Abstract
There is an emerging consensus that secondary structures of DNA have the potential for genomic instability. Palindromic AT-rich repeats (PATRRs) are a characteristic sequence identified at each breakpoint of the recurrent constitutional t(11;22) and t(17;22) translocations in humans, named PATRR22 (∼600 bp), PATRR11 (∼450 bp) and PATRR17 (∼190 bp). The secondary structure-forming propensity in vitro and the instability in vivo have been experimentally evaluated for various PATRRs that differ regarding their size and symmetry. At physiological ionic strength, a cruciform structure is most frequently observed for the symmetric PATRR22, less often for the symmetric PATRR11, but not for the other PATRRs. In wild-type E. coli, only these two PATRRs undergo extensive instability, consistent with the relatively high incidence of the t(11;22) in humans. The resultant deletions are putatively mediated by central cleavage by the structure-specific endonuclease SbcCD, indicating the possibility of a cruciform conformation in vivo. Insertion of a short spacer at the centre of the PATRR22 greatly reduces both its cruciform extrusion in vitro and instability in vivo. Taken together, cruciform extrusion propensity depends on the length and central symmetry of the PATRR, and is likely to determine the instability that leads to recurrent translocations in humans.
INTRODUCTION
The constitutional t(11;22)(q23;q11) is the most common recurrent non-Robertsonian translocations in humans. Whereas balanced carriers of the translocation usually manifest no clinical symptoms, they often have problems in reproduction, such as male infertility, recurrent abortion and the birth of offspring with chromosomal imbalance (1,2). The constitutional t(17;22)(q11;q11), which is a cause of neurofibromatosis type I (NF1; MIM# 162200) by disrupting the NF1 gene on 17q11, is another recurrent but uncommon translocation (3). Through the analysis of numerous t(11;22) and two t(17;22) cases, we have identified characteristic DNA sequences, termed palindromic AT-rich repeats (PATRRs), at the translocation breakpoints on 11q23, 17q11 and 22q11 (PATRR11, PATRR17, PATRR22) (4,5). Polymorphisms have been identified in alleles of the PATRR11 and the PATRR17 regarding their size and symmetry (4,6). The shorter alleles are generally not palindromic, and are likely to be deletion derivatives of the longer symmetric alleles. Sequence analysis of the junction fragments of forty t(11;22) and two t(17;22) families demonstrated that all the breakpoints, with only one exception, are located at the centre of symmetric PATRRs, suggesting that the centre of the palindrome is susceptible to double-strand breaks that lead to the translocation (4,7,8).
It has been suggested that palindromic sequences are unstable in vivo, as a result of their formation of hairpin and/or cruciform structures (9). If the presence of such a secondary structure in vivo is aetiologic for the translocation, there should be a correlation between the rate of translocation occurrence and the secondary structure-forming propensity among these PATRRs. In fact, there is a difference in the relative susceptibility to translocation formation among PATRRs. The t(11;22) is so frequent that a substantial number of de novo translocations are detectable in sperm from normal males (10). The t(17;22) has been reported in only two families (3,5) and de novo translocations are hardly detectable (6). The theoretically possible t(11;17) has not been identified so far. In addition, recent findings demonstrated the involvement of the PATRR22 in a non-recurrent t(4;22) and a t(1;22) (11,12). So, it is assumed that the tendency to initiate a translocation is greatest for the PATRR22, followed by the PATRR11, and lowest for the PATRR17. Consistent with this assumption, a previous study calculated the free energy differences between the intrastrand and interstrand basepairing for various PATRRs in silico and found a correlation between translocation propensity and stability of the secondary structure (12). Although the result of the in silico analysis is convincing, experimental analyses should provide further insights into the molecular characteristics of the PATRR sequences with respect to their propensity for adopting a secondary structure.
The molecular characteristics of inverted repeat sequences have been extensively studied using artificial palindromes both in vitro and in E. coli as an in vivo model system (13–15). These studies have distinguished some aspects of hairpin and cruciform structures. For example, in vitro analyses have convincingly demonstrated two distinct mechanisms for cruciform configuration formation, termed C-type and S-type. These mechanisms essentially differ in the extent of the initial denaturation, and correspond to hairpin formation on single-stranded DNA and cruciform extrusion from double-stranded DNA, respectively (13). An in vivo analysis using E. coli distinguished the two mechanisms for deletion of inverted repeats, which are likely to be mediated by hairpin and cruciform structures, respectively (16). The application of such experimental analyses to the PATRR sequences at 22q11, 11q23 and 17q12 should certainly prove useful for evaluating their secondary structure-forming capability. It has, however, been technically difficult to examine long palindromic sequences such as PATRRs, because long palindromic sequences are generally not clonable due to the difficulties with their PCR amplification and extreme instability in E. coli. In our recent studies we have successfully cloned a short allele of the PATRR11 and two alleles of the PATRR17 by using optimized PCR conditions and a specific E. coli strain, thus overcoming the aforementioned difficulties. Thus, we have demonstrated a stable cruciform configuration of these PATRRs in vitro (6,17). As a result of these studies, we have proposed that the characteristics of PATRR sequences, including a long AT-rich central region with relatively GC-rich ends, should help in forming a stable cruciform structure. This stable cruciform is likely the aetiology for the instability that leads to translocation formation in humans. To date, however, direct evidence for cruciform extrusion of PATRR sequences in vivo has not been demonstrated.
In the present study, some longer PATRRs have been successfully cloned into plasmid vectors, and the secondary structure-forming propensity of various PATRRs has been examined both in vitro and in vivo. The in vitro analysis clearly demonstrates that the different cruciform-forming propensities among various PATRRs are dependent on both length and central symmetry. The in vivo analysis demonstrates that long and symmetric PATRR sequences appear to form a cruciform structure that is susceptible to structure-specific nucleases. Our data provide a molecular basis for the palindrome-mediated instability that is likely the source of these recurrent translocations in humans.
MATERIALS AND METHODS
Bacterial strains
The SURE strain (Stratagene), whose relevant genotype concerning DNA rearrangement and deletion is recB, recJ, sbcC, umuC::Tn5, uvrC, demonstrates increased stability of palindromic sequences, and was used for cloning and propagation of plasmids. The K-12-derived E. coli strains, AB1157, RIK200, CES200, JC5519, BW1012, were obtained from the E. coli Genetic Stock Center (http://cgsc.biology.yale.edu/). The AB1157 strain was used as wild type. The additional mutations of relevant genes in each strain are as indicated elsewhere.
