of SOX9 to the wild-type and mutant COL9A1 D1/D2 elements in vitro correlated with transcriptional activity in vivo, transient transfections were performed using enhancer/reporter plasmids in RCS cells where SOX9 is endogenously expressed. To our surprise, neither the wild-type nor mutant D1/D2 elements displayed any enhancer activity in RCS cells (Figure 3).
Figure 3.
The D1/D2 element has no enhancer activity. Transient transfections were performed in RCS cells using reporter plasmids containing four tandem copies of the short D1/D2 element or a single copy of the longer M1/M2/D1/D2 enhancer. Neither wild-type D1/D2 nor corresponding mutants activated gene expression. The longer M1/M2/D1/D2 enhancer, however, was transcriptionally active in RCS cells, as previously reported (16). Graph includes data from three independent experiments, each performed in triplicate, and data was normalized to the activity of the D1/D2 element.
Although they did not contain the full-length enhancer previously identified by Zhang et al. (16), these elements did contain the same SOX9 binding site, D2, which had been identified as the critical site for activation. We hypothesized, therefore, that another critical sequence must exist in the upstream region contained in Zhang's enhancer but omitted from our D1/D2 element. To test this hypothesis, the same enhancer region as described by Zhang et al. (Figure 1C) was used to construct a single-copy enhancer/reporter plasmid, which was tested in transient transfections in RCS cells. The full-length 96-bp element (M1/M2/D1/D2) displayed significant enhancer activity (P < 0.0001), indicating that the upstream region does indeed contain additional critical sequences (Figure 3).
The upstream region was examined for possible transcription factor binding sites, and two new putative SOX9 binding sites arranged in opposite orientation to each other and separated by 4 bp were discovered. The site designated M1 was a 2-bp mismatch with the SOX consensus sequence, and the M2 site was a 1-bp mismatch (Figure 1B). EMSAs were performed using probes containing only the M1 and M2 sites to determine whether SOX9 bound in vitro to these newly identified sites. Probes used included wild-type M1/M2, as well as mutants M1*/M2 and M1/M2* (Figure 1B). The addition of anti-SOX9 antibody to the wild-type binding reaction resulted in an antibody shift indicating that SOX9 binding did occur at this pair of sites (Figure 2, lanes 7 and 8). Binding was very weak, however, compared to the D1/D2 element, and visualization of the complex required longer exposure of the X-ray film. When the M1 site was mutated, SOX9 still bound weakly to the M2 site and was supershifted by anti-SOX9 antibodies (Figure 2, lanes 9 and 10). But when the M2 site was mutated, no SOX9 binding occurred (Figure 2, lanes 11 and 12), suggesting that, as with the D1/D2 pair of sites, SOX9 binding occurs in a cooperative sequential manner with monomeric binding at the M2 site required before dimerization can occur. The M1 site is not capable of binding monomeric SOX9.
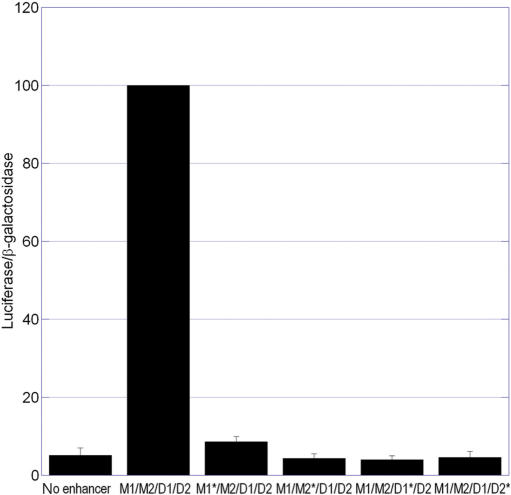
To determine whether these newly identified SOX9 binding sites were important for the transcriptional activity of the full-length M1/M2/D1/D2 enhancer, substitution mutations were introduced within each site and the mutant enhancers were tested in enhancer/reporter assays. The D1 and D2 sites were also mutated in the context of this longer enhancer (Figure 1D). Surprisingly, introducing a mutation into any one of the four putative SOX9 binding sites within the M1/M2/D1/D2 enhancer resulted in the complete elimination of enhancer activity in chondrocytic RCS cells (Figure 4).
