Abstract
Inhibitor 1 (I-1) is a protein inhibitor of protein phosphatase 1 (PP1), a major eukaryotic Ser/Thr phosphatase. Nonphosphorylated I-1 is inactive, whereas phosphorylated I-1 is a potent PP1 inhibitor. I-1 is phosphorylated in vivo on Thr35 and Ser67. Thr35 is phosphorylated by cAMP-dependent protein kinase (A kinase), and Thr35-phosphorylated I-1 inhibits PP1. Until now the kinase that phosphorylates Ser67 had not been identified and the physiological role of Ser67 phosphorylation was unknown. In this study we detected a high level of kinase activity in brain extract when a glutathione S-transferase (GST) fusion I-1 mutant containing an Ala substituted for Thr35 [GST-I-1(T35A)] was used as the substrate. GST-I-1(T35A) kinase and neuronal cdc2-like protein kinase (NCLK) in the brain extract could not be separated from each other by a series of sequential chromatographies. GST-I-1(T35A) kinase immunoprecipitated with anti-NCLK antibody from kinase-active column fractions. Purified NCLK-phosphorylated GST-I-1(T35A) and I-1 (0.7 mole of phosphate per mole of I-1). HPLC phosphopeptide mapping, amino acid sequencing, and site-directed mutagenesis determined that NCLK phosphorylates Ser67 of I-1. NCLK-phosphorylated I-1 and I-1(T35A) inhibited PP1 with IC50 values ≈9.5 and 13.8 nM, respectively. When compared, A kinase-phosphorylated I-1 was only ≈1.2 times more inhibitory than NCLK-phosphorylated I-1. Our data indicate that NCLK is a potential in vivo I-1 kinase and that Thr35 and Ser67 phosphorylation independently activate I-1.
Keywords: neuronal cdc2-like protein kinase, cAMP-dependent protein kinase
Protein phosphatase 1 (PP1) is a major eukaryotic Ser/Thr phosphatase that regulates diverse cellular processes such as glycogen metabolism, cell division, muscle contraction, signal transduction, neural functions, and RNA processing (reviews in refs. 1–5). PP1 is a 37-kDa catalytic subunit whose activity is regulated by two types of regulatory subunits: targeting subunits and inhibitory subunits. Targeting subunits confer substrate specificity and localize PP1 to various subcellular compartments (1–3). Inhibitory subunits suppress the catalytic activity of PP1 (4, 5). Inhibitor 1 (I-1), DARPP-32, and inhibitor 2 are heat-stable PP1 inhibitory subunits. DARPP-32 is an I-1 homologue found mainly in brain. I-1 and DARPP-32 require phosphorylation for the inhibitory activity, whereas inhibitor 2 inhibits PP1 without phosphorylation.
I-1 is an ≈19-kDa protein widely expressed in various mammalian tissues (4–7). It is phosphorylated on Thr35 and Ser67 in vivo (6). Thr35 is phosphorylated by cAMP-dependent protein kinase (A kinase), and this phosphorylation makes I-1 a PP1 inhibitor (6, 7). Until now an I-1 Ser67 kinase(s) was unidentified. The physiological role of Ser67 phosphorylation was also unknown. In this study, we used a glutathione S-transferase (GST) fusion I-1 mutant containing Ala in place of Thr35, GST-I-1(T35A), as the substrate and detected a high level of kinase activity in brain extract. Herein we report the identity of I-1 Ser67 kinase and describe the role of Ser67 phosphorylation on I-1 activity.
