Abstract
Complex formation between the intracellular domain of the Notch receptor (NICD) and the transcription factor CSL is indispensable for transcriptional activation. To understand how NICD displaces CSL-associated co-repressors, we have quantified the binding of different Notch1 ICD regions to a key interaction domain (the beta trefoil domain, or BTD) of human CSL. Electrophoresis, scattering, and titration calorimetry indicate that NICD and BTD combine to form a 1:1 heterodimer. Neither the Notch1 ankyrin domain (ANK) nor C-terminal region contributes binding energy towards BTD. In contrast, binding energy is attributed largely to a short segment including the conserved WFP sequence motif within the RAM region (the ~140 residue polypeptide segment N-terminal to the ANK domain); substitution of this motif substantially reduces affinity. Short (≤ 25 residues) WFP-containing peptides encoded by the four mammalian Notch genes have similar affinities to BTD; thus, activity differences between paralogues either result from other regions of NICD and CSL or from differences in interaction with downstream components. The importance of RAM was demonstrated by the ability of a short RAM peptides to dissociate NICD:CSL interaction in cellular lysates. These results support an emerging molecular mechanism for the displacement of co-repressors from DNA-bound CSL by NICD.
Keywords: CSL, Isothermal Titration Calorimetry, Notch Intracellular Domain, Notch Signaling, Transcriptional Activation
Introduction
The Notch signaling pathway mediates short-range signals involved in regulating a myriad of cellular processes.1,2 Notch signaling is unique among the major signal transduction pathways because it does not involve second messengers, and as a result, ligand mediated activation is not amplified. Activation of the Notch pathway is triggered by binding of a cell surface ligand from the DSL (Delta, Serrate and Lag2) family to specific EGF-like repeats in the extracellular domain of Notch receptors (Figure 1A). This binding is likely to induce a conformational change3–6 that exposes the juxtamembrane region to cleavage by ADAM metalloproteases.7,8 The exposed amino terminus is recognized by γ -secretase9, which cleaves Notch within its transmembrane domain leading to the dissociation of its intracellular domain (referred to as NICD) from the membrane. Liberated NICD is able to enter the nucleus, where it initiates a “transcriptional switch” by binding to a DNA-bound CSL repressor complex (CBF or RBPjκ in vertebrates, Su(H), Drosophila, Lag-1 in C. elegans) and converting it to a transcriptionally active complex (Figure 1B).10 The molecular mechanisms underlying this conversion, which involves the displacement of repressor proteins by NICD and recruitment of transcriptional activators to DNA-bound CSL, are not well understood. The biophysical and biochemical studies described here are aimed at dissecting a step central to the NICD-mediated transcriptional switch: the interaction between DNA binding protein CSL and NICD.
Figure 1.
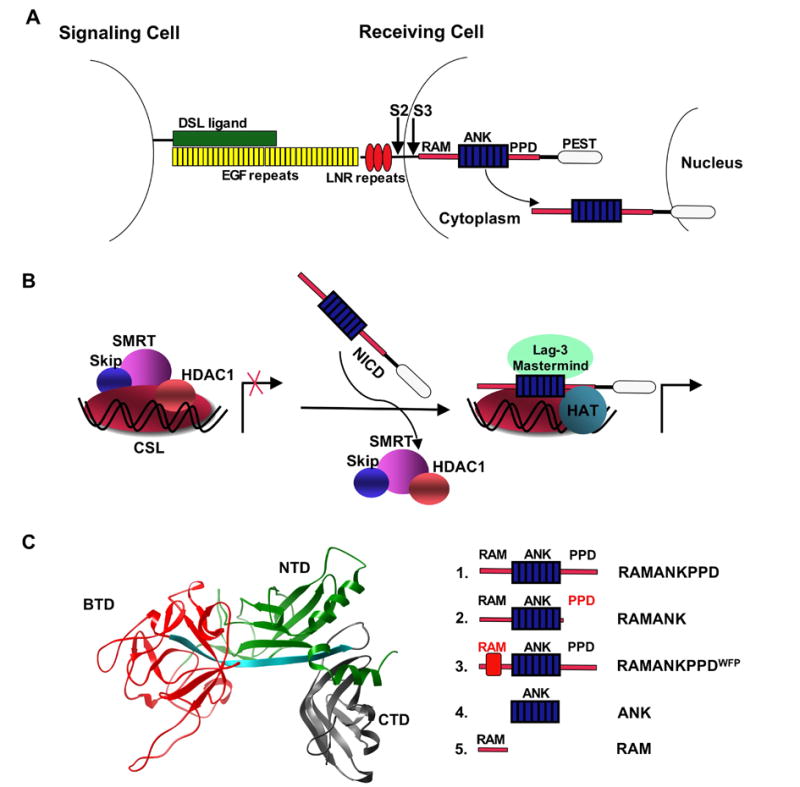
Interaction between CSL and NICD is crucial for both activation and repression of Notch target genes. (A) Activation of the Notch receptor is trigged by binding of a DSL ligand to the EGF repeats of Notch, followed by two consecutive proteolytic events (S2 and S3 arrows) that liberate NICD. (B) Cartoon representation of the Notch-mediated transcriptional switch. In the absence of NICD, CSL is found in complex with transcriptional repressor proteins. Binding of NICD to CSL allows for the displacement the repressors and recruitment of transcriptional activators. (C) Left: crystal structure of CSL homolog (Lag-1; PDB id 1TTU),21 showing the BTD domain of CSL used in this study (red). Right: schematic representation illustrating the Notch1 deletion series used in this study.
CSL proteins reside in the nucleus, where they are constitutively bound to upstream regulatory regions of Notch-regulated genes.11 In Drosophila, the CSL protein (Suppressor of Hairless) interacts with Hairless and CtBP repressor proteins.12 In vertebrates, the CSL protein (RBP-jκ ) interacts with the ubiquitous proteins SKIP and SMRT which recruit one of several abundant transcriptional co-repressor complexes.13,14 Successful signaling via the Notch receptors depends on the ability of low concentrations of NICD to displace the abundant co-repressor complexes.15,16 Once formed, the NICD:CSL complex recruits the Mastermind (MAM) protein10 and the histone acetylase (p300)17 to assemble a transcriptionally active complex (Figure 1B, reviewed by Lubman et al).18
Identification of the NICD binding sites on CSL was greatly aided by initial deletion mutagenesis studies19,20 and more recently by several crystal structures of DNA-bound CSL proteins (RBP-jκ and Lag-1) alone21 and in quaternary complex with Notch and MAM.22,23 All structures reveal that CSL is composed of three integrated domains: a N-terminal Rel homology domain (NTD), a central beta-trefoil domain (BTD) and a C-terminal Rel homology domain (CTD). The BTD domain makes extensive hydrophobic and hydrogen-bonding interactions with the N-terminal RAM (RBPjκ Associated Molecule24) region of NICD23 while the NTD and CTD make extensive electrostatic and van der Waals interactions with the more C-terminal ankyrin domain (ANK) of NICD.22,23 Co-immunoprecipitation studies suggest that although the ANK:CSL interaction involves a larger surface area than the RAM:CSL interaction, the interaction with ANK is weaker than that with RAM.25
Previous genetic and tissue culture experiments have identified an essential peptide sequence within the RAM region (WFP in mouse and human Notch 1) as being critical for the interaction of Notch with CSL. This motif is well-conserved not only in the four mammalian Notch paralogues, which have all been shown to interact with CSL,26,27 but also among Notch receptors from different species. In addition to the WFP motif, the 120 residue segment located between the transmembrane and ANK domains of the Notch protein has conserved regions that have also been implicated in the interaction with CSL. NICD fragments beginning at the C-terminal part of RAM can activate transcription28–30 and bind to CSL (RK and Eric Schroeter, unpublished observation); however, mutants lacking only the WFP motif fail to interact with CSL.15,31 The crystal structure of the Caehonrhabditis elegans CSL, Notch and Mastermind proteins on cognate DNA suggests that only 10 most membrane-proximal residues including WFP interact with the BTD domain of CSL. Both in the crystal structure and in isolation, RAM is largely unstructured (23,25 and A. Bertagna and D.B, in preparation).
