Abstract
Appropriate supply of deoxyribonucleotides by the ribonucleotide reductase (RNR) complex is essential for DNA replication and repair. One recent model for the RNR activation in Schizosaccharomyces pombe is translocation of the regulatory subunit Suc22 from the nucleoplasm to the cytoplasm. The RNR inhibitory protein Spd1, which retains Suc22 in the nucleoplasm, is rapidly degraded upon DNA-replication stress, resulting in release of Suc22 to form the active RNR complex in the cytoplasm. Here, we show that Caf1, a component of the Ccr4–Not complex, is responsible for resistance of the replication stress and control of the Suc22 translocation. Caf1 is required not only for the stress-induced translocation of Suc22 from nucleoplasm to cytoplasm but also for the degradation of nucleoplasmic Spd1. DNA-replication stress appears to allow Caf1 to interact with Suc22, resulting in release of the nucleoplasmic Spd1–Suc22 assembly. Taken together, these results suggest a novel function of Caf1 as a key regulator in the stress-induced RNR activation.
INTRODUCTION
The Ccr4–Not complex is known not only as the transcriptional factor but also as the major cytoplasmic deadenylase in Saccharomyces cerevisiae (1,2). The complex, which has been initially identified as a global regulator of transcription (3–5), consists of nine core subunits (Ccr4, Caf1/Pop2, Not1–5, Caf40 and Caf130) and additional components, such as Dbf2, Mob1, Caf4 and Caf16. Among these constituents, Ccr4 and Caf1/Pop2 have been well characterized. The Ccr4 or Caf1/Pop2 and the other proteins show distinct growth phenotypes and different binding partners (6). Ccr4 and Caf1/Pop2 also appear to function as cytoplasmic deadenylases (7). Their primary structure suggests that Ccr4 is a member of exo III family of nucleases, Mg2+-dependent endonuclease and Caf1/Pop2 is categorized as a member of DEDDh family of RNases (8). Some residues, which are crucial for exonuclease activity, are missing in ScCaf1/Pop2, although the deadenylase activity of Caf1/Pop2 is detected in vitro (9). On the other hand, it has been reported in S. cerevisiae that the Ccr4–Not complex is responsible for the sensitivity to DNA-replication stress in large-scale studies (10,11). The sensitivity appears to be dependent on the deadenylase activity of Ccr4 (12) and the transcription of RNR genes by Ccr4, Caf1/Pop2 and Not1–5 (13). However, it remains unclear whether these activities in the Ccr4–Not complex are sufficient for the stress resistance.
In response to replication stress and DNA damage, stress-response and highly conserved checkpoint pathways are activated in order to prevent genome instability. The checkpoint pathway and the supplement of dNTPs are activated in response to chemical reagents that induce DNA-replication stress and DNA damage. The S-phase DNA-replication checkpoint pathway induces cell-cycle blockage (14–16). Proteins involved in the checkpoint pathway are categorized into three groups: damage sensors, adaptors and effector kinases. In Schizosaccharomyces. pombe, Rad3 and Rad26, which belong to the phosphoinositide 3-kinase family, have been found to sense DNA damage (17,18). These sensor proteins phosphorylate Serine/Threonine-kinase adaptors, and the activated adaptor kinases in turn phosphorylate effector kinases, such as Cds1, which control further downstream targets involved in the stress response (19).
In addition to the DNA-replication checkpoint pathway, the dNTP flow is precisely controlled under DNA-replication stress and DNA damage. High-fidelity DNA replication requires an adequate supply of dNTPs (20–22). The dNTPs are synthesized from NTPs by the ribonucleotide reductase (RNR) complex, whose activity is elaborately controlled. In both S. cerevisiae and S. pombe, RNR is principally composed of two components; the large catalytic subunit Rnr1 or Rnr3 (SpCdc22) in the cytoplasm and the small regulatory subunit Rnr2–Rnr4 complex (SpSuc22) in the nucleoplasm. Multiple layers of regulation are imposed on the RNR activity. In S. pombe, Cid13 contributes to stabilize 1.9-kb Suc22 mRNA by its poly-adenylation, which proceeds only in response to DNA-replication stress (23). Furthermore, several proteins have been identified to regulate the activity and translocation of the RNR subunits. For example, Sml1 interacts with the catalytic subunit Rnr1 to inhibit the RNR activity in S. cerevisiae (24,25), and Spd1 (in S. pombe) is capable of inhibiting the RNR activity of Suc22–Cdc22 in vitro (26). In addition to the role in regulating activity of RNR, Spd1 captures the regulatory subunit Suc22 in the nucleoplasm and acts as a negative regulator for RNR in S. pombe. During S phase or DNA damage and replication stress, Spd1 is degraded by the Pcu4–Ddb1–Cop9 signalosome (CSN) complex; this releases Suc22 from the nucleoplasm to the cytoplasm, where it associates with Cdc22 to form the active complex (27). However, it is unclear how Spd1 degradation is initiated or regulated resulting in the dissociation of the nucleoplasmic Spd1–Suc22 complex in response to DNA damage or replication stress.
Here, we identified Caf1, a component of the Ccr4–Not complex, as a key regulator of Spd1 degradation in S. pombe. Caf1 is required for resistance to DNA-replication stress through the control of Suc22 translocation. Caf1 interacts with Suc22 in response to DNA-replication stress and promotes the degradation of nucleoplasmic Spd1 and the cytoplasmic translocation of Suc22. We propose that Caf1 plays an important role in regulating the RNR activity through releasing the nucleoplasmic Spd1–Suc22 assembly.
MATERIALS AND METHODS
Yeast strains
Schizosaccharomyces pombe strains were grown on YE3S (0.5% yeast extract, 2% glucose, 225 μg/ml each of adenine, leucine and uracil) for vegetative growth, or Edinburgh minimal medium (EMM) (28). DNA constructs for chromosomal disruptions and epitope tagging were made by PCR-using method and integrated by homologous recombination into the desired loci using the method described previously (29). When the kanMX6 and hygromycin cassette were used for a disruption marker, transformants were grown on YE3S plate for 1 day for integration and resistance gene expression, before plating on YE3S containing G418 or hygromycin. Some of the yeast strains used in this study are shown in Table 1.
Table 1.
Yeast strains used in this study
| Name | Relevant genotype |
|---|---|
| YSP 001 | Wild-type JY746 |
| YSP 002 | ccr4::kanr |
| YSP 003 | pan2::kanr |
| YSP 004 | parn::hygBr |
| YSP 027 | Wild-type pRep1-FLAG |
| YSP 066 | caf1::hygBr |
| YSP 069 | cds1::kanr |
| YSP 071 | rad26::kanr |
| YSP 072 | ccr4::kanr rad26::hygBr |
| YSP 086 | cid13::hygBr |
| YSP 091 | spd1-13myc::ura4+ |
| YSP 095 | caf1::hygBr cid13::kanr |
| YSP 097 | ccr4::kanr cid13::hygBr |
| YSP 102 | caf1::hygBr rad26::kanr |
| YSP 103 | caf1::hygBr cds1::kanr |
| YSP 104 | caf1::hygBr ccr4::kanr |
| YSP 106 | ccr4::kanr cds1::hygBr |
| YSP 112 | spd1::hygBr |
| YSP 131 | not4::hygBr |
| YSP 143 | ski2::hygBr |
| YSP 154 | ccr4::hygBr spd1-13myc::ura4+ |
| YSP 161 | caf1::hygBr spd1-13myc::ura4+ |
| YSP 162 | not4::hygBr spd1-13myc::ura4+ |
| YSP 172 | xrn1::hygBr |
| YSP 184 | ccr4::kanr pRep41-HA |
| YSP 185 | ccr4::kanr pRep41-HA-Ccr4 |
| YSP 188 | caf1::hygBr pRep1-FLAG |
| YSP 189 | caf1::hygBr pRep1-FLAG-Caf1 |
| YSP 190 | caf1::hygBr pRep1-FLAG-Caf1 D50A |
| YSP 195 | caf1::hygBr spd1-13myc::ura4+ pRep1-FLAG |
| YSP 196 | caf1::hygBr spd1-13myc::ura4+ pRep1-FLAG-Caf1 |
| YSP 197 | caf1::hygBr spd1-13myc::ura4+ pRep1-FLAG-Caf1 D50A |
| YSP 226 | spd1-13myc::ura4+ pRep1-FLAG |
All strains are derivatives of JY746 with the following genotype: h + ade6-M216 leu1-32 ura4-D18.