Materials and Methods
I-1 cDNA Clones and Site-Specific I-1 Mutagenesis.
pGEX-2T vectors containing human I-1 and I-1(T35A) were gifts from Shirish Shenolikar of Duke University. An I-1 mutant [I-1(S67A)] containing Ala substituted for Ser67 was constructed by PCR-based megaprimer mutagenesis (8). Two-step Pfu DNA polymerase-catalyzed PCR was carried out with human I-1 cDNA as the template. In the first step, the forward primer, 5′-CTTCCGGAATTCATGGAGCAAGACAACAGCCCCCG-3′, containing an EcoRI site (italics) and the I-1 mutagenesis reverse primer Ser67 → Ala, 5′-189CCGTTGCCGTGGCGCCATTGCCAAAG213-3′ were used. PCR (30 cycles) was carried out at 94°C for 30 sec, 55°C for 1 min, and 72°C for 40 sec, and the PCR product was purified. In the second step, the above purified PCR product was used as the forward primer and 5′-GCGGCCGCCTCGAGTCAGACCGAGTTGGCTCCCTTGG-3′ containing a XhoI site (italicized) was used as the reverse primer. PCR (30 cycles) was performed at 94°C for 30 sec, 55°C for 1 min, and 72°C for 1.5 min. The final PCR product was purified and subcloned into the EcoRI/XhoI sites of the pGEX-6P-1 vector. The recombinant plasmid was transformed into Escherichia coli DH10B first and then into BL21(DE3). The mutagenesis was confirmed by DNA sequencing.
Proteins and Peptides.
GST-I-1, GST-I-1(T35A), and GST-I-1(S67A) were purified from E. coli lysates by using glutathione-S-Sepharose affinity chromatography (7). The GST tag was removed from the various I-1 constructs by using a thrombin Cleancleave kit from Sigma and following the manufacturer's instructions. Neuronal cdc2-like kinase (NCLK) was purified from fresh bovine brain (9). Phosphorylase kinase was purified from rabbit skeletal muscle (10). PP1α was a gift from E. Y. C. Lee (New York Medical College). The catalytic subunit of A kinase, phosphorylase b, A kinase substrate kemptide (LRRASLG), and A kinase inhibitory peptide PKI (TTYADPIASGRTGRRNAIHD) were from Sigma. The making of polyclonal antibodies against cdk5 subunit of NCLK and the C terminus of mitogen-activated protein (MAP) kinase (p43Erk1) and the synthesis of the peptide substrate of NCLK (KTPKKAKKPKTPKKAKKL) have been described previously (10). Polyclonal antibodies against casein kinase 1 and casein kinase 2 were gifts from Louise Larose and Stephan Richards of McGill University.
Protein and Peptide Concentrations.
GST-I-1 and I-1 concentrations were determined by Bio-Rad protein assay using BSA as the standard. Concentrations of I-1(T35A), I-1(S67A), NCLK-phosphorylated I-1, NCLK-phosphorylated 1(T35A), A kinase-phosphorylated I-1, and NCLK/A kinase-phosphorylated I-1 were determined by Bio-Rad protein assay using I-1 as standard. Concentrations of all GST-I-1 species were determined by Bio-Rad protein assay using GST-I-1 as standard. NCLK concentration was based on its activity (9). Concentrations of A kinase, phosphorylase b, kemptide, and A kinase inhibitory peptide were based on dry weights. All other protein concentrations were determined by Bio-Rad protein assay using BSA as the standard. Concentration of NCLK substrate peptide was based on amino acid analysis.
Activity Assays.
The GST-I-1(T35A) kinase assay was initiated at 30°C by adding 10 μl of the kinase solution to 30 μl of reaction mixture containing the rest of the assay components. The final concentrations of the various assay components were 25 mM Hepes (pH 7.2), 50 mM β-glycerophosphate, 10 mM NaF, 1 mM EDTA, 1 mM DTT, 10 mM MgCl2, 0.4 mM [γ-32P]ATP, and 0.2 mg/ml GST-I-1(T35A). After 30 min, 20 μl was withdrawn from the reaction mixture, mixed with SDS/PAGE sample buffer, and electrophoresed on SDS/10% polyacrylamide gels. The GST-I-1(T35A) bands in the gel were sliced out and their radioactivities were measured in a liquid scintillation counter. NCLK activity was also assayed as above, using 50 μM peptide substrate (9). After 30 min, 10 μl of 50% trichloroacetic acid was added to the assay mixture, which was incubated at 4°C for 10 min and centrifuged. Supernatant from the assay mixture was withdrawn and analyzed by phosphocellulose strip assay (11). A kinase activity was measured as above, using kemptide as the substrate.