In addition to the RAM and ANK regions, sequence elements C-terminal to the ANK domain (referred to as the PPD region) have recently been implicated in binding to CSL.31 This region, which has not been characterized structurally, spans 50 residues downstream of the 7th ankyrin repeat and contains a ten-residue Drosophila-specific sequence insertion. In vitro co-immunoprecipitation experiments together with in vivo experiments in Drosophila embryos suggest that this region constitutes yet another binding site for CSL, and its deletion reduces Notch activity.31
Genetic and tissue culture experiments, as well as the crystal structure of the quaternary complex formed by CSL, NICD, MAM and DNA, have significantly advanced our understanding of Notch mediated transcriptional activation.23,25,32 The binding of RAM induced long-range conformational changes in CSL that may facilitate dissociation of co-repressors,23 however, several important mechanistic questions remain unanswered: What is the binding affinity of NICD to CSL? Is the affinity of NICD to the BTD domain of CSL determined exclusively by RAM or do the other two domains in NICD (ANK, PPD) contribute to this interaction? Are the three CSL binding sites on NICD acting synergistically or independently of one another, perhaps facilitating an allosteric change in CSL? To begin addressing these questions, and to identify CSL:Notch binding interfaces that are suitable for future inhibitor design, we have undertaken biochemical and biophysical studies of BTD:NICD interaction. These studies, which utilize thermodynamic, hydrodynamic and biochemical methods, comprise the first quantitative analysis of this association. We correlate the thermodynamic results presented here with the complex formation between intact CSL (RBP-jκ ) and full length NICD derived from mammalian cells by demonstrating the effectiveness of minimal BTD-binding peptides in disrupting Notch:CSL interaction. Through defining and quantifying this critical component of Notch:CSL binding, we provide a framework for a model to explain how low concentrations of mammalian Notch can effectively displace abundant co-repressor complexes.
Results
This study focuses on binding of the BTD domain of CSL to different regions of NICD (Figure 1C). The longest NICD construct examined here, referred to as RAMANKPPD contains three previously described binding regions: the RAM region,24 the ANK domain, and the PPD region located C-terminal to the ANK domain. The construct referred to as RAMANK lacks the PPD site, whereas in the RAMANKPPDWFP construct these three critical residues are substituted with alanines. This triple-substitution was previously shown to abolish NICD:CSL interaction in co-immunoprecipitation experiments and transcriptional assays.24 We used two different CSL constructs (RBP-jκ residues 161–392, and residues 161–349) because these boundaries span the BTD domain,21 and were amenable to expression and purification at sufficiently high levels for biophysical studies. In addition to the BTD, both constructs contain a long β -strand that connects the NTD, BTD, and CTD domains; furthermore, the longer of the two constructs contains two β -strands from the CTD domain of human CSL. A recent crystal structure of CSL/RAMANK/MAM on DNA shows that these β strands are critical for the interaction between the CTD of CSL and the ANK domain of Notch.23 To ensure that both constructs are properly folded, we have used far-UV CD spectroscopy to monitor secondary structure elements. The far-UV spectra of Notch and the CSL polypeptides derived from RBP-jκ are consistent with the presence of α -helical (Notch) and β -strand (CSL) structures, as expected (data not shown).
The BTD domain of CSL and the Intracellular Domain of Notch form a Heterodimeric Complex in Solution
We first used native gel electrophoresis to ask if the purified proteins form stable complexes in solution. Mixing of the BTD domain of CSL with RAMANKPPD gave rise to a third species of higher mobility in a concentration dependent manner (Figure 2A). The amount of the complex formed during titration is limited by the amount of the binding partner that is held constant. The complex also formed between the BTD and RAMANK, indicating that the PPD site is not necessary for complex formation (Figure 2B). Although the WFP to AAA substitution still permits the formation of an apparent higher molecular weight complex, the stability of this complex appears to be substantially reduced, as the unbound static component (BTD) was not significantly depleted by the excess of RAMANKPPDWFP (Figure 2C). Consistent with this interpretation, the high mobility species formed by RAMANKPPDWFP were smeared and diffuse, suggesting dissociation of the complex during electrophoresis.33 In contrast, neither BTD construct forms a complex with the isolated ANK domain (Figure 2D), as expected from the biochemical and crystallographic studies.22,23,25 Although the BTD161–392 construct contains two orphaned β -strands from the CTD domain of CSL that have been implicated in ANK binding, these structural elements are not likely to fold or support binding in the absence of the entire CTD domain. Overall, these native electrophoresis experiments indicate that the RAM region of NICD is critical for complex formation with the BTD domain of CSL in solution, and that the ANK and PPD regions of NICD are less critical for formation of this complex with BTD (if they participate at all).
Figure 2.
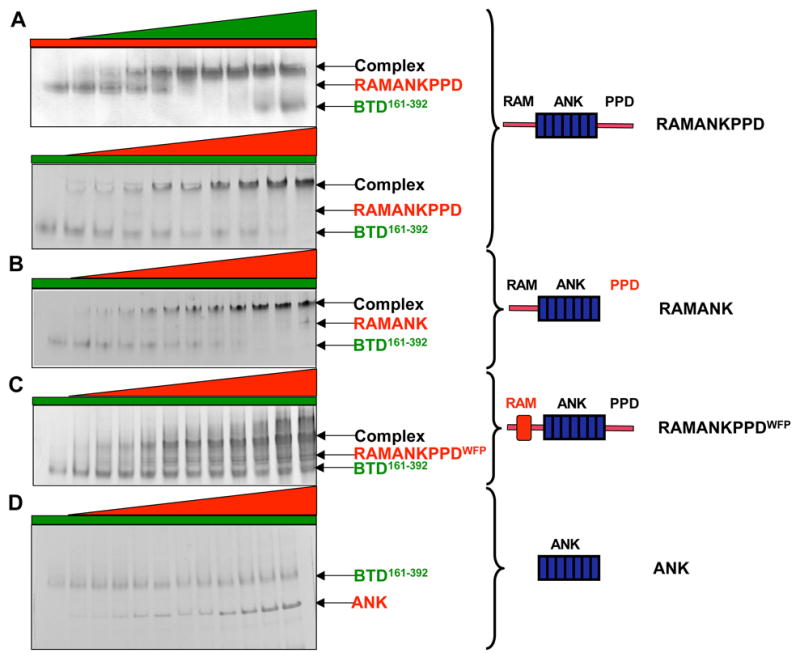
Monitoring complex formation using native gel electrophoresis. Increasing concentrations of different length Notch1 polypeptides (0, 2 μ M, 3μ M, 5μ M, 7μ M, 9μ M, 10μ M, 12μ M, 15 μ M and 20μ M) were titrated into 10μ M BTD161–392. (A) Upper panel: 10μ M of RAMANKPPD was titrated with increasing concentrations of BTD161–392; lower panel; 10μ M of BTD161–392 was titrated with increasing concentrations of RAMANKPPD. (B) 10μ M of BTD161–392 was titrated with increasing concentrations of RAMANK. (C) 10μ M of BTD161–392 was titrated with increasing concentrations of RAMANKPPDWFP. (D) 10μ M of BTD161–392 was titrated with increasing concentrations of ANK.
The BTD domain of CSL binds to Notch with 1:1 stoichiometry
To determine the stoichiometry of NICD:BTD interaction, we have used chemical cross-linking and multi-angle light scattering (MASLS). The advantage of chemical cross-linking over native gel electrophoresis is that molecular weight can be determined from mobility on SDS PAGE. Using the BS3 cross-linker, we detected a ~ 72kD band that was only observed when both CSL and RAMANKPPD or RAMANK were included (Figure 3A). This apparent size is very close to the sum of the molecular weights of RAMANK and BTD 161–349 (44 and 24kD, respectively), suggesting a 1:1 stoichiometry in solution.
Figure 3.