HU-sensitivity analysis
For spot assays, 5 μl of 10-fold serial dilutions of logarithmically growing cells were spotted onto YE3S plates containing the indicated concentrations of HU and incubated for 2–3 days at 30°C. For complementation analysis, 5 μl of 10-fold serial dilutions of logarithmically growing cells in EMM-leu medium were spotted onto EMM-leu plate containing HU and incubated for 2–3 days at 30°C.
Construction of plasmid and yeast mutants
The plasmid pRep1-His-FLAG was constructed by inserting the double-stranded oligonucleotide, annealed OLI1 (5′-CA TGG ATG ACT GGT CAT CAC CAT CAC CAT CAC GGT GAC TAC AAG GAT GAC GAT GAC AAG GGT CA-3′) and OLI2 (5′-TAT GAC CCT TGT CAT CGT CAT CCT TGT AGT CAC CGT GAT GGT GAT GGT GAT GAC CAG TCA TC-3′) into the NcoI and NdeI site of pRep1. The caf1 and ccr4 ORF was amplified by PCR from an S. pombe cDNA library and inserted into pGEM-T EASY vector (Promega). The plasmid pRep1-His-FLAG-Caf1 was constructed by inserting a sequenced clone of Caf1 in NdeI and SalI site of pRep1-His-FLAG. The plasmid pRep41-HA-Ccr4 was also constructed by inserting a sequenced clone of Ccr4 in SalI site of pRep41-HA. To isolate caf1 mutants, an error-prone PCR method was performed using rTaq polymerase in the presence of 3.0 mM Mg2+, 0.5 mM Mn2+, 0.2 mM dATP, 0.2 mM dGTP, 1.0 mM dCTP, and 1.0 mM dTTP as previously reported (30).
Protein preparation, immunoprecipitation and western blot analyses
Logarithmically growing cells (5 × 108) in the EMM selective medium or YE3S were pelleted, washed once and resuspended in 400 μl of a lysis buffer consisting of 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1 mM dithiothreitol, 0.1% NP-40 and protease inhibitors. The cells were mixed with glass beads (1 g) and disrupted by 12 cycles of vortexing for 30 s followed by incubating on ice for 1 min. The cell extracts were obtained by two consecutive runs of centrifugation (15 000 g for 10 min). Immunoprecipitation of tagged proteins was performed as follows. The extracts (350 μl) were mixed with anti-FLAG antibody conjugated-beads (M2-Agarose-AFFINITY; Sigma) or anti-HA antibody conjugated-beads (Anti-HA Affinity Matrix; Roche) and further incubated on a rotator at 4°C for 2 h. The beads were spun down and washed extensively with the lysis buffer without protease inhibitors. Proteins binding to the beads were eluted with an SDS-PAGE sample buffer by boiling for 5 min. The cell extracts (12.5 μl) and one-third of the eluted fraction were subjected to SDS-PAGE and immunoblotted with anti-Myc (9E10; 1:1000), anti-FLAG (M2; 1:1000), anti-HA (12CA5; 1:1000), anti-tubulin (gifted from K. Gull; 1:3000) monoclonal and anti-Suc22 (generous gift from Dr Masuda; 1:2000) polyclonal antibodies.
Northern blot analysis and the detection of poly(A) tails of total mRNAs
Logarithmically growing cells (5 × 108 cells) in the EMM selective medium or YE3S were removed, spun down and immediately frozen in liquid nitrogen. RNA preparation and northern blot analysis were performed as previously described (31) with minor modification. RNA was separated by 1% agarose-gel electrophoresis and transferred to the Hybond XL (Amersham Pharmacia). suc22 mRNA was detected by northern blotting using a PCR fraction of the ORF. The poly(A) length of total RNAs was analyzed as previously described (32) with some modifications. One microgram of purified total RNA was end-labeled with 5′-[32P] pCp using T4 RNA ligase. The radiolabeled mRNA was digested with RNase A and separated on 12% polyacrylamide–7.5 M urea gel by electrophoresis. The membranes and gels were exposed to phosphorImager screens followed by quantitative analysis using Molecular Dynamics software.
Microscopic analysis
Spd1-13myc and Suc22 staining was performed by the method described previously (33) with some modifications. Logarithmically growing cells (5 × 108 cells) were incubated in YE3S liquid in the presence or absence of 10 mM HU for 2 h. The cells were resuspended in 1 ml of PEM (100 mM PIPES, pH 6.9, 5 mM EGTA and 5 mM MgCl2), fixed with 3.7% para-formaldehyde for 1 h at 30°C, washed and mixed with 10 mg/ml Zymolyase 100T in PEM containing 1.2 M sorbitol for 1 h at 37°C. Spd1-13myc was detected using anti-Myc (9E10) at 1:100 and Alexa488-conjugated anti-mouse antibody (Molecular Probes) at 1:100. Suc22 was detected using anti-Suc22 antibody at 1:100 and Alexa Fluor 488-conjugated anti-rabbit antibody (Molecular Probes). The cells were also counterstained with DAPI to visualize the DNA.
