Abstract
Cholesterol synthesis in animals is controlled by the regulated transport of sterol regulatory element-binding proteins (SREBPs) from the endoplasmic reticulum to the Golgi, where the transcription factors are processed proteolytically to release active fragments. Transport is inhibited by either cholesterol or oxysterols, blocking cholesterol synthesis. Cholesterol acts by binding to the SREBP-escort protein Scap, thereby causing Scap to bind to anchor proteins called Insigs. Here, we show that oxysterols act by binding to Insigs, causing Insigs to bind to Scap. Mutational analysis of the six transmembrane helices of Insigs reveals that the third and fourth are important for Insig's binding to oxysterols and to Scap. These studies define Insigs as oxysterol-binding proteins, explaining the long-known ability of oxysterols to inhibit cholesterol synthesis in animal cells.
Keywords: cholesterol homeostasis, Scap, sterol regulatory element-binding protein pathway, COPII vesicles
The concentration of cholesterol in cell membranes is controlled by the gated transport of a membrane-bound transcription factor from the endoplasmic reticulum (ER) to the Golgi complex (1, 2). Knowledge of this regulatory system is important for several reasons: (i) it controls the ratio of cholesterol to phospholipids in cell membranes, a ratio that is crucial for cell viability and function; (ii) it is a prototype for a new type of regulatory system in which a small molecule controls the exit of a protein from the ER (3); (iii) it is also a prototype for a newly recognized mechanism of signal transduction called regulated intramembrane proteolysis (Rip) that generates regulatory proteins through proteolysis of precursor membrane proteins (4); and (iv) this regulatory system is exploited by statins, widely used drugs that lower plasma low-density lipoprotein cholesterol and prevent heart attacks (5).
The cholesterol regulatory system adjusts the synthesis and uptake of cholesterol so as to maintain a constant level of membrane cholesterol. By adjusting these processes, cells can acquire additional cholesterol during periods of rapid growth, and they can prevent toxic accumulation of cholesterol when excess is available. For >30 years, scientists have known that the cholesterol regulatory system is controlled not only by the end product, cholesterol, but also by oxysterols, which are derivatives of cholesterol with extra hydroxyl or keto groups, usually at the 7-position on the B ring or at the 24-, 25-, or 27- positions on the side chain (6–10). Oxysterols are synthesized in various tissues by specific hydroxylases. They play a role in the export of excess cholesterol from brain, lung, and other organs (11), and they also are intermediates in bile acid synthesis (12). Although oxysterols are potent feedback regulators of cholesterol homeostasis, they make up only a minute fraction of total sterols in various tissues and in blood, present at concentrations 104- to 106-fold less than that of cholesterol (11, 13).
Cholesterol and oxysterols suppress cholesterol synthesis in part by blocking the proteolytic activation of a family of membrane-bound transcription factors called sterol regulatory element-binding proteins (SREBPs). Synthesized on ER membranes, each SREBP must be transported to the Golgi complex where proteases liberate a transcriptionally active fragment, allowing it to enter the nucleus (1). There, the SREBP activates transcription of genes encoding 3-hydroxy-3-methylglutaryl CoA reductase (HMGR) and all of the other enzymes involved in the synthesis and uptake of cholesterol (14).
SREBPs are transported from ER to Golgi through the action of an escort protein called Scap, which forms a complex with SREBPs immediately after their synthesis (3). Scap contains a binding site for COPII proteins, which cluster the Scap·SREBP complex into COPII-coated vesicles (15). The vesicles then bud from ER membranes and fuse with the Golgi (16). In SRD-13A cells, which are mutant Chinese hamster ovary (CHO) cells that lack Scap, SREBPs cannot move to the Golgi, and the cells are unable to synthesize cholesterol (17). Cholesterol and oxysterols inhibit SREBP processing by blocking the Scap-mediated movement of SREBPs to the Golgi (18). Both types of sterols act by inducing Scap to bind to either of two related ER anchor proteins called Insig-1 and Insig-2 (3, 19). When Scap binds to Insig, COPII proteins can no longer bind to Scap, and hence the Scap·SREBP complex can no longer move to the Golgi (3, 15).
Although cholesterol and oxysterols both induce the Scap·Insig interaction, they do so by different mechanisms (20). In vitro binding studies show that cholesterol acts by binding to Scap (21), causing a conformational change that induces Scap to bind to Insig (20). The conformational change can be monitored by a change in the tryptic cleavage pattern of Scap. In membranes from sterol-depleted cells, Arg-505 of Scap is inaccessible to trypsin. Addition of cholesterol, either to living cells or to isolated membranes in vitro, causes Arg-505 to become exposed so that it is cleavable by trypsin (20, 22). Remarkably, oxysterols such as 25-hydroxycholesterol (25-HC) also induce Scap to bind to Insig (20), but they do not bind to Scap in vitro (21), nor do they induce a trypsin-detectable conformational change in Scap in the absence of Insigs (20). This finding led us to postulate the existence of another ER membrane protein that binds oxysterols and promotes the Scap·Insig interaction (20).
Here and in an accompanying paper (23), we show that the postulated oxysterol binding protein is Insig. Here, we prepare a purified recombinant version of Insig-2 (one of the two Insig isoforms) and use it to show that Insig-2 binds 25-HC and other oxysterols with saturation kinetics and a high degree of specificity. However, Insig-2 does not bind cholesterol. In the companion paper, we show that formation of the Scap·Insig complex can be initiated either by the binding of cholesterol to Scap or the binding of 25-HC to Insig. Independent of the initiating event, the formation of the Scap·Insig complex occludes the binding site on Scap that is recognized by COPII proteins and thereby blocks the transport of SREBPs from ER to Golgi (23).
Results
Solubilization and Purification of His10–Insig-2–Flag.
Human Insig-2 is a 225-aa protein. Most of the protein (residues 32–207) is embedded in the ER membranes as six membrane-spanning segments (see Fig. 4A). We used the baculovirus expression system in Sf-9 insect cells to express human Insig-2 with a His10-tag at the NH2 terminus and a FLAG-tag at the COOH terminus. The resulting recombinant protein is designated His10–Insig-2–FLAG. Of 20 detergents tested, the only ones that solubilized >90% of His10–Insig-2–FLAG from insect cell membranes were those belonging to the Fos–Choline series of phospholipid-like detergents. After solubilization in Fos–Choline 13 detergent, His10–Insig-2–FLAG was purified to homogeneity by using Ni-chromatography followed by gel filtration. The purified protein was monodisperse for at least 1 week when stored at 4°C at concentrations of 2–5 mg/ml. Mutant versions of His10–Insig-2–FLAG were expressed and purified in a similar fashion. For unknown reasons, we have so far been unable to produce a stable form of full length or truncated human Insig-1 despite the use of multiple epitope tags, added at either or both ends of the molecule.
