Abstract
Two classes of sterols, cholesterol and oxysterols, block export of sterol regulatory element-binding proteins (SREBPs) from the endoplasmic reticulum (ER) to the Golgi by preventing the binding of COPII-coated proteins to a hexapeptide sorting signal (MELADL) in Scap, the SREBP-escort protein. Here, we show that anti-MELADL blocks COPII binding in vitro, and microinjection of Fab anti-MELADL blocks Scap·SREBP movement in cells. Cholesterol and oxysterols block COPII binding to MELADL by binding to different intracellular receptors, cholesterol to Scap and oxysterols to Insig. Cysteine labeling shows that both binding events produce a conformational change near the MELADL sequence, abrogating COPII binding but not anti-MELADL binding. Mutagenesis experiments raise the possibility that the distance of MELADL from the ER membrane is crucial for COPII binding, and we speculate that sterols and Insig block SREBP transport by altering the location of MELADL with respect to the membrane, rendering it inaccessible to COPII proteins.
Keywords: cholesterol homeostasis, COPII vesicles, SREBP pathway, oxysterols
Cholesterol is an essential lipid of vertebrate cell membranes. Not only does it regulate membrane fluidity and integrity (1), but it also serves as a critical component of lipid rafts or caveolae, which organize signaling events (2, 3). Cholesterol homeostasis is achieved by a feedback mechanism that monitors membrane cholesterol levels and controls transcription of genes mediating cholesterol biosynthesis and uptake. Central to this regulatory system is a family of membrane-bound transcription factors termed sterol regulatory element-binding proteins (SREBPs) (4). SREBPs are synthesized in the endoplasmic reticulum (ER), and their activation depends on a polytopic membrane protein called Scap. Scap is comprised of two domains: a cytosolic COOH-terminal regulatory domain that binds to SREBPs and an NH2-terminal domain that consists of eight transmembrane helices (5).
Scap plays a dual role in cellular cholesterol homeostasis, as an escort protein and as a sterol sensor. When cellular membranes are depleted of sterols, Scap mediates the clustering of SREBP in coat protein complex II (COPII)-coated vesicles. The vesicles carry the Scap·SREBP complex to the Golgi complex where the SREBP is cleaved sequentially by two proteases to release the transcriptionally active basic helix–loop–helix leucine zipper fragment. The released fragment travels to the nucleus, where it activates many genes involved in cholesterol synthesis and uptake (6). When cholesterol builds up in ER membranes, the sterol binds to Scap, altering its conformation and causing the Scap·SREBP complex to bind to Insig-1 or Insig-2, which are closely related polytopic membrane proteins (5). When the Scap·SREBP complex binds to Insig, the complex can no longer be incorporated into COPII-coated vesicles (7). The ER-retained SREBP is not processed, and transcription of the target genes declines. As a consequence, cholesterol synthesis and uptake fall.
The vesicles that carry SREBPs to the Golgi (7–9) are typical COPII-coated vesicles that have been studied widely in yeast and animal cells (10, 11). The COPII coat consists of five proteins: Sar-1, Sec23/Sec24 complex, and Sec13/Sec31 complex. Newly synthesized membrane proteins and secreted proteins destined for export (called cargo proteins) are clustered into COPII-coated vesicles in a reaction initiated when an ER resident protein, Sec12, stimulates the exchange of GTP for GDP on Sar-1. This exchange triggers the binding of Sar-1-GTP to ER membranes. There, the Sar-1-GTP complex provides a platform for recruitment of Sec23/Sec24, followed by Sec13/31. The Sec23/24 complex selects certain membrane cargo proteins that have sequences recognized by Sec24, which thereby clusters them into budding vesicles (11–14).
In the case of Scap, the binding site for Sec24 has been identified as a hexapeptide sequence, Met–Glu–Leu–Ala–Asp–Leu (MELADL), located in the cytoplasmic loop between transmembrane helices 6 and 7 (7). Sterols trigger the binding of Scap to Insig-1 or Insig-2, and this binding somehow precludes the binding of Sec24 to MELADL (7). How Insig blocks the binding of COPII proteins to the MELADL sequence in Scap is currently unknown.
In the current studies, we have sought to determine how Insig blocks the binding of COPII proteins to Scap's MELADL sequence. The results indicate that Insig does not occlude the MELADL sequence. Rather, Insig appears to cause a conformational change that may move the MELADL sequence closer to the membrane, and this change sequesters MELADL from Sec24. We show that this MELADL change can be triggered either by the binding of cholesterol to Scap or by the binding of oxysterols, such as 25-hydroxycholesterol (25-HC), to Insig (see accompanying paper, ref. 15). These studies provide a mechanistic explanation for the control of cholesterol metabolism in mammalian cells.
Results
Inhibition of Scap Transport by Anti-MELADL.
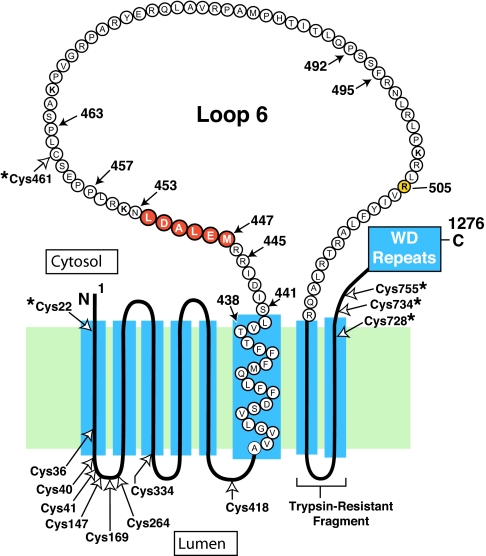
Fig. 1 shows the membrane domain of hamster Scap with emphasis on the 78-aa cytoplasmic loop (loop 6) between transmembrane helices 6 and 7. The hexapeptide MELADL, on the NH2-terminal side of loop 6, is crucial for binding COPII proteins and for the ER-to-Golgi transport of Scap·SREBP (7). Following the membrane domain of Scap, there is a sequence of 546 aa comprising multiple WD repeats that binds SREBPs (16).
Fig. 1.
Amino acid sequence and topology of the membrane domain of hamster Scap. The cytoplasmic loop between transmembrane helices 6 and 7 is referred to as loop 6, and it is postulated to extend from Ser-441 to Arg-518. Amino acids 447–452 (highlighted in red) constitute the hexapeptide sequence MELADL. Filled arrows denote amino acids that were mutated in the current experiments. Open arrows denote all of the cysteine residues in the membrane domains of Scap; those cysteines marked by asterisks (*) were changed to alanines to create the Cys(−) construct used as the parent plasmid for the experiments in Fig. 4. Arg-505 (highlighted in yellow) denotes a cholesterol-induced, trypsin-sensitive cleavage site (20, 21). Trypsin-resistant fragment denotes the sequence between membrane helices 7 and 8 (amino acids 540–707) that is detected by IgG-R139 (33).