Construction of the PATRR plasmids
The plasmids containing the polymorphic PATRR11s (accession numbers AF391128 and AF391129) and the PATRR17s (accession numbers AB195812 and AB195813) were constructed by PCR amplification and TA cloning into pBluescript II (Stratagene) and pT7-blue (Novagen) as previously described (4,6). Although, to date, the authentic PATRR22 has not been cloned due to its localization in an unclonable region in the human genome (18), the PATRR22 is likely to be a nearly perfect palindrome of ∼590 bp as deduced from the junction fragment sequence of the derivative 11 (der(11)) and 22 (der(22)) of the t(11;22) (4). For experimental analysis, we created two artificial constructs of the PATRR22s, the PATRR22-pal (palindromic, P) and the PATRR22-quasi (quasipalindromic, Q), by ligating two PATRR22-derived sequences of the t(11;22) junction fragment using a SnaBI site that is present at the centre of the original PATRR11. The PATRR22-quasi was created simply by ligating the proximal fragment of the der(22) and the distal fragment of the der(11), but contains an asymmetric 9 bp at the centre due to the PATRR11-derived sequences. The PATRR22-pal is a completely perfect palindrome, and was produced by ligating, in opposite directions, two identical distal fragments of the PATRR22 derived from the der(11) with flanking, non-palindromic sequences unchanged. The exact sequence of the PATRR22s used in this study is available in DDBJ (accession numbers AB261544 and AB261545). Pairs of plasmids with inserts in both orientations were made for each of the PATRRs, and designated as “p” (proximal) and “d” (distal), indicating which end of the PATRR sequence is closest to the plasmid's origin of replication. While most of the PATRR inserts were preserved in the SURE strain, some PATRR sequences, such as the PATRR11-long and the PATRR17-long, were unstable even in this strain. In these cases, clones having relatively low levels of deletion were selected and used for further analyses.
Cruciform extrusion assay
PATRR-containing plasmids readily formed a cruciform structure when prepared using an alkaline lysis protocol (6,17). Cruciform-free plasmids were successfully obtained using a non-denaturing, Triton-lysis method as previously described (17). In brief, E. coli cells from a 50-ml culture were dissolved with 10 ml of lysis buffer containing 50 mM Tris–HCl (pH 7.5), 5% sucrose, 1.5 mg/ml lysozyme, 0.1 M EDTA, 25 μg/ml RNase A and 0.75% Triton X-100. The plasmids were extracted without the use of phenol, and purified using an ion-exchange column (QIAGEN). Aliquots of plasmid DNA were precipitated with 2-propanol and stored at −30°C until used in an experiment. All of the procedures were performed at 4°C in a cold room to avoid spontaneous cruciform formation during the procedure. To induce cruciform formation, plasmids were incubated overnight at room temperature or for 30 min at 37°C in 10 mM Tris–HCl (pH 7.5), 0.1 mM EDTA with various NaCl concentrations (0–150 mM). As a negative control, plasmids were incubated in 10 mM Tris–HCl (pH 7.5), 0.1 mM EDTA and 100 mM NaCl for 30 min on ice. The plasmids were cooled on ice before electrophoresis at 50 V for ∼4 h in a 0.9% agarose gel at 4°C. The gel was stained with ethidium bromide and photographed using the ImageMaster VDS system (Amersham Pharmacia Biotech). Band intensities were quantified using NIH image 1.62 software. Treatment of the plasmids with T7 endonuclease I (New England Biolabs) was performed using 5 units of the enzyme in 20 μl of NEB2 buffer for 40 min on ice. This condition was found to minimize additional cruciform extrusion during digestion. The digested plasmids were purified by phenol/chloroform extraction and ethanol precipitation, and treated with the appropriate restriction enzymes to excise the PATRR-containing fragments, and then subjected to 2% agarose gel electrophoresis.
Analysis of instability in E. coli
Competent E. coli cells were transformed with plasmids by electroporation and plated on LB agar plates containing ampicillin. Colonies were cultured in 2 ml LB broth and plasmids were prepared using an automatic DNA isolation system (PI-200, Kurabo). Insert size was determined by restriction enzyme digestion and electrophoresis in a 2% agarose gel. For analysis of deletions of the PATRR22-pal and -quasi in various E. coli strains, insert fragments were amplified by colony PCR using previously described conditions (6), and subjected to electrophoresis in 2% agarose gels. Deletion products of the PATRRs were sequenced using the BigDye Terminator Cycle Sequencing Ready Reaction Kit Ver. 3 (Applied Biosystems).
RESULTS
Cruciform extrusion assays of various PATRRs in vitro
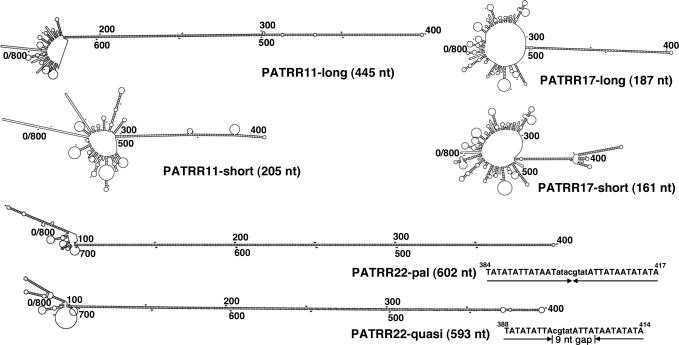
We have previously cloned the PATRR sequences on human chromosomes 11 and 17 using an optimized PCR method (4,6). The PATRR11 and the PATRR17 manifest length polymorphisms; PATRR11-long (445 bp) and PATRR11-short (205 bp), and PATRR17-long (187 bp) and PATRR17-short (161 bp). The long and short PATRRs are different not only with respect to their length, but also their symmetry. The long types of both PATRR11 and PATRR17 are nearly perfect palindromes, whereas the short versions have asymmetric regions around their centres. The authentic PATRR22 has not yet been cloned to date, but is presumed to be a nearly perfect palindrome of ∼590 bp based on junction fragment sequences of the der(11) and the der(22) of the t(11;22) (4,12). For experimental analysis in the present study, we prepared two artificial PATRR22 constructs; one is a completely perfect palindrome (PATRR22-pal, 602 bp), and the other is a quasipalindrome having a short spacer at the centre and four mismatched bases near the centre (PATRR22-quasi, 593 bp). The possible secondary structures formed by these PATRRs were analysed using the DNA mfold server (http://www.bioinfo.rpi.edu/applications/mfold/old/dna/), and they are schematically shown in Figure 1.
Figure 1.
Potential secondary structures formed by single-stranded PATRR sequences. Eight-hundred nucleotides containing each PATRR at the centre are analysed by mfold to demonstrate the putative stem-loop structures formed by intrastrand basepairing. The most prominent stem-loop structure for each sequence is formed by every PATRR sequence, whose length is indicated in parentheses. With regard to the PATRR22-pal and the PATRR22-quasi, sequences around the centre are indicated. The lowercase sequences are PATRR11-derived sequences in the junction fragment of the t(11;22) that provide the SnaBI site used to ligate the proximal and distal arms of the PATRR22-derived sequence. The PATRR22-pal is a perfect palindrome, whereas the PATRR22-quasi is a quasipalindrome having a spacer of 9 nt at the centre and four mismatched bases at 23, 27, 28 and 29 nt away from the hairpin tip.