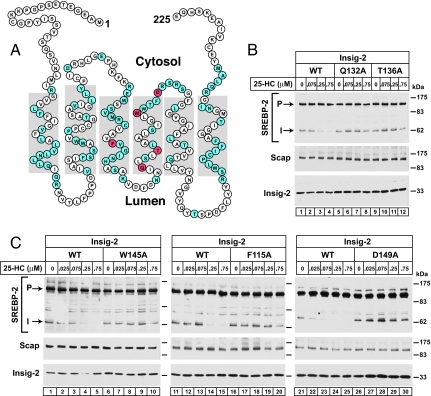
Fig. 4.
Alanine scanning mutagenesis of human Insig-2 reveals five amino acid residues crucial for 25-HC mediated inhibition of SREBP-2 processing in insect cells. (A) Amino acid sequence and predicted topology of human Insig-2. The topology is based on that of Insig-1 (26). Amino acids in blue denote residues that, when mutated to alanine, did not interfere with 25-HC-mediated inhibition of SREBP-2 cleavage. Amino acids in red denote residues that when mutated to alanine created resistance to 25-HC mediated inhibition of SREBP-2 cleavage. (B) Immunoblot analysis of SREBP-2 cleavage. Drosophila S2 cells were transfected with the following plasmids per dish: 0.4 μg of pDS-HSV-SREBP-2, 0.05 μg of pAc-Scap, and 0.05 μg of either wild-type pAc–Insig-2–Myc or the indicated mutant. On day 2, the cells were switched to medium F containing the indicated concentration of 25-HC. After incubation for 6 h at 23°C, cells were harvested and whole cell lysates were subjected to SDS/PAGE and immunoblot analysis with anti-HSV-IgG (against SREBP-2), IgG-9D5 (against Scap), and IgG-9E10 (against Insig-2). P and I denote the uncleaved precursor and cleaved intermediate forms of SREBP-2, respectively.
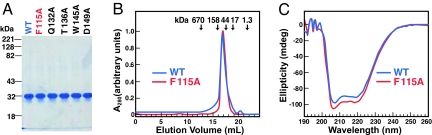
Fig. 1A shows the migration of purified His10–Insig-2–FLAG (wild-type and indicated mutant versions) on SDS/PAGE, as visualized by Coomassie blue staining. Gel filtration studies of His10–Insig-2–FLAG in Fos–Choline detergent micelles showed that the protein eluted as an 80-kDa species (blue curve in Fig. 1B). The calculated molecular mass of a His10–Insig-2–FLAG monomer is 30 kDa and that of a Fos–Choline 13 micelle is ≈25 kDa (21), and thus we suspect that the 80-kDa species represents a dimer. Fig. 1C shows the far UV circular dichroism (CD) spectrum of His10–Insig-2–FLAG (blue curve) in Fos–Choline 13 micelles. The CD spectrum suggests a highly helical protein with characteristic minima at ≈206 and 222 nm (24, 25). This result is consistent with previous data showing that the bulk of Insig proteins resides within the ER membrane (26), where it likely forms α-helices. The gel filtration profiles and CD spectra of all five mutant forms of His10–Insig-2–FLAG studied in this paper were similar to those of the wild-type protein. The data for one of the mutants, His10–Insig-2(F115A)–FLAG, are shown in the red curves in Fig. 1 B and C.
Fig. 1.
Characterization of purified His10–Insig-2–FLAG. (A) Coomassie staining. Recombinant Insig-2 proteins were purified in two steps as described in Materials and Methods. Five micrograms of wild-type His10–Insig-2–FLAG or the indicated mutant version were subjected to 10% SDS/PAGE, and the proteins were visualized with Coomassie brilliant blue R-250 stain (Bio-Rad). Molecular masses of protein standards are indicated. (B) Gel filtration chromatography of purified proteins. Buffer A (100 μl) containing 6 nmol of either His10–Insig-2–FLAG or His10–Insig-2–FLAG(F115A) was loaded onto a Tricorn 10/300 Superose 6 column (Amersham) and chromatographed at a flow rate of 0.4 ml/min. Absorbance at 280 nm was monitored continuously to identify His10–Insig-2–FLAG (blue) or His10-Insig-2(F115A)-FLAG (red). Standard molecular mass markers (thyroglobulin, Mr 670,000; γ globulin, Mr 158,000; ovalbumin, Mr 44,000; myoglobin, Mr 17,000; and vitamin B12, Mr 1350) were chromatographed on the same column (arrows). The apparent molecular mass of His10–Insig-2–FLAG is 80 kDa. (C) Circular dichroism spectroscopy. Circular dichroism of 3 μM His10–Insig-2–FLAG or His10–Insig-2–FLAG(F115A) in buffer A was measured on an Aviv 62DS spectrometer using a 2-mm path length cuvette. The average of 10 spectra is shown.
Differential Binding Specificity for Insig-2 and Scap.
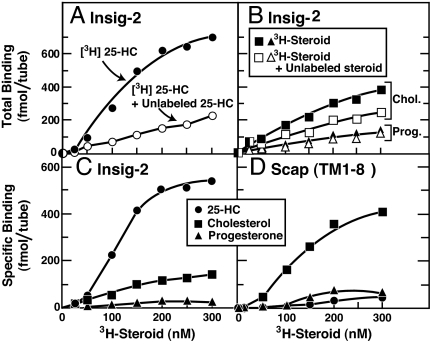
We used an assay developed in earlier work on Scap (21) to investigate the direct binding of 3H-labeled cholesterol, 25-HC, and progesterone to His10–Insig-2–FLAG. As described in this earlier work, the [3H]steroids were solubilized in Fos–Choline 13, the same detergent used for solubilizing and purifying His10–Insig-2–FLAG. Fig. 2A shows a saturation curve for binding of [3H]25-HC to His10–Insig-2–FLAG. The binding was saturable, reaching a half-maximal value at ≈100 nM as judged from multiple experiments. Binding was reduced in the presence of excess of unlabeled 25-HC. In contrast, as shown in Fig. 2B, the binding of [3H]cholesterol to His10–Insig-2–FLAG was not saturable and was reduced only slightly by an excess of unlabeled cholesterol. The specific binding of [3H]25-HC to His10–Insig-2–FLAG (obtained by subtracting the binding value in the absence of the respective unlabeled sterol from that in its presence) was ≈5-fold greater than for [3H]cholesterol (Fig. 2C). [3H]Progesterone showed no specific binding (Fig. 2 B and C). In studies not shown, we found that equilibrium binding of [3H]25-HC was achieved after 1–2 h, the binding was optimal at a pH of 6.5–8.0, and there was no significant enhancement of binding by changes in ionic strength, addition of metal ions, or addition of crude lipid extracts from bovine liver or chicken egg solubilized in Fos–Choline 13 micelles. For unknown reasons, maximal binding was less than stoichiometric. At saturation, one molecule of [3H]25-HC bound to ≈40 His10–Insig-2–FLAG dimers (or 80 monomers).
Fig. 2.