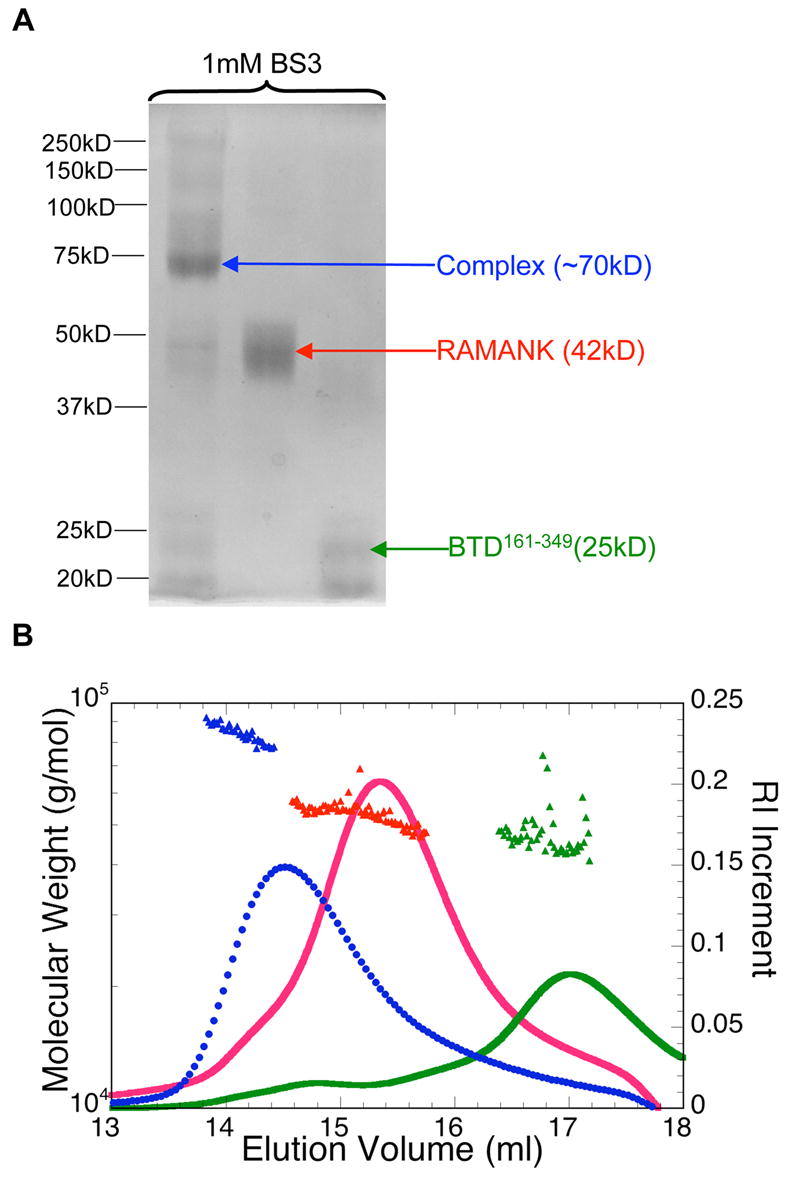
Notch1 RAMANK and BTD161–349 form 1:1 heterodimer. (A) Migration profiles on 12% SDS PAGE of BTD161–349, RAMANK and 1:1 complex (10μ M protein) in the presence of 1 mM chemical cross-linker BS3. (B) Association in solution of BTD161–349 and RAMANK detected by multi-angle static light scattering was resolved either separately (green and red) or together (blue) on a size exclusion column, and the total protein concentration in the elution was monitored by differential refractometry (right axis). This signal was combined with signals from multiple scattering detectors to determine weight-averaged molecular weights (filled triangles, left axis).
We also examined the molecular weight of RAMANK, BTD161–349, and a 1:1 mixture of the two proteins using size exclusion chromatography coupled with MASLS (Figure 3B). This technique provides the weight-averaged molecular weight at each point of the chromatographic elution profile 34. At 1:1 molar ratio, RAMANK and BTD elute together in a single peak with the molecular weight of 75 ± 1.6kD (Figure 3B). Interestingly, MASLS detected no association between RAMANKPPDWFP and BTD. This is consistent with a decreased affinity suggested from native gel electrophoresis; the lack of detection of any dimeric species by MASLS suggests a more complete dissociation on the gel filtration matrix than in a cross-linked gel cage. Likewise, a mixture of the ANK domain with BTD elutes as two separate peaks at monomer molecular weight, with no evidence of complex formation (data not shown). The failure to form complexes of higher molecular weight when either component is in large stoichiometric excess indicates that the RAMANK:BTD complex is limited to a 1:1 stoichiometry.
The RAM domain is the sole binding determinant of the BTD:Notch interaction
To quantitatively dissect the contribution of the different regions of NICD towards binding to the BTD, we examined the binding of RAMANKPPD, RAMANK, RAMANKPPDWFP, and a collection of peptides of varying lengths centered on the WFP motif of the RAM region, to the BTD domain of CSL using isothermal titration calorimetry (ITC). ITC directly measures the heat of association of a titrant (NICD fragments) with its binding partner (BTD), providing a direct determination of the association constant (Ka), the stoichiometry (n), and the enthalpy of binding (ΔH°b). The free energy and entropy of binding are then calculated from Ka and ΔHb using well-known relations:
Figure 4A shows baseline-corrected heat peaks resulting from substoichiometric additions of RAM to the BTD domain. Figures 4B and 4C display representative integrated ITC heats of titration of BTD161–349 with RAMANKPPD, RAMANK, RAM and RAM-derived peptides. RAMANKPPD binds to BTD with a relatively high affinity (Kd=200nM) and binding enthalpy (−9.3 kcal/mole). RAMANK binds to BTD with an affinity nearly identical to that of RAMANKPPD, suggesting that the PPD region does not contribute to the stability of NICD:BTD interaction (Table 1). The full length RAM polypeptide (120 residues) binds to the BTD domain of CSL with the same affinity as RAMANK, indicating that the RAM region is the only element within NICD contributing to the affinity of interaction with the BTD domain. Moreover, the similar affinities of RAMANK and the RAM polypeptides suggest that the interaction of the RAM domain on the isolated BTD domain is not affected by covalent linkage to the ANK domain.
Figure 4.
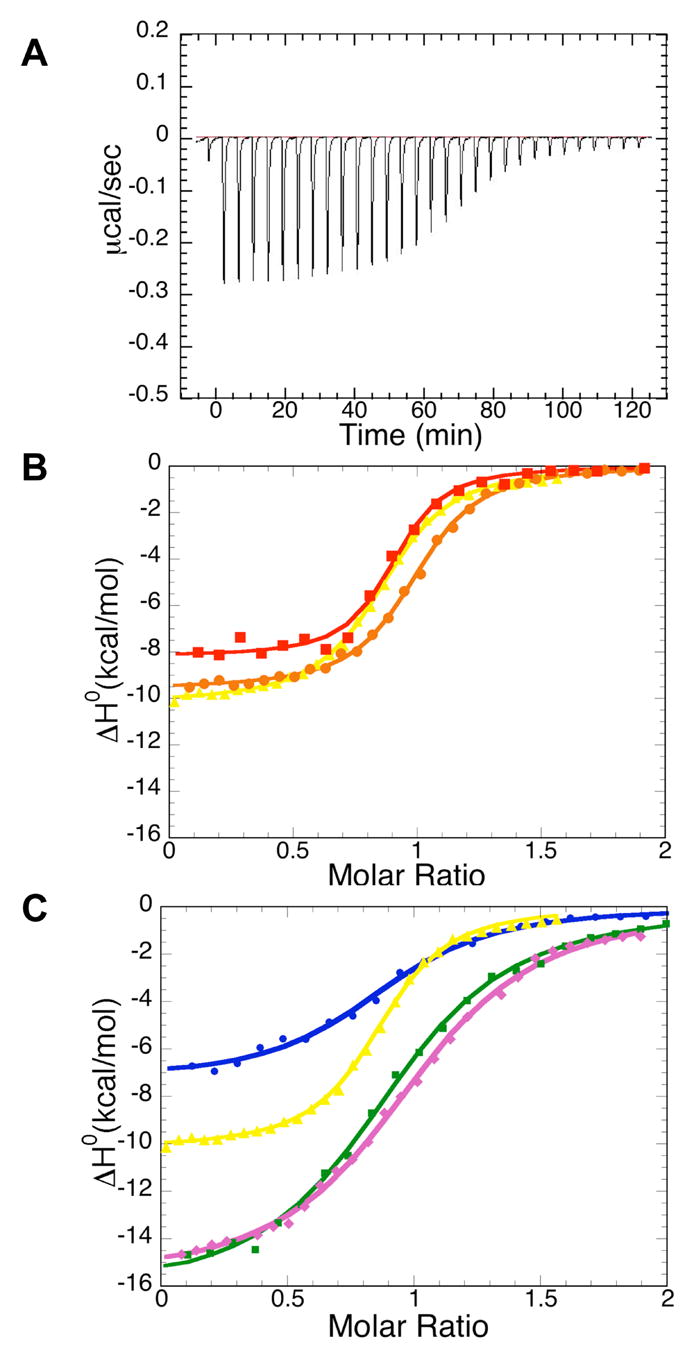
Isothermal calorimetric titrations for binding of the BTD161–349 domain to Notch1 polypeptides. (A) Raw heat signal of an isothermal titration of BTD161–349 with the RAM domain of Notch. The power output is shown as a function of time (seconds). 10μ l increments of 0.12mM RAM were injected into a 1.35 ml cell containing 10μ M BTD161–349. (B) Integrated data for titrations of RAMANKPPD (red filled squares), RAMANK (brown filled circles) and RAM (yellow filled triangles) with the BTD161–349. (C) Integrated data for titrations of RAM (yellow filled triangles), N1RAM13 peptide (purple filled diamonds), N1RAM16 (blue filled circles) and N1RAM22 (green filled squires) with BTD161–349. Solid lines represent nonlinear least-squares fits to the data. All titrations shown here were performed at 30 ° C in 20mM HEPES pH7.5, 100mM NaCl, 1mM EDTA and 2mM BME.