Figure 4.
Mutation of any one of the four SOX sites in the M1/M2/D1/D2 element abolishes enhancer activity in RCS cells. Transient transfections were performed in RCS cells using reporter plasmids containing the wild-type enhancer (M1/M2/D1/D2) or mutant enhancers (M1*/M2/D1/D2, M1/M2*/D1/D2, M1/M2/D1*/D2 and M1/M2/D1/D2*). Results were normalized to the activity of the wild-type enhancer. Each panel contains data from three independent experiments, performed in triplicate.
To verify that the activity of the M1/M2/D1/D2 enhancer was the result of SOX9 transactivation, enhancer/reporter assays were performed in 10T1/2 cells, which do not express SOX9. The wild-type M1/M2/D1/D2 enhancer showed a nonsignificant level of transcriptional activity (P = 0.053) in the absence of SOX9, and none of the four mutants was significantly different from the wild-type level of activity in the absence of SOX9 (P > 0.165) (Figure 5, white bars). Cotransfection with the SOX9 expression plasmid, however, strongly activated the wild-type M1/M2/D1/D2 enhancer (P < 0.0001). Individual mutation of the M1, M2 or D1 sites reduced activation to between 30 and 50% of wild type, and mutation of the D2 site eliminated activation by SOX9 (Figure 5, grey bars), demonstrating that each of the four sites participates in transactivation by SOX9.
Figure 5.
SOX9 overexpression activates the M1/M2/D1/D2 enhancer in 10T1/2 cells. Transient transfections were performed in 10T1/2 cells using reporter plasmids containing the wild-type enhancer (M1/M2/D1/D2) or mutant enhancers (M1*/M2/D1/D2, M1/M2*/D1/D2, M1/M2/D1*/D2 and M1/M2/D1/D2*). Cells were cotransfected with the SOX9 or the ▵66–75 SOX9 mutant expression plasmid. Results were normalized to the activity of the wild-type enhancer cotransfected with SOX9.
To examine the importance of SOX9 dimerization for activation of the full-length M1/M2/D1/D2 enhancer, the ▵66–75 mutant of SOX9 was used in EMSAs and enhancer/reporter assays. As previously shown, SOX9 that is lacking amino acids 66–75 within the dimerization domain is incapable of DNA-dependent dimerization and is unable to activate enhancers that require dimeric SOX9 binding (15). Elements M1/M2, D1/D2 and M1/M2/D1/D2 were used as probes in EMSA with ▵66–75 SOX9 (Figure 6). With each probe, the dimeric SOX9 band was lost and the intensity of the monomeric band increased when ▵66–75 was substituted for SOX9 in the binding reaction (Figure 6, lanes 4, 9 and 14).
Figure 6.
The ▵66–75 SOX9 mutant cannot dimerize at M1/M2/D1/D2 SOX9 binding sites. EMSA was performed using the M1/M2, D1/D2 or M1/M2/D1/D2 elements as probes and either SOX9 or ▵66–75 mutant SOX9 as indicated. The ▵66–75 SOX9 mutant bound to these probes only as a monomer. Lanes 1, 6 and 11 contain the in vitro protein synthesis reaction blank control. The three panels all contain lanes from the same EMSA experiment, but the panel on the left was exposed longer because SOX9 binding to the M1/M2 probe was very weak compared to the other two probes. *denotes a nonspecific band.