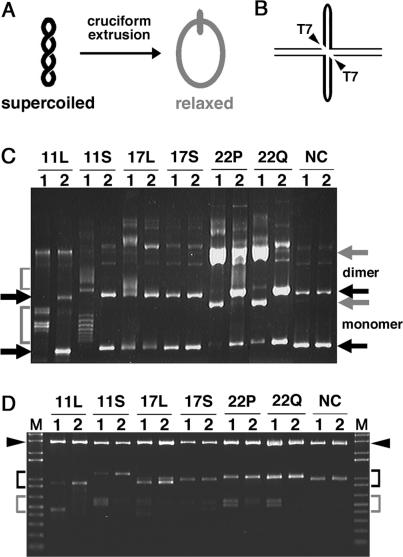
Palindrome-containing supercoiled plasmid is known to be relaxed when a cruciform is formed by the palindromic DNA sequence (Figure 2A). Consistent with that finding, we previously demonstrated that the PATRR11-short-containing plasmid undergoes a conformational change, which results in a mobility shift on agarose gel electrophoresis (17). We employed a non-denaturing, Triton-lysis method for the preparation of cruciform-free plasmids. The plasmids prepared using this technique are mostly supercoiled upon preparation (Figure 2C, condition 2, black arrows). After incubation at room temperature for 16 h, all of the PATRR-containing plasmids, except for the PATRR17-short, exhibited the following electrophoretic mobility shifts (Figure 2C, condition 1). With plasmids containing PATRR11-long and -short, the shifted bands are seen as a ladder and a smear, which reflect the various linking numbers of the plasmids (Figure 2C, gray brackets). A partial mobility shift of the PATRR17-long plasmid was evident only with the dimeric plasmid (Figure 2C, 17L, condition 1). In contrast to the results seen for the PATRR11s and the PATRR17-long, the shifted bands for the PATRR22-pal and -quasi plasmids appear as single bands migrating at the position of the open circular form (Figure 2C, gray arrows), indicating that the cruciform-forming palindrome sequence is long enough to fully relax the plasmids. Theoretically, transition of about 300 bp of the inverted repeat sequence into a cruciform structure would be sufficient for full relaxation of the ∼4 kb plasmids, which contain, in general, 15–25 superhelical turns in E. coli. This is because one superhelical turn is relaxed for every 10.5 bp of DNA that adopts a cruciform configuration (19). Two clusters of shifted bands are observed in the PATRR11-long plasmids (Figure 2C, 11L, condition 1). They are likely to represent two different stable conformations on the basis of previous observations using two-dimensional gel electrophoresis that demonstrated multiple configurations adopted by the PATRR11-short plasmid (17). Indeed, there is no sequence difference of the PATRRs between the two band clusters (data not shown). In contrast, no mobility shift was observed in a plasmid with non-palindromic sequence (Figure 2C, NC).
Figure 2.
Electrophoresis mobility shifts caused by cruciform extrusion in closed circular plasmids containing PATRR sequences. (A) Schematic representation of the topological change of circular plasmid DNA caused by cruciform formation. Negative supercoiling is relaxed if a cruciform is formed in the supercoiled plasmid DNA. The degree of relaxation is dependent on the length of the extrusion. (B) Diagram of T7 endonuclease I digestion. If the PATRR sequences (thick lines) adopt a cruciform structure, they will be cut into two fragments by a diagonal cleavage with the T7 endonuclease I. (C) PATRR-containing plasmids prepared by the Triton-lysis method were dissolved in TE (pH 7.5) and incubated at room temperature for 16 h (condition 1), or were dissolved in TE with 100 mM NaCl (pH 7.5) on ice without incubation (condition 2) before the electrophoresis in 0.9% agarose gel. Electrophoretic mobility shifts are observed as ladder bands (brackets) with plasmids containing the PATRR11-long (11L), -short (11S) and the PATRR17-long (17L), or observed as a single band (gray arrows) with plasmids containing the PATRR22-pal (22P) and the PATRR22-quasi (22Q). No mobility shift was observed with plasmids containing the PATRR17-short (17S) and non-palindrome sequence (NC). The positions of monomeric and dimeric supercoiled plasmid DNAs are indicated with black arrows. (D) Plasmids dissolved in TE (pH 7.5) and incubated at room temperature for 16 h (condition 1) and those dissolved in NEB2 buffer on ice without incubation (condition 2) were subjected to T7 endonuclease I digestion on ice, and followed by appropriate restriction enzyme digestion. Vectors and intact PATRR-containing inserts are indicated with arrowheads and black brackets, respectively. T7 endonuclease I-digested, PATRR-containing fragments are indicated with gray brackets. The expected sizes of intact fragments (and T7 endonuclease I-digested fragments) are as follows: PATRR11-long, 900 bp (452 and 448 bp); PATRR11-short, 1140 bp (609 and 531 bp); PATRR17-long, 1009 bp (567 and 442 bp); PATRR17-short, 975 bp (554 and 421 bp); PATRR22-pal, 1044 bp (556 and 488 bp) and PATRR22-quasi, 1035 bp (552 and 483 bp).
To confirm the existence of a cruciform structure in relaxed plasmids, their sensitivity to T7 endonuclease I was examined. T7 endonuclease I is capable of recognizing and cleaving the four-way junction of a cruciform structure. If the cruciform structure formed by PATRRs is digested by the nuclease, the PATRR-containing DNA fragment will be cut into two fragments (Figure 2B). The plasmids prepared in two conditions as in Figure 2C were digested with T7 endonuclease I in conditions that minimize additional cruciform extrusion during incubation. As a result, although digestion was likely to be partial, T7 endonuclease I-digested fragments were clearly detected in condition 1 of the plasmids containing PATRR11-long and -short, and PATRR22-pal and -quasi, but were hardly detectable in condition 2 (Figure 2D, 11L, 11S, 22P and 22Q). For the PATRR17-long plasmids, slight bands were preferentially detected in condition 1 (Figure 2D, 17L). Although very weak bands were also observed for the PATRR17-short plasmids, there was no difference between the two conditions (Figure 2D, 17S). No digested band was detected in the plasmid with non-palindromic sequence (Figure 2D, NC). These results convincingly demonstrate the correlation between mobility shifts of plasmids (Figure 2C) and the existence of a cruciform structure (Figure 2D) for each PATRR-containing plasmid, supporting the assumption that the relaxation of the plasmids is due to cruciform formation. In addition, the sizes of the T7 endonuclease I-digested fragments were consistent with cleavage of the expected cruciform structures whose intrastrand basepairing is formed as shown in Figure 1. Taken together, these results demonstrate that PATRR-containing plasmids, except for the PATRR17-short, prefer to adopt a cruciform configuration at low ionic strength at room temperature.