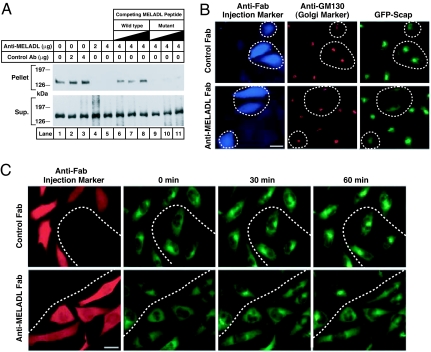
To further explore the function of the MELADL sequence, we produced an affinity-purified polyclonal rabbit antibody directed against a 16-aa synthetic peptide corresponding to residues 446 to 461, which includes the MELADL sequence (residues 447–452). For simplicity, we call this antibody anti-MELADL. Fig. 2A shows an experiment in which we tested the ability of anti-MELADL to block the binding of COPII proteins to Scap in membranes. For this purpose, we used SRD-13A cells, a line of mutant CHO cells that does not produce Scap, owing to mutations in both copies of the Scap gene (17). The SRD-13A cells were depleted of sterols by a brief incubation with hydroxypropyl-β-cyclodextrin (HPCD), which removes cholesterol from the plasma membrane (18). Before the experiment, the SRD-13A cells were transfected with plasmids encoding wild-type Scap, Insig-1, and SREBP-2. After sterol depletion, the cells were harvested and microsomal membranes were incubated with the indicated amount of anti-MELADL or an irrelevant antibody. As a further control, we blocked the interaction of anti-MELADL with Scap by adding various amounts of the 16-aa synthetic peptide containing the wild-type MELADL sequence (lanes 6–8) or the identical peptide with AAAAAA substituted for MELADL (lanes 9–11). After incubation on ice for 30 min, we added recombinant GST-tagged Sar-1 and FLAG-tagged Sec23/Sec24. After incubation for 15 min at 28°C, the membranes were solubilized in detergent and incubated with FLAG antibody beads to pull down the Sec23/Sec24 complex. The supernatant and pellet fractions were subjected to SDS/PAGE and blotted with an antibody against Scap. In the absence of anti-MELADL, the COPII proteins pulled down Scap (Fig. 2A, lane 1). Preincubation with anti-MELADL blocked the COPII pull down of Scap (lanes 4, 5). Preincubation with the control antibody had no effect (lanes 2, 3). The inhibiting effect of anti-MELADL was blocked by inclusion of an excess of the wild-type peptide containing MELADL (lanes 6–8), but not the mutant peptide with the 6-alanine substitution (lanes 9–11). These data show that the binding of the antibody to the MELADL sequence prevents COPII protein binding to Scap in vitro.
Fig. 2.
Anti-MELADL inhibits Scap transport from ER to Golgi. (A) Binding of COPII proteins to Scap in vitro. On day 0, Scap-deficient SRD-13A cells were set up in 100-mm dishes. On day 2, cells were transfected with 2 μg of pTK-SREBP-2, 0.4 μg of pCMV–Scap, and 0.2 μg of pCMV-Insig-1-Myc. Twelve hours later, cells were switched to sterol-depleting medium containing 1% HPCD and incubated for 1 h at 37°C. Cells were then switched to the same medium without HPCD. After 3 h at 37°C, cells were harvested, and 150 μg of microsomal membranes (16,000 × g pellet) were incubated, in a final volume of 0.3 ml Buffer B, with the indicated amount of affinity-purified control anti-T7 tag or anti-MELADL antibody in absence or presence of increasing amounts (0.2, 0.5, and 1.0 mg) of a 16-aa synthetic peptide corresponding to residues 446–461 of Scap and containing wild-type (lanes 6–8) or a mutant MELADL sequence substituted with AAAAAA (lanes 9–11). After a 30-min incubation on ice, we added 10 μg of a recombinant mutant of GST-Sar-1(H79G; GTPase-defective) and 10 μg of recombinant Flag-Sec23/24 in the presence of 0.5 mM sodium GTP. The Scap/COPII complex was precipitated with anti-FLAG. The resulting supernatant (Sup.) and pellet (5% of Sup.) fractions were subjected to 8% SDS/PAGE and immunoblot analysis with IgG-R139 (anti-Scap). (B) Immunofluorescence. On day 0, CHO/pGFP-Scap cells were set up on 12-mm coverslips as described in Materials and Methods. On day 2, cells were switched to microinjection medium supplemented with 5% FCS, after which Fab fragments (0.2 mg/ml) of either anti-MELADL or control antibody (IgG-R139) were microinjected into the cytoplasm. After 1-h incubation at 37°C, cells were switched to sterol-depleting medium containing 1% HPCD without sterols and incubated for 1 h at 37°C. Cells were then fixed for 15 min in 3.7% formaldehyde in PBS at room temperature and permeabilized for 10 min in methanol at −20°C. Cells were stained with monoclonal IgG-GM130 (1.2 μg/ml, anti-GM130) and visualized with Alexa Fluor 594 goat anti-mouse IgG (1:300 dilution, GM130), and Alexa Fluor 350 goat anti-rabbit IgG (1:200 dilution, microinjected Fab fragments). GFP-Scap was visualized directly. (Scale bar, 25 μm.) (C) Time-lapse imaging. On day 0, CHO/pGFP-Scap cells were set up on 18-mm coverslips as described in Materials and Methods. On day 1, cells were switched to medium A with 5% LPDS and 1% HPCD in the presence of sterols (10 μg/ml 25-HC and 10 μg/ml cholesterol in 0.2% ethanol) and incubated for 16 h. On day 2, cells were refed with microinjection medium with 5% LPDS and 1% HPCD in the presence of sterols, after which Fab fragments of either anti-MELADL or control antibody (IgG-R139) were microinjected into the cytoplasm (34). After 30 min (zero time), the coverslip was mounted into a square holder (Ludin chamber) and positioned on the microscope stage, after which the cells were switched to sterol-depleting imaging medium, and images were collected every 5 min for 75 min. Note that only images taken at 0, 30, and 60 min are presented. During the experiment, cells were kept at 37°C on a microscope enclosed in an environmental chamber. After imaging, the coverslip chamber was removed from the stage after which the cells were fixed in the chamber, permeabilized as described above, and stained with Alexa Fluor 594 conjugated goat anti-rabbit IgG (1:200 dilution). To identify injected cells, the coverslip chamber was mounted back on the microscope stage, which was repositioned to the stored x–y coordinates on the stage. This allowed us to image the same field of cells as detected during live cell imaging. (Scale bar, 25 μm.)
To determine whether anti-MELADL blocks Scap transport in intact cells, we microinjected the Fab fragment of anti-MELADL into the cytoplasm of Scap-deficient SRD-13A cells that were stably transfected with a plasmid encoding GFP-Scap (8). We studied ER-to-Golgi transport of GFP-Scap using fluorescence microscopy (Fig. 2B). The cells were depleted of sterols by incubation with medium containing HPCD. Under these sterol-depleting conditions, GFP-Scap displayed strong juxtanuclear staining (Fig. 2B Right), which coincided with structures stained by antibodies to the Golgi resident protein GM130 (Fig. 2B Center), confirming that GFP-Scap had moved to the Golgi. In cells microinjected with Fab fragments of anti-MELADL, the concentration of GFP-Scap in the Golgi was lost, and the protein displayed a diffuse, reticular pattern corresponding to the ER (Fig. 2B Lower Right). Microinjection of Fab fragments of a control antibody did not block the Golgi localization of GFP-Scap (Fig. 2B Upper Right).