RESULTS
Caf1 and Ccr4 contribute to protect against DNA-replication stress by interacting with RNR-activation pathway
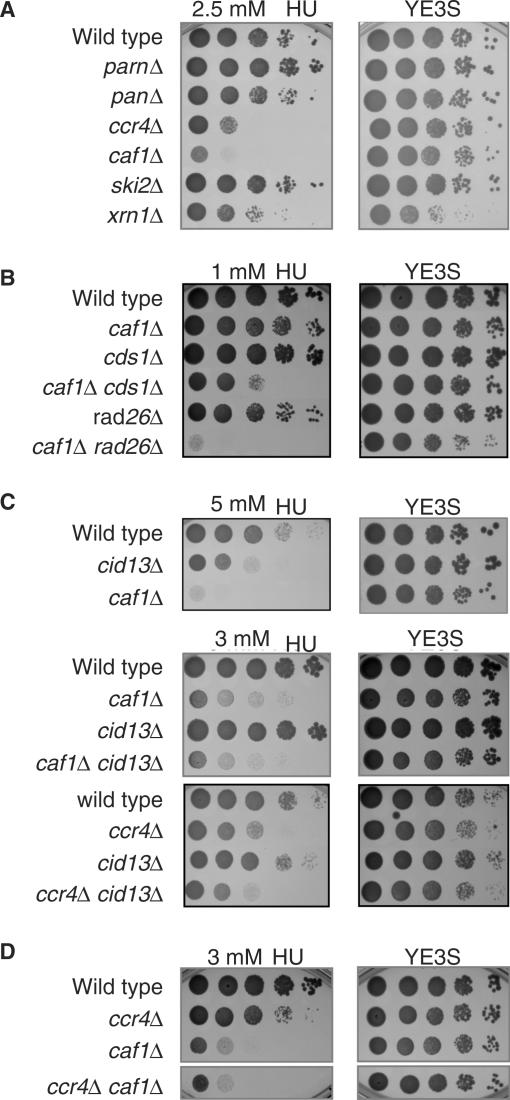
It has recently been reported that several components of mRNA-decay machinery are required for efficient cell-cycle progression after DNA damage and during replication stress in various species including S. cerevisiae (10,11,34). To clarify the importance of the decay machinery toward the stress responses in S. pombe, we initially investigated whether the disruption of genes encoding mRNA-specific exonucleases show high sensitivity to stress stimuli. For the analysis, hydroxyurea (HU), which impairs DNA replication by inhibiting RNR activity (35,36), was used as the stress stimulus. In accordance to previous work in S. cerevisiae, ccr4Δ and caf1Δ cells in fission yeast also exhibited high sensitivity to HU (Figure 1A). In contrast, the deletion of other possible deadenylases (Parn and Pan2) or a component of the 3′–5′ helicase (Ski2) did not impair the cell growth with HU, though cells lacking the 5′-exonuclease Xrn1 exhibited non-selective growth defect under the present conditions (37). Thus, among various mRNA-specific exonucleases, Caf1 and Ccr4 appear to have unique properties in terms of recovery from the DNA-replication stress.
Figure 1.
Caf1 and Ccr4 contribute to protect against DNA-replication stress by interacting with RNR-activation pathway. Ten-fold serial dilutions of logarithmically growing cells, wild type (YSP001) parnΔ (YSP004), pan2Δ (YSP003), ccr4Δ (YSP002), caf1Δ (YSP066), ski2Δ (YSP143) and xrn1Δ (YSP172) for (A), wild type (YSP001), caf1Δ (YSP066), cds1Δ (YSP069), caf1Δ cds1Δ (YSP103), rad26Δ (YSP071) and caf1Δ rad26Δ (YSP102) for (B), wild type (YSP001), cid13Δ (YSP086), caf1Δ (YSP066), caf1Δ cid13Δ (YSP095), ccr4Δ (YSP002) and ccr4Δ cid13Δ (YSP097) for (C) and ccr4Δ (YSP002), caf1Δ (YSP066) and ccr4Δ caf1Δ (YSP104) for (D) were spotted on YE3S plates containing the indicated concentrations of HU. The plates were photographed after 2 or 3 days for growth at 30°C.
Various pathways are activated in response to HU, including DNA-structure checkpoint and synthesis of dNTPs by RNR activation. To test whether Caf1 and Ccr4 are involved in these pathways, we carried out genetic interaction experiments. Double mutants lacking Caf1 or Ccr4 and well-characterized components of the above pathways were constructed and analyzed for their sensitivity to HU. As shown in Figure 1B, double mutants, caf1Δ with additional deletion of the checkpoint gene CDS1 (cds1Δ) or RAD26 (rad26Δ), showed marked increases in sensitivity to HU compared to either single mutant. Such synergistic phenotypes were also observed in double mutants of ccr4Δ and cds1Δ or rad26Δ (data not shown), suggesting that Caf1 and Ccr4 might act in a pathway(s) distinct from the DNA checkpoint. The possible involvement of Caf1 and Ccr4 in the dNTP-synthesis pathway was next investigated. We compared the sensitivity of caf1Δ cid13Δor ccr4Δ cid13Δ double mutant with the respective single mutants, since HU inhibits RNR consisting of Suc22 and Cdc22 in the fission yeast and stimulates the poly-adenylation by the cytoplasmic poly(A) polymerase Cid13 of specific mRNAs coding stress-inducible genes, such as 1.9-kb suc22 mRNA (23). As shown in Figure 1C, the sensitivity of these double mutant cells was equivalent to the single caf1Δ or ccr4Δ mutant, suggesting that Caf1 and Ccr4 act potentially in a step of the RNA activation pathway distal to Cid13. Although the phenotypes observed between caf1Δ and ccr4Δ cells were quite similar to each other, there was no synergistic enhancement of HU sensitivity in caf1Δ ccr4Δ double mutant cells (Figure 1D). Thus, both Caf1 and Ccr4 appear to contribute to protect against the HU-induced replication stress probably through the stabilization or translocation of a component(s) of the RNR complex.
Isolation of Caf1 mutants that have a defect in HU-induced stress response
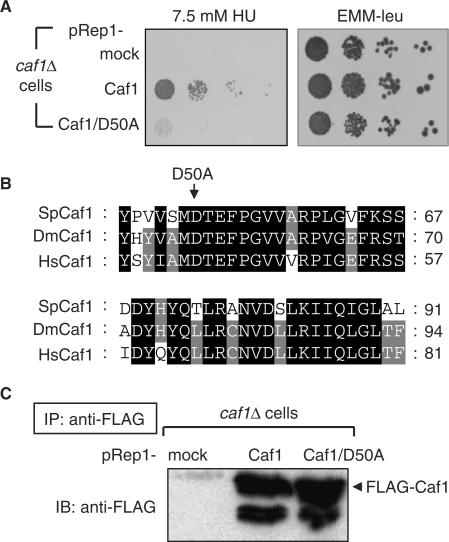
To better understand what biochemical properties of Caf1 and Ccr4 are relevant to the high sensitivity to HU stress, we focused on Caf1 showing more potent phenotype and isolated its point mutants by using the error-PCR method. One allele, which has a D50A mutation, was isolated as a mutant lacking the high sensitivity to the replication stress (Figure 2A), and this mutation is located in a region of Caf1 that is highly conserved from yeast to human (Figure 2B). Interestingly, the D50 residue is one of two residues known to be involved in the catalytic action of other members of the RNase D family and has been shown to be critical for both S. cerevisiae and mammalian Caf1 deadenylase activities in vitro (38–40). To examine the expression of this mutant protein, immunoprecipitation and western-blotting assays were performed. The protein level of FLAG-Caf1 under the nmt1 promoter was almost the same between the mutant and wild-type cells, indicating that the D50A mutation does not disrupt the stability of Caf1 (Figure 2C). As will be described below, this Caf1 mutant exhibits unique biochemical properties in the HU-induced RNR pathway.
Figure 2.
Isolation of Caf1 mutants that have defects in the HU-induced stress response. (A) Ten-fold serial dilutions of logarithmically growing cells, caf1Δ pRep1 (YSP188), caf1Δ pRep1-Caf1 (YSP189) and caf1Δ pRep1-Caf1/D50A (YSP190), were spotted on YE3S plates containing 7.5 mM HU and EMM-leu. (B) Sequence alignment of CAF1 was performed with Clustal W: the most conserved amino acid residues are highlighted with a black background. Sp, Schizosaccharomyces pombe; Dm, Drosophila melanogaster; Hs, Homo sapiens. (C) Lysates of logarithmically growing cells (YSP188, YSP189 and YSP190) were immunoprecipitated with anti-FLAG beads, and the precipitated fractions were subjected to SDS-PAGE and immunoblotted with anti-FLAG for caf1-FLAG.