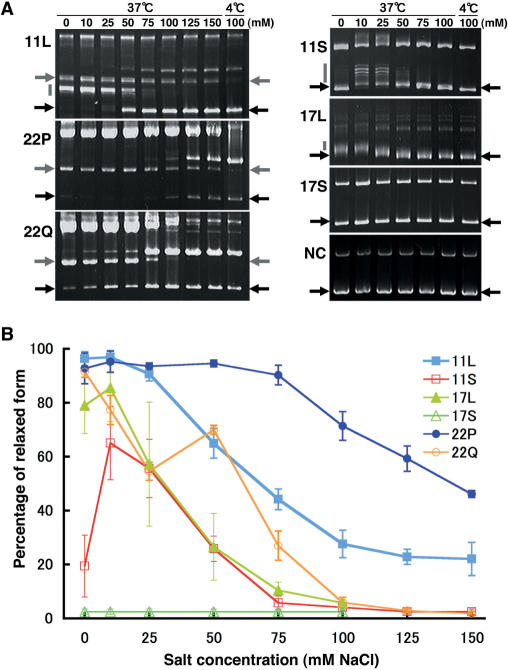
We analysed the cruciform extrusion propensities of various PATRRs by incubating the plasmids in various salt concentrations at 37°C. Electrophoretic mobility shifts were observed with all PATRR-containing plasmids except for the PATRR17-short when incubated in relatively low salt concentration buffers (0–50 mM NaCl) (Figure 3A). Quantitative analysis of the ratio of the relaxed to the supercoiled plasmid bands demonstrated that secondary structures were formed most efficiently by the PATRR11-long and the PATRR22-pal. Secondary structure is less frequently adopted for the PATRR11-short, the PATRR17-long and the PATRR22-quasi, and not at all by the PATRR17-short (Figure 3B). In contrast, only the PATRR11-long and the PATRR22-pal showed significant cruciform extrusion among various PATRRs in physiologic salt concentrations (75–150 mM NaCl) (Figure 3A). Interestingly, the PATRR22-pal and -quasi exhibited striking difference in their cruciform extrusion abilities at relatively high salt concentrations. This result points out the importance of perfect symmetry at the centre for cruciform extrusion under physiologic conditions. Quantitative analysis at high salt concentration clearly demonstrated that the propensity for cruciform extrusion was highest with the PATRR22-pal, followed by the PATRR11-long and lowest with the PATRR17-long among the three symmetric PATRRs (Figure 3B). This result is quite consistent with previous in silico studies (12), and indicates the striking correlation between cruciform-forming propensity and putative translocation susceptibility in humans.
Figure 3.
Effects of salt concentration on cruciform extrusion from various PATRR-containing plasmids. (A) Gel images showing electrophoretic mobility shifts after incubation in TE buffer containing various salt concentrations. Black arrows indicate the positions of supercoiled plasmids. Gray arrows and gray bars indicate the mobility shifts of plasmids due to relaxation by cruciform formation. Fully relaxed forms (gray arrows) were observed in plasmids containing the PATRR11-long (11L), the PATRR22-pal (22P) and the PATRR22-quasi (22Q) and partially relaxed forms (gray bars) were observed in plasmids containing the PATRR11-long (11L), the PATRR11-short (11S) and the PATRR17-long (17L). At relatively low concentrations of NaCl (0–50 mM), most PATRR-containing plasmids, except for the PATRR17-short (17S), exhibited mobility shifts. In contrast, only the plasmids containing the PATRR11-long (11L) and the PATRR22-pal (22P) exhibited significant mobility shifts at relatively physiologic salt concentrations (75–150 mM). The two clusters of bands in the PATRR11-long (11L) exhibit different salt concentration dependency. (B) Quantitative analysis of cruciform-forming ratios in PATRR-containing plasmids incubated at 37°C with various salt concentrations. The band intensities were quantified using NIH image, and the percentages of the shifted bands were plotted against the concentration of NaCl. Data represent mean ± SD of three independent assays.
Instability of various PATRRs in E. coli
To obtain evidence for secondary structure formation of the PATRRs in vivo, we examined instability characteristics of the PATRRs in E. coli. A previous study has demonstrated two distinct mechanisms for palindrome-stimulated deletions; replication slipped mispairing and single-strand annealing following cruciform structure cleavage (16). The former mechanism is independent of the hairpin endonuclease SbcCD and preferentially occurs on a lagging strand that stalls at a hairpin structure formed on the single-stranded template during DNA replication. In contrast, the latter mechanism is largely dependent on SbcCD activity, and is assumed to be provoked by double-strand cleavage of a cruciform structure because the deletion is independent of the direction of replication. Thus, by examining deletions of PATRR-containing plasmids, it should be clear whether the deletion was caused by a hairpin or cruciform structure in vivo based on dependency on SbcCD activity and the direction of replication.
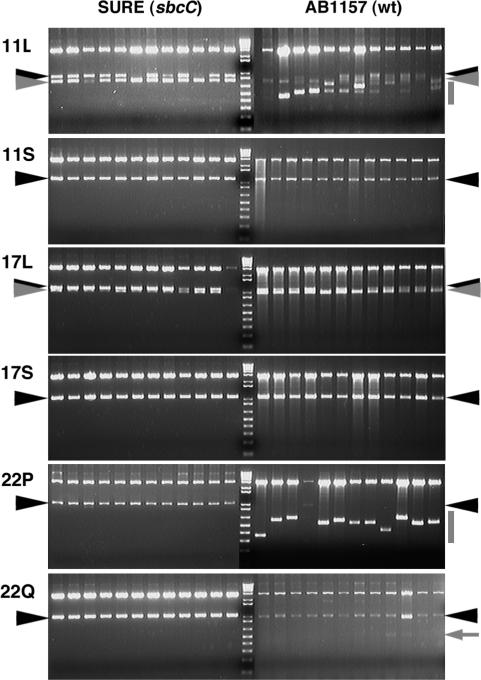
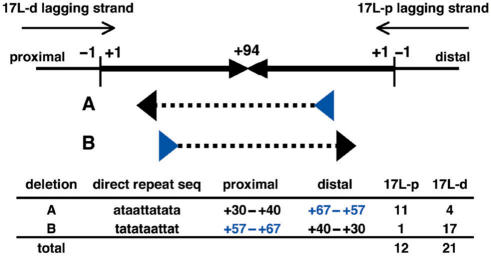
We first evaluated SbcCD-independent deletion in the SURE strain, which has a mutation in the SbcC gene. When multiple independent clones were examined, PATRR11-long and PATRR17-long underwent partial, short deletions of the same length in the SURE strain (Figure 4, gray arrowheads), whereas other PATRRs were stable. We then analysed the replication direction dependency of these deletions by comparing two plasmids with the PATRR17-long sequence inserted in opposite directions (Figure 5, 17L-p and 17L-d). Similar deletions were observed with the two constructs (data not shown). Sequence analysis of the deletion products identified two types of mirror image rearrangement with respect to the direction of replication that were mediated by a direct repeat consisting of 11 nt (Figure 5). The frequency of the two deletions was clearly biased by the replication direction of the plasmids. The location of the direct repeats was consistent with preferential occurrence of the slippage on the lagging strand based on the knowledge that the ‘donor repeat’ (Figure 5, blue arrowheads) is located closer to the centre than the ‘target repeat’ (Figure 5, black arrowheads) for a slippage (9). These results indicate that deletion of the PATRR17-long and perhaps the PATRR11-long are potentially mediated by replication slipped mispairing at the hairpin structure formed on the lagging strand template.