To see if transcriptional activation could occur in the absence of SOX9 dimerization, enhancer/reporter assays were performed using ▵66–75 mutant SOX9 overexpression in 10T1/2 cells. The M1/M2/D1/D2 enhancer and each of the four mutant enhancer elements were tested. The ▵66–75 mutant SOX9 produced approximately half as much activation of the M1/M2/D1/D2 enhancer as wild-type SOX9, indicating that dimerization is required for full activation. Interestingly, however, when tested with enhancers containing mutations in the M1, M2, D1 or D2 sites individually, the ▵66–75 SOX9 mutant protein was not less active than wild-type SOX9 (Figure 5, black bars).
To determine whether the spacing between the two pairs of SOX9 binding sites was a significant factor in enhancer activation, RCS cells were transfected with mutant enhancer/reporter plasmids containing two different 4-bp insertion mutations between the two paired SOX9 binding sites (M1/M2/A*/D1/D2 and M1/M2/B*/D1/D2). Two different insertions were used in order to verify that the specific sequence of the inserted DNA had no inherent activity. These insertions increased the space between the two dimeric binding sites from 14 to 18 bp and thus offset the two pairs of binding sites by nearly half a turn of the DNA helix. The presence of either insert resulted in complete inactivation of the enhancer, suggesting that the SOX9 dimers bound at the two pairs of sites must interact, either directly or indirectly, to form a complex that results in transcriptional activation (Figure 7).
Figure 7.
Insertion of 4 bp between the two SOX9 binding site pairs eliminates enhancer activity. Transient transfections were performed in RCS cells using reporter plasmids containing the wild-type enhancer (M1/M2/D1/D2) or enhancers containing one of two different 4-bp insertions between the two SOX9 binding site pairs (M1/M2/A*/D1/D2 and M1/M2/B*/D1/D2). Both insertions eliminated enhancer activity. Graph includes data from three independent experiments, each performed in triplicate, and data was normalized to the activity of the wild-type enhancer.
Although our results had shown that cotransfection with SOX9 was sufficient to activate the M1/M2/D1/D2 enhancer in 10T1/2 cells, and mutation of any one of the four SOX consensus sites reduced responsiveness to SOX9, we wondered whether other SOX family proteins besides SOX9 might bind to some of the sites and cooperate with SOX9 to regulate transcription. The proteins l-SOX5 and SOX6 have been shown to cooperate with SOX9 to activate the type II collagen gene, Col2a1, through binding to a chondrocyte-specific enhancer with multiple SOX consensus sequences (17). The loss of both l-Sox5 and Sox6 results in severe cartilage abnormalities in a dual knock-out mouse, so we wondered if l-Sox5 and Sox6 might also help activate the COL9A1 M1/M2/D1/D2 enhancer in cartilage cells (18). Plasmids expressing l-Sox5 and Sox6 were included in addition to the SOX9 expression plasmid in M1/M2/D1/D2 enhancer/reporter assays in 10T1/2 cells, to see if either protein could regulate transcriptional activity independently or in cooperation with SOX9. Neither l-Sox5 nor Sox6 had any effect on the level of activation produced by SOX9 (data not shown), suggesting that SOX9 binds to all four SOX consensus sequences in the COL9A1 M1/M2/D1/D2 enhancer and that l-Sox5 and Sox6 do not play a role in activating this enhancer.
EMSAs were performed to determine whether the same mutations that eliminated M1/M2/D1/D2 enhancer activity in enhancer/reporter assays also prevented SOX9 binding in vitro. Inclusion of anti-SOX9 antibodies demonstrated that although binding was reduced, some SOX9 still bound the enhancer when the M1, M2 or D1 sites were mutated (Figure 8). Mutation of the D2 site, however, eliminated SOX9 binding, suggesting that SOX9 must bind first to the D2 site before the rest of the SOX9 complex can assemble. Taken together with the enhancer/reporter assay results presented in Figure 4, these results indicate that even though some SOX9 complex assembly can occur when the M1, M2 or D1 sites are mutated, the partially assembled SOX9 complexes are completely inactive for the purposes of transcriptional activation in chondrocytic cells.