Steroid specificity of binding to Insig-2 (A–C) and Scap(TM1–8) (D). (A and B) Total binding of [3H]steroids to Insig-2. Each assay tube, in a total volume of 100 μl of buffer A, contained 400 nM His10–Insig-2–FLAG (40 pmol), 25 mM phosphocholine chloride, and varying concentrations of the indicated [3H]steroid [[3H]25-HC (152 dpm/fmol) (A), [3H]cholesterol (120 dpm/fmol) (B), or [3H]progesterone (215 dpm/fmol) (B)] in the absence (filled symbols) or presence (open symbols) of the respective unlabeled steroid at a final concentration of 5 μM. After incubation for 4 h at room temperature, bound [3H]steroids were measured as described in Materials and Methods. Each data point is the average of duplicate assays and represents the total binding without subtraction of any blank values. (C) Specific binding of [3H]steroids to Insig-2. These data are replotted from A and B. (D) Specific binding of [3H]steroids to Scap(TM1–8). Each assay was carried out as described in A and B except that the tubes contained 120 nM His10–Scap(TM1–8) (12 pmol) instead of His10–Insig-2–Flag. Each data point is the average of duplicate assays. Specific binding in C and D was calculated by subtracting the binding value in the presence of unlabeled steroid from that in its absence.
For comparative purposes, Fig. 2D shows specific binding of [3H]25-HC, [3H]cholesterol, and [3H]progesterone to His10–Scap(TM1–8). High-affinity, saturable binding to His10–Scap(TM1–8) was observed with [3H]cholesterol, but not with [3H]25-HC or [3H]progesterone. At saturation, one cholesterol molecule bound to ≈6 His10–Scap(TM1–8) tetramers.
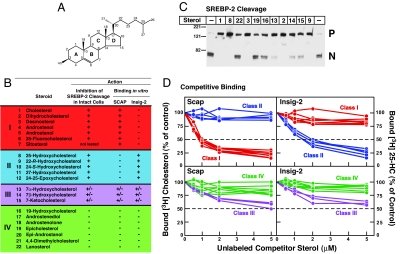
To more completely define the differential sterol specificity for Insig-2 and Scap, we used a competitive binding assay. Fig. 3A shows cholesterol with each ring and carbon atom labeled. Fig. 3B lists the 22 sterols that were studied. The sterols are divided into four classes. class I sterols have an intact steroid nucleus (3β-hydroxyl group and no additional methyl groups) and no hydroxyl or epoxy groups on their iso-octyl side chains (1–7). The iso-octyl side chains of class I sterols include those that are unsaturated (3), acylated (7), or fluorinated (6). Two of these sterols have no side chain at all (4, 5). Two of these sterols lack a C5–C6 double bond (2, 5). Class II sterols have the cholesterol backbone and a hydroxyl or epoxy group on their intact iso-octyl side chains (8–12). Class III sterols have a hydroxyl or keto group at the 7 position on the B ring (13–15). Class IV sterols have modifications to the steroid nucleus other than saturation of the C5–C6 double bond (16–22).
Fig. 3.
Differential sterol specificity for Insig-2 and Scap: comparison of binding in vitro and inhibition of SREBP-2 cleavage in mammalian cells. (A) Chemical structure of cholesterol. (B) List of sterols tested and their actions in inhibiting SREBP-2 cleavage in intact cells and in competing with the binding of [3H]cholesterol to Scap and of [3H]25-HC to Insig-2. The degree of effect (maximal, intermediate, or minimal) is denoted by +, +/−, and −, respectively. For assays of SREBP-2 cleavage, 11 sterols (each at a final concentration of 20 μM in a 1:12 molar ratio with MCD) were tested in the current study (see C). The data for all other steroids have been reported (20, 27). Sterol 7 (sitosterol) could not be solubilized in a 1:12 molar ratio with MCD. For binding assays, all 22 sterols were tested in the current study; representative competition curves are shown in D and E. Trivial sterol names and their standard nomenclature (other than for cholesterol and its derivatives) are as follows: dihydrocholesterol for 5α-cholestan-3β-ol (2); desmosterol, 5,24-cholestadien-3β-ol (3); androstenol, 5-androsten-3β-ol (4); androstanol, 5α-androstan-3β-ol (5); sitosterol, 5-cholesten-24β-ethyl-3β-ol (7); 17-androstenediol, 5-androsten-3β,17β-diol (17); androstenolone, 5-androsten-3β-ol-17-one (18); epicholesterol, 5-cholesten-3α-ol (19); epi-androstanol, 5α-androstan-3α-ol (20); and lanosterol, 8,24,(5α)-cholestadien-4,4,14α-trimethyl-3β-ol (22). (C) Immunoblot analysis of SREBP-2 cleavage in mammalian cells. On day 2, CHO-K1 cells were incubated for 1 h in medium D containing 1% HPCD and then switched to medium D containing the indicated sterol (20 μM) complexed to MCD. After incubation for 6 h, cells were harvested, and total cell extracts were immunoblotted with IgG-7D4 (anti-SREBP-2). The precursor and mature nuclear form of SREBP-2 are denoted by P and N, respectively. (D) Competitive binding of [3H]cholesterol to Scap (Left) and [3H]25-HC to Insig-2 (Right). Each assay tube, in a total volume of 100 μl of buffer A, contained 25 mM phosphocholine chloride, 120 nM His10–Scap(TM1–8) or 400 nM His10–Insig-2–FLAG as indicated, 100 nM [3H]cholesterol (120 dpm/fmol) or 100 nM [3H]25-HC (152 dpm/fmol) as indicated, and varying concentration of the indicated unlabeled sterol. After incubation for 4 h at room temperature, bound [3H]cholesterol or [3H]25-HC was measured as described in Materials and Methods. Each data point is the average of duplicate assays and represents the amount of [3H]cholesterol or [3H]25-HC bound relative to that in the control tube, which contained no unlabeled sterol. In a typical competition experiment, six to nine unlabeled sterols were tested, two of which were always cholesterol and 25-HC. “100% of control” values for [3H]cholesterol binding (Left) ranged from 209 to 263 fmol per tube in four experiments. “100% of control” values for [3H]25-HC binding (Right) ranged from 199 to 261 fmol per tube in four experiments. All unlabeled sterols were tested in duplicate in two or more experiments with similar results.
We first tested the ability of all four classes of sterols to inhibit SREBP-2 cleavage when added to sterol-deprived CHO-K1 cells. The degree of inhibition (maximal, intermediate, or minimal) is denoted by +, +/−, and −, respectively. Fig. 3C shows a representative experiment in which we assayed 11 sterols for their ability to inhibit SREBP-2 cleavage when delivered to CHO-K1 cells in a 1:12 complex with MCD at a sterol concentration of 20 μM. Maximal inhibitory effects were consistently seen with class I and II sterols. These included cholesterol (1), dihydrocholesterol (2), desmosterol (3), 25-HC (8), and 22-R-hydroxycholesterol (9). Intermediate inhibitory effects were seen with class III sterols: 7α-hydroxycholesterol (13), 7β-hydroxycholesterol (14), and 7-ketocholesterol (15). Minimal inhibitory effects were seen with class IV sterols: 19-hydroxycholesterol (16), epi-cholesterol (19), and lanosterol (22). We were unable to solubilize sitosterol (7) in a 1:12 molar ratio with MCD. The data for the remaining sterols have been reported (20, 27), and are summarized in Fig. 3B.