Table 1.
Calorimetric data for binding of Notch polypeptides to the BTD domain of CSL
| BTD domain Of | Ligand | K (M−1) | Kd (μM) | ΔG° (kcal/mol) | ΔH° (kcal/mol) | TΔS° (kcal/mol) |
|---|---|---|---|---|---|---|
| BTD 161–349 | RAMANKPPD | 4.3( ± 2.5) x106 | 0.23 | −9.2 ± 0.4 | −9.0 ± 0.6 | 1.2 ± 0.5 |
| BTD161–349 | RAMANK | 4.9( ± 0.5) x 106 | 0.20 | −9.2 ± 0.2 | −9.4 ± 0.4 | −0.2 ± 0.6 |
| BTD161–349 | RAM | 6.48( ± 2.3) x106 | 0.15 | −9.4 ± 0.25 | −10.9 ± 0.8 | −1.5 ± 0.6 |
| BTD161–349 | N1RAM13 | 8.8( ± 1.4) x105 | 1.0 | −8.2 ± 0.1 | −16 ± 0.5 | −7.8 ± 0.6 |
| BTD161–349 | N1RAM16 | 9.9( ± 0.45) x105 | 1.1 | −8.2 ± 0.2 | −8.4 ± 1.0 | 0.2 ± 0.7 |
| BTD161–349 | N1RAM22 | 1.5( ± 0.3) x106 | 0.8 | −8.6 ± 0.1 | −14.6 ± 0.8 | −6 ± 0.8 |
| BTD161–349 | N2RAM23 | 2.0( ± 0.4) x 106 | 0.5 | −8.7 ± 0.1 | −9.4 ± 0.7 | −0.7 ± 0.6 |
| BTD161–349 | N3RAM25 | 6.3( ± 0.1) x105 | 1.6 | −8.0 ± 0.1 | −12.4 ± 1.0 | −4.4 ± 0.7 |
| BTD161–349 | N4RAM25 | 1.4 ( ± 0.7) X106 | 0.7 | −8.5 ± 0.5 | −11.2 ± 1.0 | −2.7 ± 0.8 |
Reported values are the mean of at least three independent titrations. Uncertainties in K, ΔG° and ΔH° represent the standard deviations of multiple experiments. Uncertainty in TΔS° was calculated by standard error propagation.
To further dissect the boundaries of the BTD binding motif within the RAM region, we synthesized a series of shorter peptides centered on the Notch1 WFP motif and examined their ability to bind BTD (Figure 4C and Table 1). The longest peptide (N1RAM22, 22 residues; Figure 5A) has a modestly decreased affinity towards BTD (Δ ΔG°=0.6 kcal/mol, relative to the 120 residue RAM construct). A peptide with a C-terminal truncation (N1RAM13, 13 residues; Figure 5A) produces only a slight (0.4 kcal/mol) additional decrease in binding free energy compared to N1RAM22 (Table 1). N-terminal extension by three residues (N1RAM16, Figure 5A) has no effect on binding free energy compared to this minimal 13 residue peptide (Table 1). As expected, an inverted, 22 residue peptide control (InvN1RAM22) does not associate with BTD in the ITC experiment. ITC also confirmed that the interaction between BTD and the RAMANKPPDWFP protein is very weak, since the heat peaks observed for that titration did not deviate significantly from the baseline (data not shown). Combined, these observations confirm that 13 membrane-proximal residues that span the WFP motif of RAM are critical for the interaction with BTD domain of CSL.
Figure 5.
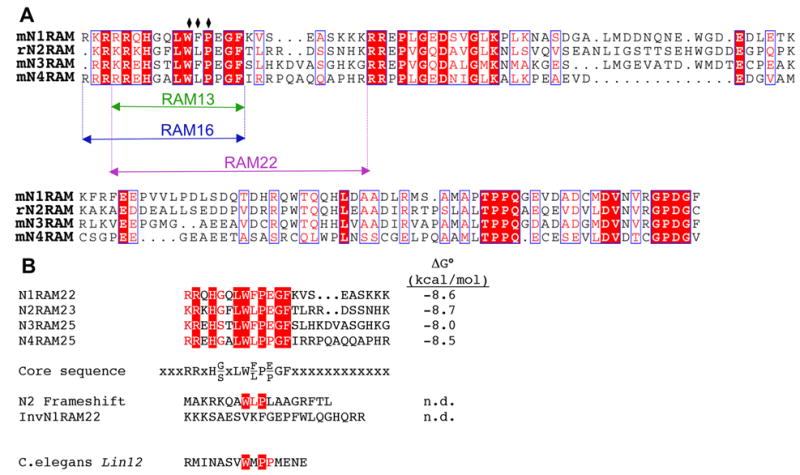
Sequences of RAM regions from Notch genes. (A) Alignment of the four mouse Notch paralogues from the juxtamembrane region to the segment flanking the first ankyrin repeat. Identities are outlined in red boxes; similarities are outlined in white boxes. N- and C-terminal boundaries of the 13,16 and 22 residue Notch1 peptides used in this study are colored green, blue and magenta, respectively. (B) Peptides used to compare affinities of the four mammalian Notch WFP segments with BTD. Binding free energies (from ITC) are given to the right. The consensus sequence is defined by sequence similarity among the four Notch peptides, which all bind with similar affinity; as such, not all residues in this consensus necessarily contribute to binding free energy. Positions marked with x’s show no conservation; although these sequences do not contribute to binding free energy, variation at these positions appears to contribute to variations in enthalpy and entropy. Neither the frameshifted Notch2 sequence nor the inverse N1RAM22 show any detectable binding in the ITC.
Although the free energies of binding of the 120 residue RAM polypeptide to BTD are similar to those of the short peptides, enthalpies of binding differ substantially, with N1RAM13 and N1RAM22 producing a larger heat release upon binding (Δ ΔH°=−7 kcal/mol). In contrast, N1RAM16 has a binding enthalpy similar to the 120 residue RAM polypeptide (Table 1, Figure 6), suggesting that the three N-terminal residues included in N1RAM16 contribute to both binding entropy and enthalpy, with little net contribution to free energy. Consistent with cross-linking and MASLS experiments described earlier, all the NICD constructs that interact with the BTD domain show a 1:1 stoichiometry in the ITC.
Figure 6.