Fig. 2C shows an experiment designed to demonstrate the rapid movement of GFP-Scap immediately after sterol depletion. To begin the experiment, cells were cultured in medium containing 25-HC and cholesterol, a mixture that causes Scap to be retained in the ER (19). The cells were microinjected with Fab fragments of anti-MELADL or a control Fab, and they were switched to sterol-depleting imaging medium that contained HPCD. Under these conditions, the HPCD triggers the movement of Scap to the Golgi (8). The injected cells were visualized by fixation, permeabilization, and incubation with a fluorescent anti-Fab antibody; they appeared red in the fluorescence micrographs. At zero min of this experiment, GFP-Scap was localized to the ER in a diffuse, reticular pattern. After 30 min of sterol depletion, GFP-Scap had already moved to the Golgi (Fig. 2C Upper), and remained there at 60 min. In the cells that were injected with Fab fragments of anti-MELADL, transport of GFP-Scap from ER to Golgi was abolished (Fig. 2C Lower). Transport was unaffected in the cells that were not injected.
Insig Prevents Binding of COPII Proteins, but Not Anti-MELADL, to Scap.
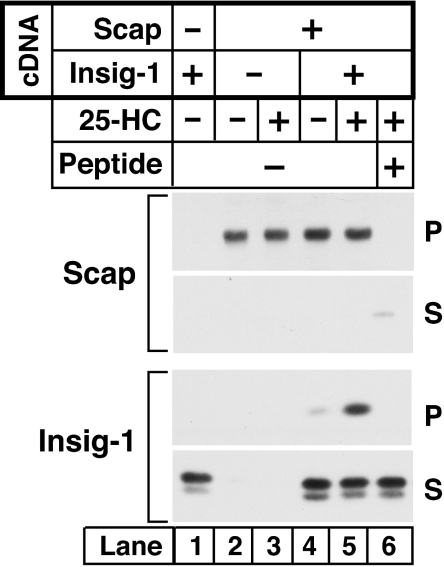
In the presence of Insig-1 or Insig-2, cholesterol or oxysterols cause Scap to bind to Insig, and this prevents binding of COPII proteins to Scap (7). Inasmuch as the MELADL sequence is required for COPII binding, the question arises as to whether Insig physically occludes the MELADL sequence, or whether it prevents COPII binding by some other means. To answer this question, we tested whether Insig binding prevents anti-MELADL from binding to Scap. SRD-13A cells were transfected with plasmids encoding Scap and SREBP-2 with or without Insig-1. The membranes were isolated and incubated with affinity-purified anti-MELADL, and the immune complexes were solubilized in detergent and precipitated with Protein A/G beads. In the absence of 25-HC and Insig-1, anti-MELADL immunoprecipitated all of the Scap (Fig. 3Upper, lane 2). Addition of the MELADL-containing peptide blocked this precipitation (Fig. 3 Upper, lane 6). Precipitation of Scap remained complete, even when 25-HC and Insig-1 were present (lane 5). Under these conditions, anti-MELADL pulled down Insig-1 (Fig. 3 Lower, lane 5), showing formally that Scap can bind anti-MELADL and Insig-1 at the same time. Thus, Insig-1 does not completely occlude the MELADL sequence, and it must block COPII binding by some other mechanism.
Fig. 3.
Binding of Insig-1 to Scap does not block binding of antibody to the MELADL sequence. On day 0, Scap-deficient SRD-13A cells were set up in 60-mm dishes. On day 2, cells were transfected with 2 μg of pTK-SREBP-2 (all dishes) and 0.2 μg of pCMV–Scap and 0.05 μg of pCMV-Insig-1-Myc as indicated. Twelve hours after transfection, cells were switched to sterol-depleting medium containing 1% HPCD and incubated at 37°C for 1 h. The cells were then incubated with sterol-depleting medium (without HPCD) in the absence or presence of 1 μg/ml of 25-HC. After incubation for 3 h at 37°C, the cells were harvested, and aliquots of microsomal membranes (150 μg), in a final volume of 0.3 ml of Buffer B, were mixed with 3 μg of anti-MELADL antibody in the absence or presence of 0.5 mg of the MELADL-containing synthetic peptide as indicated. After incubation for 1 h at 4°C, the microsomal membranes were pelleted by centrifugation, solubilized in Buffer D (50 mM Hepes-KOH, pH 7.2/150 mM KCl/1 mM MgCl2, and a mixture of protease inhibitors; see Materials and Methods) supplemented with 0.1% Nonidet P-40, and incubated with Protein A/G beads for 30 min at 4°C. The resulting supernatant (S) and pellet (P) (10% of S) fractions were subjected to 8% SDS/PAGE and immunoblot analysis with IgG-9D5 (anti-Scap) and IgG-9E10 (anti-Insig-1).
Conformational Changes in Scap Induced by Insig and Sterols.
If Insig-1 does not physically occlude the MELADL sequence, perhaps it alters the conformation of Scap in such a way that the spatially restricted COPII proteins no longer have access to the MELADL site. Recent studies showed that addition of cholesterol to isolated cell membranes causes a conformational change in loop 6 of Scap, as detected by exposure of a previously buried arginine (Arg-505) to cleavage by trypsin (20, 21). Arg-505 is located in the COOH-terminal portion of loop 6, whereas the MELADL sequence is in the NH2-terminal portion (Fig. 1). To determine whether sterols also induce a conformational change in the NH2-terminal portion of loop 6, we used mPEG-MAL-5000, a high molecular weight membrane-impermeable agent that forms a covalent bond with exposed cysteines, thereby causing proteins to migrate with higher molecular weight on SDS/PAGE (22). The first step was to prepare a plasmid encoding the truncated membrane domain of Scap (residues 1–767) in which all five cytoplasmically disposed cysteines were changed to alanine (see Fig. 1). We designate this plasmid as pCMV–Scap(1–767;Cys−). We then introduced a single cysteine residue at one of several positions near the MELADL sequence at the NH2-terminal end of loop 6. CMV-Scap(1–767;Cys−) and all of the single-cysteine proteins retained their ability to bind to Insig-1 in a sterol-dependent manner (data not shown).