We then tested these same sterols for their ability to compete for the binding of [3H]cholesterol to His10–Scap(TM1–8) or the binding of [3H]25-HC to His10–Insig-2–FLAG. Competition curves for all 22 sterols listed in Fig. 3B are shown in Fig. 3D for Scap(TM1–8) binding (Left) and Insig-2 binding (Right). Here, a remarkable reciprocity was observed. Class I sterols (shown in red) competed with [3H]cholesterol for binding to Scap, whereas class II sterols (shown in blue) competed with [3H]25-HC for binding to Insig-2. Class III sterols (shown in lavender) had weak affinity for both His10–Scap(TM1–8) and His10–Insig-2–FLAG. Class IV sterols (shown in green), which have minimal to no effect on inhibition of SREBP-2 cleavage, did not compete for binding to either recombinant protein.
In other studies not shown, three other steroids, the two major bile acids, 3α,7α,12α-trihydroxy-5β-cholanic acid (cholic acid) and 3α,7α-dihydroxy-5β-cholanic acid (chenodeoxycholic acid), and the hypocholesterolemic agent, (3α,4α,5α)-4-(2-propenyl-cholestan-3α-ol) (LY295427) (28, 29), did not compete for the binding of either [3H]cholesterol to Scap or [3H]25-HC to Insig-2 when each was tested at 5 μM (i.e., 50-fold above the concentration of radioactive ligand).
Identification of Amino Acids in Insig-2 Required for 25-HC Binding.
To determine the residues in Insig-2 that are important for its binding to 25-HC, we subjected the protein molecule to alanine-scanning mutagenesis. Fig. 4A shows the amino acid sequence and the predicted topology of human Insig-2. The topology is based on the demonstrated topology of the closely related Insig-1 (26). Comparison of the Insig-2 membrane-spanning sequences from nine vertebrate species reveals an identity of 73%, whereas there is lesser identity in the sequences of the nonmembrane regions, which compose 11–22% of the molecule. In a subset of five mammalian species, the membrane regions are virtually the same (97% identity). Thus, we focused our mutagenesis studies on this highly conserved membrane region of human Insig-2, which is also 85% identical to that of human Insig-1 (30).
Insect cells provide a convenient model system to study mutations in Insig-2 because they do not contain a recognizable Insig-2 or Insig-1 gene and because it is possible to reconstitute 25-HC regulation by coexpressing mammalian SREBP-2, Scap, and Insig-2 (31). As shown in Fig. 4B, when human SREBP-2, hamster Scap, and human Insig-2 were coexpressed in Drosophila S2 cells, we observed the processing of SREBP-2 from its precursor form to a membrane-bound cleaved fragment whose size corresponded to the product of cleavage by the Golgi-resident Site-1 protease (S1P) of Drosophila. S1P cleaves SREBPs in the luminal loop between the two membrane-spanning helices. The NH2-terminal fragment remains membrane bound by virtue of its single transmembrane helix. In mammalian cells, the intermediate fragment is immediately cleaved by Site-2 protease (S2P), which cuts within the transmembrane helix, thereby releasing the NH2-terminal fragment into the cytosol (1). Drosophila S2P does not recognize mammalian SREBP-2, and thus the intermediate fragment remains membrane bound. We refer to this band as the intermediate fragment (31, 32). Addition of 25-HC to the triply transfected Drosophila cells led to a nearly complete inhibition of cleavage to the intermediate form (Fig. 4 B, lanes 1–4, and C, lanes 1–5, 11–15, and 21–25) (31).
We carried out alanine-scanning to search for point mutations that compromised the ability of Insig-2 to block SREBP-2 cleavage in a 25-HC-dependent manner. Of 81 amino acid residues that were mutated to alanine, 76 substitutions (shaded blue in Fig. 4A) did not interfere with 25-HC mediated inhibition of SREBP-2 cleavage. The other five substitutions (shaded red in Fig. 4A) created resistance to 25-HC mediated inhibition of SREBP-2 cleavage (Fig. 4B, lanes 5–12, and C, lanes 6–10, 16–20, and 26–30). Four of the amino acids (Q132, T136, W145, and D149) are in the fourth transmembrane helix, and a helical wheel analysis predicts that Q132 and T136 lie on the opposite face of the helix from D149 and W145. The fifth amino acid (F115) lies in the middle of the third transmembrane helix.
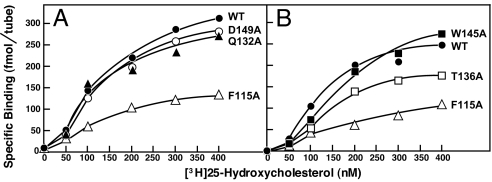
These five mutations could block 25-HC regulation by interfering with the ability of Insig-2 to bind to 25-HC or the ability of the Insig-2·25-HC complex to bind to Scap, or both. To test whether 25-HC binding was affected, we prepared baculoviruses encoding these five mutant versions of His10–Insig-2–FLAG and purified the recombinant proteins as described earlier. The physical properties of these mutant proteins were similar to those of the wild-type version (see Fig. 1). Fig. 5 A and B shows saturation curves for specific binding of [3H]25-HC to His10–Insig-2–FLAG (wild-type and five mutant versions). The maximum specific binding of [3H]25-HC to His10–Insig-2(F115A)–FLAG and His10–Insig-2(T136A)–FLAG was reduced by ≈70% and ≈35%, respectively, compared with the wild-type version. The other three mutants (Q132A, W145A, and D149A) bound [3H]25-HC normally. The D149A mutant was shown to be defective in its binding to Scap in the presence of 25-HC (33). It is likely that the Q132A and W145A mutants also are unable to bind to Scap even though they bind 25-HC.
Fig. 5.
[3H]25-HC binding to Insig-2 mutants. Each assay tube, in a total volume of 100 μl of buffer A, contained 400 nM His10–Insig-2–FLAG (wild-type or the indicated mutant version), 25 mM phosphocholine chloride, and the indicated concentration of [3H]25-HC (152 dpm/fmol). After incubation for 4 h at room temperature, specifically bound [3H]25-HC was measured as described in Fig. 2C. Each data point is the average of duplicate assays.