Neither increase of 1.9-kb suc22 mRNA level nor deadenylation of mRNA poly(A) tails by Caf1 contributes to protect against the HU-induced replication stress
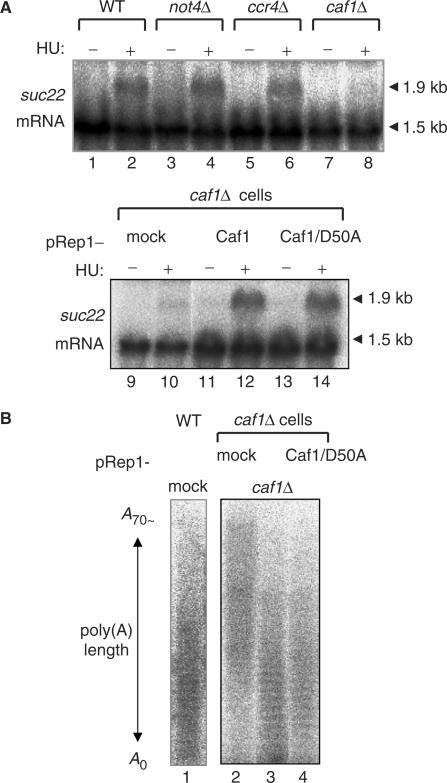
In S. cerevisiae, mutations of Ccr4–Not complex impair the transcription of mRNAs coding RNR genes in response to HU (13). In accordance to previous work (41), HU induced the 1.9-kb suc22 mRNA in S. pombe wild-type cells (Figure 3A, lane 2). The increase of suc22 mRNA level was still observed in not4Δ (lane 4) and ccr4Δ (lane 6) cells, but it was totally abolished in caf1Δ cells (lane 8). Moreover, the defect of suc22 mRNA increase in caf1Δ cells was compensated not only by the introduction of wild-type Caf1 (lane 12) but also by the Caf1/D50A mutant (lane 14). It thus appeared that Caf1/D50A mutation is still capable of inducing the suc22 mRNA and that the ability of Caf1 to induce the 1.9-kb suc22 mRNA is not responsible for the HU-high sensitivity observed in caf1Δ and ccr4Δ cells.
Figure 3.
Neither increase of 1.9-kb suc22 mRNA level nor deadenylation of mRNA poly(A) tails by Caf1 contributes to the HU-induced stress-response pathway. (A) Logarithmically growing cells, wild type (YSP001), not4Δ (YSP131), ccr4Δ (YSP002), caf1Δ (YSP066), caf1Δ pRep1 (YSP188), caf1Δ pRep1-Caf1 (YSP189) and caf1Δ pRep1-Caf1/D50A (YSP190), were incubated in the presence (+) and absence (−) of 10 mM HU for 2 h at 30°C. RNA preparation and northern blotting analysis were performed as described in the Materials and Methods section. Membranes were exposed to phosphorImager screens and followed by the quantitative analysis of suc22 mRNAs using Molecular Dynamics software. (B) Total RNA (1 μg) were purified from the above cells (YSP027, YSP188, YSP189 and YSP190) and end-labeled with 5′-[32P] pCp using T4 RNA ligase. The radiolabeled mRNAs were digested with RNase A and separated with 12% polyacrylamide–7.5 M urea gel electrophoresis.
In S. cerevisiae, Caf1/Pop2 has been reported as a cofactor of Ccr4 deadenylase. To examine the deadenylase activity in caf1Δ cells, the poly(A)-tail regions of total mRNAs were isolated, and the length of poly(A) tails was examined. As shown in Figure 3B, longer poly(A) tails were observed in caf1Δ cells (lane 2) compared to wild-type cells (lane 1). The poly(A) length was much shortened by the introduction of wild-type Caf1 into the mutant cells (lane 3). Interestingly, the Caf1/D50A mutant was also capable of shortening the poly(A) length (lane 4), suggesting that the deadenylation-supportive activity of Caf1 is not involved in the HU-induced stress-response pathway. Altogether, these results raise the possibility that the Caf1/D50A mutation disrupts a cryptic biochemical activity of Caf1 that is distinct from the previously identified properties, increase of suc22 mRNA level or deadenylation.
Caf1 is required for HU-induced cytoplasmic translocation of Suc22
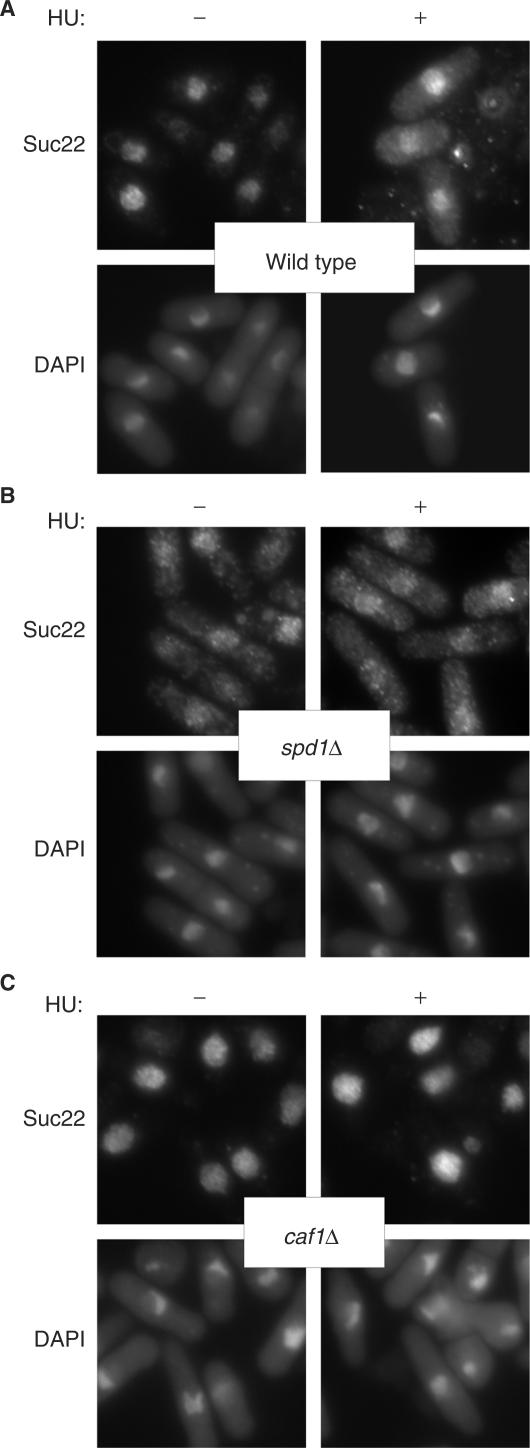
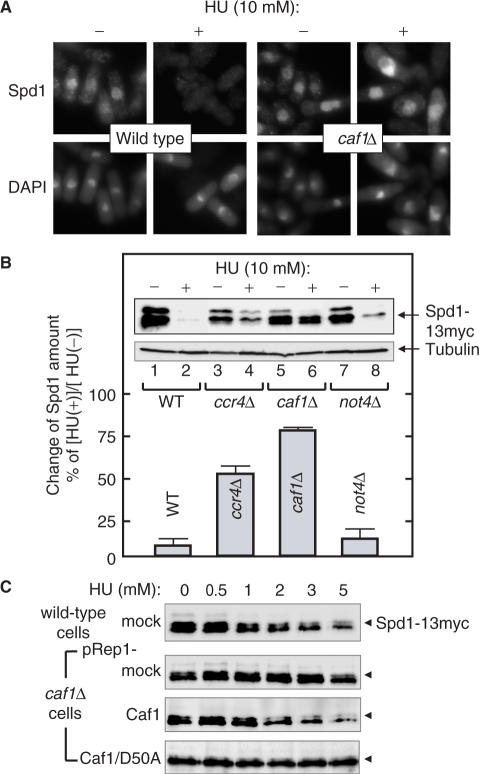
The RNR activity is stimulated through the interaction of Suc22 with Cdc22 in the cytoplasm. However, translocation of Suc22 into cytoplasm is inhibited by nuclear Spd1 through the formation of a nucleoplasmic Spd1–Suc22 assembly. In S phase, degradation of Spd1 via the Pcu4–Ddb1–CSN complex leads Suc22 to be exported to cytoplasm (27). To explore the undescribed function of Caf1 in HU-induced stress response, we next investigated subcellular localization of the components of RNR pathway (Figure 4). In the absence of the replication stress, Suc22 was localized in the nucleoplasm of both wild-type and caf1Δ cells. In contrast, Suc22 dispersed into the cytoplasm in spd1Δ cells is probably due to the loss of the ability to retain the nucleoplasmic Suc22. After HU treatment, the cytoplasmic signal of Suc22 was detected in wild type cells. However, the localization of Suc22 was not altered in caf1Δ cells even after the HU treatment. These results indicate that Caf1 is required for HU-induced cytoplasmic translocation of Suc22.