Figure 4.
Instabilities of PATRR-containing plasmids in E. coli. The expected fragment sizes containing the intact PATRR sequences are as follows: PATRR11-long, 900 bp; PATRR11-short, 1140 bp; PATRR17-long, 1093 bp; PATRR17-short, 1067 bp; PATRR22-pal, 1044 bp and PATRR22-quasi, 1035 bp. The positions of the intact bands are indicated with black arrowheads. Twelve randomly selected independent clones are represented for each plasmid and each strain. In both SURE and AB1157 strain, the PATRR11-long (11L) and the PATRR17-long (17L) exhibited the same-sized deletions of ∼150 and ∼100 bp, respectively (gray arrowheads). Prominent deletions of various sizes occurred in plasmids containing the PATRR11-long (11L) and the PATRR22-pal (22P) specifically in AB1157 strain (gray bars). The remarkable instability is consistent with their cruciform-forming propensity at physiological conditions in vitro.
Figure 5.
Sequence analysis of SbcC-independent deletions of the PATRR17-long. The head-to-head arrows represent the palindromic sequence of the PATRR17-long. Numbers with plus (+1 to +94) indicate the position of bases from the end of the palindrome (vertical bars) in both the proximal and distal arms. Larger numbers indicate positions closer to the centre. Thin arrows represent the direction of lagging strand synthesis in two different plasmids containing the same PATRR sequence in opposite directions (17L-p and 17L-d). Deletions (dashed lines) are flanked by a pair of direct repeats (arrowheads). A and B are the same deletions with opposite direction, and their frequencies are biased with the direction of replication.
We next analysed the instability of the PATRRs in the wild-type E. coli strain AB1157 to evaluate SbcCD-dependent deletions by comparing with the results obtained in the SURE strain. With regard to hairpin-mediated deletions of the PATRR11-long and the PATRR17-long, similar-sized deletions were also observed in the AB1157 strain (Figure 4, gray arrowheads). More importantly, the wild type strain-specific, extensive deletions were observed in most clones of the PATRR22-pal, and to a lesser extent in the clones of the PATRR11-long (Figure 4, gray bars), but not in the clones of the other PATRRs. In contrast to the PATRR22-pal, the PATRR22-quasi was quite stable, and only a subtle deletion was infrequently observed in the wild-type E. coli (Figure 4, arrow). The different levels of instability among these PATRRs are in good correlation with their ability to adopt a cruciform configuration under physiologic conditions in vitro (Figure 3). These results suggest that wild-type-specific instability is caused by the cruciform structures formed in vivo that could act as a substrate for the SbcCD nuclease. Further analyses of SbcCD-dependent deletions were performed only with the PATRR22-pal but not with the PATRR11-long. This was because the rearranged products of the PATRR11-long were complicated and most sustained their palindromic nature even after deletion, which made analysis by sequencing impossible (data not shown).
SbcC-dependent instability of the PATRR22s in E. coli
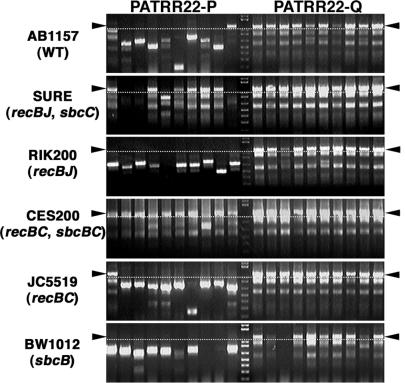
To confirm the SbcC-dependency of the PATRR22-pal deletions, we examined the instability of the PATRR22-pal and -quasi in various E. coli strains by the colony PCR method for rapid and sensitive analysis. The PATRR22-pal were extensively deleted in a wild-type strain (Figure 6, AB1157), but were kept intact in most, but not all, colonies in two independent SbcC mutant strains (Figure 6, SURE and CES200). The other relevant mutations that are in the SURE and CES200 strains, such as recB, recC, recJ and sbcB, did not inhibit the deletions as demonstrated by the use of the other mutant strains (Figure 6, RIK200, JC5519 and BW1012). This result suggests that the hairpin endonuclease activity of SbcCD is crucial for the instability of PATRR sequences, as has been demonstrated for the instability of short artificial palindromes (16). Furthermore, the PATRR22-quasi was not conspicuously deleted in the E. coli strains examined (Figure 6).
Figure 6.
Deletion of the PATRR22-pal requires SbcC activity. The SbcC-dependency of the PATRR22-pal deletions was evaluated using various E. coli strains. Relevant genotypes for each strain are indicated in parentheses. The insert size of PATRR22s was examined by colony PCR amplification. The bands above the dotted lines represent intact inserts. The PATRR22-pal was extensively deleted in the wild-type strain (AB1157), but was remarkably kept intact in two independent SbcC mutant strains (SURE and CES200). The PATRR22-quasi was stable in all strains examined. Premature termination products of PCR reactions were observed as smaller-sized bands when the template DNA retained its palindromic nature.
Sequence analysis of the PATRR22-pal deletions
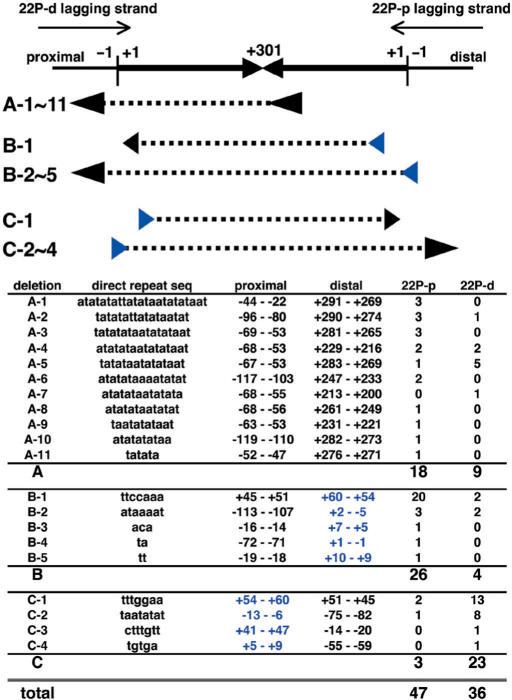
To further characterize the SbcCD-dependent deletions of the PATRR22-pal, the influence of replication direction was analysed using two plasmids with opposite replication directions in the AB1157 strain. As a result, we found that rearranged products could be largely divided into two types; one is significantly dependent on replication direction (Figure 7, types B and C) and the other is relatively independent (Figure 7, type A). Replication direction-dependency for the SbcCD-dependent deletions was rather unexpected, as a previous study demonstrated that such deletions were independent of replication direction (16). The most frequent deletions among types B and C were mediated by the same direct repeats in opposite directions (Figure 7, B-1 and C-1). In all the types B and C deletions, the putative donor repeats on the lagging strand were located around the start of the palindromic sequence, and were more internal than the target repeats (Figure 7, B and C series). These results indicate that types B and C deletions are likely to be mediated by a slipped-replication-mispairing mechanism. In contrast to types B and C deletions, type A deletions were relatively independent of replication direction, and occurred more asymmetrically, i.e. near the centre and the outside of the palindrome (Figure 7, type A). The localization of one deletion end near the centre of the palindrome is in agreement with putative DNA cleavage at the tips of the cruciform structure by the SbcCD endonuclease followed by asymmetric resection of the DNA ends (16). Thus, the type A deletion is likely to be caused by cleavage of the cruciform structure followed by a single-strand annealing mechanism. The assumption of an essential difference between the two types of deletions would be further supported by differences in the direct repeat lengths; the type A deletions utilized longer direct repeats (6–23 bp) than the types B and C deletions (2–7 bp). The length of the direct repeats mediating the latter deletions was consistent with a previous report showing the utilization of short direct repeats (3–8 bp) for replication slippage (20). Although direct evaluation was impossible, the PATRR11-long may also form a cruciform structure in vivo that is responsible for its SbcCD-dependent deletion in wild-type E. coli. Taken together, these results provide the first experimental evidence for cruciform structure formation by long, symmetric PATRR sequences in vivo. This cruciform structure was extremely unstable in wild-type E. coli because of its susceptibility to structure-specific nuclease.