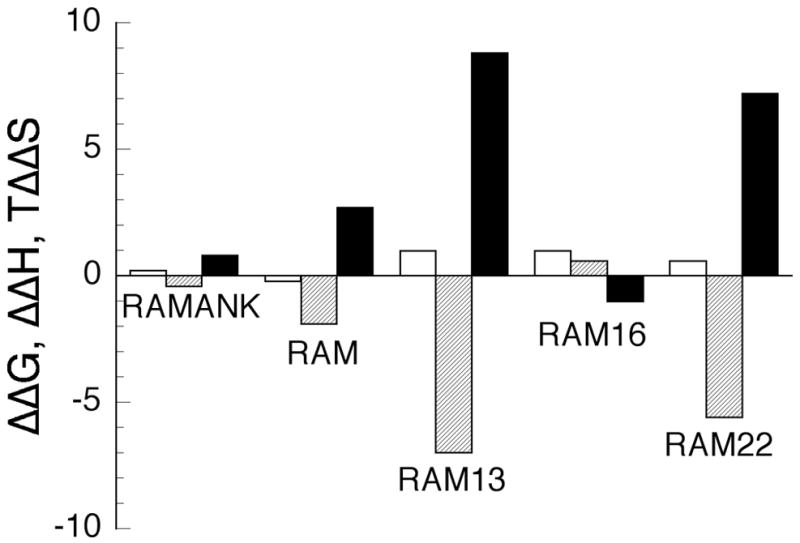
Binding energetics of Notch1 polypeptide variants. Shown are the differences in the ΔG ° (open), ΔH ° (cross-hatched) and TΔS ° (solid) between RAMANKPPD and the shorter Notch polypeptides. Values for Δ ΔG ° , Δ ΔH ° and TΔ ΔS ° are given at 30 ° C.
Binding energies of RAM peptides from the four Notch paralogues
Unlike CSL, mammals have four different Notch genes, which when tested in parallel within the same cell-based transcription assays display different activities.35–37 As the RAM region of NICD has previously been shown to be critical for interaction with CSL,24,25,38 and our studies demonstrate the interaction of Notch1 with the BTD domain to be restricted to a short segment surrounding the WFP motif of RAM, divergence within the RAM region seems likely to account for the differences among the four Notch paralogues (Figure 5B).
To test whether differences in activities among the four Notch genes reflect differences in the affinities of respective RAM regions with BTD, we compared the binding affinities of peptides containing the WFP regions of the four mouse Notch receptors. Peptides were synthesized to be analogous to N1RAM22, which showed the highest affinity of the three Notch1 RAM peptides we examined (Table 1). Because there are insertions of 1–3 residues C-terminal to the WFP region of Notch2-Notch4, these peptides were slightly longer than N1RAM22 (Figure 5B). Although the Notch3 peptide interaction with BTD may be slightly weaker than for the other three peptides (see N3RAM25, Table 1), correlating with its poor activation potential, binding free energies of the four peptides are remarkably similar to one another.
The finding that these four peptides bind with such similar affinities indicates that any specificities arising from differences in NICD:CSL interaction likely involves other regions of Notch. One likely candidate region for such affinity differences is the ANK domain, which shows substantial surface sequence variation39, and may result in differences in affinity toward the NTD and CTD domains of CSL. Consistent with this, Beatus et al. have found that differences in transcriptional activation between Notch1 and Notch3 are determined by the ANK domains.40 Moreover, the similarities of the binding affinities of these four Notch peptides further define the consensus sequence determinants for BTD binding surrounding the WFP motif (Figure 5B). Although structural analysis of the C.elegans lin12 RAM region bound to the BTD domain of lag1 might be expected to further refine this binding sequence, the sequence similarity of the mammalian paralogues to the lin12 RAM sequence is limited, making this comparison tenuous. Finally, although the sequence differences among the four Notch RAM peptides do not affect free energies of binding, the binding enthalpies and entropies vary significantly (see discussion).
The lack of detectable binding by ITC of RAMANKPPDWFP confirms the importance of the WFP sequence for the RAM:BTD interaction. As all four of the Notch paralogues contain close approximations to this motif (WLP in Notch2 and Notch4), it might be expected that this short tripeptide sequence would sufficient for the ~8 kcal/mol binding free energy of these peptides and longer constructs. This is not, however, the case, as demonstrated by the lack of interaction of BTD with a variant of the mouse Notch2 RAM peptide that results from what was likely a sequencing error (N2 frameshift, Figure 5B).1 Like the inverted Notch1 sequence, this frameshifted sequence shows no heat peaks upon injection into BTD in the ITC, although it contains a WLP tripeptide sequence. This observation confirms that sequences outside of the W(F/L)P sequence (for Notch2, four and three residues in the N- and C-terminal direction, respectively) also contribute significantly to binding.
Notch1 RAM peptides inhibit NICD1:CSL interaction in cellular extracts
The experiments above that map the NICD:BTD interactions to a short segment of RAM were performed with a bacterially produced BTD domain, not the entire CSL protein. To demonstrate the relevance of this interaction in the context of the full-length CSL protein and in a more complex cellular environment, we asked if the Notch1 RAM peptides could compete with NICD1 for binding to full length CSL in cellular extracts. To evaluate binding, we used an immunodepletion strategy. Human HEK293 Cells were transfected either with a 6MT-NICD1 (overexpressing a Myc-tagged version of the entire mouse Notch-1 intracellular domain) or a 3xFlag-RBP-jκ (expressing the human CSL orthologue) expression vector (see methods); cell lysates containing each of the two constructs were mixed and were immediately challenged with increasing concentrations of RAM peptide. We then asked whether the Notch1 RAM peptides could block association between NICD1 and RBP-jκ by measuring the amounts of NICD1 remaining in the supernatant after clearing the mixture by FLAG immunoprecipitation.
Myc immunoreactivity in the supernatant increased with increasing concentration of Notch1 RAM peptide, but not control peptide (Fig. 7A). Consistent with the ITC-derived binding affinity, the effectiveness N1RAM22 in disrupting the NICD1:RPBjκ interaction was modestly but reproducibly greater than that of N1RAM13 at low concentration (Fig 7A). When FLAG antibodies are used on extracts containing NICD1 lacking the RAM domain (6MT-NICD1 RAM), NICD was not depleted (Fig. 7, lane 14). We also examined whether Notch1 RAM peptides were effective in converting pre-assembled NICD:CSL complexes to dissociated polypeptides by adding RAM peptides to lysates from cells cotransfected with NICD1 and RBP expression vectors. These RAM peptides are also effective at displacing NICD1 from preformed complexes with RBP (Figure 7B) as when added during assembly. This finding suggests that RAM-derived peptide fragments may be able to actively reverse Notch signaling.
Figure 7.
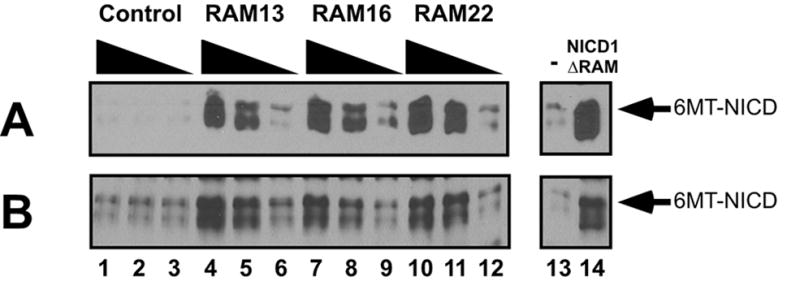
Notch1 RAM peptides inhibit NICD:CSL association in cellular extracts. The effects of different RAM peptides on 6MT-NICD1:3XFLAG-RBPjκ (the mammalian CSL orthologues) interactions were assessed by immunodepletion. Shown are the amounts of 6MT-NICD1 remaining in the supernatants (analyzed by 9E10/MYC Western blots) after FLAG immunoprecipitation. (A) Lysates from cells expressing 6MT-NICD1 were combined with lysates from cells expressing 3XFlag-RBPjκ , and were immediately exposed to increasing concentrations (0.2, 1, 5 μ M) of different Notch1 RAM peptides (Lanes 1–12). (B) Lysates from cells co-expressing 6MT-NICD1 and 3XFlag-RBPjκ were pooled and similarly treated with different RAM peptides. As controls, immunodepletion assays were also performed without peptide (Lane 13–14) and with 6MT-NICD RAM expression vector (Lane 14).
Discussion
The ability of Notch to switch CSL from transcriptional repression to activation is a central control point in the Notch signaling pathway.19 Molecular, genetic, cell-biological, biochemical and structural studies observed that this switch involves multiple binding sites both on NICD and CSL. The thermodynamic and hydrodynamic approaches used here lay out a quantitative foundation for dissecting this important switch. It is well-established that the integrity of the NICD:CSL complex relies on the presence of the WFP residues of the RAM domain.23–25,38 Here we have quantified the binding energy of NICD to a single domain in CSL (BTD), and dissected NICD to determine the relative contributions of different regions within NICD to this interaction.
