Abstract
Quantitative measurements of regional and tissue specific concentrations of brain metabolites were measured in elderly subjects using multislice proton magnetic resonance spectroscopic imaging (1H MRSI). Selective k-space extrapolation and an inversion-recovery sequence were used to minimize lipid contamination and linear regression was used to account for partial volume problems. The technique was applied to measure the concentrations of N-acetyl aspartate (NAA), and creatine (Cr)- and choline (Cho)-containing compounds in cortical gray and white matter, and white matter lesions of the frontal and the parietal lobe in 40 normal elderly subjects (22 females and 18 males, 56–89 years old, mean age 74 ± 8). NAA was about 15% lower in cortical gray matter and 23% lower in white matter lesions when compared to normal white matter. Cr was 11% higher in cortical gray matter than in white matter, and also about 15% higher in the parietal cortex than in the frontal cortex. Cho was 28% lower in cortical gray matter than in white matter. Furthermore, NAA and Cr changes correlated with age. In conclusion, regional and tissue differences of brain metabolites must be considered in addition to age-related changes when interpreting 1H MRSI data.
Keywords: magnetic resonance spectroscopic imaging, quantification, aging, gray and white matter, white matter lesions
Most magnetic resonance spectroscopy (MRS) studies of the human brain have used single-volume localization methods that measure concentrations of N-acetyl aspartate (NAA), creatine (Cr)- and choline (Cho)-containing compounds, and other metabolites in one operator-specified location. Although these methods are generally easy to implement, they provide limited information about the regional distribution of the brain metabolites. In contrast, MRS imaging (MRSI) techniques acquire spectra simultaneously over a wide brain region, enabling the generation of maps of metabolite distributions. However, 1H MRSI is technically more challenging. For brain studies, care must be taken to account for an intense signal from extracranial lipids that may otherwise distort weak metabolite resonances, especially in brain regions close to the skull. To avoid distortion from lipids, many 1H MRSI studies have incorporated volume preselection with point-resolved spectroscopy (PRESS) (1) or stimulated echo acquisition modes (STEAM) (2) to limit the observed region to within the brain. 1H MRSI without volume preselection enabling full brain coverage was first demonstrated by Dynn et al. (3), using outer-volume suppression (OVS) pulses to reduce the lipid signal within the skull region. However, OVS pulses may also partially saturate brain tissue adjacent to the skull, complicating metabolite quantification from this region. Lipid reduction without OVS pulses was accomplished using inversion recovery methods that null the lipid signal during data acquisition (4,5) or data processing methods using lipid selective k-space extrapolation (6,7). The first goal of this study was therefore to develop a multislice 1H MRSI method with minimal lipid contamination, using a combination of lipid nulling and k-space extrapolation to avoid OVS pulses and improve the accuracy of NAA measurements with 1H MRSI.
Because many 1H MRSI voxel locations in the brain include cerebrospinal fluid (CSF), which contributes no observable signal, it is necessary to account for the relative inclusion of CSF in order to obtain metabolite concentrations in the tissue. Furthermore, because signal contributions may arise from both gray and white matter tissue, it is critical to differentiate between metabolite changes of gray and white matter, and other tissue types. One approach for differentiation is the use of linear regressions to predict the relationship between metabolite intensity changes and gray/white matter variations in MRSI voxels (4,8-12). However, most previous MRSI studies that used linear regression averaged metabolite concentrations over different lobes of the brain, ignoring regional variations. In addition, tissues other than gray and white matter—such as white matter lesions, which occur frequently in the aged brain—were ignored or not determined. Therefore, a second goal in this study was to develop an approach for obtaining metabolite concentrations of gray and white matter, as well as of white matter lesions in different lobes of the brain using linear regression.
Before MRSI results can be interpreted properly for pathological abnormalities, the metabolite patterns must first be established in normal brain. The concentrations of NAA, Cr, and Cho in gray and white matter of normal brain have been studied by a number of investigators using linear regression. However, the results were not always in agreement—especially with respect to NAA. While several studies (8-10,13) reported higher NAA in gray matter than in white matter; others (4,11) found the opposite. Therefore, the third goal of this study was to apply the new techniques to measure metabolite concentrations in different regions and tissues of the brain of normal elderly subjects.
METHODS
The committee of human research at the University of California–San Francisco (UCSF) approved the study, and written informed consent was obtained from each subject before participation.
Study Population
This study included 40 (22 females and 18 males) healthy elderly subjects between 56 and 89 years of age, mean age 74 ± 8 years. The subjects were recruited from the UCSF Institute of Aging, the UCSF memory clinic, and the University of California–Davis Alzheimer's Center. The subjects received the standard neurological examinations at the centers, and were considered cognitively normal for their age group. Furthermore, a neuroradiologist read the MR images of the subjects to exclude major neuropathology, such as brain tumors and cortical and subcortical infarcts. Subjects with white matter lesions or marked brain atrophy were not excluded from the study.
Structural MRI and 1H MRSI Acquisition
The subjects were scanned on a 1.5T VISION™ MR system (Siemens Inc., Iselin, NJ). A vacuum-molded head holder (Vac-Pac, Olympic Medical, Seattle, WA) was employed to restrict head movements. Structural MRI data were acquired using a double spin echo sequence (DSE) with TR/TE1/TE2 = 2500/20/80 msec timing, 1.0 × 1.4 mm2 in-plane resolution, and about 50 contiguous 3-mm-thick axial slices oriented along the optic nerve as seen from a midsection scout MRI. In addition, a volumetric magnetization-prepared rapid gradient echo (MPRAGE) sequence was acquired, with TR/TE/TI = 13.5/7/300 msec timing, 15° flip angle, 1.0 × 1.0 mm2 in-plane resolution, and 1.4-mm-thick coronal partitions, oriented orthogonal to the DSE image plane. Proton density and T2-weighted images from DSE and T1-weighted images from MPRAGE were used together for MRI tissue segmentation with software developed in-house and described previously (14).