Figure 8.
The D2 site is required for SOX9 binding to the M1/M2/D1/D2 enhancer in vitro. EMSAs were performed using SOX9 made in vitro and wild-type or mutant enhancer probes as indicated. Anti-SOX9 antibody supershifted the wild-type enhancer complex (lane 2). It also supershifted weaker complexes formed with enhancers bearing mutations at the M1, M2 and D1 sites, indicating that SOX9 can still bind these mutated enhancers in vitro (lanes 4, 6 and 8). Mutation of the D2 site, however, abolished SOX9 binding (lanes 9 and 10).
DISCUSSION
This work has resulted in the discovery of a new type of enhancer element through which the chondrogenic transcription factor SOX9 can activate transcription. Previously discovered enhancer elements from the Col11a2, COL27A1, Col2a1, Col9a2 and CD-RAP genes each contain two SOX9 binding sites arranged in opposite orientation to each other and separated by 3 or 4 bp (8–12). Mutation of either site in a pair inactivates those enhancers, suggesting that dimeric SOX9 binding is required for transcriptional activation of these genes. The COL9A1 enhancer, in contrast, contains four SOX9 binding sites arranged in two pairs.
This two-pair arrangement initially suggested the possibility of two independent enhancer elements. A shorter element (D1/D2) containing only one of the two pairs (including the site that had previously been shown to be important for COL9A1 enhancer activity), however, was completely inactive in RCS cells. Even when multimerized to four tandem copies containing a total of eight SOX9 binding sites arranged in four pairs, this element was inactive. In contrast, a single copy of the longer M1/M2/D1/D2 element containing all four SOX9 binding sites arranged in two pairs showed enhancer activity in chondrocytic cells and was activated by SOX9 in 10T1/2 cells. Activity, therefore, is not simply a function of the number of SOX9 binding sites. Instead, each of the four sites contributes some essential function.
The importance of each individual SOX9 binding site to the activity of the COL9A1 enhancer was demonstrated by generating targeted mutations at each of the four sites. If activation of the enhancer were accomplished by the independent contributions of four SOX9 monomers or by two independently functioning SOX9 dimers, mutation of any one site would have produced an intermediate level of transcriptional activation resulting from SOX9 binding at the three remaining monomeric binding sites or the one intact dimeric binding site. Instead, the mutation of any one site completely inactivated the enhancer in chondrocytic cells, demonstrating that the four SOX9 binding sites function cooperatively and that neither dimer has the ability to activate transcription independently in chondrocytic cells.
Mutation of any single SOX9 site completely eliminated enhancer activity in chondrocytic RCS cells, but the M1, M2 and D1 mutant enhancers were still partially activated in 10T1/2 cells by overexpression of SOX9, probably because the levels of SOX9 were unnaturally high. When the Δ66–75 SOX9 dimerization mutant was overexpressed in 10T1/2 cells, we found that activation of the four mutant enhancers was the same as with wild-type SOX9, suggesting that elimination of any individual SOX9 binding site in the enhancer has the same effect as inactivating the SOX9 dimerization domain. This result provided additional support for the hypothesis that dimerization of SOX9 at each pair of sites is required for full transcriptional activation of the M1/M2/D1/D2 enhancer in chondrocytic cells.
To determine whether the two individual SOX9 dimers interact with one another, 4 bp insertions (nearly one-half turn of the double helix) were generated to change the spacing from 14 to 18 bp between the two pairs of sites. These insertions inactivated the M1/M2/D1/D2 enhancer, even though all four of the SOX9 binding site sequences were unaltered. This result suggested that SOX9 dimers that are bound at the two pairs of sites must interact with each other, either directly or through contact with an additional transcription factor, and that disrupting their spatial arrangement prevented this interaction from occurring. Changing the spacing between the two pairs of sites prevented the formation of the functional transcriptional activating complex.