Our findings have implications for several aspects of the transcriptional switch mechanism. First, the submicromolar binding affinity between BTD and the RAM region of the receptor may help to make NICD a potent antagonist of the transcriptional co-repressors, facilitating the transcriptional switch. Although the free energy of interaction is moderate compared to other protein-protein interactions that have been determined among transcription factors (Table S1), it is in a similar range to that of the NF-κ B p50/p50 homodimer, another eukaryotic transcription factor Although the identical affinity of RAM binding to BTD compared with longer constructs that include the ANK domain demonstrates that the ANK domain does not contribute to the binding affinity toward BTD, it is likely that affinity of NICD with full length CSL is increased by interaction between the ANK domain and the NTD and CTD domains of CSL; this interaction was recently demonstrated crystallographically.22,23 Second, these studies indicate that the PPD site does not contribute to the binding affinity to BTD; like ANK, PPD may interact with a region of CSL located outside of the BTD. Third, the similar affinities of all four mammalian Notch WFP peptides indicates that differences in transcriptional activation among the Notch paralogues resides outside this region. Again, it is possible that these differences result from differences in the ANK domains, either in their interactions with CSL or with a nearby factor in the activation complex. Alternatively, differences in activation may result from differences in intramolecular interactions involving the WFP motifs and a distal region of NICD. Fourth, this affinity range predicts that at the endogenous (low) nuclear NICD concentrations associated with Notch signaling, NICD will partition among all putative partners according to their relative affinity, rather than saturating all available partners. For NICD to have a significant impact on p50 localization for example41, it will have to have a higher affinity to p50 than 0.2μ M.
Energetics of the NICD:BTD interaction
The quantitative ITC results presented here demonstrate that the determinants of NICD:BTD binding energy reside entirely within the RAM region. Moreover, we find that short WFP-containing RAM peptides which overlap the 20 residue region of RAM defined crystallographically in an NICD:CSL complex,23 bind BTD with similar (albeit slightly lower) affinity to the full-length RAM polypeptide.
In addition to providing free energies of binding, the ITC measurements allow dissection of binding energy into enthalpic and entropic components (Figure 6). While both RAMANKPPD and RAMANK bind BTD with relatively modest enthalpy changes, the three RAM-derived peptides vary greatly in their enthalpies and entropies of binding. Both the 13 and 22 residue peptides, which share a common N-terminus (see Figure 5A) bind BTD with large favorable enthalpies and compensating unfavorable entropies (Figure 6). In contrast, 16 residue peptide, which shares a common C-terminus with N1RAM13 but contains three additional N-terminal residues, binds BTD with an enthalpy and entropy much closer to that seen for RAM and the longer NICD constructs (Figure 6).
Although a number of molecular interactions can contribute to these enthalpy and entropy differences, it is clear that the N-terminal extension in N1RAM16 (and in RAM), which consists of three positively charged residues, play a direct role in this enthalpy/entropy difference. One of these residues, Arg1750 has been shown to interact with negatively charged patch of the BTD domain in the crystal structure of Notch/CSL/MAM.23 It is possible that partial desolvation of these basic residues accompanies binding, and that this desolvation is electrostatically unfavorable but is compensated by increased entropy in the released waters. As is often observed with molecular interactions, these entropy and enthalpy changes are equal and offsetting, leaving the free energy of binding unchanged. Likewise, the substantial differences in enthalpies and entropies of binding of N1RAM22 to BTD compared with the three analogous peptides from Notch2-Notch4 are offsetting, resulting in similar free energies of binding of all these peptides. These results suggest that that the affinity of RAM to BTD is determined by a core binding sequence including the WFP motif (Figure 5B), and that distal sequence elements contribute in modest, subtle ways.
Mechanistic implications
The ability of the isolated RAM region to enhance the transcriptional activity of the otherwise inactive RBP-jκ (the human CSL orthologue) constructs highlighted the importance of this region in canonical Notch signaling.42 Recently, two landmark papers describe the Notch/CSL/MAM/DNA complex at the atomic level.22,23 Comparing the crystals suggests that the RAM:BTD interaction may trigger long-range allosteric changes in CSL.22,23,32
Our biophysical studies support this model by demonstrating that the RAM domain contributes substantially to the binding energy of the Notch:CSL interaction. Our biochemical experiments demonstrate that the RAM domain alone convert a pre-formed cellular Notch:CSL complex to dissociated subunits, either through a concerted mechanism or by shifting a dynamic equilibrium. It is possible that RAM binding to the BTD domain is an early step in the transcriptional switch from repression to activation; this is especially likely if the RAM-binding region of CSL is not directly involved in co-repressor association.14 Early RAM:BTD interaction may then facilitate allosteric changes that lower the binding affinity of bound co-repressor proteins to CSL, facilitating their dissociation, permitting ANK domain association with CSL and subsequent recruitment of MAM.25
Implications for targeted therapeutics against aberrant Notch signaling
Perturbations in Notch signaling are often associated with disease.2 Notch was first implicated in human disease by its involvement in T-cell acute lymphoblastic leukemia (T-ALL).43 Recently it was demonstrated that 50% of T-ALL patients acquire sporadic mutations in the Notch1 locus.5 Several other tumors are thought to involve Notch signaling.44 Targeting aberrant Notch signaling is currently approached through inhibition of Notch proteolysis; however, targeting of Notch interactions with effector proteins is also being considered: a peptide encompassing amino acids 13–74 of the transcriptional activator MAM was sufficient to inhibit the growth of Notch-induced leukemia cell lines.45 Perhaps an approach more amenable to small molecule design would be to directly target the critical RAM:BTD binding interface. Our results indicate that 13 residues surrounding the WFP motif retain most of the binding free energy for RAMANKPPD:BTD interaction and successfully converts a preformed Notch:CSL complex isolated from human cells to dissociated polypeptides, making this peptide a promising starting point for small-molecule inhibitors that directly interfere with protein-protein interactions.
Materials and Methods
Subcloning
A fragment of mouse Notch-1 protein encoding RAM domain, seven ankyrin repeats and 80 residues C-terminal to ankyrin repeats (RAMANKPPD; residues 1749 through 2194) was amplified using polymerase chain reaction (PCR). The PCR product was ligated into the T7 expression vector pProEX HTc (Invitrogen; Carlsbad, CA) in-frame with an N-terminal His-tag and a TEV protease cleavage site. The same approach was used to clone ANK domain of Notch, encompassing residues 1872 through 2105. The N-terminal boundary of the ANK construct does not include the capping helix recently described in the crystal structure of Notch/CSL/MAM complex on DNA.23The remaining Notch polypeptides used in this study were made by introducing a stop codon at various places in the RAMANKPPD construct through QuickChange Mutagenesis Kit (Invitrogen). The RAMANK polypeptide spans residues 1749 through 2114 of Notch-1, whereas the RAM polypeptide spans residues 1749 through 1868. To generate the 6MT-NICD1 expression construct, full length mouse Notch 1 cDNA was amplified by PCR and subcloned into the pCS2+6MT vector.
A fragment of isoform 1 of human isoform1 RBP-jκ, the human CSL orthologue, encoding BTD domain and part of CTD domain (residues 161-392; BTD161–392) was amplified using PCR from a plasmid containing cloned full length human RBP-jκ cDNA (a gift from Alan Israel). The resulting PCR product was cloned into pPro EX HTc as described for Notch constructs. Midway through this study, we made a BTD construct lacking additional β -strands from the CTD domain (BTD161–349) by introducing stop codon at residue 350. The full length RBPjκ was subcloned into the p3XFLAG-CMV7 vector (Sigma) to generate the 3XFLAG-RBP-jκ expression construct.