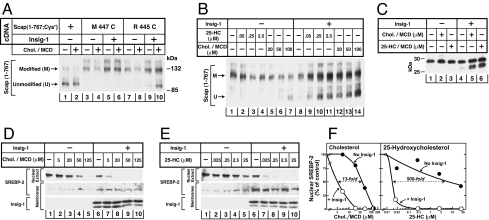
Fig. 4A shows an experiment in which we used SRD-15 cells, a line of mutant CHO cells that are deficient in both Insig-1 and Insig-2 (23). The cells were transfected with plasmids encoding single-cysteine versions of Scap (1–767) and then incubated with or without cholesterol delivered in methyl-β-cyclodextrin (MCD). Sealed membrane vesicles were isolated and then treated with mPEG-MAL-5000, after which the reaction was quenched with DTT. The proteins were solubilized, subjected to SDS/PAGE, and immunoblotted with anti-Scap. In cells expressing Scap(1–767;Cys−), mPEG-MAL-5000 did not react with the cysteine-deficient Scap, and only the unmodified protein was seen in the SDS/PAGE (lower band in Fig. 4A, lanes 1, 2). Introduction of a single cysteine at residue 447 rendered the protein susceptible to mPEG-MAL-5000 modification as indicated by a slowed migration on SDS/PAGE (Fig. 4A, lanes 3–6). Incubation of the cells with cholesterol did not alter the susceptibility of Cys-447 to modification, regardless of whether Insig-1 was coexpressed. This finding indicates that Cys-447 is exposed to the cytoplasm in both conformations of Scap. A different result was observed when the cysteine was located two residues closer to the membrane, i.e., at residue 445 (Fig. 4A, lanes 7–10). In extracts from sterol-depleted cells, this cysteine was modified by mPEG-MAL-5000 (Fig. 4A, lanes 7 and 9). When the cells were incubated with 20 μM cholesterol in MCD in the absence of Insigs, Cys-445 continued to be modified (Fig. 4A, lane 8). However, a substantial fraction of this protein became resistant to modification when cholesterol was added to cells that expressed Insig-1 (Fig. 4A, lane 10). These findings suggest that cholesterol caused a conformational change in Scap such that the amino acid at residue 445 became resistant to modification. At 20 μM cholesterol, this change was detected only when Insig-1 was present.
Fig. 4.
Sterol-mediated conformational change in region of MELADL sequence of Scap. (A) mPEG-MAL labeling after sterol treatment of intact cells with cholesterol. On day 0, Insig-1- and Insig-2-deficient SRD-15 cells were set up in 60-mm dishes. On day 1, cells were transfected with 2 μg of a plasmid encoding mutant version of Scap(1–767;Cys−) with either no cytoplasmically oriented cysteines or with a single cysteine at the indicated residue. The cells also received 0.5 μg of pCMV–Insig1–T7 as indicated. Twelve hours after transfection, cells were switched to sterol-depleting medium containing 1% HPCD for 1 h at 37°C. Cells then received sterol-depleting medium (without HPCD) with 25 μg/ml ALLN in absence or presence of 20 μM cholesterol [delivered in MCD at a cholesterol:MCD ratio (wt/wt) of 1:10]. After 3 h at 37°C, cells were harvested for preparation of sealed microsomal membrane vesicles, which were then treated with 2 mM mPEG-MAL-5000 to modify cytoplasmically exposed cysteines as described in Materials and Methods. The membranes were pelleted, solubilized, and subjected to 7% SDS/PAGE and immunoblotted with IgG-R139 (anti-Scap). (B) mPEG-MAL labeling after treatment of intact cells with 25-HC or cholesterol. On day 0, Insig-1- and Insig-2-deficient SRD-15 cells were set up in 60-mm dishes. On day 1, cells were transfected with 2.5 μg of pTK-HSV-SREBP-2 and 2 μg of pCMV–Scap(1–767;Cys−;R445C). Cells were also transfected with 0.5 μg of pCMV–Insig1–T7 as indicated. Twelve h after transfection, cells were incubated with sterol-depleting medium containing 1% HPCD for 1 h at 37°C. Cells were then switched to sterol-depleting medium (without HPCD) with the indicated concentration of either 25-HC (dissolved in ethanol) or cholesterol (delivered in MCD). After incubation for 6 h (ALLN at 25 μg/ml added 3 h before harvest), cells were harvested, and sealed microsomal membrane vesicles were prepared and treated with 2 mM mPEG-MAL-5000 as described above. The modified membranes were subjected to 7% SDS/PAGE and immunoblot analysis with IgG-R139 (anti-Scap). (C) Trypsin cleavage of Scap after treatment of intact cells with cholesterol or 25-HC. On day 0, Scap-deficient SRD-13A cells were set up in 60-mm dishes. On day 2, cells were transfected with 2 μg of pTK-HSV-SREBP-2 and 1.25 μg of pCMV–Scap in the absence or presence of 0.6 μg pCMV-Insig-1-Myc. Twelve hours after transfection, cells were switched to sterol-depleting medium containing 1% HPCD. After incubation for 1 h at 37°C, cells were incubated with sterol-depleting medium (without HPCD) containing no sterols, 50 μM of either cholesterol or 25-HC [delivered in MCD at a sterol/MCD ratio (wt/wt) of 1:10]. After incubation for 3 h, cells were harvested, and aliquots of microsomal membranes (100 μg) were treated sequentially with 2 μg of trypsin (30°C, 30 min) and 625 units of PNGase F (37°C, 12 h) and then subjected to 12% Tris-tricine SDS/PAGE and immunoblot analysis with IgG-R139 (anti-Scap). (D and E) SREBP-2 cleavage after treatment of cells with cholesterol (D) or 25-HC (E). SRD-15 cells were set up, transfected, and treated with the indicated concentration of cholesterol (delivered in MCD) or 25-HC (dissolved in ethanol) as in B. Nuclear extract and membrane fractions were prepared and subjected to 8% SDS/PAGE and immunoblot analysis with anti-HSV IgG (anti-SREBP-2) and anti-T7 IgG (anti-Insig-1) as indicated. (F) Densitometric quantification of the bands in D and E. The intensity of the cleaved nuclear form of SREBP-2 in the absence of sterol treatment was arbitrarily set at 100%.
As discussed above, cholesterol and oxysterols such as 25-HC both block SREBP processing. In previous studies we showed that Scap binds cholesterol, but it does not bind oxysterols (24). In a companion paper to the current one, we show that Insigs bind oxysterols such as 25-HC, but they do not bind cholesterol (15). The question then arose as to whether 25-HC, by binding to Insigs, can produce the same conformational change at residue 445 in Scap that cholesterol causes by binding directly to Scap. To test this hypothesis, we transfected the cDNA encoding Scap(1–767;Cys−;R445C) into SRD-15 cells (Fig. 4B). The cells were then incubated with the indicated amount of 25-HC dissolved in ethanol or cholesterol complexed with MCD, after which membrane vesicles were isolated and subjected to cysteine modification. In cells lacking Insigs, the sterol-depleted membranes showed only an upper band consistent with complete modification of Cys-445 (Fig. 4B, lane 1). Addition of 100 μM cholesterol blocked this modification, causing the appearance of the lower band (Fig. 4B, lane 7). The presence of Insig-1 markedly enhanced the cholesterol effect with a lower concentration of cholesterol (20 μM) producing an appreciable amount of unmodified Scap (1–767) (Fig. 4B, lane 12). In the absence of Insig-1, incubation of cells with 25-HC up to 2.5 μM had no effect (lanes 2–4). In cells expressing Insig-1, addition of as little as 0.05 μM 25-HC led to an appreciable fraction of unmodified Scap (1–767) (Fig. 4B, lanes 9–11). These data indicate that high concentrations of cholesterol can induce a conformational change in Scap in the absence of Insigs and that this effect is enhanced by Insig-1. In contrast, 25-HC causes very little conformational change in Scap in the absence of Insigs. In experiments not shown, we found that 25-HC delivered in MCD caused no conformation change in Scap even at the highest concentration tested (100 μM). In the presence of Insig-1, 25-HC becomes extremely potent in altering the conformation of Scap. Thus, the 25-HC effect shows a greater dependence on Insig-1 than does the cholesterol effect.