The in vitro DNA–protein binding data presented here demonstrates that even though the short D1/D2 element is transcriptionally inactive, SOX9 is capable of binding to it as a monomer or dimer in vitro. SOX9 can bind to the short D1/D2 probe as a monomer at the D2 site, which is a perfect consensus binding site, even when the D1 site is mutated. When the D2 site is mutated, in contrast, SOX9 does not bind as a monomer to the intact D1 site. This suggests that a SOX9 monomer must first bind the D2 site, and then a second SOX9 molecule can bind at the D1 site and dimerize with the first. EMSA results indicate that SOX9 binding at the M1/M2 pair of sites alone is very weak, and it is likely that the SOX9 dimer bound at the M1/M2 pair of sites is stabilized by its interaction with the D1/D2 SOX9 dimer in the full-length enhancer. Consistent with this sequential cooperative binding hypothesis, the D2 site mutation is the only mutation that prevented all SOX9 binding to the full-length M1/M2/D1/D2 COL9A1 enhancer probe in EMSA. It is also the only mutation that made the enhancer completely nonresponsive to SOX9 overexpression in 10T1/2 cells. We suggest that mutation of the D2 primary SOX9 binding site prevented the initial SOX9–DNA interaction, and without this primary interaction, subsequent binding of SOX9 to the D1, M2 and M1 sites could not occur. It follows then that when SOX9 was overexpressed in 10T1/2 cells, the enhancer activity that was observed even when the M1, M2 or D1 sites were mutated was probably produced by monomeric binding of SOX9 at the D2 site. Importantly, however, such activation of the mutant enhancers occurred only in 10T1/2 cells with artificially high levels of SOX9, and never in chondrocytic cells with physiologically relevant levels of SOX9. In RCS cells, binding of both SOX9 dimers is essential for enhancer activity.
The in vitro DNA–protein binding data also emphasizes another important point: binding of SOX9 to an element in vitro does not necessarily mean that the element will be transcriptionally active in vivo. Transcription factors must not only bind the DNA but also interact with one another, as well as with any required cofactors in order for productive transactivation to occur. In the case of the COL9A1 enhancer, it is possible that cofactors are required to coordinate the interaction between the two SOX9 dimers. It is also possible that the two SOX9 dimers interact directly with one another. Regardless of how this interaction takes place, the work presented here demonstrates that the interaction of proteins bound at four interdependent SOX9 binding sites is essential for the activity of the COL9A1 enhancer element. This represents a novel mechanism for transcriptional activation by SOX9.
ACKNOWLEDGEMENTS
We thank Dr. Benoit de Crombrugghe of the M.D. Anderson Cancer Center in Houston, TX, for the RCS cells and 10T1/2 cells, and Dr. Veronique Lefebvre of the Cleveland Clinic in Cleveland, OH, for the p89Col2a1Bs and SOX9-pcDNA-5′-UT plasmids. This work was supported by NIAMS/NIH grant #AR48839 to L.C.B. Funding to pay the Open Access publication charge was provided by Brigham Young University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Harley VR, Clarkson MJ, Argentaro A. The molecular action and regulation of the testis-determining factors, SRY (sex-determining region on the Y chromosome) and SOX9 [SRY-related high-mobility group (HMG) box 9] Endocr. Rev. 2003;24:466–487. doi: 10.1210/er.2002-0025. [DOI] [PubMed] [Google Scholar]