Expression and Purification of Notch and RBPjκ polypeptides
E. coli strain BL21*(DE3) was transformed with either Notch or RBP-jκ constructs and was grown at 37°C in the presence of 0.1mg/ml of ampicillin to an OD of 0.7–0.8. Protein expression was induced by adding IPTG to 1 mM, and cells were grown for 4 hours at 37°C. Cells were collected by centrifugation and were frozen at –80 ° C for at least an hour. Cell pellets were re-suspended in 50ml of lysis buffer containing 50mM Tris pH 8.0, 300mM NaCl, 10% Glycerol, 2mM β -mercaptoethanol (BME), and an EDTA-free protease inhibitor tablet (Roche Biosciences). Cells were lysed using a French press. Lysates were cleared by centrifugation at 15,000 rpm in a JA-20 rotor. The soluble portion of the cleared lysate was loaded onto -NTA column (Qiagen; Valencia, CA) equilibrated with lysis buffer and eluted with 300mM of imidazole. Fractions were pooled and dialyzed against buffer containing 25mM Tris pH 8.0, 100mM NaCl and 2mM BME. After dialysis, material was then loaded onto a Hi-Trap Q column (Amersham Biosciences AB) and eluted with a linear gradient of 100mM-1M NaCl. These purification steps yielded purities >95%, as determined using SDS-PAGE. Occasionally, gel filtration chromatography was performed as a final purification step. Yields for the purified polypeptides, per liter of bacterial culture, were as follows: RAMANKPPD=~6mg/L; RAMANK=~10mg/L; RAMANKPPDWFP=~5mg/L; RAM=~2mg/L; RBPjκ 161–392=~1.5mg/L; RBPjκ 161–349=~2.5mg/L. Proteins were concentrated and flash frozen for future experiments.
Isothermal Titration Calorimetry
Titrations were carried out using a VP-ITC micro-calorimeter from Microcal (Northhampton, MA). Notch polypeptides, synthetic peptides, and BTD polypeptides were dialyzed against 20mM HEPES pH 7.5, 100mM NaCl, 1mM EDTA and 2mM BME for at least 48 hours prior to experiments. For all titrations, BTD polypeptides were loaded in the calorimetry cell (10–25 μ M) and were titrated with Notch polypeptides (0.1–0.2 mM syringe concentration) at 30°C. The concentration of the Notch and BTD polypeptides was determined spectroscopically using their calculated extinction coefficients which are: ε 280(RAMANKPPD)=34,280 M−1 cm−1, ε 280(RAMANK)=33,120M−1cm−1,ε 280(RAMANKPPDWFP)=28,590M−1cm−1,ε 280(ANK)=15,930 M−1 cm−1, ε 280(RAM)=22,190 M−1 cm−1, ε 280(RBPjκ 161–392)=31,150 M−1 cm−1 ε280(RBPjκ 161–349)=25,460 M−1 cm−1. Concentration of the all of the peptides used in this study was determined using extinction coefficient of Trp ε 280(Trp)=5600 M−1 cm−1. Data were analyzed using ORIGIN software 46. All titrations could be fitted to a 1:1 binding model with a stoichiometry of binding (n) being between 0.8 to 1. Calorimetric parameters are reported as a mean value of at least three independent experiments with reported uncertainty being the standard deviation of multiple experiments. The c-value, defined to be the product of protein concentration and association constant, was between 5 and 200 for all experiments.
Cross-linking Experiments
Cross-linking of RAMANK to the BTD domain of RBPjκ was performed in using the bi-functional cross-linker bis(sulphosuccinimidyl)suberate (BS3; Pierce Biotechnology, Rockford IL). Reactions included 20mM HEPES pH 7.5, 150mM NaCl and 2mM BME. Initial experiments were conducted with range of protein (from 5 to 25μ M) and cross-linker concentrations (from 0.5 to 5mM). These experiments provided an optimal range of concentrations for both BTD and RAMANK and minimized non-specific cross-linking. The final concentration of both proteins in Figure 3 is 10μ M, final cross-linker concentration was 1.0mM. Reactions were incubated at 25°C for 30 min, and were terminated by the addition of SDS sample buffer. The cross-linked proteins were analyzed by 12 % SDS-PAGE under reducing conditions and visualized using by Coomassie staining.
MASLS
Static light scattering measurements were performed at 25°C using a three-angle light scattering detector (Wyatt MiniDAWN; Wyatt Technologies, Santa Barbara, CA) and a differential refractive index detector (Wyatt Optilab DSP; Wyatt Technologies) running in-line with an HPLC system. Samples were dialyzed overnight at 4°C against running buffer (150 mM NaCl, 2mM BME, either 20 mM Tris at pH 8.0 or 20 mM HEPES at pH 7.5 and 0.01% NaN3). 50 μ l samples were injected onto a G3000PWXL (TosoHaas, Montgomeryville, PA) size exclusion column equilibrated with running buffer at loading concentrations between 1.0 and 3.5 mg/ml (approximately 80 to 200μ M protein). Data acquisition and analysis were performed using ASTRA 4.0 software (Wyatt Technologies).
Native Gel Electrophoresis
BTD polypeptides were incubated with NICD polypeptides at 4°C for at least 1 h. Complexes were resolved on 10–20% native polyacrylamide gels (Invitrogen) and were visualized by staining with Simply Blue (Invitrogen).
Peptide Synthesis and Purification
RAM-derived peptides were synthesized using a Protein Technologies Symphony Quartet Peptide Synthesizer. Peptide identities were verified by mass using a Finnigan LCQ Deca ion-trap mass spectrometer equipped with an electrospray ionization source (Thermo Finnigan, San Jose, CA). Peptides were purified using an HPLC system equipped with a C18 semi-preparative column (Grace-Vydac; Hesperia, CA) and purified with linear acetonitrile gradient (0–50%). Fractions containing pure peptide were lyophilized and stored at −20 ° C.
Immunodepletion assays
HEK293 cells were grown at 37ºC and 5% CO2 in DMEM containing 10% FBS. Cells were transfected either with pCS2+6MT-NICD1 or pCMV-3XFlag-RBPjκ or co-transfected with both expression vectors using FuGENE reagent (Roche; Nutley, NJ). 40–44 hours after transfection, cells were lysed in cold CoIP buffer (25 mM HEPES, pH 7.4, 1% Nonidet P-40, 200 mM KCl, 20 mM NaF, 0.2 mM EGTA 27) supplemented with complete protease inhibitor cocktail (Roche). In one experiment, lysates from cells expressing 6MT-NICD1 alone and 3XFlag-RBPjκ alone were pooled and immediately aliquotted among microfuge tubes containing increasing concentrations of the different RAM peptides. In a separate experiment, lysates from cells co-expressing 6MT-NICD1 and 3XFlag-RBPjκ were pooled and similarly challenged with the different RAM peptides. To assess the effects of the RAM peptides on NICD1-RBPjκ association, we immunoprecipitated the lysates with anti-FLAG M2 antibodies (Sigma) and then measured the amounts of NICD1 remaining in the supernatant by monitoring Myc immunoreactivity by SDS-PAGE/Western blotting. Western analyses with polyclonal anti-FLAG antibodies (Sigma) indicated that immunoprecipitation of 3XFlag-RBPjκ was complete under these conditions (data not shown).
Supplementary Material
Acknowledgments
We thank Dr. Tamara Hendrickson, Jennifer Meitzler and Megan Ehrenwerth for help with peptide synthesis, and gratefully acknowledge the support of an Instrumentation Development Grant (0500580 to Dr. Hendrickson) from the National Science Foundation Division of Biological Infrastructure for Peptide Synthesis instrumentation. We thank members of Barrick and Kopan laboratories for technical advice and support. We thank Chin-Tong Ong for the 6MT-NICD1 construct as well as Mary Blandford and Gabriel Rice for technical assistance. OL was supported by a Dermatology training grant and Dimitri D’Arbeloff post-doctoral fellowship, and by a training grant from the US. Department of Energy (DE-FG0204ER25626). This work was supported by NIH grants GM55479 to RK, and GM60001 to DB.