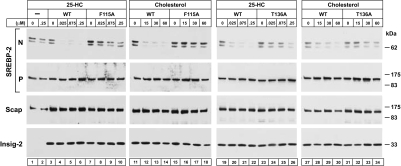
The F115A and T136A mutants of Insig-2 are partially defective in their ability to bind 25-HC. The question arose as to whether these mutant proteins could bind to Scap in its cholesterol-induced conformation. To test this possibility functionally, we compared the ability of 25-HC and cholesterol to suppress SREBP-2 cleavage in Scap-deficient hamster cells (SRD-13A cells) that were transfected with plasmids encoding SREBP-2, Scap, and wild-type or mutant Insig-2. The immunoblots in Fig. 6 show that either 25-HC (lanes 3–6 and 19–22) or cholesterol (lanes 11–14 and 27–30) inhibited SREBP-2 cleavage in cells expressing wild-type Insig-2. In cells expressing either mutant Insig-2(F115A) or Insig-2(T136A), there was resistance to inhibition of SREBP-2 cleavage by both 25-HC (lanes 7–10 and 23–26, respectively) and cholesterol (lanes 15–18 and 31–34, respectively). Considered together with the direct binding data in Fig. 5, these findings suggest that the F115A and T136A mutations alter two functions of Insig-2: binding to 25-HC and binding to Scap in its cholesterol-induced conformation.
Fig. 6.
Failure of cholesterol and 25-HC to inhibit SREBP-2 processing in mammalian cells expressing Insig-2 mutants. On day 0, Scap-deficient SRD-13A cells were set up in medium C at 3.5 × 105 cells per 60-mm dish. On day 2, each dish of cells was transfected in medium B with 2 μg of pTK-HSV-SREBP-2, 0.1 μg of pCMV-Scap, and one of the following pCMV–Insig-2–Myc plasmids: wild-type, 0.34 μg; F115A mutant, 0.64 μg; or T136A mutant, 0.34 μg. After incubation for 16 h at 37°C, the cells were switched to medium D containing 1% HPCD for 1 h, after which the cells were washed twice with PBS and switched to medium D containing the indicated amounts of cholesterol (complexed to MCD) or 25-HC (in ethanol). After incubation for 6 h at 37°C, the cells were harvested, fractionated, and subjected to SDS/PAGE and immunoblot analysis with anti-HSV-IgG (against SREBP-2), IgG-9D5 (against Scap), and IgG-9E10 (against Insig-2). N and P denote the cleaved nuclear and precursor forms of SREBP-2, respectively.
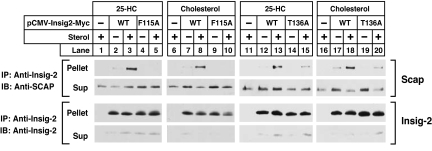
The experiments in Fig. 7 were designed to confirm the conclusions regarding Scap·Insig binding through use of coimmunoprecipitation. For this purpose, we transfected plasmids encoding Scap and Myc-tagged wild-type or mutant versions of Insig-2 into Scap-deficient SRD-13A cells. After incubation in the presence or absence of 25-HC or cholesterol, cells were harvested, and the cell lysates were incubated with anti-Myc to precipitate Insig-2. Supernatants and pellets were subjected to SDS/PAGE, and Insig-2 and Scap were visualized by immunoblotting. As shown in Fig. 7, Scap was coimmunoprecipitated with wild-type Insig-2 in the presence of 25-HC (lanes 3 and 13) or cholesterol (lanes 8 and 18), but not in the absence of either sterol (lanes 2, 7, 12, and 17). Scap was not coimmunoprecipitated with Insig-2(F115A) regardless of the presence of 25-HC (lanes 4 and 5) or cholesterol (lanes 9 and 10). The less severe Insig-2(T136A) mutant retained some ability to interact with Scap in the presence of cholesterol (lane 20) or 25-HC (lane 15).
Fig. 7.
Mutant Insig-2s do not bind to Scap in presence of 25-HC or cholesterol. On day 0, Scap-deficient SRD-13A cells were set up in medium C at 3.5 × 105 cells per 60-mm dish. On day 2, each dish of cells was transfected in medium B with 0.1 μg of pCMV-Scap and one of the following pCMV–Insig-2–Myc plasmids: wild-type, 0.16 μg; F115A mutant, 0.32 μg; or T136A mutant, 0.16 μg. After incubation for 6 h at 37°C, the cells were switched to medium D for 16 h, then switched to medium D containing 1% HPCD for 1 h, after which the cells were washed twice with PBS and switched to medium D containing no sterols, 0.075 μM 25-HC (in ethanol), or 30 μM cholesterol (complexed to MCD), as indicated. After incubation for 6 h at 37°C, cells were harvested, lysed, and immunoprecipitated with polyclonal anti-Myc to precipitate Insig-2. Pellets and supernatants (2.5:1 ratio) were subjected to SDS/PAGE and immunoblot analysis. Sup, supernatant fraction; IP, immunoprecipitate; IB, immunoblot.
Discussion
The current experiments define Insig-2 as a membrane-bound oxysterol-binding protein with specificity for cholesterol derivatives that possess hydroxyl groups on the side chain. This finding appears to unravel the conundrum raised by previous studies showing that oxysterols could block SREBP cleavage by promoting Scap/Insig binding, and yet oxysterols did not bind to Scap (20, 21). Instead, the data now show that oxysterols bind to Insigs. Inasmuch as we were unable to purify human Insig-1 in a stable form, all of the current experiments were conducted with human Insig-2. However, we believe that the conclusions apply equally to human Insig-1, whose membrane domain is 85% identical to that of human Insig-2 (26, 30). Previous studies have shown that 25-HC indeed induces the binding of Insig-1 to Scap (20). Moreover, each of the five amino acids that we show to be crucial for Insig-2 function are conserved in both Insig-1 and Insig-2 from nine vertebrate species, including lamprey, puffer fish, and zebrafish. One of these residues (D149 in Insig-2) is equivalent to D205 in Insig-1. Previous studies showed that D205 of Insig-1 is crucial for its function in blocking SREBP processing (34).
The reciprocity between the sterol-binding properties of Scap and Insig-2 is striking (Figs. 2 and 3). Scap recognizes the tetracyclic steroid nucleus and the 3β-hydroxyl group of cholesterol. It binds the sterol equally well even when it lacks a side chain. However, Scap does not bind a sterol that contains a polar group such as a hydroxyl group on the side chain. In contrast, Insig-2 binding absolutely requires the sterol side chain with a hydroxyl group that can be located at positions 22, 24, 25, or 27. Insig-2 binding also requires the steroid nucleus, as evidenced by the observation that binding of [3H]25-HC was not inhibited by mimics of the oxygenated iso-octyl side chain of 25-HC, such as 2-octanol and 3,7-dimethyl-3-octanol (data not shown). The contrasts between the binding properties of Scap and Insig-2 suggest that Scap may recognize cholesterol in its usual orientation in the membrane, i.e., when its 3β-hydroxyl is exposed at the surface and its side chain is buried in the hydrophobic bilayer. It is likely that a hydroxylated side chain would prevent such membrane insertion (35, 36), and hence Insig-2 would more likely recognize a sterol that lies on the surface of a membrane with its hydroxylated side chain exposed. It is unlikely that Insig-2 binds directly to the hydroxylated side chain because the hydroxyl group can be on any one of several different positions. Rather, it may be that the hydroxyl group orients the sterol in relation to the lipid bilayer so that Insig-2 can recognize it. It is also possible that Insig-2 interacts indirectly with the hydroxyl group on an oxysterol through water-mediated interactions as in the case of Osh4, a yeast oxysterol-binding protein whose crystal structures has been determined with bound ligand (37). Whatever the mechanism of binding, it is noteworthy that all sterols that completely inhibit SREBP processing bind to either Scap or Insig, but not to both (Fig. 3B).