Figure 4.
Caf1 is required for the cytoplasmic translocation of Suc22 in response to DNA-replication stress. Logarithmically growing cells, wild type (A, YSP001), spd1Δ (B, YSP112) and caf1Δ (C, YSP066), were incubated in the presence and absence of 10 mM HU for 2 h at 30°C. Immunofluorescence-staining images of Suc22 were obtained with an anti-Suc22 antibody.
Caf1 and Ccr4 are required for HU-induced nucleoplasmic reduction of Spd1
The data presented above predict that Caf1 is required for the cytoplasmic translocation of Suc22 in response to the replication stress by regulating the localization or the degradation of Spd1. To examine these possibilities, we investigated subcellular localization of Spd1 after HU treatment in wild-type and caf1Δ cells (Figure 5A). Under no-stress conditions, Spd1 was localized in the nucleoplasm of both cells. In response to HU, the signal of Spd1 disappeared from the nucleoplasm in wild-type cells, but was still present in caf1Δ cells. Thus, the stress-induced Spd1 degradation is impaired in caf1Δ cells. To confirm the role of Caf1 in the nucleoplasmic reduction of Spd1, we determined the quantity of Spd1 in various cells before and after HU treatment. The expression level of spd1 mRNA was almost unchanged among these cells (data not shown). As shown in Figure 5B, Spd1 was detected as multiple bands in untreated wild-type cells (lane 1), and these bands disappeared almost completely after incubation of the cells with 10 mM HU (lane 2). Figure 5C (first panel) shows the concentration-dependent effect of HU: there was a progressive decrease in the amount of Spd1, as the concentration of HU was increased (0–5 mM). The HU treatment also reduced the amount of Spd1 in not4Δ cells (Figure 5B, lane 8). However, the HU-induced reduction of Spd1 was markedly inhibited in caf1Δ cells (lane 6). Such inhibition was also observed in ccr4Δ cells to a lesser extent (lane 4), and this may be related to the observation that ccr4Δ cells were less sensitive to HU (Figure 1A). The HU-induced reduction of Spd1 in wild-type or not4Δ cells and its diminishment in caf1Δ or ccr4Δ cells were still apparently observed in the presence of a protein synthesis inhibitor (50 μg/ml cycloheximide, data not shown). This excludes the possibility that the reduction of Spd1 might be due to changes in de novo protein synthesis of Spd1.
Figure 5.
Caf1 is required for HU-induced degradation of Spd1. (A) Logarithmically growing cells, spd1-13Myc (YSP091) and caf1Δ spd1-13Myc (YSP161) were incubated in the presence and absence of 10 mM HU for 2 h at 30°C. Immunofluorescence-staining images of Spd1 were obtained with an anti-Myc antibody. (B) Logarithmically growing cells, spd1-13Myc (YSP091), ccr4Δ spd1-13Myc (YSP154), caf1Δ spd1-13Myc (YSP161) and not4Δ spd1-13Myc (YSP162), were incubated in the presence (+) and absence (−) of 10 mM HU for 2 h. Extracts from the cells were blotted with an anti-tubulin antibody as an internal control. The extracts were also incubated with anti-Myc antibody, and the immunoprecipitated fractions were subjected to western blot analysis for the detection of Spd1. Three independent experiments were performed, and the HU-induced reduction of the Spd1 bands was quantitated by an imaging analyzer LAS1000. The results of one representative set are also shown in the inset. (C) Logarithmically growing cells, spd1-13Myc pRep1 (YSP226), caf1Δ spd1-13Myc pRep1 (YSP195), caf1Δ spd1-13Myc pRep1-Caf1 (YSP196) and caf1Δ spd1-13Myc pRep1-Caf1/D50A (YSP197), were incubated with the indicated concentrations of HU for 2 h at 30°C, and Spd1 was detected as described in (B).
We next investigated whether the defect of Spd1 reduction in caf1Δ cells is compensated with the wild type and D50A mutant of Caf1 (Figure 5C). As expected, HU-induced Spd1 reduction was clearly restored by the expression of wild-type Caf1 under the nmt1-promoter (third panel). In contrast, the Caf1/D50A mutant failed to promote the reduction of Spd1 (fourth panel). Since the Caf1/D50A mutant still retains the ability to induce 1.9-kb suc22 mRNA and shorten poly(A) tails (Figure 3), the additional biochemical activity of Caf1 is likely to be involved in the Spd1 turnover. Thus, this novel activity defective in Caf1/D50A appears to be responsible for the mutant phenotype that lacks the ability to complement HU-induced growth defect. Taken together, these observations further reinforce the notion that Caf1 and Ccr4 are involved in the HU-induced degradation of Spd1 leading to the activation of the RNR pathway.