Figure 7.
Sequence analysis of the PATRR22-pal deletions in AB1157 strain. The head-to-head arrows represent the palindromic sequence of the PATRR22-pal. Numbers with a plus (+1 to +301) indicate the position of bases from the end of the palindrome (vertical bars) in both the proximal and distal arms, while numbers with a minus indicate the positions of bases outside the PATRR. The larger plus numbers indicate positions closer to the centre (i.e. the palindrome is labelled from +1 to +301 and back to +1). Thin arrows represent the direction of lagging strand synthesis in the PATRR22-pal-pBluescript II-KS (22P-d) and -SK (22P-p) plasmids. Deletion types are classified by the positions of direct repeat pairs (arrowheads) that flank the deleted sequences (dashed lines). One of a pair is located near the centre in the type A deletions, and is located at the distal and proximal ends in the type B and type C deletions, respectively. The different distribution of direct repeats supports the existence of different putative mechanisms for these deletions.
DISCUSSION
In this study, we evaluated the hairpin and cruciform structure formation of various PATRR sequences in vitro and in vivo. In vitro analysis was useful to compare the secondary structure-forming propensities of the PATRRs. Previous studies have characterized two distinct mechanisms for cruciform formation in vitro, termed C-type and S-type (13). The C-type extrusion mechanism, which includes extensive denaturation of an inverted repeat-containing DNA region (up to hundreds of basepairs) followed by hairpin formation in the single-stranded sequence, occurs preferentially with AT-rich sequences in solutions of very low ionic strength. In contrast, in physiologic salt concentration, cruciform extrusion can occur only by the S-type mechanism, which includes an initial opening of only 8–10 bp at the centre, nucleation of intrastrand basepairing, followed by branch migration to the fully extruded cruciform. Thus, cruciform structure formation in vitro is likely to be mediated by hairpin structure formation in a low-salt environment, while in physiologic salt conditions, it is likely to be mediated by cruciform extrusion from the double-stranded DNA. Our in vitro analysis demonstrated that a ‘hairpin structure’ could be formed by most PATRRs based on the results at low-salt concentrations. In contrast, the propensity for cruciform extrusion from double-stranded DNA was quite different among the PATRRs. Quantitative analysis demonstrated that cruciform-extruding propensity was highest with PATRR22-pal, followed by PATRR11-long, and negligible with PATRR17-long among the three symmetric PATRRs. This difference in propensity is consistent with the putative translocation susceptibility of these PATRRs estimated by free energy as well as by observations of the actual translocation frequency in humans (12). Taken together, in vitro analysis demonstrated that the propensity for cruciform extrusion, rather than that for hairpin formation, is in good agreement with the putative instability of various PATRR sequences in humans.
The mechanism of palindrome-mediated instability in vivo has been extensively characterized in E. coli as a model system. Very early studies on the DNA secondary structure of inverted repeat sequence suggested the possibility that its instability was due to cruciform formation in vivo (21). However, initial studies failed to demonstrate the existence of such a structure in vivo, and argued against the phenomenon based on the kinetic barrier to cruciform formation from double-stranded DNA (22). Besides a cruciform structure, a hairpin structure is another type causing instability of inverted repeat sequences as evidenced by replication-dependent deletions (23). From a kinetic viewpoint, formation of a hairpin structure at an inverted repeat is favoured when DNA become single stranded during replication. Although there is a presumption that cruciform extrusion from double-stranded DNA would be unfavourable, a series of studies using specifically designed inverted repeat sequences demonstrated that highly AT-rich sequences, in particular alternating (AT)n sequence, have no discernible kinetic barrier to cruciform extrusion in vitro (14,24,25), and form cruciform structures in E. coli (19,26). Through these and other studies, it has been established that AT-richness around the centre is a prerequisite for cruciform extrusion in vivo. In this context, all of the PATRR sequences are sufficiently AT-rich at their centres (only 1–3 GCs within the central 20 bp).
In a recent study, instability has been postulated to occur by two distinct mechanisms mediated by hairpin and cruciform structures, respectively (16). In E. coli, a hairpin structure could be formed by an inverted repeat sequence on the lagging strand template when it becomes single-stranded during replication (9,23). This hairpin structure will cause deletion by replication slippage that includes polymerase pausing, dissociation and re-annealing of the synthesizing strand to a downstream region using short direct repeats (27). In our in vivo experiments, only the PATRR11-long and the PATRR17-long underwent deletion by replication slippage in the SURE strain. It is puzzling why the other PATRRs, especially the PATRR22-pal and -quasi, were quite stable in the SURE strain despite their potential hairpin structure formation. We are assuming that a paucity of deletions is more likely to reflect the absence of available direct repeats for slippage than the inability to form a hairpin structure in vivo, because the location of the direct repeats is critical for efficient slippage (20,28). In this context, our results indicate that even long perfect palindromes, such as the PATRR22s, can be preserved in plasmids in the SURE strain if no direct repeat is available to allow for slippage.