Abbreviations Used
- NICD
Notch receptor intracellular domain
- ANK
Ankyrin domain of the Notch receptor
- RAM
segment of the Notch receptor N-terminal to the ANK domain (RBP-associated molecule)
- CSL
transcription factor downstream of the Notch receptor
- BTD
beta trefoil domain of CSL
- ITC
isothermal titration calorimetry
Footnotes
This sequence, which is derived from the NCBI database, contains a WLP sequence but differs significantly from ensembl mouse Notch2 sequence as well as from and human Notch2. It appears that this sequence resulted from a sequencing error in which a two BP insertion 5’ and a one base insertion 3’ to the the divergent sequence produced a local frameshift (supplemental Figure 1). With these changes the NCBI can be converted to the rat, human, and the ensembl mouse Notch2 sequence. Note that a WLP sequence in the frameshifted Notch2 NCBI sequence is fortuitously coded within the alternate frame.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Gridley T. Notch signaling and inherited disease syndromes. Hum Mol Genet 12 Spec No. 2003;1:R9–13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Irizarry C, Carpenter AC, Weng AP, Pear WS, Aster JC, Blacklow SC. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol Cell Biol. 2004;24:9265–73. doi: 10.1128/MCB.24.21.9265-9273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vooijs M, Schroeter EH, Pan Y, Blandford M, Kopan R. Ectodomain shedding and intramembrane cleavage of mammalian Notch proteins is not regulated through oligomerization. J Biol Chem. 2004;279:50864–73. doi: 10.1074/jbc.M409430200. [DOI] [PubMed] [Google Scholar]
- 5.Weng AP, Aster JC. Multiple niches for Notch in cancer: context is everything. Curr Opin Genet Dev. 2004;14:48–54. doi: 10.1016/j.gde.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Vardar D, North CL, Sanchez-Irizarry C, Aster JC, Blacklow SC. Nuclear magnetic resonance structure of a prototype Lin12-Notch repeat module from human Notch1. Biochemistry. 2003;42:7061–7. doi: 10.1021/bi034156y. [DOI] [PubMed] [Google Scholar]
- 7.Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 8.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–16. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 9.Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, LaPlant Q, Ball H, Dann CE, 3rd, Sudhof T, Yu G. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005;122:435–47. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 10.Fryer CJ, Lamar E, Turbachova I, Kintner C, Jones KA. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 2002;16:1397–411. doi: 10.1101/gad.991602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell. 2004;16:509–20. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Morel V, Lecourtois M, Massiani O, Maier D, Preiss A, Schweisguth F. Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr Biol. 2001;11:789–92. doi: 10.1016/s0960-9822(01)00224-x. [DOI] [PubMed] [Google Scholar]
- 13.Zhou S, Hayward SD. Nuclear localization of CBF1 is regulated by interactions with the SMRT corepressor complex. Mol Cell Biol. 2001;21:6222–32. doi: 10.1128/MCB.21.18.6222-6232.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou S, Fujimuro M, Hsieh JJ, Chen L, Miyamoto A, Weinmaster G, Hayward SD. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC To facilitate NotchIC function. Mol Cell Biol. 2000;20:2400–10. doi: 10.1128/mcb.20.7.2400-2410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–6. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 16.Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, Kopan R. Collaboration between discrete protein domains and binding site architecture on the promoter determine target selectivity of vertebrate notch proteins. J Biol Chem. 2005 doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- 17.Oswald F, Tauber B, Dobner T, Bourteele S, Kostezka U, Adler G, Liptay S, Schmid RM. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol Cell Biol. 2001;21:7761–74. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lubman OY, Korolev SV, Kopan R. Anchoring notch genetics and biochemistry; structural analysis of the ankyrin domain sheds light on existing data. Mol Cell. 2004;13:619–26. doi: 10.1016/s1097-2765(04)00120-0. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD. Truncated mammalian Notch1 activates CBF1/RBPJκ -repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–9. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tani S, Kurooka H, Aoki T, Hashimoto N, Honjo T. The N- and C-terminal regions of RBP-J interact with the ankyrin repeats of Notch1 RAMIC to activate transcription. Nucleic Acids Res. 2001;29:1373–80. doi: 10.1093/nar/29.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovall RA, Hendrickson WA. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. Embo J. 2004;23:3441–51. doi: 10.1038/sj.emboj.7600349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–83. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 23.Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;124:985–96. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 24.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Curr Biol. 1995;5:1416–23. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 25.Nam Y, Weng AP, Aster JC, Blacklow SC. Structural requirements for assembly of the CSL.intracellular Notch1.Mastermind-like 1 transcriptional activation complex. J Biol Chem. 2003;278:21232–9. doi: 10.1074/jbc.M301567200. [DOI] [PubMed] [Google Scholar]
- 26.Kato H, Sakai T, Tamura K, Minoguchi S, Shirayoshi Y, Hamada Y, Tsujimoto Y, Honjo T. Functional conservation of mouse Notch receptor family members. FEBS Lett. 1996;395:221–4. doi: 10.1016/0014-5793(96)01046-0. [DOI] [PubMed] [Google Scholar]
- 27.Saxena MT, Schroeter EH, Mumm JS, Kopan R. Murine notch homologs (N1-4) undergo presenilin-dependent proteolysis. J Biol Chem. 2001;276:40268–73. doi: 10.1074/jbc.M107234200. [DOI] [PubMed] [Google Scholar]
- 28.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–8. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 29.Kopan R, Nye JS, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120:2385–96. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 30.Nofziger D, Miyamoto A, Lyons KM, Weinmaster G. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 1999;126:1689–702. doi: 10.1242/dev.126.8.1689. [DOI] [PubMed] [Google Scholar]
- 31.Le Gall M, Giniger E. Identification of two binding regions for the suppressor of hairless protein within the intracellular domain of Drosophila notch. J Biol Chem. 2004;279:29418–26. doi: 10.1074/jbc.M404589200. [DOI] [PubMed] [Google Scholar]
- 32.Barrick D, Kopan R. The Notch transcription activation complex makes its move. Cell. 2006;124:883–5. doi: 10.1016/j.cell.2006.02.028. [DOI] [PubMed] [Google Scholar]
- 33.Fried MG, Daugherty MA. Electrophoretic analysis of multiple protein-DNA interactions. Electrophoresis. 1998;19:1247–53. doi: 10.1002/elps.1150190804. [DOI] [PubMed] [Google Scholar]
- 34.Wyatt P. Light scattering and the absolute characterization of macromolecules. Analytica chimica Acta. 1993;272:1–40. [Google Scholar]
- 35.Beatus P, Lundkvist J, Oberg C, Lendahl U. The notch 3 intracellular domain represses notch 1-mediated activation through Hairy/Enhancer of split (HES) promoters. Development. 1999;126:3925–35. doi: 10.1242/dev.126.17.3925. [DOI] [PubMed] [Google Scholar]
- 36.Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–81. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 37.Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, Kopan R. Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J Biol Chem. 2006;281:5106–19. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- 38.Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–41. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 39.Lubman OY, Kopan R, Waksman G, Korolev S. The crystal structure of a partial mouse Notch-1 ankyrin domain: repeats 4 through 7 preserve an ankyrin fold. Protein Sci. 2005;14:1274–81. doi: 10.1110/ps.041184105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beatus P, Lundkvist J, Oberg C, Pedersen K, Lendahl U. The origin of the ankyrin repeat region in Notch intracellular domains is critical for regulation of HES promoter activity. Mech Dev. 2001;104:3–20. doi: 10.1016/s0925-4773(01)00373-2. [DOI] [PubMed] [Google Scholar]
- 41.Shin HM, Minter LM, Cho OH, Gottipati S, Fauq AH, Golde TE, Sonenshein GE, Osborne BA. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. Embo J. 2006;25:129–38. doi: 10.1038/sj.emboj.7600902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurooka H, Kuroda K, Honjo T. Roles of the ankyrin repeats and C-terminal region of the mouse Notch1 intracellular region. Nucleic Acids Research. 1998;26:5448–5455. doi: 10.1093/nar/26.23.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–61. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 44.Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307:1904–9. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 45.Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC, Aster JC. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol. 2003;23:655–64. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–7. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 47.Phelps CB, Sengchanthalangsy LL, Huxford T, Ghosh G. Mechanism of I kappa B alpha binding to NF-kappa B dimers. J Biol Chem. 2000;275:29840–6. doi: 10.1074/jbc.M004899200. [DOI] [PubMed] [Google Scholar]
- 48.Schon O, Friedler A, Bycroft M, Freund SM, Fersht AR. Molecular mechanism of the interaction between MDM2 and p53. J Mol Biol. 2002;323:491–501. doi: 10.1016/s0022-2836(02)00852-5. [DOI] [PubMed] [Google Scholar]
- 49.Paal K, Baeuerle PA, Schmitz ML. Basal transcription factors TBP and TFIIB and the viral coactivator E1A 13S bind with distinct affinities and kinetics to the transactivation domain of NF-kappaB p65. Nucleic Acids Res. 1997;25:1050–5. doi: 10.1093/nar/25.5.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desrosiers DC, Peng ZY. A binding free energy hot spot in the ankyrin repeat protein GABPbeta mediated protein-protein interaction. J Mol Biol. 2005;354:375–84. doi: 10.1016/j.jmb.2005.09.045. [DOI] [PubMed] [Google Scholar]
- 51.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26:2887–900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