Multislice 1H MRSI (TR/TE = 1800/135 msec, 30-min total acquisition) was acquired from two 15-mm-thick slices aligned parallel to the axial-oblique DSE images, including slice-selective inversion-recovery (TI = 170 msec) to null the lipid signal and chemical shift-selected (CHESS) water suppression (2). k-Space sampling was accomplished with 36 × 36 circularly bounded encoding steps across a 2802 mm2 field of view (FOV), yielding a nominal voxel size of about 0.9 ml. The center of the lowest MRSI slice was positioned to be immediately below the superior aspect of the corpus callosum, and the second slice was about 22 mm more superior. This configuration covered large areas of the left and right, and frontal and parietal lobes with little inclusion of the occipital and temporal lobes and subcortical regions. Together, structural MRI and multislice 1H MRSI, including adjustments for CHESS water suppression, required about 50 min of acquisition time.
MRI Segmentation
First-pass segmentation of whole-brain MRI data into the primary tissue categories of gray and white matter and CSF was performed by an automated procedure based on K-means cluster analysis, as described previously with minor modifications (14). Second-pass, operator-assisted segmentation further classified white matter into normal white matter (WM) and white matter lesions (WML), and gray matter into cortical gray matter (cGM) and subcortical gray matter. Finally, the hemispheric and Rolandic fissures were marked on MRI approximately over the region covered by the MRSI slices to mark the boundaries between the right and left cerebral hemispheres, and frontal and posterior lobes. One trained operator (D.L.A.) performed semiautomated segmentation, and one experienced radiologist (A.A.C.) marked the MRI data.
Spectral Processing
The 1H MRSI data were zero-padded to 64 × 64 points in the spatial domain and 1024 points in the spectral domain. Before Fourier reconstruction, the time domain data were 2 Hz Gaussian filtered and deconvolved using a Hamming filter to eliminate residual water resonances. No filters were applied along the spatial domain. Reduction of spurious resonances from extracranial lipids was accomplished by selective k-space extrapolation, as described by Haupt et al. (7). A fully automated spectral-fitting software package (15) was employed to estimate the peak areas of NAA, Cr, and Cho. This software utilizes a parametric model based on a priori information of spectral metabolite patterns and a nonparametric baseline model based on wavelets to fit the data. Quality control was ensured by rejecting peaks that had larger than 12 Hz linewidth at half-peak height or fits with residual sum squares that were outside the upper 95 percentile distribution of residuals from all fits. Typically, no more than 10% of the MRSI data from each subject were rejected by this method. Finally, the peak areas were corrected for receiver gain and expressed relative to the intensity of median ventricular CSF of each subject, as measured from proton density MRI. The coefficient of variation (standard deviation divided by the mean value) for ventricular CSF intensities of the subjects as measured from proton density MRI was 2.5%. All metabolite concentrations are expressed in arbitrary units.
MRI and 1H MRSI Coanalysis
The next goal of the analysis was to estimate tissue weight WTissue (in units of volume) of each tissue type (i.e., cGM, subcortical gray matter, WM, WML, CSF, and nonbrain tissue) in each of the MRSI voxels using information from the segmented MR images. First, the stack of segmented MR images was aligned with the MRSI slices, using slice position and orientation information, and assuming that there was no head movement between MRI and MRSI scans. Then the tissue weights were computed (Eq. [1]) by convolving each tissue map of the segmented MR images with the discrete transform of the MRSI spatial response function (Eq. [2]) and MRSI slice profile, including corrections for chemical shift displacement and coil sensitivity (determined from phantom studies):
| [1] |
| [2] |
Here, is the estimated cGM weight associated with NAA in the nth MRSI voxel with coordinates (xn,yn,zn). IcGM(x′j,y′k,z′l) represents the cGM map of the segmented MR images, which is unity when MRI indicates cGM, and 0 for other tissue types at coordinates (x′j,y′k,z′l). SRF(x,y) is the 2D spatial response function of MRSI, arising from the two phase-encoding directions, and(x′j − xn,y′k − yn). are the x and y distances (in millimeters) from the center of the nth MRSI voxel to the segmented MRI pixel (j,k). The characteristic form of SRF (16) for values specific to this study is given in Eq. [2] for one dimension of 36 MRSI phase-encoding steps over the FOV of 2802 mm2. P(z′l, − znNAA) represents the MRSI slice profile as a function of the distance between the z coordinates of the nth MRSI and lth MRI voxel, and is the correction for coil sensitivity, where smax is the maximum sensitivity and is the sensitivity profile of the coil. indicates a slight spatial displacement that is associated with the chemical shift frequency of NAA. Summations are performed over all N × N pixels and M slices of the segmented MRI data. Finally, was classified as belonging to either the left or right frontal lobe, or the left or right parietal lobe, based on a computerized comparison of the voxel position with respect to the marks of the hemispheric and Rolandic fissures on MRI. Similarly, tissue weights of other tissue types can be calculated by substituting the corresponding tissue maps for IcGM(x′j,y′k,z′l), and tissue weights associated with Cr and Cho can be calculated by substituting znCr and znCho for znNAA in Eq. [1].
The next goal of the analysis was the formulation of a reliable method for the estimation of NAA, Cr, and Cho concentrations (hereafter termed [NAA], [Cr], and [Cho]) in cortical gray and white matter for each of the four brain regions (left frontal, right frontal, left parietal, and right parietal) of each subject. With the assumption that such levels exist, and that cortical gray and white matter give rise to most of the MRSI signal, the following simplified model for the measured signal intensity Y (Y = corrected peak areas of NAA, Cr, or Cho) in the nth MRSI voxel was applied:
| [3] |
Here, n runs through all voxels in one of the four lobe regions of each subject. XcGM and XWM represent the signal contributions from cortical gray and white matter, respectively, and are synonymous with concentrations in this context. WncGM and WnWM are the weights of cortical gray and white matter in the voxels, respectively. is the signal component from spurious contributions of other tissue types, such as WML and subcortical gray matter. The bias term is, by hypothesis, a function of the weights WncGM and WnWM, because the lower their sum for a voxel the more likely it is that WML and subcortical gray matter are contributing to the signal. A major exception to this is when the rest of the voxel contains CSF or nonbrain tissue, as both contribute nothing to the signal. The error term represents simply the error from estimations of the metabolite signal itself and is assumed to be reasonably Gaussian distributed. The model was fit using a multiterm linear regression algorithm (SPLUS, Mathsoft Inc, Seattle, WA) to obtain estimations for XcGM and XWM. Furthermore, the accuracy for XcGM and XWM was determined, especially given the possibility of bias induced by other tissue types, using the width of 95% confidence intervals of the regression fits. These confidence intervals were constructed by refitting XcGM and XWM from a set of 500 resample observations using a general nonparametric bootstrap procedure (SPLUS). Signal contributions corresponding to the bias term in Eq. [3] were discriminated by comparing mean and median values of the confidence intervals, according to bias = (mean − median)/mean. For bias values larger than ±0.05, some MRSI voxels with high tissue weights of WML or subcortical gray matter were removed before refitting the regressions until a bias of less than or equal to ±0.05 was obtained.