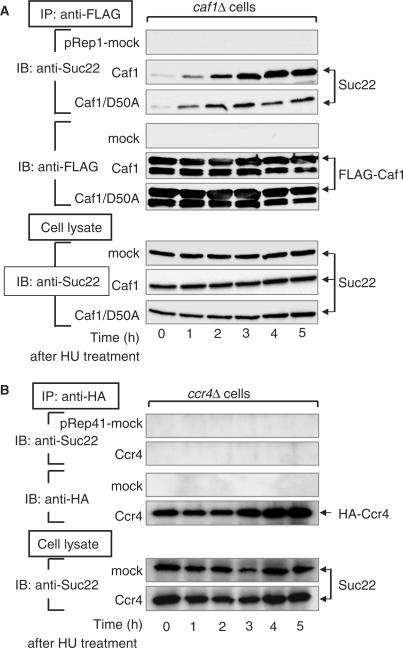
HU stress allows Caf1 to interact with Suc22 in the RNR pathway
To further investigate the role of Caf1 and Ccr4 in the degradation of Spd1, we examined whether Caf1 and/or Ccr4 are capable of interacting physically with the RNR component Suc22. When FLAG-tagged Caf1 that had been expressed in caf1Δ cells was immunoprecipitated with an anti-FLAG antibody, no apparent band of Suc22 was observed in the Caf1-immunoprecipitated fraction (Figure 6A, 0-h time). However, Suc22 appeared to interact with Caf1 when the cells had been incubated with HU. The amount of Suc22 that interacted with Caf1 increased progressively with the incubation times and reached a plateau at 4-h incubation, though total protein levels of Suc22 and Caf1 in the cell lysates were constant during the incubation. The same experiments were performed with HA-tagged Ccr4 that had been expressed in ccr4Δ cells. However, Suc22 was not observed in the Ccr4-immunoprecipitated fraction after HU treatment (Figure 6B). The physical association observed between Suc22 and Caf1 was not due to an artifact resulting from aggregation of the immunoprecipitated materials, since the immunoprecipitated fractions exhibited essentially the same patterns of protein distribution upon SDS-PAGE analysis (data not shown). These results indicate that there is a separation of Suc22 and Caf1 in the absence of HU stress and that HU stress allows Caf1 to interact with Suc22. Interestingly, when the Caf1/D50A mutant had been expressed in caf1Δ cells, HU-dependent interaction of the mutant Caf1 with Suc22 was markedly reduced compared with wild-type Caf1 (Figure 6A). We thus conclude that Caf1, in concert with Ccr4, induces the Spd1–Suc22 disassembly to stimulate dNTP synthesis in the recovery from DNA-replication stress. Although more-detailed biochemical analysis is required, it is very likely that Caf1 tightly associates with Suc22 to release and/or degrade the inhibitory Spd1 in a manner dependent on Ccr4 and that this novel Caf1 function is impaired by D50A mutation.
Figure 6.
HU stress allows Caf1 to interact with Suc22 in the RNR pathway. Logarithmically growing cells, caf1Δ pRep1 (YSP188), caf1Δ pRep1-Caf1 (YSP189) and caf1Δ pRep1-Caf1/D50A (YSP190) for (A) and ccr4Δ pRep41 (YSP184) and ccr4Δ pRep41-Ccr4 (YSP185) for (B), were incubated with 10 mM HU for the indicated times at 30°C. Extracts from the cells were incubated with an anti-FLAG (A) or an anti-HA (B) antibody, and the immunoprecipitated fractions were subjected to western-blot analysis for the detection of Suc22 and Caf1 or Ccr4 as described in the Materials and Methods section. The whole cell lysates were also subjected to western blot analysis for the detection of Suc22.
DISCUSSION
We have shown that Caf1, a component of the Ccr4–Not complex, plays an important role in the mechanisms for resistance to DNA-replication stress and in the proper supply of dNTPs to maintain genome stability and DNA replication in S. pombe. Although Caf1 has been characterized as a multi-functional component that is largely involved in the transcription of RNR genes and the regulation of Ccr4 deadenylase, the defects observed in the present caf1Δ cells are not solely explicable by either of the two major pathways. Caf1 deletion impairs not only the stress-induced translocation of Suc22 into cytoplasm but also the nucleoplasmic degradation of Spd1. The Caf1/D50A mutant isolated in the present study also reinforces the novel role of Caf1 in the RNR pathway. The Caf1 mutant, that fails to restore the growth-defect phenotype of caf1Δ cells, still has the ability to increase 1.9-kb suc22 mRNA level and shorten poly(A) tails as the wild type. However, the Caf1 ability to support the stress-induced degradation of Spd1 is completely abolished by the D50A mutation. Taken together, interaction of Caf1 with Suc22 leading to Spd1–Suc22 disassembly is necessary for the protection against DNA-replication stress.
Caf1 as a multi-functional component of the Ccr4–Not complex
Each component of the Ccr4–Not complex has its own biochemical role(s) and seems to coordinate the activity of other components. In addition, some components confer distinct biochemical functions independently from the whole complex. For examples, Ccr4–Caf1 (Pop2) and Not4 have deadenylation and ubiquitination activities, respectively (42–44). This is also true in stress responses and in Spd1 degradation. We observed that ccr4Δ and caf1Δ cells show high sensitivity to HU (Figure 1A), though not4Δ cells do not (data not shown). The HU-induced degradation of Spd1 was markedly impaired in ccr4Δ and caf1Δ cells, though Spd1 was degraded normally in not4Δ cells (Figure 5B). However, Caf1 was required for the increase of 1.9-kb suc22 level, while Ccr4 and Not4 were not (Figure 3A). Suc22 appeared to interact with Caf1, not with Ccr4 after HU treatment (Figure 6). It is interesting that distinct components in the complex are required for the respective biochemical reactions even though they are all triggered by the same reagent, HU. The Ccr4–Not complex has been found in several species as a large complex, 1 or 1.9 MDa, in addition to their constituent subunits (5,6). The spectrum of the Ccr4–Not components associated with the platform protein Not1 may be altered rapidly in response to external conditions as well as cell-cycle progression. The function of each member is possibly distinct and drastically dependent on their constituent members and/or the complex conformation. One possible model is that Caf1 in the context of the Ccr4–Not complex receives a signal following HU stress and Caf1 is then released to bind Suc22. Characterization of components in each form of the Ccr4–Not complex should help to explain their diverse biochemical versatility.
As described above, one of the outstanding features in caf1Δ cells is the lack of the HU-dependent increase of 1.9-kb suc22 mRNA level (Figure 3A). It has been shown that Cid13 stabilizes the suc22 mRNA by its poly-adenylation upon DNA-replication stress and stimulates RNR activity (23). We have shown that caf1Δ cid13Δ double mutant cells are not synergistically sensitive to HU. This indicates that Caf1 and Cid13 act potentially in the same pathway initiated by DNA-replication stress. The key to the non-synthetic phenotype in caf1Δ and cid13Δ cells is the increase of 1.9-kb suc22 mRNA level. Therefore, Suc22 mRNA may be first induced by Caf1, and its poly-adenylation subsequently takes place by Cid13. Alternatively, Caf1 may poly-adenylate 1.9-kb suc22 mRNA cooperatively with Cid13. Since there are many other genes that are regulated by Caf1 upon DNA-replication stress, Caf1 may induce and stabilize stress-response genes in cooperation with Cid13. Thus, the stabilization of stress-induced mRNAs closely correlates with the absence of phenotypic enhancement in caf1Δ cid13Δ double mutant cells.
Proper dNTP pools ensure genome stability and DNA replication
It has been shown that dNTP pools are elevated upon DNA damage, for example 7-fold in S. cerevisiae and 2-fold in S. pombe (26,45). However, excessive dNTP pools cause genomic mutation (46), and reduced dNTP pools affect genome stability (45,47). In mammalian cells, constitutive RNR activation through inhibition of the RB pathway transforms cells into carcinoma (48). Moreover, dNTP imbalances also induce increased frequency of replication frame-shift mutations to a significant extent (46,49). These data indicate that tight control of dNTP pools is necessary to maintain complete fidelity of DNA replication. Similarly, the increase of unavailable nucleotides or the intermediates are also harmful to the regulation of genome stability and induce aberrant DNA replication. Indeed, we have observed the accumulation of abnormal nucleotides in caf1Δ cells under logarithmic growing conditions and a slight defect in the cell proliferation (data not shown). Thus, Caf1 may have an important role in the maintenance of proper dNTP pools upon DNA damage via its interaction with Suc22.