As discussed above, the cholesterol-induced conformational change at the COOH-terminal end of loop 6 in Scap is manifested by exposure of Arg-505 to cleavage by trypsin (20, 25). To detect this change, sealed membrane vesicles are isolated from sterol-depleted cells and treated with trypsin. The proteins are subjected to SDS/PAGE and blotted with IgG-R139, a polyclonal antibody to the loop of Scap between membrane helices 7 and 8, which is intraluminal and therefore protected from trypsin (see Fig. 1). When blotted with IgG-R139, sterol-depleted membranes show a trypsin-protected fragment with an apparent molecular mass of ≈28 kDa. When cholesterol is added to intact cells or to membranes in vitro, Arg-505 becomes exposed to trypsin, and a smaller band of ≈27 kDa is observed.
Fig. 4C shows an experiment designed to determine whether the sterol-induced conformational change in the NH2-terminal end of loop 6 (detected by mPEG-MAL-5000 modification as in Fig. 4B) correlates with the sterol-induced conformational change in the COOH-terminal end of this loop (detected by trypsin digestion). For this purpose, we transfected Scap-deficient SRD-13A cells with wild-type Scap in the absence or presence of transfected Insig-1, after which the cells were incubated with either cholesterol or 25-HC delivered in MCD. We then harvested the cells, isolated membrane vesicles, and subjected them to trypsin treatment. In the absence of Insig-1, treatment of the intact cells with either cholesterol or 25-HC at 50 μM yielded only the 28-kDa upper band (Fig. 4C, lanes 1–3), indicating that Arg-505 was not accessible to trypsin (20, 25). In the presence of Insig-1, both cholesterol and 25-HC produced a clear bottom band of 27 kDa, consistent with cleavage at arginine-505 (Fig. 4C, lanes 5 and 6). Considered together with the experiment in Fig. 4B, these data indicate that the cholesterol and 25-HC induced conformational change at residue 445 in loop 6 of Scap occurs concomitantly with the aforementioned conformational change at residue 505 in the same loop. Both changes are strongly influenced by Insig.
To determine whether the Insig-dependent, sterol-induced conformational change in the region of the MELADL sequence correlates with the block in SREBP-2 processing, SRD-15 cells were transfected with a plasmid encoding HSV-tagged SREBP-2 with or without a plasmid encoding Insig-1 and incubated for 6 h in the presence of varying amounts of cholesterol complexed with MCD (Fig. 4D) or with 25-HC in ethanol (Fig. 4E). Membrane fractions and nuclear extracts were subjected to SDS/PAGE and blotted with antibodies against the NH2-terminal HSV tag on SREBP-2 or against Insig-1. In the absence of Insig-1, SREBP-2 cleavage was abolished by 125 μM cholesterol as indicated by a disappearance of SREBP-2 from the nuclear fraction (Fig. 4D Upper, lane 5). In the presence of Insig-1, the effect of cholesterol was enhanced (Fig. 4D Upper, lanes 7–10). In the absence of Insig-1, 25-HC had only a modest effect in reducing SREBP-2 cleavage, even at levels as high as 25 μM (Fig. 4E Upper, lanes 1–5). In contrast, in the presence of Insig-1, addition of 25-HC at a 1,000-fold lower concentration (25 nM) decreased SREBP-2 processing by 80% (Fig. 4E Upper, lanes 6–10). Fig. 4F shows densitometric quantification of the amount of nuclear SREBP-2 as a function of cholesterol or 25-HC concentrations. In this experiment Insig-1 increased the sensitivity to cholesterol by 13-fold (Fig. 4F Left). In 3 other experiments not shown, Insig-1 increased cholesterol sensitivity by 7- to 20-fold (average of 13-fold). Insig-1 increased the sensitivity to 25-HC by a much higher amount: 500-fold as shown in Fig. 4F Right. In three other experiments not shown, Insig-1 shifted the sensitivity 160- to 700-fold (average of 390-fold). Thus, inhibition of SREBP-2 processing by 25-HC shows a greater dependence on Insig-1 than does the inhibition by cholesterol. These findings are consistent with the results of the cysteine modification assays, which show greater dependence on Insig-1 for the 25-HC effect on Scap conformation as compared with the cholesterol effect (Fig. 4B).
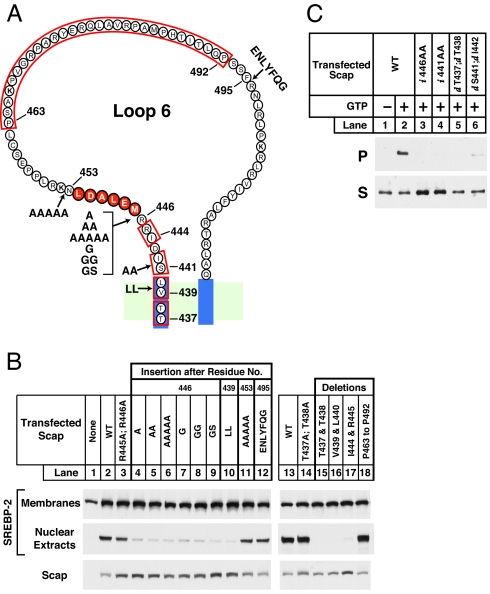
Amino Acid Insertions and Deletions Between MELADL and the Membrane Alter Scap Transport.
To determine whether the distance between the MELADL sequence and the membrane is important for the binding of COPII proteins, we constructed a series of insertion or deletion mutants (Fig. 5). We introduced the mutant plasmids into Scap-deficient SRD-13A cells by transfection, together with a plasmid encoding epitope-tagged SREBP-2. After depleting the cells of cholesterol, we measured the amount of cleaved nuclear SREBP-2 by immunoblotting of nuclear extracts. These data show that insertion of various residues between the MELADL sequence and the membrane decreased SREBP-2 processing. These insertions consisted of a single alanine (A), two alanines (AA), five alanines (AAAAA), one glycine (G), two glycines (GG), or glycine–serine (GS) between residues 446 and the MELADL sequence (Fig. 5B, lanes 4–9). A similar result was observed when we inserted two leucines between residues 439 and 440 (lane 10), which are predicted to lie within the membrane. All of these insertions are predicted to move the MELADL sequence further from the membrane. In contrast, insertion of residues on the COOH-terminal side of the MELADL sequence, such as five alanines (AAAAA) after residue 453 or a TEV protease cleavage site (ENLYFQG) after residue 495, did not affect SREBP-2 processing (lanes 11 and 12). Amino acid deletions within the membrane (T437 and T438 or V439 and L440) or between MELADL and the membrane (I444 and R445) led to a profound reduction in SREBP-2 processing (lanes 15–17). These deletions are predicted to move the MELADL sequence closer to the membrane. In contrast, large deletions on the COOH-terminal side of MELADL, such as removal of 30 aa from residue 463 to residue 492, had no effect on SREBP-2 processing (lane 18). Although insertions and deletions between MELADL and the membrane reduce SREBP-2 cleavage, substitution mutations in this region such as R445A;R446A (lane 3) or T437A;T438A (lane 14) did not abolish SREBP-2 processing. None of these mutations altered the level of expression of Scap, as determined by immunoblotting (Fig. 5B). These data raise the possibility that the distance between MELADL and the membrane is crucial in allowing COPII proteins to transport the Scap·SREBP-2 complex from ER to Golgi. Moving the MELADL sequence either closer to or further from the membrane impairs the function of the MELADL sequence.