Finally, metabolite intensities of WML were calculated from a voxel j with maximum weight WWML and minimal contributions from cortical and subcortical gray matter in each subject. Such a condition can usually be found, because white matter lesions appear frequently within white matter regions at some distance to gray matter. This leads to the simplified expression for the signal component from WML:
| [4] |
Here, Yj, WWM, and error have the same meanings as in Eq. [3]. Xwm is the estimated signal component from white matter, obtained from the regression analysis according to Eq. [3]. (A similar analysis can be performed for subcortical gray matter; but this was not carried out in this study because the MRSI data included very little subcortical gray matter due to the MRSI slice positions). All together, MRI segmentation, automated spectral processing, and bootstrap procedures required about 4 hr computation time per subject (on a 500 MHz Pentium processor) and an additional 2 hr for operator-interactive processes, such as the marking of white matter lesions and hemispheric and Rolandic fissures on the MRI data.
Statistics
Differences between measurements with and without k-space extrapolation were evaluated using paired t-tests. Differences of metabolite concentrations between cortical gray and white matter and regional variations were tested using repeated measure analysis of variance (rmANOVA, two tissue types by four regions), followed by post hoc Scheffe tests. Differences of metabolite concentrations between WML and white matter were evaluated using t-tests. Pearson correlation coefficients were used to determine metabolite changes with age.
RESULTS
In Fig. 1 are shown representative metabolite images of NAA, Cr, and Cho from an 80-year-old healthy male subject. Also shown are representative spectra from the frontal and parietal surface cortex, sampled at a location in close proximity to the skull. Spectral fits are superimposed on the raw 1H MR spectra. In Fig. 2 are shown representative regression plots for NAA, Cr, and Cho intensity changes as a function of cortical gray matter weights in MRSI voxels from the left frontal lobe of the same subject. Table 1 lists the confidence intervals at the 95% level for estimations of the metabolite concentrations with linear regression, separately for each metabolite and each brain region. The 95% confidence intervals, which were obtained from bootstrapping, are expressed in percent of the metabolite mean values from all subjects. This shows that confidence intervals for [NAA], [Cr], and [Cho] were smaller (and thus, accuracy was higher) for white matter than for cortical gray matter (F(1,301) = 160; P < 0.001). Similarly, confidence intervals were smaller and accuracy higher for the frontal lobe than for the parietal lobe (F(3,301) = 20; P < 0.001). The 95% confidence intervals ranged from about 6% for [NAA] of white matter to about 32% for [Cho] of cortical gray matter.
FIG. 1.
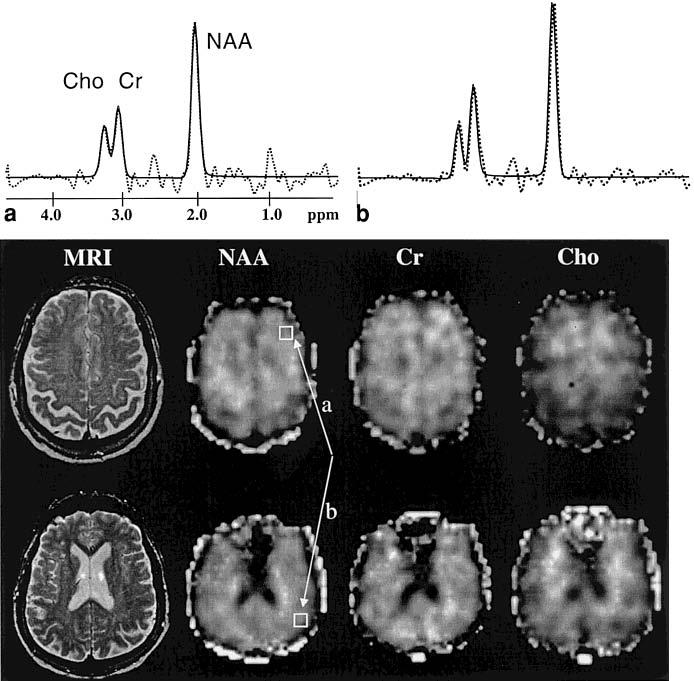
Metabolite images of NAA, Cr, and Cho, and the corresponding anatomical images from a 75-year-old male subject. Also shown are representative spectra from a location in the frontal (a) and parietal (b) cortex, as indicated in the NAA images. Dashed lines represent raw spectral data; solid lines represent results from the automated spectral fit.
FIG. 2.