Possible roles of Caf1 in promotion of Spd1–Suc22 disassembly
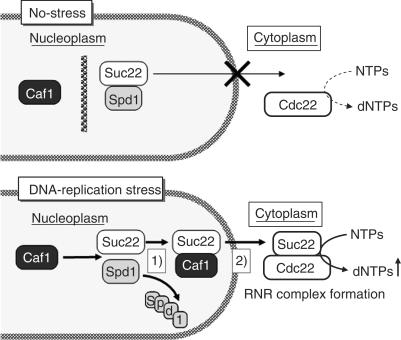
As discussed above, the RNR activity is regulated not only by the transcription of their constituent subunits but also by the subcellular translocation of the regulated subunits. Our present results address the mechanism of this translocation (Figure 7). There may be at least two possible mechanisms, by which Caf1 promotes the Spd1–Suc22 disassembly that involves the degradation of Spd1 (scheme 1) and the translocation of Suc22 from nucleoplasm to cytoplasm (scheme 2). First, Caf1 interacts with Suc22 in a manner competitive with Spd1, which facilitates the Pcu4–Ddb1–CSN-induced degradation of Spd1 and the release of Suc22 into cytoplasm. Thus, Caf1 may have a transporter role in the Suc22 translocation. Alternatively, Caf1 may function as an activator protein associating with a component(s) of the Pcu4–Ddb1–CSN complex to enhance its degradation activity. These two models are not mutually exclusive. Our findings that Caf1 progressively associates with Suc22 after HU treatment and that the stress-induced Spd1 degradation requires Caf1 may be consistent with the first transporter model. On the other hand, a physical interaction analysis shows that Caf1 interacts with Csi1, a subunit of the CSN complex (50). Moreover, we detected a possible interaction between Caf1 and Csn1 by immunoprecipitation assay (data not shown). Immunofluorescence observations revealed that Caf1 localizes both in the nucleoplasm and in the cytoplasm and that HU exposure does not apparently alter the Caf1 localization. These observations may support the second activator model.
Figure 7.
Possible models for the DNA-replication stress-induced translocation of Suc22. Under no-stress conditions, there is a compartment between Caf1 and the Suc22–Spd1 complex in the nucleoplasm. When cells sense DNA-replication stress, Caf1 interacts with Suc22 to facilitate the degradation of Spd1 (scheme 1) and the translocation of Suc22 from nucleoplasm to cytoplasm (scheme 2). See Discussion for further explanation.
Each organism may have evolved distinct mechanisms to localize the regulatory subunits of RNR. In mammalian cells, the regulatory subunit p53R2 localizes in the cytoplasm under normal conditions and relocates to the nucleoplasm in response to DNA damage (51). However, in yeast cells, the regulatory subunits (Suc22 or Rnr2–Rnr4 complex) localize in the nucleoplasm under the standard conditions and enter the cytoplasm upon DNA damage. The apparent reason for the differential translocation mechanisms between yeast and mammals remains unknown. In this regard, Lee and Elledge point out that the different amounts of DNA have to be replicated between the two species (52). Consistently, the efficiency of replication rate is possibly important. The balance between the length of S phase (approximately 40 min and 8 h for yeast and mammal, respectively) and the DNA amounts to be replicated (mammalian genome is 250 times longer than yeast) suggests that mammalian cells replicate DNA about 20 times faster than yeast. We speculate that the efficiency may explain the difference in translocation mechanism.
Mechanisms of nucleoplasmic anchoring of RNR regulatory subunits in yeast
It has been recently reported in S. cerevisiae that Wtm1, containing WD40 repeats, functions to anchor the regulatory subunit Rnr2–Rnr4 in the nucleoplasm (52,53). It is not known whether a WD40-dependent anchoring system is also conserved in S. pombe. In this regard, one of the Caf1-interacting proteins, Caf4, has a WD40 repeat, although Caf1 itself does not. It is thus tempting to speculate that Caf4 anchors Suc22 in the nucleoplasm to inhibit the cytoplasmic release of the regulatory subunit under no-stress conditions. Alternatively, such a WD40-containing protein may be involved in the cytoplasmic translocation of Suc22 and/or the recruitment of Spd1 to the Pcu4–Ddb1–CSN complex in S. pombe. A WD40 repeat-containing protein, Cdt2, which regulates the degradation of Spd1 via Pcu4–Ddb1–CSN ubiquitin ligase, might be one of the potential candidates (54). It would be interesting to determine whether WD40-dependent mechanisms are conserved across species, and whether there is a cytoplasmic anchoring protein for the regulatory subunit p53R2 in mammals. The understanding of RNR translocation mechanisms will help to explain the precise timing of the elevation of dNTP flows for DNA replication. These important issues are currently under investigation in our laboratory.
ACKNOWLEDGEMENTS
We thank Dr Eric Witze (University of Colorado), Dr Masamitsu Fukuyama and Satoshi Kofuji in this laboratory for their comments and charitable supports for this report. We are most grateful to Drs Hirohisa Masuda and Yasushi Hiraoka (KARC, NiCT) for generous gift of anti-Suc22 antibody. This work was supported in part by research grants from the Scientific Research Funds of the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government. Funding agency is the Scientific Research Funds of the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government and Japan Society for the Promotion of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Collart M. Global control of gene expression in yeast by the Ccr4-Not complex. Gene. 2003;313:1–16. doi: 10.1016/s0378-1119(03)00672-3. [DOI] [PubMed] [Google Scholar]
- 2.Denis C, Chen J. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 2003;73:221–250. doi: 10.1016/s0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- 3.Denis CL. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics. 1984;108:833–844. doi: 10.1093/genetics/108.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collart MA, Struhl K. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 1994;8:525–537. doi: 10.1101/gad.8.5.525. [DOI] [PubMed] [Google Scholar]
- 5.Liu HY, Badarinarayana V, Audino DC, Rappsilber J, Mann M, Denis CL. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17:1096–1106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol. Cell. Biol. 1999;19:6642–6651. doi: 10.1128/mcb.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 8.Moser MJ, Holley WR, Chatterjee A, Mian IS. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daugeron MC, Mauxion F, Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett CB, Lewis LK, Karthikeyan G, Lobachev KS, Jin YH, Sterling JF, Snipe JR, Resnick MA. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 2001;29:426–434. doi: 10.1038/ng778. [DOI] [PubMed] [Google Scholar]