One caveat to the current studies is the low stoichiometry of 25-HC binding to Insig-2 in the in vitro binding assay. Gel filtration suggests that purified Insig-2 exists as a dimer in Fos–Choline 13 micelles. At saturation, we calculated that one 25-HC molecule was bound for every 40 dimers. It is likely that this inefficiency relates to the physical configuration of the Insig-2 protein in the micelles. This configuration only partially approximates the configuration of Insig-2 in a planar lipid bilayer. We also noted that detergent-solubilized Insig-2 was stable for only a few days. Thereafter, the binding activity declined even further. So far, experiments designed to reconstitute Insig-2 into lipid bilayers have failed, but efforts to overcome this obstacle are being pursued.
The alanine-scanning mutagenesis studies reveal that the third and fourth membrane spanning helices of Insig-2 play a role in 25-HC binding as well as Scap binding. Two mutations in these helices (F115A and T136A) reduced the amount of [3H]25-HC binding (Fig. 5). Interestingly, the reduction occurred in Bmax without an apparent change in affinity. This raises the possibility that these two mutations alter the configuration of Insig-2 in the micelle so that fewer binding sites are exposed. These mutations also reduced the ability of Insig-2 to bind to Scap, even when Scap was induced to assume its binding-competent configuration through the binding of cholesterol (Fig. 7). Other mutations in the fourth membrane spanning helix reduced the ability of Insig to bind to Scap without reducing its ability to bind to 25-HC. As expected, all of these mutations impaired the ability of both cholesterol and 25-HC to block SREBP-2 cleavage in intact cells. Taken together, these data suggest that the third and fourth membrane helices of Insig-2 play a primary role in Insig binding to Scap and a lesser role in its binding to 25-HC.
Insig proteins also bind in a sterol-dependent manner to HMGR, the rate-limiting enzyme of the cholesterol biosynthesis pathway (38). Formation of the HMGR·Insig complex leads to ubiquitination and degradation of HMGR, whereas formation of the Scap·Insig complex leads to ER retention of SREBP (3). There is partially overlapping sterol specificity for HMGR·Insig and Scap·Insig complex formation. Oxysterols (such as 25-HC) can induce formation of both HMGR·Insig and Scap·Insig complexes. Cholesterol, which binds to Scap and induces Scap·Insig complex formation, has no effect on HMGR·Insig complex formation (27). On the other hand, lanosterol (a cholesterol precursor) can stimulate formation of HMGR·Insig complex (27), although it has no effect on Scap·Insig complex formation as inferred from its inability to produce a conformational change in Scap (22) and its inability to block SREBP processing (27). Based on the current studies, it is tempting to speculate that the oxysterol-bound form of Insig (the common denominator to both systems) can form a complex with HMGR just as it can with Scap, whereas lanosterol acts by binding to HMGR much like cholesterol binds directly to Scap. It is possible that Insig, upon activation by oxysterol, can bind other proteins as well.
In an accompanying paper, we show that the conformational change in Scap is similar whether it is elicited when Scap binds to cholesterol or when Scap binds to the Insig·25-HC complex (23). In both cases, this conformational change precludes the binding of COPII proteins to Scap, and this appears to explain the inhibition of Scap · SREBP transport from ER to Golgi. Thus, vertebrate cells have two sterol sensors for the SREBP pathway, Scap and Insig, which together enable them to turn off cholesterol synthesis in response to the buildup of either cholesterol or its hydroxylated derivatives.
Materials and Methods
Solubilization of Steroids.
Stock solutions of steroids in detergent were prepared as described (21). An ethanol solution containing 4–5 nmol of [3H]25-hydroxycholesterol was evaporated to dryness on the sides of a microcentrifuge tube in a SpeedVac centrifuge. One milliliter of Buffer A [see supporting information (SI) Materials and Methods] was then added to the tube, after which it was agitated for 4 h on a vortex mixer and centrifuged at 20,000 × g for 30 min, all at room temperature. As determined by scintillation counting, the concentration of solubilized [3H]25-HC in the supernatant ranged from 2 to 3 μM. Stock solutions of [3H]cholesterol and [3H]progesterone were prepared in a similar manner. Saturated solutions of unlabeled sterols in Buffer A were prepared by evaporating 200 nmol of ethanolic sterol solution in a microcentrifuge tube. The dried sterol was solubilized by the same procedure as outlined above. Concentrations were determined by adding tracer amounts of 3H-labeled sterols. The choice of the labeled tracer sterol was based on structural similarity to the bulk sterol, i.e., [3H]cholesterol for sterols 1–7 and 19–20, [3H]25-hydroxycholesterol for sterols 8–18, and [3H]lanosterol for sterols 21–22 (see Fig. 3B).
Purification of Recombinant Scap and Insig-2.
The following recombinant baculoviruses were constructed in pFastBacHTa expression vector (Invitrogen) as described (21): His10–Scap(TM1–8) encoding the eight transmembrane region of wild-type hamster Scap with an NH2-terminal His10-tag, and His10–Insig-2–FLAG encoding wild-type or mutant versions of human Insig-2 with an NH2-terminal His10-tag and a COOH-terminal FLAG-tag. Recombinant proteins were overexpressed in Sf9 insect cells and purified in Buffer A using nickel chromatography and gel filtration as described (21). Protein concentrations were measured by using a BCA kit (Pierce).
[3H]Steroid Binding Assay.
Binding of [3H]steroids to His10–Scap(TM1–8) and His10–Insig-2–FLAG was measured by using procedures described in ref. 21. In the standard assay, binding reactions were set up at room temperature. Each reaction, in a final volume of 100 μl of buffer A, contained either 1 μg (120 nM final concentration) of purified His10–Scap(TM1–8) or 1.2 μg (400 nM) of purified wild-type or mutant versions of His10–Insig-2–FLAG, 10–400 nM [3H]steroid, and 25 mM phosphocholine chloride. Phosphocholine chloride increases binding of sterols to His10–Scap(TM1–8) by ≈3-fold and to His10–Insig-2–FLAG by ≈1.5-fold. After incubation for 4 h at room temperature, the mixture was passed through a column packed with 0.3 ml of Ni-NTA agarose beads (Qiagen). Each column was then washed for ≈15 min with 7 ml of buffer A in the case of His10–Scap(TM1–8) or buffer B in the case of His10–Insig-2–FLAG. The choice of wash buffers is important in lowering nonspecific binding. The protein-bound [3H]steroid was eluted with 250 mM imidazole and measured by scintillation counting. For competition experiments with unlabeled sterols, the standard assays were carried out in the presence of Buffer A containing the indicated unlabeled sterol (0–5 μM).