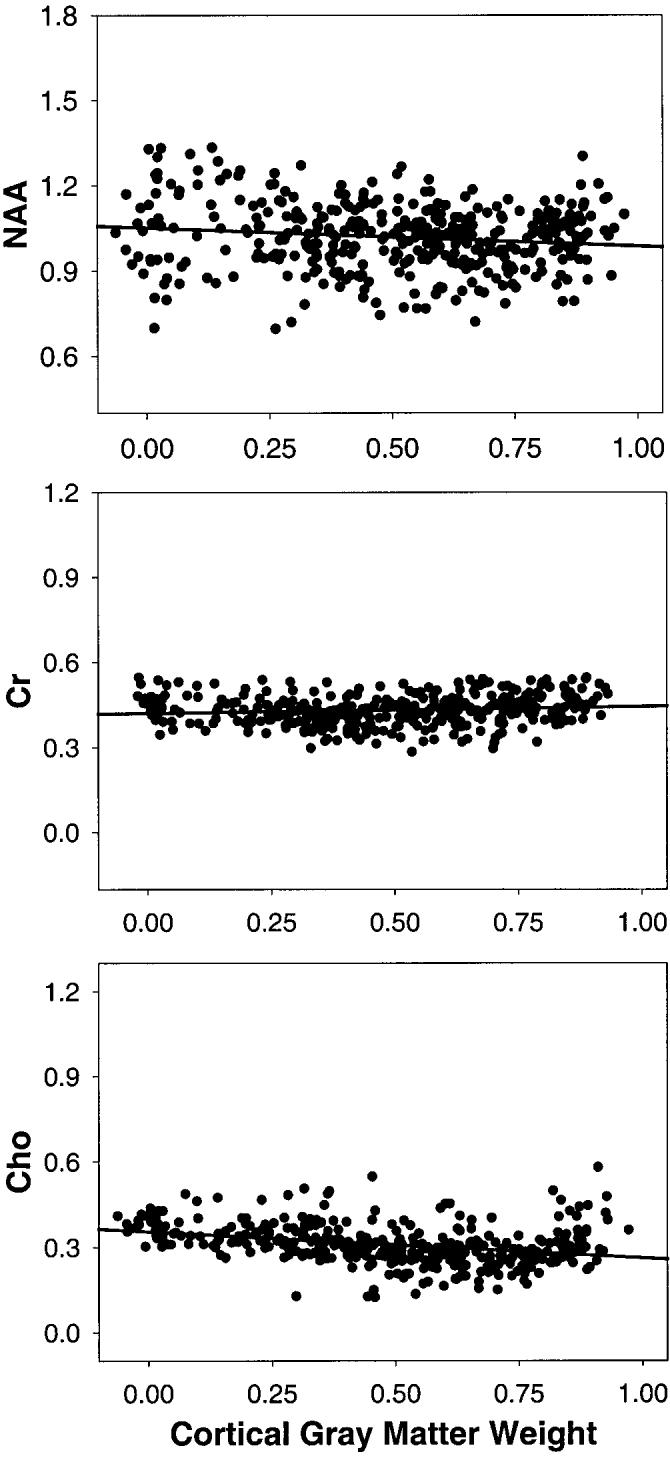
Regression plots of NAA, Cr, and Cho intensity changes (in arbitrary units) against cortical gray matter weight in MRSI voxels. Cortical gray matter weight is defined as the ratio of cortical gray matter to total brain tissue in a voxel. Its value is unity when all tissue in a voxel is cortical gray matter.
Table 1.
Confidence Intervals at the 95% Level for Estimations of the Concentrations of NAA, Cr, and Cho, Obtained From 500 Resampled Regressions by Bootstrapping*
| Metabolite tissue type | Brain lobes |
|||
|---|---|---|---|---|
| Frontal right | Frontal left | Parietal right | Parietal left | |
| NAA | ||||
| Cortical gray | 10.7 ± 3.3 | 10.8 ± 3.3 | 12.6 ± 4.1 | 14.8 ± 4.0 |
| White | 6.6 ± 2.0 | 7.0 ± 2.7 | 8.7 ± 3.4 | 8.9 ± 3.0 |
| Cr | ||||
| Cortical gray | 15.4 ± 3.7 | 14.7 ± 4.2 | 20.1 ± 4.5 | 20.1 ± 4.3 |
| White | 8.2 ± 3.0 | 8.6 ± 2.7 | 11.2 ± 3.5 | 11.7 ± 3.7 |
| Cho | ||||
| Cortical gray | 19.7 ± 5.5 | 19.5 ± 4.0 | 31.7 ± 6.0 | 30.1 ± 6.1 |
| White | 11.2 ± 3.6 | 11.7 ± 3.1 | 16.8 ± 5.8 | 19.2 ± 6.6 |
In percent of the metabolite mean values from all subjects.
The effect of lipid k-space extrapolation in addition to lipid nulling on the accuracy for determination of [NAA] is illustrated in Table 2. This analysis was not performed for Cr and Cho because no difference was anticipated. The data are from the left frontal lobe of 20 subjects who were processed with and without k-space extrapolation, and with identical conditions otherwise. This shows that when k-space extrapolation was performed in addition to lipid nulling, [NAA] values were 10% (P < 0.005, paired t-test) lower in cortical gray matter and 8% (P < 0.005) lower in white matter than with lipid nulling alone. Furthermore, the confidence intervals at 95% became narrower (thus accuracy improved) with k-space extrapolation by 44% for [NAA] of cortical gray matter and by 37% for [NAA] of white matter. Similar results were obtained for the other brain lobes. This implies that although lipid contamination was already reduced with a 135-msec spin-echo time and lipid nulling, it was removed further with the application of k-space extrapolation.
Table 2.
NAA Concentrations and Confidence Intervals (CI)* at the 95% Level for MRSI Data With and Without Lipid k-Space Extrapolation
| Tissue | NAA | CI |
|---|---|---|
| Cortical gray matter | ||
| With extrapolation | 0.72 ± 0.11 | 9.1 ± 1.7 |
| Without extrapolation | 0.79 ± 0.12 | 13.2 ± 2.8 |
| % difference | 10a | 44a |
| White matter | ||
| With extrapolation | 0.89 ± 0.09 | 6.7 ± 1.6 |
| Without extrapolation | 0.96 ± 0.11 | 9.2 ± 1.9 |
| % difference | 8a | 37a |
In percent of the mean NAA values in cortical gray and white matter.
P < 0.005, paired t-tests.
In Table 3 are listed the [NAA], [Cr], and [Cho] values of cortical gray and white matter for the left and right frontal and parietal lobes. For [NAA], there was a significant difference in metabolite concentration between tissue types (F (3,301) = 65.88; P < 0.0001), with [NAA] being about 15% lower in cortical gray matter than in white matter, but with no regional effect (F (3,301) = 1.02; P = 0.38) or tissue-by-region interaction (F (3,301) = 1.72; P = 0.16). For [Cr], there was a significant tissue (F (3,301) = 17.38; P < 0.0001) and region effect (F (3,301) = 6.46; P = 0.0002), but no tissue-by-region interaction (F (3,301) = 1.16; P = 0.32). [Cr] was about 11% higher in cortical gray matter than in white matter, and about 15% higher in the parietal cortex than in the frontal cortex (Scheffe, P < 0.001). There was no significant left-to-right difference for [Cr] and no significant regional difference for [Cr] of white matter. For [Cho], there was a significant tissue effect (F (3.298) = 148.71; P < 0.0001), but no regional effect (F (3,298) = 0.48; P = 0.69) or tissue-by-region interaction (F (3,298) = 1.92; P = 0.12). [Cho] was 28% lower in cortical gray matter than in white matter.