Fig. 5.
Amino acid insertions or deletions adjacent to MELADL sequence alter Scap function. (A) Amino acid sequence of loop 6 of Scap, showing sites of insertions (arrows) and deletions (red box). (B and C) On day 0, Scap-deficient SRD-13A cells were set up in 60-mm dishes. (B) SREBP-2 cleavage. On day 2, cells were transfected with 2.5 μg of pTK-SREBP-2 and 0.2 μg of the indicated wild-type or mutant version of pCMV–Scap. Twelve hours after transfection, cells were switched to sterol-depleting medium with 1% HPCD and incubated for 1 h at 37°C. Cells were then incubated with sterol-depleting medium (without HPCD) for 3 h at 37°C and harvested for preparation of nuclear extract and membrane fractions. These fractions were subjected to 8% SDS/PAGE and immunoblot analysis with anti-HSV IgG (anti-SREBP-2) and IgG-R139 (anti-Scap) as indicated. (C) Flag-Sec23 pulldown. On day 2, cells were transfected with 0.4 μg of wild-type or the indicated mutant version of pCMV–Scap (i, insertion; d, deletion). Twelve hours after transfection, cells were treated the same as in B and harvested. Microsomal membranes (150 μg) were analyzed for Scap binding to COPII proteins using the Flag-Sec23 pull-down assay as described in Fig. 2A. Supernatant (S) and pellet (P) (5% of S) fractions were subjected to 8% SDS/PAGE and immunoblot analysis with IgG-R139 (anti-Scap).
To confirm that the insertions and deletions blocked SREBP-2 processing by preventing COPII protein binding to Scap, we measured the ability of FLAG-Sec23/24 to bind Scap in vitro (Fig. 5C). In the presence of GTP, FLAG-Sec23 bound to wild-type Scap, and a fraction of the Scap was found in the pellet fraction after precipitation with anti-FLAG (lane 2). Insertion of two alanines at positions 446 or 441 between MELADL and the membrane led to a marked decrease in FLAG-Sec23 binding (lanes 3 and 4). A similar reduction was observed when we deleted pairs of residues between MELADL and the membrane (437 plus 438 or 441 plus 442) (lanes 5 and 6). Thus, moving MELADL with respect to the membrane reduced the ability of COPII proteins to bind to the MELADL sequence in vitro.
Discussion
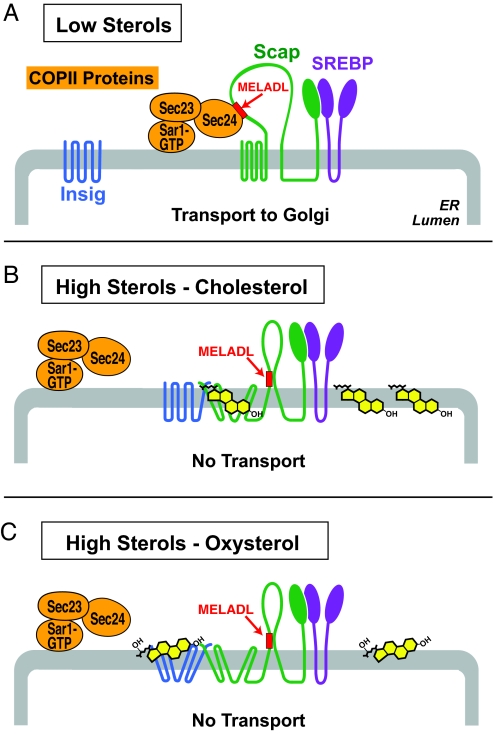
The current results, together with those in the accompanying paper (15), indicate that animal cells evolved distinct but convergent mechanisms to facilitate the control of cholesterol metabolism by two classes of sterols: cholesterol and oxysterols. This control mechanism is illustrated schematically in Fig. 6. The pivotal event is the sterol-regulated binding of the Sec23/Sec24 complex of COPII coat proteins to the MELADL sequence in Scap, an event that is required in order for the Scap·SREBP complex to exit the ER. Cholesterol and oxysterols both block COPII protein binding by causing Insigs to bind to Scap. However, they do so by different mechanisms. Cholesterol binds to Scap, triggering Scap to bind to Insigs. Oxysterols bind to Insigs, triggering Insigs to bind to Scap. In both cases, the end result is a conformational change in the cytoplasmic loop 6 of Scap that prevents the COPII proteins from gaining access to the MELADL sequence.
Fig. 6.
Differential mechanisms by which cholesterol and oxysterols trigger Insig binding to Scap, which in turn abrogates COPII binding to the MELADL sorting signal in Scap, inhibiting the ER-to-Golgi transport of SREBPs.
The crucial role of the MELADL sequence in ER-to-Golgi transport of Scap was demonstrated visually in Fig. 2, which shows that microinjection of a Fab fragment of anti-MELADL abolishes Scap movement to the Golgi. When sterols trigger the binding of Insigs to Scap, the COPII proteins can no longer bind to the MELADL sequence (7). Nevertheless, the MELADL sequence is still accessible to anti-MELADL (Fig. 3). We speculate that this difference indicates that the MELADL sequence is not covered up, but rather that its conformation may be changed or may be moved to a location where the Sec24/23 complex can no longer have access, but where the antibody retains access. Because the Sec23/24 heterodimer is in a complex with membrane-bound GST-Sar-1, it seems likely that the binding site on Sec24 is located at a fixed distance from the membrane, and it is possible that the Insig-dependent conformational change in Scap may move the MELADL sequence with respect to the membrane so that it is no longer in range of the binding site on Sec24.
The hypothesis regarding the importance of juxtamembrane spacing of MELADL was supported when we altered the distance between the MELADL sequence and the membrane by making a series of insertion and deletion mutations (Fig. 5A). The insertion of even a single alanine or glycine between MELADL and the membrane abolished SREBP transport and processing, as did the deletion of various pairs of residues (Fig. 5B). Small insertions and deletions also blocked the binding of COPII proteins in vitro (Fig. 5C). Substitution mutations in this region had no effect on SREBP processing (Fig. 5B, lanes 3 and 14), suggesting that the length of this segment, rather than its absolute structure, was crucial for COPII binding. In contrast to the profound effects of length alterations between MELADL and the membrane, even large deletions on the COOH-terminal side of MELADL had no effect on transport (Fig. 5 A and B), further supporting the notion that the spacing between MELADL and the membrane is crucial.