In contrast to a hairpin structure-mediated mechanism, a cruciform structure-mediated deletion is assumed to be dependent on a structure-specific nuclease, and not dependent on a replication event (16). Among the various PATRRs, SbcCD-dependent deletion was observed most prominently with the PATRR22-pal, to a lesser extent with the PATRR11-long, and not with the other PATRRs. The differences in stability of these PATRRs correlate well with their cruciform extrusion propensity in vitro, which has also been demonstrated with a set of artificial long palindromes in lambda phage (15). Furthermore, sequence analysis of the PATRR22-pal demonstrated that some deletions (type A) are likely to be mediated by double-strand breakage of a cruciform structure based on their lack of dependence on the direction of replication and their putative central cleavage. This speculation seems to be credible because of the remarkable contrast to the other distinct deletions (types B and C) observed for the same PATRR. The types B and C deletions of PATRR22-pal are also SbcCD-dependent, but are likely to be mediated by a replication slippage mechanism based on their dependency on replication direction and the position of the direct repeats. The reason that these replication slippages occur only rarely in the sbcC mutant strains remains elusive. As a possible explanation, in the palindromes that do not have a direct repeat available for slippage like the PATRR22, the cleavage of the hairpin structure by the SbcCD activity might allow slippage between the cryptic direct repeats that are not available with the intact hairpin structure, possibly by allowing for the resumption of DNA synthesis and annealing to a relatively distant position.
Another significant thing that we have demonstrated is the importance of central symmetry for the instability of a PATRR sequence in vivo based on the striking differences between the PATRR22-pal and the PATRR22-quasi. In particular, is the case of the PATRR22-quasi. It is almost the same as the PATRR22-pal in length and AT-richness, but has a 9-bp spacer at the centre and four mismatched bases near the centre. It could neither form a cruciform structure at physiologic conditions in vitro nor cause instability in wild-type E. coli. In the kinetics of cruciform extrusion, there are two critical steps, central denaturation and followed by intrastrand basepairing. The former step is largely due to the central AT-richness as discussed earlier, and thus is not different between the two PATRR22s. Therefore, the central symmetry, which is critical for the initiation of intrastrand basepairing, is likely responsible for the remarkable differences of SbcCD-dependent instability between the two PATRR22s. In conjunction with the fact that the longer palindrome sequence exhibits greater instability amongst the symmetric PATRRs, not only the initial denaturation, but also the following intrastrand basepairing is a critical determinant for cruciform formation in vivo that leads to instability. This is also likely to be the case for instability of the PATRR sequences in humans, because our recent study demonstrated reduced susceptibility of asymmetric PATRR11 alleles to the recurrent 11;22 translocation by comparing de novo translocation frequency among various PATRR11 alleles (29). The critical dependency of translocation on the presence of central symmetry may further support the possibility that a cruciform structure, rather than a hairpin structure, provides the substrate for frequent recurrent t(11;22) translocations, because hairpin formation efficiency is less likely to depend on central symmetry, as has been demonstrated in previous studies (19) and in our in vitro experiments. In addition, the PATRR22-quasi, which is likely to form a hairpin structure in vivo, underwent only a subtle deletion in wild-type E. coli as shown in Figure 4. This result suggests that a hairpin structure is much more stable than a cruciform structure in vivo with regard to its susceptibility to a structure-specific nuclease. Of course, this speculation does not exclude the possibility that hairpin-mediated double-strand breaks lead to a translocation, as has been successfully demonstrated in a yeast model (30). However, the hypothetical greater instability of a cruciform structure than a hairpin structure may be a plausible explanation for the striking difference between the frequency of the t(11;22) and the t(17;22), because the PATRR17, in contrast to the PATRR11 and the PATRR22, is likely to only form a hairpin structure and not a cruciform in vivo as demonstrated in the present study.
Although a cruciform structure in vivo is likely to be aetiologic for palindrome-mediated recurrent translocations, no direct evidence has demonstrated the presence of such a configuration in eukaryotic chromatin. The energetically unfavourable structure will require a sufficient negative superhelicity for stabilizing the structure, while the existence of such a level of negative supercoiling has not been proven. We have previously reported male meiosis-specific occurrence of this translocation in humans (10), suggesting that a meiosis-specific physiological event might be involved in a mechanism of cruciform extrusion and/or the structure-dependent instability. During the process of spermatogenesis, a series of unique chromatin remodeling occurs related to homologous pairing and recombination, transcriptional regulation and chromatin compaction (31). Among factors involved in chromatin remodeling, ATP-dependent chromatin remodeling factors, which contain a Swi2/Snf2-family ATPase subunit, were shown to have the ability to generate unconstrained negative supercoiling, leading to cruciform extrusion of palindromic DNA in chromatin in vitro (32). Although it has not been studied yet, the specific chromatin remodeling activity during spermatogenesis might provide a mechanism to induce cruciform extrusion in vivo. Allowing that a cruciform structure exists in vivo, the exact mechanism for the instability of a cruciform structure in humans is also totally unknown. There are two physiological types of DNA breakage that could provide a reasonable candidate substrate for generation of double-strand breaks leading to the translocation in meiotic nuclei. One is the double-strand breaks that are required for meiotic recombination, and the other lies in the resolution of Holliday junctions that are formed as intermediates of homologous recombination, and are analogous to cruciform structures. The breakpoints of human t(11;22)s localize at the centre of the palindrome, which could represent the tip of the hairpin when a PATRR sequence forms such a structure. The ‘centre-break mechanism’ has also been demonstrated in the mammalian germ line as a result of a long palindromic transgene in mice (33). In this context, the Mre11–Rad50–Nbs1 complex, the eukaryotic homolog of the bacterial hairpin nuclease SbcCD, would represent a candidate to be involved in PATRR-mediated translocations, as this complex is also likely to play a role in meiotic recombination (34). We also postulate the possible involvement of a diagonal cleavage of the cruciform structure, like Holliday junction resolution (35). Such diagonal cleavage will produce hairpin-structured ends, which might be a target for a specialized form of non-homologous end joining such as that observed with the rearrangement of immunoglobulin and T-cell receptor genes (36). This hypothesis is intriguing because it could provide a good explanation for enigmatic PATRR-specific translocations (12).