Cell Culture.
CHO-K1 cells and SRD-13A cells (a Scap null mutant clone derived from γ-irradiated CHO-7 cells; ref. 39) were grown in monolayer at 37°C in an atmosphere of 8–9% CO2 and maintained in medium B and medium C, respectively. Stock cultures of Drosophila S2 cells were maintained in medium E at 23°C as described (31).
Analysis of SREBP-2 Processing in Insect Cells.
On day 0, Drosophila S2 cells were set up for experiments in Drosophila-SFM medium (Invitrogen) at a density of 6 × 105 per 37-mm dish and cultured at 23°C. Several hours later, the cells were transfected with 6 μl per dish of Cellfectin reagent (Invitrogen) in Drosophila-SFM medium according to the manufacturer's instructions. The total amount of DNA was adjusted to 1 μg per dish by supplementing with pAc5.1/V5-HisB mock vector. After overnight incubation, cells were refed with 3 ml per dish of medium F. On day 2, the cells were refed with 3 ml per dish of medium F containing various amounts of 25-HC in ethanol (final concentration of added ethanol was 0.03% vol/vol). All control cultures received ethanol (0.03%). After incubation for 6 h, cells were harvested by scraping in PBS. For preparation of whole-cell lysates, the cell pellets from two replicate dishes were combined and solubilized in Buffer C. The insoluble residue was removed by centrifugation for 10 min at 20,000 × g at 4°C. An aliquot of the supernatant solution (100 μl) was mixed with 25 μl of 5× SDS loading buffer, and aliquots were subjected to 8% SDS/PAGE and analyzed by immunoblotting.
Analysis of SREBP-2 Processing in Mammalian Cells.
On day 0, CHO-K1 cells were set up for experiments in medium B at 7 × 105 cells per 100-mm dish and cultured at 37°C. On day 2, the cells were switched to medium D containing 1% HPCD for 1 h. The cells were then washed twice with PBS and switched to medium D containing 20 μM concentrations of various sterols complexed in a 1:12 molar ratio with MCD. Stock solutions of sterol–MCD complexes at a final sterol concentration of 2.5 mM were prepared as described (22). After incubation for 6 h, the cells were washed once with PBS, then treated with 300 μl of Buffer D and scraped into 1.5-ml tubes. The cells were passed through a 22-gauge needle 15 times and then agitated for 10 min. The protein concentration of each cell extract was measured (BCA kit), after which an aliquot of cell extract (25 μg) was mixed with an equal volume of Laemmli sample buffer (Bio-Rad), heated for 10 min at 95°C, and then subjected to 8% SDS/PAGE and immunoblot detection of SREBP-2.
Transient Transfection and Fractionation of SRD-13A Cells.
On day 0, SRD-13A cells were set up in medium C at 3.5 × 105 cells per 60-mm dish and cultured at 37°C. On day 2, cells were transfected in medium B with the indicated plasmids by using FUGENE 6 reagent (Roche Applied Science) according to the manufacturer's instructions. The amount of plasmid DNA in each dish was adjusted to 0.75–2.75 μg by addition of pcDNA3 mock vector. After incubation for 16 h, the cells were switched to medium D containing 1% HPCD for 1 h. Cells were then washed twice with PBS and switched to medium D containing various amounts of cholesterol (complexed in a 1:12 molar ratio with MCD) or 25-HC (from 1 mg/ml stocks in ethanol). After incubation for 6 h, duplicate or triplicate dishes of cells were harvested and pooled for measurement of SREBP-2 cleavage and detection of Scap·Insig-2 complex formation.
SREBP-2 Cleavage.
Nuclear extracts and 20,000 × g membrane fractions from duplicate dishes of transiently transfected SRD-13A cells (see above) were prepared as described (33) and subjected to 8% SDS/PAGE and immunoblot analysis (33).
Detection of Scap·Insig-2 Complex by Immunoprecipitation.
Pooled cell pellets from triplicate dishes of transiently transfected SRD-13A cells (see above) were resuspended in 0.5 ml of Buffer E. Cell lysates were passed through a 22-gauge needle 15 times, extracted by rotating for 1 h at 4°C, and clarified by centrifugation at 20,000 × g for 15 min. The lysates were precleared by incubation for 1 h at 4°C with 20 μg of an irrelevant rabbit polyclonal antibody (IgG fraction) together with 40 μl of Protein A/G agarose beads (Santa Cruz Biotechnology). Precleared lysates were rotated for 16 h at 4°C with 15 μg of polyclonal anti-Myc together with 40 μl of protein A/G agarose beads. After centrifugation at 200 × g for 3 min, the resulting supernatants were mixed with 5× SDS loading buffer. The pelleted beads were washed three times (10 min each at 4°C) with 0.7 ml of Buffer E and resuspended in 100 μl of Buffer D containing 1 mM sodium EDTA and 1 mM sodium EGTA, and mixed with 5× SDS loading buffer. Immunoprecipitated material was eluted by boiling and collected by centrifugation. Supernatant and pellet fractions were then subjected to 10% SDS/PAGE and immunoblot analysis.
Immunoblot Analysis.
After SDS/PAGE, the proteins were transferred to Hybond-C extra nitrocellulose filters (Amersham). The filters were incubated at room temperature with the following primary antibodies: 0.5 μg/ml anti-HSV IgG (Novagen); 1 μg/ml of IgG-9E10, a mouse monoclonal antibody against c-Myc (19); 10 μg/ml of IgG-9D5, a mouse monoclonal antibody against hamster Scap (40); 10 μg/ml of IgG-7D4, a mouse monoclonal antibody against the NH2 terminus of hamster-SREBP-2 (41). Bound antibodies were visualized by chemiluminescence (Super Signal Substrate; Pierce) using a 1:5,000 dilution of donkey anti-mouse IgG (Jackson ImmunoResearch) conjugated to horseradish peroxidase. Filters were exposed to Kodak X-Omat Blue XB-1 film at room temperature for 1–60 s.
Other Materials and Methods.
Reagents, buffers, tissue culture media, and expression plasmids are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank our colleagues Jin Ye, Russell DeBose-Boyd, and Li-Ping Sun for helpful discussions; Lisa Beatty, Angela Carroll, and Marissa Hodgin for invaluable help with tissue culture; and Debra Morgan and Mathew Francis for excellent technical assistance. This work was supported by National Institutes of Health Grant HL20948 and grants from the Perot Family Foundation. A.R. was the recipient of a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research.