- 11.Hanway D, Chin JK, Xia G, Oshiro G, Winzeler EA, Romesberg FE. Previously uncharacterized genes in the UV- and MMS-induced DNA damage response in yeast. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10605–10610. doi: 10.1073/pnas.152264899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Traven A, Hammet A, Tenis N, Denis CL, Heierhorst J. Ccr4-not complex mRNA deadenylase activity contributes to DNA damage responses in Saccharomyces cerevisiae. Genetics. 2005;169:65–75. doi: 10.1534/genetics.104.030940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulder KW, Winkler GS, Timmers HT. DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4-Not complex. Nucleic Acids Res. 2005;33:6384–6392. doi: 10.1093/nar/gki938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murakami H, Nurse P. DNA replication and damage checkpoints and meiotic cell cycle controls in the fission and budding yeasts. Biochem. J. 2000;349:1–12. doi: 10.1042/0264-6021:3490001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osborn AJ, Elledge SJ, Zou L. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 2002;12:509–516. doi: 10.1016/s0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- 16.Lambert S, Watson A, Sheedy DM, Martin B, Carr AM. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 17.Al-Khodairy F, Fotou E, Sheldrick KS, Griffiths DJ, Lehmann AR, Carr AM. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol. Biol. Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unsal-Kacmaz K, Makhov AM, Griffith JD, Sancar A. Preferential binding of ATR protein to UV-damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6673–6678. doi: 10.1073/pnas.102167799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boddy MN, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 20.Weinberg G, Ullman B, Martin DW., Jr Mutator phenotypes in mammalian cell mutants with distinct biochemical defects and abnormal deoxyribonucleoside triphosphate pools. Proc. Natl. Acad. Sci. U.S.A. 1981;78:2447–2451. doi: 10.1073/pnas.78.4.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunz BA, Kohalmi SE, Kunkel TA, Mathews CK, McIntosh EM, Reidy JA. International commission for protection against environmental mutagens and carcinogens. deoxyribonucleoside triphosphate levels: a critical factor in the maintenance of genetic stability. Mutat. Res. 1994;318:1–64. doi: 10.1016/0165-1110(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 22.Meuth M. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp. Cell Res. 1989;181:305–316. doi: 10.1016/0014-4827(89)90090-6. [DOI] [PubMed] [Google Scholar]
- 23.Saitoh S, Chabes A, McDonald WH, Thelander L, Yates JR, Russell P. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell. 2002;109:563–573. doi: 10.1016/s0092-8674(02)00753-5. [DOI] [PubMed] [Google Scholar]
- 24.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 25.Chabes A, Domkin V, Thelander L. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem. 1999;274:36679–36683. doi: 10.1074/jbc.274.51.36679. [DOI] [PubMed] [Google Scholar]
- 26.Hakansson P, Dahl L, Chilkova O, Domkin V, Thelander L. The Schizosaccharomyces pombe replication inhibitor Spd1 regulates ribonucleotide reductase activity and dNTPs by binding to the large Cdc22 subunit. J. Biol. Chem. 2006;281:1778–1783. doi: 10.1074/jbc.M511716200. [DOI] [PubMed] [Google Scholar]
- 27.Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev. 2003;17:1130–1140. doi: 10.1101/gad.1090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno A, Navarro E, Senent F, Baeza A, Miro C, del Rio M. Short and medium effects on the environment of Valencia, Spain, of the Chernobyl nuclear plant accident. Bull. Environ. Contam. Toxicol. 1991;46:14–21. doi: 10.1007/BF01688249. [DOI] [PubMed] [Google Scholar]
- 29.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 30.Oki M, Noguchi E, Hayashi N, Nishimoto T. Nuclear protein import, but not mRNA export, is defective in all Saccharomyces cerevisiae mutants that produce temperature-sensitive forms of the Ran GTPase homologue Gsp1p. Mol. Gen. Genet. 1998;257:624–634. doi: 10.1007/s004380050690. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi S, Araki Y, Sakuno T, Katada T. Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′-to-5′ mRNA decay in yeast. EMBO J. 2003;22:3951–3959. doi: 10.1093/emboj/cdg374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sachs AB, Davis RW. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 33.Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- 34.Westmoreland TJ, Marks JR, Olson JA, Jr, Thompson EM, Resnick MA, Bennett CB. Cell cycle progression in G1 and S phases is CCR4 dependent following ionizing radiation or replication stress in Saccharomyces cerevisiae. Eukaryot. Cell.z. 2004;3:430–446. doi: 10.1128/EC.3.2.430-446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krakoff IH, Brown NC, Reichard P. Inhibition of ribonucleoside diphosphate reductase by hydroxyurea. Cancer Res. 1968;28:1559–1565. [PubMed] [Google Scholar]
- 36.Wang PJ, Chabes A, Casagrande R, Tian XC, Thelander L, Huffaker TC. Rnr4p, a novel ribonucleotide reductase small-subunit protein. Mol. Cell. Biol. 1997;17:6114–6121. doi: 10.1128/mcb.17.10.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szankasi P, Smith GR. Requirement of S. pombe exonuclease II, a homologue of S. cerevisiae Sep1, for normal mitotic growth and viability. Curr. Genet. 1996;30:284–293. doi: 10.1007/s002940050134. [DOI] [PubMed] [Google Scholar]
- 38.Thore S, Mauxion F, Seraphin B, Suck D. X-ray structure and activity of the yeast Pop2 protein: a nuclease subunit of the mRNA deadenylase complex. EMBO Rep. 2003;4:1150–1155. doi: 10.1038/sj.embor.7400020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viswanathan P, Ohn T, Chiang, yc, Chen J, Denis CL. Mouse CAF1 can function as a processive deadenylase/3′-5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J. Biol. Chem. 2004;279:23988–23995. doi: 10.1074/jbc.M402803200. [DOI] [PubMed] [Google Scholar]
- 40.Bianchin C, Mauxion F, Sentis S, Seraphin B, Corbo L. Conservation of the deadenylase activity of proteins of the Caf1 family in human. RNA. 2005;11:487–494. doi: 10.1261/rna.7135305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris P, Kersey PJ, McInerny CJ, Fantes PA. Cell cycle, DNA damage and heat shock regulate suc22+ expression in fission yeast. Mol. Gen. Genet. 1996;252:284–291. doi: 10.1007/BF02173774. [DOI] [PubMed] [Google Scholar]
- 42.Chen J, Chiang YC, Denis CL. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albert TK, Hanzawa H, Legtenberg YI, de Ruwe MJ, van den Heuvel FA, Collart MA, Boelens R, Timmers HT. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 2002;21:355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 46.Bebenek K, Roberts JD, Kunkel TA. The effects of dNTP pool imbalances on frameshift fidelity during DNA replication. J. Biol. Chem. 1992;267:3589–3596. [PubMed] [Google Scholar]
- 47.Holmberg C, Fleck O, Hansen HA, Liu C, Slaaby R, Carr AM, Nielsen O. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev. 2005;19:853–862. doi: 10.1101/gad.329905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Angus SP, Wheeler LJ, Ranmal SA, Zhang X, Markey MP, Mathews CK, Knudsen ES. Retinoblastoma tumor suppressor targets dNTP metabolism to regulate DNA replication. J. Biol. Chem. 2002;277:44376–44384. doi: 10.1074/jbc.M205911200. [DOI] [PubMed] [Google Scholar]
- 49.Lu Q, Zhang X, Almaula N, Mathews CK, Inouye M. The gene for nucleoside diphosphate kinase functions as a mutator gene in Escherichia coli. J. Mol. Biol. 1995;254:337–341. doi: 10.1006/jmbi.1995.0620. [DOI] [PubMed] [Google Scholar]
- 50.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 52.Lee YD, Elledge SJ. Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes Dev. 2006;20:334–344. doi: 10.1101/gad.1380506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Z, An X, Yang K, Perlstein DL, Hicks L, Kelleher N, Stubbe J, Huang M. Nuclear localization of the Saccharomyces cerevisiae ribonucleotide reductase small subunit requires a karyopherin and a WD40 repeat protein. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1422–1427. doi: 10.1073/pnas.0510516103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu C, Poitelea M, Watson A, Yoshida SH, Shimoda C, Holmberg C, Nielsen O, Carr AM. Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4-Ddb1-CSN ubiquitin ligase. EMBO J. 2005;24:3940–3951. doi: 10.1038/sj.emboj.7600854. [DOI] [PMC free article] [PubMed] [Google Scholar]