Table 3.
Concentrations* of NAA, Cr, and Cho in Cortical Gray Matter and White Matter of the Left and Right, Frontal and Parietal Lobes
| Metabolite/tissue type | Brain lobes |
|||
|---|---|---|---|---|
| Frontal right | Frontal left | Parietal right | Parietal left | |
| NAA | ||||
| Cortical gray | 0.85 ± 0.12 | 0.83 ± 0.12 | 0.84 ± 0.18 | 0.82 ± 0.17 |
| White | 1.00 ± 0.14 | 0.96 ± 0.13 | 1.01 ± 0.16 | 0.97 ± 0.16 |
| Cr | ||||
| Cortical gray | 0.41 ± 0.08 | 0.40 ± 0.07 | 0.46 ± 0.11 | 0.47 ± 0.11 |
| White | 0.37 ± 0.05 | 0.39 ± 0.06 | 0.40 ± 0.09 | 0.41 ± 0.09 |
| Cho | ||||
| Cortical gray | 0.28 ± 0.07 | 0.28 ± 0.07 | 0.26 ± 0.10 | 0.26 ± 0.13 |
| White | 0.36 ± 0.07 | 0.38 ± 0.06 | 0.37 ± 0.06 | 0.38 ± 0.07 |
In arbitrary units.
In Table 4 are listed the metabolite concentrations in WML tissue for 10 subjects who presented MRSI voxels with at least 25% or more WML tissue content. The other subjects had either no WML or very small areas of WML that filled MRSI voxels less than 25%. When compared to normal white matter, [NAA] of WML was significantly reduced by 21% (P < 0.03, two-sided t-test). Similarly, [Cr] and [Cho] were also lower in WML than in normal white matter. But the differences were not significant.
Table 4.
Concentrations* of NAA, Cr, and Cho in White Matter Lesions (WML) and Normal White Matter
| Tissue/region | NAA | Cr | Cho |
|---|---|---|---|
| WML | 0.84 ± 0.23 | 0.36 ± 0.10 | 0.37 ± 0.09 |
| Normal white | 1.07 ± 0.18 | 0.41 ± 0.04 | 0.40 ± 0.11 |
| % difference | −21a | −12 | −8 |
In arbitrary units.
P < 0.03 by two-tailed t-test.
Age effects were determined over the age range of 56 to 89 years. Metabolite concentrations of cortical gray matter were not significantly correlated with age. In white matter, however, [NAA] (r 5.35, P = 0.04) and [Cr] (r = .52, P = 0.01) increased with increasing age, while [Cho] changes and age were not significantly correlated (r = .18, P = 0.1). [NAA] and [Cr] changes as a function of age are depicted in Fig. 3. There were no significant regional differences for age-related changes of [NAA] (F (3,145) = 0.19; P = 0.5) or [Cr] (F (3,145) = 1.03; P = 0.4).
FIG. 3.
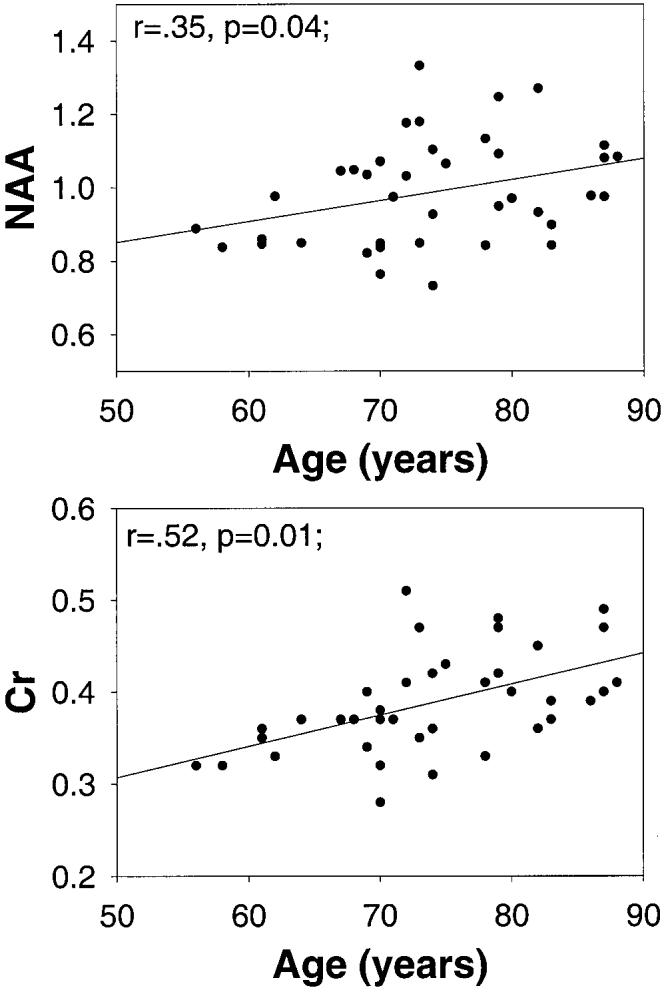
Changes of NAA and Cr concentrations (in arbitrary units) in white matter as a function of age.
DISCUSSION
In this work, a robust procedure is described for quantification of metabolites in gray and white matter, and WML of the frontal and parietal lobes of the brain. Selective k-space extrapolation was used in addition to lipid signal nulling to minimize lipid contamination, and linear regression was used to compensate for partial volume problems in 1H MRSI. Our main findings were: 1) Compared to white matter, [NAA] was about 15% lower in cortical gray matter and 23% lower in white matter lesions. 2) [Cr] was about 11% higher in cortical gray than in white matter, and also about 15% higher in parietal cortex compared to frontal cortex. 3) [Cho] was about 28% lower in cortical gray matter than in white matter. Finally, [NAA] and [Cr] of white matter increased with increasing age.