ACKNOWLEDGEMENTS
The authors wish to thank E. Hosoba for technical assistances and Dr A. Iizuka-Kogo for helpful discussions. This work was supported by Grants-in-Aid for Scientific Research and 21st Century COE program from the Ministry of Education, Science, Sports and Culture of the Japanese Government (H.K.). This work was partially supported by funds from CA39926 from the United States National Institutes of Health (B.S.E.). Funding to the pay the Open Access publication charge was provided by Daiko Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shaikh TH, Budarf ML, Celle L, Zackai EH, Emanuel BS. Clustered 11q23 and 22q11 breakpoints and 3:1 meiotic malsegregation in multiple unrelated t(11;22) families. Am. J. Hum. Genet. 1999;65:1595–1607. doi: 10.1086/302666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zackai EH, Emanuel BS. Site-specific reciprocal translocation, t(11;22) (q23;q11), in several unrelated families with 3:1 meiotic disjunction. Am. J. Med. Genet. 1980;7:507–521. doi: 10.1002/ajmg.1320070412. [DOI] [PubMed] [Google Scholar]
- 3.Kehrer-Sawatzki H, Häussler J, Krone W, Bode H, Jenne DE, Mehnert KU, Tümmers U, Assum G. The second case of a t(17;22) in a family with neurofibromatosis type 1: sequence analysis of the breakpoint regions. Hum. Genet. 1997;99:237–247. doi: 10.1007/s004390050346. [DOI] [PubMed] [Google Scholar]
- 4.Kurahashi H, Emanuel BS. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Mol. Genet. 2001;10:2605–2617. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- 5.Kurahashi H, Shaikh T, Takata M, Toda T, Emanuel BS. The constitutional t(17;22): another translocation mediated by palindromic AT-rich repeats. Am. J. Hum. Genet. 2003;72:733–738. doi: 10.1086/368062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inagaki H, Ohye T, Kogo H, Yamada K, Kowa H, Shaikh TH, Emanuel BS, Kurahashi H. Palindromic AT-rich repeat in the NF1 gene is hypervariable in humans and evolutionarily conserved in primates. Hum. Mutat. 2005;26:332–342. doi: 10.1002/humu.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edelmann L, Spiteri E, Koren K, Pulijaal V, Bialer MG, Shanske A, Goldberg R, Morrow BE. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am. J. Hum. Genet. 2001;68:1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tapia-Páez I, Kost-Alimova M, Hu P, Roe BA, Blennow E, Fedorova L, Imreh S, Dumanski JP. The position of t(11;22)(q23;q11) constitutional translocation breakpoint is conserved among its carriers. Hum. Genet. 2001;109:167–177. doi: 10.1007/s004390100560. [DOI] [PubMed] [Google Scholar]
- 9.Leach DR. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- 10.Kurahashi H, Emanuel BS. Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat. Genet. 2001;29:139–140. doi: 10.1038/ng1001-139. [DOI] [PubMed] [Google Scholar]
- 11.Nimmakayalu MA, Gotter AL, Shaikh TH, Emanuel BS. A novel sequence-based approach to localize translocation breakpoints identifies the molecular basis of a t(4;22) Hum. Mol. Genet. 2003;12:2817–2825. doi: 10.1093/hmg/ddg301. [DOI] [PubMed] [Google Scholar]
- 12.Gotter AL, Shaikh TH, Budarf ML, Rhodes CH, Emanuel BS. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum. Mol. Genet. 2004;13:103–115. doi: 10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murchie AI, Lilley DM. The mechanism of cruciform formation in supercoiled DNA: initial opening of central basepairs in salt-dependent extrusion. Nucleic Acids Res. 1987;15:9641–9654. doi: 10.1093/nar/15.23.9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng GX, Sinden RR. Effect of base composition at the center of inverted repeated DNA sequences on cruciform transitions in DNA. J. Biol. Chem. 1988;263:5356–5361. [PubMed] [Google Scholar]
- 15.Davison A, Leach DR. The effects of nucleotide sequence changes on DNA secondary structure formation in Escherichia coli are consistent with cruciform extrusion in vivo. Genetics. 1994;137:361–368. doi: 10.1093/genetics/137.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bzymek M, Lovett ST. Evidence for two mechanisms of palindrome-stimulated deletion in Escherichia coli: single-strand annealing and replication slipped mispairing. Genetics. 2001;158:527–540. doi: 10.1093/genetics/158.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurahashi H, Inagaki H, Yamada K, Ohye T, Taniguchi M, Emanuel BS, Toda T. Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations. J. Biol. Chem. 2004;279:35377–35383. doi: 10.1074/jbc.M400354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurahashi H, Shaikh TH, Hu P, Roe BA, Emanuel BS, Budarf ML. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum. Mol. Genet. 2000;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- 19.Zheng GX, Kochel T, Hoepfner RW, Timmons SE, Sinden RR. Torsionally tuned cruciform and Z-DNA probes for measuring unrestrained supercoiling at specific sites in DNA of living cells. J. Mol. Biol. 1991;221:107–122. doi: 10.1016/0022-2836(91)80208-c. [DOI] [PubMed] [Google Scholar]
- 20.Pinder DJ, Blake CE, Lindsey JC, Leach DR. Replication strand preference for deletions associated with DNA palindromes. Mol. Microbiol. 1998;28:719–727. doi: 10.1046/j.1365-2958.1998.00831.x. [DOI] [PubMed] [Google Scholar]
- 21.Lilley DM. In vivo consequences of plasmid topology. Nature. 1981;292:380–382. doi: 10.1038/292380a0. [DOI] [PubMed] [Google Scholar]
- 22.Courey AJ, Wang JC. Cruciform formation in a negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983;33:817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- 23.Trinh TQ, Sinden RR. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature. 1991;352:544–547. doi: 10.1038/352544a0. [DOI] [PubMed] [Google Scholar]
- 24.Greaves DR, Patient RK, Lilley DM. Facile cruciform formation by an (A-T)34 sequence from a Xenopus globin gene. J. Mol. Biol. 1985;185:461–478. doi: 10.1016/0022-2836(85)90064-6. [DOI] [PubMed] [Google Scholar]
- 25.McClellan JA, Lilley DM. A two-state conformational equilibrium for alternating (A-T)n sequences in negatively supercoiled DNA. J. Mol. Biol. 1987;197:707–721. doi: 10.1016/0022-2836(87)90477-3. [DOI] [PubMed] [Google Scholar]
- 26.McClellan JA, Boublíková P, Palecek E, Lilley DM. Superhelical torsion in cellular DNA responds directly to environmental and genetic factors. Proc. Natl. Acad. Sci. USA. 1990;87:8373–8377. doi: 10.1073/pnas.87.21.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viguera E, Canceill D, Ehrlich SD. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 2001;20:2587–2595. doi: 10.1093/emboj/20.10.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosche WA, Trinh TQ, Sinden RR. Differential DNA secondary structure-mediated deletion mutation in the leading and lagging strands. J. Bacteriol. 1995;177:4385–4391. doi: 10.1128/jb.177.15.4385-4391.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato T, Inagaki H, Yamada K, Kogo H, Ohye T, Kowa H, Nagaoka K, Taniguchi M, Emanuel BS, et al. Genetic variation affects de novo translocation frequency. Science. 2006;311:971. doi: 10.1126/science.1121452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 31.Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- 32.Havas K, Flaus A, Phelan M, Kingston R, Wade PA, Lilley DM, Owen-Hughes T. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell. 2000;103:1133–1142. doi: 10.1016/s0092-8674(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 33.Akgün E, Zahn J, Baumes S, Brown G, Liang F, Romanienko PJ, Lewis S, Jasin M. Palindrome resolution and recombination in the mammalian germ line. Mol. Cell Biol. 1997;17:5559–5570. doi: 10.1128/mcb.17.9.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farah JA, Cromie G, Steiner WW, Smith GR. A novel recombination pathway initiated by the Mre11/Rad50/Nbs1 complex eliminates palindromes during meiosis in Schizosaccharomyces pombe. Genetics. 2005;169:1261–1274. doi: 10.1534/genetics.104.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurahashi H, Inagaki H, Ohye T, Kogo H, Kato T, Emanuel BS. Palindrome-mediated chromosomal translocations in humans. DNA Repair (Amst) 2006;5:1136–1145. doi: 10.1016/j.dnarep.2006.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]