- 2.Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyer J, Sudbeck P, Held M, Wagner T, Schmitz ML, Bricarelli FD, Eggermont E, Friedrich U, Haas OA, et al. Mutational analysis of the SOX9 gene in campomelic dysplasia and autosomal sex reversal: lack of genotype/phenotype correlations. Hum. Mol. Genet. 1997;6:91–98. doi: 10.1093/hmg/6.1.91. [DOI] [PubMed] [Google Scholar]
- 5.Foster JW. Mutations in SOX9 cause both autosomal sex reversal and campomelic dysplasia. Acta Paediatr. Jpn. 1996;38:405–411. doi: 10.1111/j.1442-200x.1996.tb03515.x. [DOI] [PubMed] [Google Scholar]
- 6.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 7.Bridgewater LC, Lefebvre V, de Crombrugghe B. Chondrocyte-specific enhancer elements in the Col11a2 gene resemble the Col2a1 tissue-specific enhancer. J. Biol. Chem. 1998;273:14998–15006. doi: 10.1074/jbc.273.24.14998. [DOI] [PubMed] [Google Scholar]
- 8.Bridgewater LC, Walker MD, Miller GC, Ellison TA, Holsinger LD, Potter JL, Jackson TL, Chen RK, Winkel VL, et al. Adjacent DNA sequences modulate Sox9 transcriptional activation at paired Sox sites in three chondrocyte-specific enhancer elements. Nucleic Acids Res. 2003;31:1541–1553. doi: 10.1093/nar/gkg230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins E, Moss JB, Pace JM, Bridgewater LC. The new collagen gene COL27A1 contains SOX9-responsive enhancer elements. Matrix Biol. 2005;24:177–184. doi: 10.1016/j.matbio.2005.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies SR, Li J, Okazaki K, Sandell LJ. Tissue-restricted expression of the Cdrap/Mia gene within a conserved multigenic housekeeping locus. Genomics. 2004;83:667–678. doi: 10.1016/j.ygeno.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Zhou G, Lefebvre V, Zhang Z, Eberspaecher H, de Crombrugghe B. Three high mobility group-like sequences within a 48-base pair enhancer of the Col2a1 gene are required for cartilage-specific expression in vivo. J. Biol. Chem. 1998;273:14989–14997. doi: 10.1074/jbc.273.24.14989. [DOI] [PubMed] [Google Scholar]
- 12.Bernard P, Tang P, Liu S, Dewing P, Harley VR, Vilain E. Dimerization of SOX9 is required for chondrogenesis, but not for sex determination. Hum. Mol. Genet. 2003;12:1755–1765. doi: 10.1093/hmg/ddg182. [DOI] [PubMed] [Google Scholar]
- 13.Xie WF, Zhang X, Sakano S, Lefebvre V, Sandell LJ. Trans-activation of the mouse-cartilage-derived retinoic-acid-sensitive protein gene by Sox9. J. Bone. Miner. Res. 1999;14:757–763. doi: 10.1359/jbmr.1999.14.5.757. [DOI] [PubMed] [Google Scholar]
- 14.Bell DM, Leung KK, Wheatley SC, Ng LJ, Zhou S, Ling KW, Sham MH, Koopman P, Tam PP, et al. SOX9 directly regulates the type-II collagen gene [see comments] Nat. Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- 15.Sock E, Pagon RA, Keymolen K, Lissens W, Wegner M, Scherer G. Loss of DNA-dependent dimerization of the transcription factor SOX9 as a cause for campomelic dysplasia. Hum. Mol. Genet. 2003;12:1439–1447. doi: 10.1093/hmg/ddg158. [DOI] [PubMed] [Google Scholar]
- 16.Zhang P, Jimenez SA, Stokes DG. Regulation of human COL9A1 gene expression. Activation of the proximal promoter region by SOX9. J. Biol. Chem. 2003;278:117–123. doi: 10.1074/jbc.M208049200. [DOI] [PubMed] [Google Scholar]
- 17.Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L -Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefebvre V, Behringer RR, de Crombrugghe B. L-Sox5, Sox6 and Sox9 control essential steps of the chondrocyte differentiation pathway. Osteoarthritis Cartilage. 2001;9(Suppl A):S69–5. doi: 10.1053/joca.2001.0447. [DOI] [PubMed] [Google Scholar]