A number of previous 1H MRS studies have measured metabolite concentrations in gray and white matter. The majority employed single voxel MR spectroscopy (for review see Ref. 17), selecting a region that contained primarily gray or white matter. However, as these voxels were usually large (2–18cc), they were likely heterogeneous with respect to tissue composition; thus, any differences that were found must be less than the true differences. These studies were also limited due to the small number of data samples obtained from each subject (18). To overcome these problems, several investigators (4,8-13) incorporated tissue-segmented MRI data and linear regression into the MRSI analysis, similar to the approach described here. However, our proposal provides several improvements over previous studies using regression methods. First, we reduced lipid contamination of 1H MR spectra by employing k-space extrapolation (7) in addition to lipid signal nulling. This improved the accuracy for determination of [NAA], presumably because spurious resonances from extracranial lipids were further reduced, which helped spectral fitting. Although the results may be different for other automated spectral-fitting programs, this finding implies that lipid k-space extrapolation improves 1H MRSI measurements of the brain. Second, we measured metabolite concentrations of gray and white matter separately in the left and right frontal and parietal lobes, and observed some regional differences. Although other studies have used 3D 1H MRSI (9,10), which provided better brain coverage than the present multislice study, metabolite concentrations were averaged over the regions, ignoring regional variations. Noworolsky et al. (13) differentiated between the frontal lobe and occipital lobe using surface coils for data acquisition, but required comprehensive data processing to compensate for coil inhomogeneity. Third, we accounted for signal contributions from WML and estimated metabolite concentrations of WML. Most previous studies using regression analysis ignored signal contributions from tissue other than gray or white matter. Finally, we evaluated accuracy of the metabolite estimations in each brain region. We found marked differences among [NAA], [Cr], and [Cho], as well as between gray and white matter and the frontal and parietal lobe. Overall, accuracy was highest for [NAA] of white matter in the frontal lobe and lowest for [Cho] of gray matter in the parietal lobe. Higher accuracy for [NAA] than for [Cho] or [Cr] is probably due to a frequently higher peak intensity of NAA than of Cr or Cho. Higher accuracy for white matter than for cortical gray matter is probably due to a better tissue-fill factor for voxels in white matter regions compared to voxels in cortical regions. Higher accuracy for frontal lobe than parietal lobe data is probably due a larger number of MRSI samples for the regression from the frontal lobe compared to the smaller parietal lobe. These differences should be considered when analyzing MRSI data.
Our finding of lower [NAA] (by about 15%) in cortical gray matter than in white matter is consistent with similar results by Hetherington et al. (4) and Lundbom et al. (11). However, it should be emphasized that the magnitude of this difference is on the order of the measurement accuracy for [NAA], as determined by 95% confidence intervals. Other studies (8-10,12,13) reported higher [NAA] in gray matter relative to white matter between 6% (9) and 46% (8) with linear regression. One possible explanation for the difference is the variation due to T1 and T2 relaxations. In particular, our results are slightly weighted by T1, which is induced with slice-selective inversion for lipid signal nulling, in addition to T2. Lundborn et al. (11) found also lower [NAA] in gray matter than in white matter in the absence of complications with T1 and at a longer echo time than in this study. Furthermore, if our results were corrected for differential (gray and white matter specific) T1 and T2 relaxations (using T1/T2 values reported by Kreis et al. (19) and following computations similar to those outlined by Noworolski et al. (13)), [NAA] would still be about 10% lower in gray matter than in white matter. Taken together, this implies that T1/T2 relaxation alone does not explain the difference. Another possible explanation is differences due to aging. This is further substantiated by our observation that [NAA] of white matter (but not of cortical gray matter) increased slightly from age 56 to 89, reducing [NAA] of cortical gray matter relative to white matter at older ages. Whether the gray/white matter [NAA] ratio changes with age is controversial. Pfefferbaum et al. (10) found no age effect, while others reported a marked reduction of [NAA] in gray matter and no changes in white matter (9,11), decreasing the gray/white matter [NAA] ratio with increasing age. Finally, there are a variety of substantial technical differences between this study and other studies that could be responsible for the disparate findings for [NAA], such as: 1) sampling spectra from the middle of the brain only (8,12); 2) using outer volume suppression for lipid reduction (13); 3) fitting magnitude spectra without baseline correction (9,10); 4) using noncontiguous MRI data for segmentation (9,10); and 5) computing tissue compartments of the voxels with incomplete correction for the MRSI point spread function (9,10). Although autopsy 1H MRS studies (20) reported 50% higher [NAA] in gray matter compared to white matter, it is not clear whether a similar ratio would apply to in vivo conditions.
In agreement with many 1H MRSI studies (4,8,10-13), but not all (21), we found higher [Cr] in cortical gray matter (by about 11%) than in white matter. This is in good agreement with previous MRS autopsy studies that reported higher [Cr] in gray matter than white matter by 16% (22) to 25% (20). In addition, we found about 15% higher [Cr] in parietal cortical gray matter than in frontal gray matter. Regional differences for Cr have been measured previously with single-voxel MRS (23). Although Noworolski et al. (13) found no significant regional variation for [Cr], they examined different brain regions than we did in this study, which makes a comparison difficult. Nevertheless, our findings imply that caution should be applied when using Cr as an internal reference to express metabolite changes in MRS.
Our finding of lower [Cho] in gray matter than in white matter is in agreement with several previous 1H MRSI regression studies (10,11,13). However, our result of a 28% difference between gray and white matter [Cho] is substantially higher than the 9% difference in elderly controls reported by Pfefferbaum et al. (10). Several other MRSI studies (4,8,12) reported no significant gray/white difference for [Cho]. The markedly lower measurement accuracy for [Cho] compared with [NAA] and [Cr] could explain the disparate findings for [Cho].
Another finding was a 21% reduction of [NAA] in WML when compared with normal white matter, and no significant change of [Cho] and [Cr]. There have been few previous reports of metabolite changes in WML of normal elderly with 1H MRSI (24,25). These earlier studies found reduced NAA/Cho and higher Cho/Cr ratios in regions with WML, suggesting that the changes were primarily associated with increased [Cho]. Our results suggest that reduced [NAA] in the absence of substantial [Cr] and [Cho] changes may have been primarily responsible for these earlier reports of metabolite variations in WML.