The suggestion that Insigs move the MELADL sequence out of reach of the COPII proteins is supported by the cysteine modification experiments of Fig. 4. These experiments show that a cysteine introduced at residue 445 between MELADL and the membrane becomes less accessible to covalent modification in the presence of Insigs plus sterols, suggesting that it is buried by a conformational change, perhaps one that moves it closer to the membrane. Previous studies have shown that cholesterol can bind to Scap in vitro (24) and produce a conformational change even in the absence of Insigs (25). Large amounts of cholesterol can also produce this conformational change when added to cells deficient in Insigs. However, the change becomes markedly more sensitive to cholesterol when Insigs are present (Fig. 4B). This finding suggests that the cholesterol-induced conformational change may be reversible and that the subsequent binding of Insigs shifts the equilibrium toward the cholesterol-bound state, thereby increasing the apparent affinity for cholesterol.
A different scenario applies in the case of oxysterols such as 25-HC. In contrast to cholesterol, oxysterols do not bind to Scap in vitro (15, 24). On the other hand, oxysterols, but not cholesterol, bind to Insigs (15). The notion that 25-HC acts by binding to Insigs is supported by the finding that 25-HC has very little ability to cause a conformational change in Scap or to block SREBP processing in Insig-deficient cells (Fig. 4). When SREBP-2 cleavage is measured in intact cells, the presence of Insig-1 increases the sensitivity to cholesterol by 13-fold, but it increases the sensitivity to 25-HC by 500-fold, which is nearly an all-or-none effect (Fig. 4F).
The MELADL sequence differs from other sorting signals that have been shown to be essential for COPII binding and ER exit in coated vesicles (11, 13, 26). It is not known whether these previously recognized sequences must lie at a fixed distance from the ER membrane. It is possible that nature has placed this constraint only on Scap so that it can be exploited to allow sterol regulation. Further studies of regulated ER exit of Scap·SREBP and other cargo protein should clarify this important general question.
Previous cross-linking studies of COPII proteins to artificial membrane surfaces by Matsuoka et al. (27) and crystallographic data of the Sec23/24-Sar-1 prebudding complex by Bi et al. (28) have convincingly shown that Sec23/24 is closely associated with the lipid bilayer. An important test of our hypothesis that the distance of the MELADL sequence from the membrane is crucial for sterol-mediated regulation of COPII accessibility of Scap will be whether the MELADL region of Scap can be cross-linked to membrane lipids in a cholesterol (or oxysterol) and Insig-dependent manner.
As a final comment, it is noteworthy that the molecular switch that controls sterol metabolism in animal cells (reviewed in the accompanying paper, ref. 15) is a simple hexapeptide targeting signal (MELADL) in a single membrane protein (Scap). For >30 years, scientists have known that cellular cholesterol synthesis and uptake are inhibited in feedback fashion by one of two classes of sterol molecules, either the end-product itself (cholesterol) or a hydroxylated derivative of cholesterol (oxysterol) (29–31). The latter are formed in cells through the actions of one of several hydroxylases (32). From the data in the current and companion papers, we now know that each of these classes of sterols initiates its regulatory action by binding to different intracellular receptors: cholesterol to Scap and oxysterol to Insig. Thereafter, their actions converge. Both ligands cause Insig to bind to Scap, and this produces a single conformational change that switches the MELADL sorting signal in Scap to a new location with respect to the ER membrane, thereby precluding COPII protein binding. It remains to be seen whether transport of other cargo proteins from ER to Golgi is regulated by shifts in the location of their COPII regulatory sequences.
Materials and Methods
Cultured Cells and Transfection.
SRD-13A cells, a Scap-deficient cell line derived from CHO-7 cells (17), were grown in monolayer at 37°C in 8–9% CO2 in medium A supplemented with 5% (vol/vol) FCS, 1 mM sodium mevalonate, 20 mM sodium oleate, and 5 μg/ml cholesterol. On day 0, cells were set up for experiments in the above medium at a density of either 3.25 × 105 cells per 60-mm dish or 7 × 105 cells per 100-mm dish. On day 2, cells were transfected with various plasmids in medium A supplemented with 5% FCS using FuGENE 6 according to the manufacturer's protocol (Roche Diagnostics). SRD-15 cells, an Insig-1/Insig-2 deficient cell line (23), were maintained in medium A supplemented with 5% (vol/vol) lipoprotein-deficient serum (LPDS), 10 μm of SR-12813, and 1 μg/ml 25-HC. On day 0, cells were set up for experiments in medium A supplemented with 5% LPDS at a density of 5 × 105 cells per 60-mm dish. On day 1, cells were transfected with various plasmids in medium A supplemented with 5% LPDS, 5 μM compactin, and 50 μM sodium mevalonate using the same transfection reagent as above. For each transfection, the total amount of DNA was adjusted to 3 μg per dish by addition of pcDNA3 mock vector. Conditions of incubation after transfection are described in figure legends.
Immunofluorescence.
On day 0, stably transfected CHO/pGFP-Scap cells (8) were set up on 12- to 18-mm coverslips in 60-mm dishes at a density of 3.5 × 105 cells per dish in medium A supplemented with 5% FCS. On day 1 (Fig. 2C) or day 2 (Fig. 2B), cells were used for experiments as described in the figure legends. Fluorescence analysis was performed using a Zeiss Axiovert 200M microscope (Zeiss, Thornwood, NY) and a Plan-Neofluar ×40/1.3 N.A. DIC oil immersion objective (Zeiss). Images were captured on an Orca 285 CCD camera (Hamamatsu, Hamatsu City, Japan) using the software package Openlab 4.02 (Improvision, Coventry, U.K.).
Recombinant Proteins and Microsomal Membranes.
Recombinant hamster GST-tagged-Sar-1(H79G) and the dimeric complex of Flag-tagged mouse Sec23A and His-tagged human Sec24C (Sec23/Sec24) were expressed and purified, and the hamster microsomal membranes (16,000 × g pellet) were prepared as described (7).
Flag-Sec23 Pull-Down Assay.
The assay was carried out as described (7) with a single modification: After solubilizing COPII·Scap complex in 0.5% (wt/vol) digitonin, the supernatant was incubated with anti-Flag M2 Affinity gel (20 μl in a final volume of 0.3 ml) instead of GSH-Sepharose beads.
Trypsin Cleavage of Scap.
Aliquots (100 μg) of sealed membrane vesicles (20,000 × g pellet) from transfected SRD-13A cells were subjected to trypsin digestion, followed by electrophoresis on 12% Tris-tricine gels as described (25).
Chemical Modification of Scap.
Microsomal membranes from two 60-mm dishes of SRD-13A cells expressing Scap(1–767;Cys−) were pooled and incubated for 15 min at 20°C with 2–3 mM mPEG-MAL-5000 in a final volume of 0.2 ml of Buffer B (50 mM Hepes-KOH, pH 7.2/250 mM sorbitol/70 mM potassium acetate/5 mM sodium EGTA/1.5 mM magnesium acetate, and a mixture of protease inhibitors containing 10 μg/ml leupeptin, 10 μg/ml pepstatin A, 6 μg/ml aprotinin, and 25 μg/ml ALLN). The reaction was terminated by addition of DTT at a final concentration of 10 mM. After incubation for 15 min at 4°C, the membranes were pelleted, solubilized in Buffer C (10 mM Tris·HCl, pH 7.4/100 mM NaCl/1 mM sodium EDTA/1% SDS), subjected to SDS/PAGE, and blotted with polyclonal IgG-R139 (anti-Scap).