Abbreviations
- ER
endoplasmic reticulum
- SREBP
sterol regulatory element-binding protein
- HMGR
3-hydroxy-3-methylglutaryl CoA reductase
- 25-HC
25-hydroxycholesterol
- HPCD
hydroxypropyl-β-cyclodextrin
- MCD
methyl-β-cyclodextrin.
Footnotes
This Feature Article is part of a series identified by the Editorial Board as reporting findings of exceptional significance.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 6496.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700899104/DC1.
References
- 1.Brown MS, Goldstein JL. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 2.Edwards PA, Tabor D, Kast HR, Venkateswaran A. Biochim Biophys Acta. 2000;1529:103–113. doi: 10.1016/s1388-1981(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein JL, DeBose-Boyd RA, Brown MS. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 4.Brown MS, Ye J, Rawson RB, Goldstein JL. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 5.Matsuda M, Korn BS, Hammer RE, Moon Y.-A., Komuro R, Horton JD, Goldstein JL, Brown MS, Shimomura I. Genes Dev. 2001;15:1206–1216. doi: 10.1101/gad.891301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandutsch AA, Chen HW. J Biol Chem. 1973;248:8408–84l7. [PubMed] [Google Scholar]
- 7.Brown MS, Goldstein JL. J Biol Chem. 1974;249:7306–73l4. [PubMed] [Google Scholar]
- 8.Goldstein JL, Faust JR, Brunschede GY, Brown MS. In: Lipids, Lipoproteins, and Drugs. Kritchevsky D, Paoletti R, Holmes WL, editors. New York: Plenum; 1975. pp. 77–84. [Google Scholar]
- 9.Kandutsch AA, Chen HW, Heiniger H-J. Science. 1978;201:498–501. doi: 10.1126/science.663671. [DOI] [PubMed] [Google Scholar]
- 10.Chang T-Y, Limanek JS. J Biol Chem. 1980;255:7787–7795. [PubMed] [Google Scholar]
- 11.Bjorkhem I. J Clin Invest. 2002;110:725–730. doi: 10.1172/JCI16388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell DW. Biochim Biophys Acta. 2000;1529:126–135. doi: 10.1016/s1388-1981(00)00142-6. [DOI] [PubMed] [Google Scholar]
- 13.Lund EG, Diczfalusy U. Methods Enzymol. 2003;364:24–37. doi: 10.1016/s0076-6879(03)64002-4. [DOI] [PubMed] [Google Scholar]
- 14.Horton JD, Goldstein JL, Brown MS. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L-P, Li L, Goldstein JL, Brown MS. J Biol Chem. 2005;280:26483–26490. doi: 10.1074/jbc.M504041200. [DOI] [PubMed] [Google Scholar]
- 16.Lee MCS, Miller EA, Goldberg J, Orci L, Schekman R. Annu Rev Cell Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 17.Rawson RB, Cheng D, Brown MS, Goldstein JL. J Biol Chem. 1998;273:28261–28269. doi: 10.1074/jbc.273.43.28261. [DOI] [PubMed] [Google Scholar]
- 18.Nohturfft A, Yabe D, Goldstein JL, Brown MS, Espenshade PJ. Cell. 2000;102:315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- 19.Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 20.Adams CM, Reitz J, DeBrabander JK, Feramisco JD, Brown MS, Goldstein JL. J Biol Chem. 2004;279:52772–52780. doi: 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- 21.Radhakrishnan A, Sun L-P, Kwon HJ, Brown MS, Goldstein JL. Mol Cell. 2004;15:259–268. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 22.Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL. Mol Cell. 2002;10:237–245. doi: 10.1016/s1097-2765(02)00591-9. [DOI] [PubMed] [Google Scholar]
- 23.Sun L-P, Seemann J, Goldstein JL, Brown MS. Proc Natl Acad Sci USA. 2007;104:6519–6526. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tinoco I, Jr, Woody RW. J Chem Phys. 1963;38:1117–1125. [Google Scholar]
- 25.Chen Y-H, Yang JT, Chau KH. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 26.Feramisco JD, Goldstein JL, Brown MS. J Biol Chem. 2004;279:8487–8496. doi: 10.1074/jbc.M312623200. [DOI] [PubMed] [Google Scholar]
- 27.Song B-L, Javitt NB, DeBose-Boyd RA. Cell Metab. 2005;1:179–189. doi: 10.1016/j.cmet.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Lin H-S, Rampersaud AA, Archer RA, Pawlak JM, Beavers LS, Schmidt RJ, Kauffman RF, Bensch WR, Bumol TF, Apelgren LD, et al. J Med Chem. 1995;38:277–288. doi: 10.1021/jm00002a010. [DOI] [PubMed] [Google Scholar]
- 29.Janowski BA, Shan B, Russell DW. J Biol Chem. 2001;276:45408–45416. doi: 10.1074/jbc.M108348200. [DOI] [PubMed] [Google Scholar]
- 30.Yabe D, Goldstein JL, Brown MS. Proc Natl Acad Sci USA. 2002;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobrosotskaya I, Goldstein JL, Brown MS, Rawson RB. J Biol Chem. 2003;278:35837–35843. doi: 10.1074/jbc.M306476200. [DOI] [PubMed] [Google Scholar]
- 32.Seegmiller AC, Dobrosotskaya I, Goldstein JL, Ho YK, Brown MS, Rawson RB. Dev Cell. 2002;2:229–238. doi: 10.1016/s1534-5807(01)00119-8. [DOI] [PubMed] [Google Scholar]
- 33.Gong Y, Lee JN, Brown MS, Goldstein JL, Ye J. Proc Natl Acad Sci USA. 2006;103:6154–6159. doi: 10.1073/pnas.0601923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong Y, Lee JN, Lee PCW, Goldstein JL, Brown MS, Ye J. Cell Metab. 2006;3:15–24. doi: 10.1016/j.cmet.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 35.Kauffman JM, Westerman PW, Carey MC. J Lipid Res. 2000;41:991–1003. [PubMed] [Google Scholar]
- 36.Meaney S, Bodin K, Diczfalusy U, Bjorkhem I. J Lipid Res. 2002;43:2130–2135. doi: 10.1194/jlr.m200293-jlr200. [DOI] [PubMed] [Google Scholar]
- 37.Im YJ, Raychaudhuri S, Prinz WA, Hurley JH. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sever N, Yang T, Brown MS, Goldstein JL, DeBose-Boyd RA. Mol Cell. 2003;11:25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 39.Rawson RB, DeBose-Boyd RA, Goldstein JL, Brown MS. J Biol Chem. 1999;274:28549–28556. doi: 10.1074/jbc.274.40.28549. [DOI] [PubMed] [Google Scholar]
- 40.Sakai J, Nohturfft A, Cheng D, Ho YK, Brown MS, Goldstein JL. J Biol Chem. 1997;272:20213–20221. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- 41.Yang J, Brown MS, Ho YK, Goldstein JL. J Biol Chem. 1995;270:12152–12161. doi: 10.1074/jbc.270.20.12152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.