Finally, we found for [NAA] and [Cr] (but not for [Cho]) of white matter an increase with age. Although significant, the [NAA] increase with age was modest and less prominent than the [Cr] changes. The function of NAA is still controversial, presumably reflecting the number or density of neurons, including axons and/or neuronal/axonal metabolism. Since the number and metabolism of axons are not expected to increase with age, it is unlikely that these factors are responsible for the [NAA] increase in white matter. However, increased axonal density due to decreased brain water (intra- or extracellular) could explain the NAA changes. Another explanation is prolonged T1 and/or T2 relaxation of white matter NAA at older ages. In contrast to NAA, increased Cr concentrations with age in white matter have been reported by several studies (10,26,27). Our measurement of [Cr] increase with age could be due to a combination of prolonged T1/T2 relaxations and/or higher Cr concentration. Our finding of stable [Cho] is in agreement with several MRS studies (27-29). However, two more recent studies (10,26) reported increases of white matter [Cho] with age. Again, the low accuracy of Cho measurements may be responsible for discrepant findings. The significance of [Cr] and [Cho] changes in white matter with age is still not clear and may reflect both neuronal and nonneuronal tissue deterioration, including demyelination of axons, as observed in post mortem MRI and histological studies of normal aging (30). Nevertheless, the results imply that matching groups by age is essential for 1H MRSI studies.
There were several limitations to our study. First, measurements for obtaining relaxation values for the metabolites could not be performed due to prohibitively long acquisition times. Therefore, corrections for T2 and particularly T1 were not applied, and absolute metabolite concentrations could not be determined. Second, linear regression was performed separately for each brain lobe, ignoring contributions from bordering regions. A more rigorous approach for linear regression, which includes contributions from bordering regions, is possible when the segmented MRI data incorporates regional information in addition to tissue segmentation alone. This allows regression of metabolite intensity changes simultaneously against both tissue type and regional variations. We did not perform such an analysis because regional information could not easily be incorporated into MRI segmentation at the time of this study. Third, frontal and parietal cortical gray matter was not further differentiated into gray matter of the sensory and the motor cortex. Therefore, metabolite values for cortical gray matter may be skewed to the extent that the sensory and motor cortexes have different metabolite concentrations than the frontal and the parietal cortexes.
In summary, an MRSI method has been described for measurements of regional and tissue variations of metabolites, which demonstrated metabolite differences among regions and tissue types in the brain of normal elderly controls. This information is essential when interpreting 1H MRSI data of brain disorders in the elderly, such as Alzheimer's disease, vascular dementia, and Parkinson's disease.
ACKNOWLEDGMENTS
We thank Dr. Owen Wolkowitz of the UCSF Center for Aging, and Dr. William Jagust of UC–Davis for referral and neurological evaluation of the elderly controls. We are indebted to Dr. David Norman for clinical screening of MRI scans.
Footnotes
Grant sponsor: NIH; Grant numbers: AG10897; AG12435.
REFERENCES
- 1.Bottomley PA. Spatial localization in NMR spectroscopy in vivo. Ann N Y Acad Sci. 1987;508:333–348. doi: 10.1111/j.1749-6632.1987.tb32915.x. [DOI] [PubMed] [Google Scholar]
- 2.Haase A, Frahm J. Multiple chemical-shift-selective NMR imaging using stimulated echoes. J Magn Reson. 1985;64:94–102. [Google Scholar]
- 3.Duyn JH, Gillen J, Sobering G, van Zijl PC, Moonen CT. Multisection proton MR spectroscopic imaging of the brain. Radiology. 1993;188:277–282. doi: 10.1148/radiology.188.1.8511313. [DOI] [PubMed] [Google Scholar]
- 4.Hetherington HP, Pan JW, Mason GF, Adams D, Vaughn MJ, Twieg DB, Pohost GM. Quantitative 1H spectroscopic imaging of human brain at 4.1 T using image segmentation. Magn Reson Med. 1996;36:21–29. doi: 10.1002/mrm.1910360106. [DOI] [PubMed] [Google Scholar]
- 5.Spielman D, Meyer C, Macovski A, Enzmann D. 1H spectroscopic imaging using a spectral-spatial excitation pulse. Magn Reson Med. 1991;18:269–279. doi: 10.1002/mrm.1910180203. [DOI] [PubMed] [Google Scholar]
- 6.Hu X, Patel MS, Garwood M, Ugurbil K, Merkle H, Grad J, Stillman AE. Reduction of lipid contamination in proton chemical shift imaging by a Bayesian reconstruction. Proceedings of the ISMRM. 1992:4253. [Google Scholar]
- 7.Haupt CI, Schuff N, Weiner MW, Maudsley AA. Removal of lipid artifacts in 1H spectroscopic imaging by data extrapolation. Magn Reson Med. 1996;35:678–687. doi: 10.1002/mrm.1910350509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doyle TJ, Bedell BJ, Narayana PA. Relative concentrations of proton MR visible neurochemicals in gray and white matter in human brain. Magn Reson Med. 1995;33:755–759. doi: 10.1002/mrm.1910330603. [DOI] [PubMed] [Google Scholar]
- 9.Lim KO, Spielman DM. Estimating NAA in cortical gray matter with applications for measuring changes due to aging. Magn Reson Med. 1997;37:372–377. doi: 10.1002/mrm.1910370313. [DOI] [PubMed] [Google Scholar]