Immunoblot Analysis.
SDS/PAGE gels were calibrated with molecular weight markers (Bio-Rad, 18–220 kDa). Primary antibodies were used at the following concentrations: polyclonal IgG-R139 (anti-Scap), 3 μg/ml; monoclonal IgG-9D5 (anti-Scap), 5 μg/ml; monoclonal IgG-HSV, 67 ng/ml; monoclonal anti-T7 IgG, 0.5 μg/ml; and monoclonal IgG-9E10 (anti-Myc), 2 μg/ml. Two secondary antibodies (horseradish peroxidase-conjugated, affinity-purified donkey anti-mouse IgG and anti-rabbit IgG; Jackson ImmunoResearch) were each used at a dilution of 1:5,000. All filters were exposed to Kodak X-Omat Blue XB-1 film for 1–60 s.
Other Materials and Methods.
Reagents, Antibodies, Expression Plasmids, and Tissue Culture Media are described in supporting information (SI) Materials and Methods.
Supplementary Material
Acknowledgments
We thank our colleagues Arun Radhakrishnan, Jin Ye, and Russell DeBose-Boyd for helpful suggestions; and Lisa Beatty, Angela Carroll, and Marissa Infante for invaluable help with tissue culture. This research was supported by National Institutes of Health Grant HL20948 and grants from the Perot Family Foundation. J.S. is supported by the University of Texas Southwestern Endowed Scholars Program.
Abbreviations
- SREBP
sterol regulatory element-binding protein
- ER
endoplasmic reticulum
- 25-HC
25-hydroxycholesterol
- HPCD
hydroxypropyl-β-cyclodextrin.
Footnotes
This Feature Article is part of a series identified by the Editorial Board as reporting findings of exceptional significance.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 6496.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700907104/DC1.
References
- 1.Devaux PF. Curr Opin Struct Biol. 1993;3:489–494. [Google Scholar]
- 2.Simons K, Ikonen E. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 3.Anderson RGW. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 4.Brown MS, Goldstein JL. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstein JL, DeBose-Boyd RA, Brown MS. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 6.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Proc Natl Acad Sci USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun L-P, Li L, Goldstein JL, Brown MS. J Biol Chem. 2005;280:26483–26490. doi: 10.1074/jbc.M504041200. [DOI] [PubMed] [Google Scholar]
- 8.Nohturfft A, Yabe D, Goldstein JL, Brown MS, Espenshade PJ. Cell. 2000;102:315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- 9.Espenshade PJ, Li W-P, Yabe D. Proc Natl Acad Sci USA. 2002;99:11694–11699. doi: 10.1073/pnas.182412799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antonny B, Schekman R. Curr Opin Cell Biol. 2001;13:438–443. doi: 10.1016/s0955-0674(00)00234-9. [DOI] [PubMed] [Google Scholar]
- 11.Barlowe C. Cell. 2003;114:395–397. doi: 10.1016/s0092-8674(03)00650-0. [DOI] [PubMed] [Google Scholar]
- 12.Aridor M, Weissman J, Bannykh SI, Nuoffer C, Balch WE. J Cell Biol. 1998;141:61–70. doi: 10.1083/jcb.141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mossessova E, Bickford LC, Goldberg J. Cell. 2003;114:483–495. doi: 10.1016/s0092-8674(03)00608-1. [DOI] [PubMed] [Google Scholar]
- 14.Lee MCS, Miller EA, Goldberg J, Orci L, Schekman R. Annu Rev Cell Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 15.Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Proc Natl Acad Sci USA. 2007;104:6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakai J, Nohturfft A, Cheng D, Ho YK, Brown MS, Goldstein JL. J Biol Chem. 1997;272:20213–20221. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- 17.Rawson RB, DeBose-Boyd RA, Goldstein JL, Brown MS. J Biol Chem. 1999;274:28549–28556. doi: 10.1074/jbc.274.40.28549. [DOI] [PubMed] [Google Scholar]
- 18.Kilsdonk EPC, Yancey PG, Stoudt GW, Bangerter FW, Johnson WJ, Phillips MC, Rothblat GH. J Biol Chem. 1995;270:17250–17256. doi: 10.1074/jbc.270.29.17250. [DOI] [PubMed] [Google Scholar]
- 19.Nohturfft A, DeBose-Boyd RA, Scheek S, Goldstein JL, Brown MS. Proc Natl Acad Sci USA. 1999;96:11235–11240. doi: 10.1073/pnas.96.20.11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL. Mol Cell. 2002;10:237–245. doi: 10.1016/s1097-2765(02)00591-9. [DOI] [PubMed] [Google Scholar]
- 21.Adams CM, Goldstein JL, Brown MS. Proc Natl Acad Sci USA. 2003;100:10647–10652. doi: 10.1073/pnas.1534833100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Gall S, Neuhof A, Rapoport T. Mol Biol Cell. 2004;15:447–455. doi: 10.1091/mbc.E03-05-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee PCW, Sever N, DeBose-Boyd RA. J Biol Chem. 2005;280:25242–25249. doi: 10.1074/jbc.M502989200. [DOI] [PubMed] [Google Scholar]
- 24.Radhakrishnan A, Sun L-P, Kwon HJ, Brown MS, Goldstein JL. Mol Cell. 2004;15:259–268. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 25.Adams CM, Reitz J, DeBrabander JK, Feramisco JD, Brown MS, Goldstein JL. J Biol Chem. 2004;279:52772–52780. doi: 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- 26.Bonifacino JS, Glick BS. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 27.Matsuoka K, Schekman R, Orci L, Heuser JE. Proc Natl Acad Sci USA. 2001;98:13705–13709. doi: 10.1073/pnas.241522198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bi X, Corpina RA, Goldberg J. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- 29.Kandutsch AA, Chen HW. J Biol Chem. 1973;248:8408–84l7. [PubMed] [Google Scholar]
- 30.Brown MS, Goldstein JL. J Biol Chem. 1974;249:7306–73l4. [PubMed] [Google Scholar]
- 31.Chang T-Y, Limanek JS. J Biol Chem. 1980;255:7787–7795. [PubMed] [Google Scholar]
- 32.Russell DW. Biochim Biophys Acta. 2000;1529:126–135. doi: 10.1016/s1388-1981(00)00142-6. [DOI] [PubMed] [Google Scholar]
- 33.Nohturfft A, Brown MS, Goldstein JL. J Biol Chem. 1998;273:17243–17250. doi: 10.1074/jbc.273.27.17243. [DOI] [PubMed] [Google Scholar]
- 34.Seemann J, Jokitalo EJ, Warren G. Mol Biol Cell. 2000;11:635–645. doi: 10.1091/mbc.11.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.