- 10.Pfefferbaum A, Adalsteinsson E, Spielman D, Sullivan EV, Lim KO. In vivo spectroscopic quantification of the N-acetyl moiety, creatine, and choline from large volumes of brain gray and white matter: effects of normal aging. Magn Reson Med. 1999;41:276–284. doi: 10.1002/(sici)1522-2594(199902)41:2<276::aid-mrm10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 11.Lundbom N, Barnett A, Bonavita S, Patronas N, Rajapakse J, Tedeschi, Di Chiro G. MR image segmentation and tissue metabolite contrast in 1H spectroscopic imaging of normal and aging brain. Magn Reson Med. 1999;41:841–845. doi: 10.1002/(sici)1522-2594(199904)41:4<841::aid-mrm25>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 12.McLean MA, Woermann FG, Barker GJ, Duncan JS. Quantitative analysis of short echo time 1H-MRSI of cerebral gray and white matter. Magn Reson Med. 2000;44:401–411. doi: 10.1002/1522-2594(200009)44:3<401::aid-mrm10>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 13.Noworolski SM, Nelson SJ, Henry RG, Day MR, Wald LL, Star-Lack J, Vigneron DB. High spatial resolution 1H-MRSI and segmented MRI of cortical gray matter and subcortical white matter in three regions of the human brain. Magn Reson Med. 1999;41:21–29. doi: 10.1002/(sici)1522-2594(199901)41:1<21::aid-mrm5>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 14.Tanabe JL, Amend D, Schuff N, DiSclafani V, Ezekiel F, Norman D, Fein G, Weiner MW. Tissue segmentation of the brain in Alzheimer disease. Am J Neuroradiol. 1997;18:115–123. [PMC free article] [PubMed] [Google Scholar]
- 15.Soher BJ, Young K, Govindaraju V, Maudsley AA. Automated spectral analysis III: application to in vivo proton MR spectroscopy and spectroscopic imaging. Magn Reson Med. 1998;40:822–831. doi: 10.1002/mrm.1910400607. [DOI] [PubMed] [Google Scholar]
- 16.Mareci TH, Brooker HR. Essential considerations for spectral localization using indirect gradient encoding of spatial information. J Magn Reson. 1991;92:229–246. [Google Scholar]
- 17.Ross BD, Bluml S, Cowan R, Danielsen E, Farrow N, Gruetter R. In vivo magnetic resonance spectroscopy of human brain: the biophysical basis of dementia. Biophys Chem. 1997;68:161–172. doi: 10.1016/s0301-4622(97)00032-x. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Li SJ. Differentiation of metabolic concentrations between gray matter and white matter of human brain by in vivo 1H magnetic resonance spectroscopy. Magn Reson Med. 1998;39:28–33. doi: 10.1002/mrm.1910390107. [DOI] [PubMed] [Google Scholar]
- 19.Kreis R, Ernst T, Ross BD. Absolute quantitation of water and metabolites in the human brain. II. Metabolite concentrations. J Magn Reson Ser B. 1993;102:9–19. [Google Scholar]
- 20.Petroff OAC, Spencer DD, Jeffrey R, Alger JR, Prichard JW. High-filed proton magnetic resonance spectroscopy of human cerebrum obtained during surgery for epilepsy. Neurology. 1989;39:1197–1202. doi: 10.1212/wnl.39.9.1197. [DOI] [PubMed] [Google Scholar]
- 21.Soher BJ, van Zijl PC, Duyn JH, Barker PB. Quantitative proton MR spectroscopic imaging of the human brain. Magn Reson Med. 1996;35:356–363. doi: 10.1002/mrm.1910350313. [DOI] [PubMed] [Google Scholar]
- 22.Petroff OA, Pleban LA, Spencer DD. Symbiosis between in vivo and in vitro NMR spectroscopy: the creatine, N-acetylaspartate, glutamate, and GABA content of the epileptic human brain. Magn Reson Imaging. 1995;13:1197–1211. doi: 10.1016/0730-725x(95)02033-p. [DOI] [PubMed] [Google Scholar]
- 23.Pouwels PJ, Frahm J. Differential distribution of NAA and NAAG in human brain as determined by quantitative localized proton MRS. NMR Biomed. 1997;10:73–78. doi: 10.1002/(sici)1099-1492(199704)10:2<73::aid-nbm448>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 24.Sappey-Marinier D, Calabrese G, Hetherington HP, Fisher SN, Deicken R, Van Dyke C, Fein G, Weiner MW. Proton magnetic resonance spectroscopy of human brain: applications to normal white matter, chronic infarction, and MRI white matter signal hyperintensities. Magn Reson Med. 1992;26:313–327. doi: 10.1002/mrm.1910260211. [DOI] [PubMed] [Google Scholar]
- 25.Constans JM, Meyerhoff DJ, Gerson J, MacKay S, Norman D, Fein G, Weiner MW. H-1 MR spectroscopic imaging of white matter signal hyperintensities: Alzheimer disease and ischemic vascular dementia. Radiology. 1995;197:517–523. doi: 10.1148/radiology.197.2.7480705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leary SM, Brex PA, MacManus DG, Parker GJ, Barker GJ, Miller DH, Thompson AJ. A (1)H magnetic resonance spectroscopy study of aging in parietal white matter: implications for trials in multiple sclerosis. Magn Reson Imaging. 2000;18:455–459. doi: 10.1016/s0730-725x(00)00131-4. [DOI] [PubMed] [Google Scholar]
- 27.Saunders DE, Howe FA, van den Boogaart A, Griffiths JR, Brown MM. Aging of the adult human brain: in vivo quantitation of metabolite content with proton magnetic resonance spectroscopy. J Magn Reson Imaging. 1999;9:711–716. doi: 10.1002/(sici)1522-2586(199905)9:5<711::aid-jmri14>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 28.Charles HC, Lazeyras F, Krishnan KR, Boyko OB, Payne M, Moore D. Brain choline in depression: in vivo detection of potential pharmacodynamic effects of antidepressant therapy using hydrogen localized spectroscopy. Prog Neuropsychopharmacol Biol Psych. 1994;18:1121–1127. doi: 10.1016/0278-5846(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 29.Chang L, Ernst T, Poland RE, Jenden DJ. In vivo proton magnetic resonance spectroscopy of the normal aging human brain. Life Sci. 1996;58:2049–2056. doi: 10.1016/0024-3205(96)00197-x. [DOI] [PubMed] [Google Scholar]
- 30.Scheltens P, Barkhof F, Leys D, Wolters EC, Ravid R, Kamphorst W. Histopathologic correlates of white matter changes on MRI in Alzheimer's disease and normal aging. Neurology. 1995;45:883–888. doi: 10.1212/wnl.45.5.883. [DOI] [PubMed] [Google Scholar]