To generate Fig. 3 and Table 3, phosphorylated I-1 was prepared as above except the concentration of I-1 was 0.6 mg/ml. When used, the A kinase concentration was 25 μg/ml. After 12 h at 30°C, the reaction mixture was dialyzed against 50 mM Tris⋅HCl, pH 7.4/1 mM EDTA/1 mM DTT. NCLK-phosphorylated I-1(T35A) was also prepared as above. NCLK/A kinase-phosphorylated I-1 was prepared as above by phosphorylating I-1 with NCLK, followed by A kinase, for 6 h each. NCLK-phosphorylated I-1, NCLK-phosphorylated I-1(T35A), A kinase-phosphorylated I-1, and NCLK/A kinase-phosphorylated I-1 were prepared at the same time under identical conditions and were dialyzed in the same vessel. The amounts of phosphate in NCLK-phosphorylated I-1, NCLK-phosphorylated I-1(T35A), A kinase-phosphorylated I-1, and NCLK/A kinase-phosphorylated I-1 were 0.7, 0.5, 0.8, and 1.4 mole of phosphate per mole of inhibitor protein, respectively. Similarly, the amounts of phosphate in NCLK-phosphorylated GST-I-1, NCLK-phosphorylated GST-I-1(T35A), and A kinase-phosphorylated GST-I-1 were 0.5, 0.3, and 0.8 mole of phosphate per mole of inhibitor protein, respectively.
Figure 3.
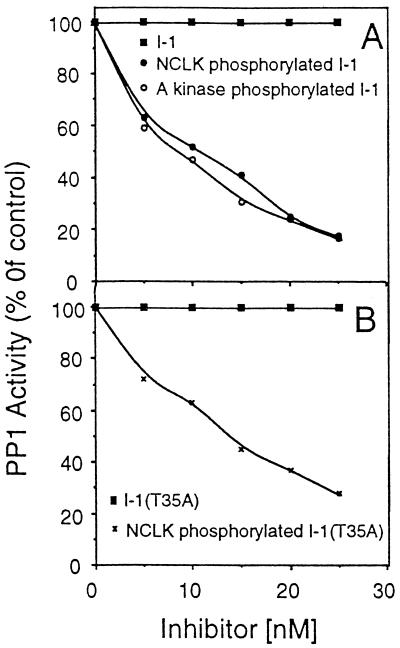
PP1 inhibition by A kinase-phosphorylated I-1 and by NCLK-phosphorylated I-1 and I-1(T35A). PP1 activity was assayed in the presence of indicated concentrations of the indicated inhibitor. After 30 min, aliquots were withdrawn and PP1 activity was analyzed. All of the assays were carried out at the same time under identical conditions.
Table 3.
Comparison of PP1 inhibitory activities of various I-1 species
| Inhibitor | IC50, nM |
|---|---|
| I-1 | No inhibition |
| A kinase-phosphorylated I-1 | 7.5 ± 0.49 |
| NCLK-phosphorylated I-1 | 9.5 ± 0.93 |
| NCLK/A kinase-phosphorylated I-1 | 4.8 ± 0.18 |
| I-1(T35A) | No inhibition |
| NCLK-phosphorylated I-1(T35A) | 13.8 ± 1.5 |
| GST-I-1 | No inhibition |
| A kinase-phosphorylated GST-I-1 | 55 ± 4 |
| NCLK-phosphorylated GST-I-1 | 74 ± 7 |
| GST-I-1(T35A) | No inhibition |
| NCLK-phosphorylated GST-I-1(T35A) | 149 ± 12 |
PP1 activity in the presence of various concentrations of indicated I-1 species was monitored and plotted as in Fig. 3. IC50 (concentration of inhibitor required for half-maximal inhibition) was determined from the plot. Values are mean of three independent determinations.
PP1 activity was assayed as described (7) in a 30-μl reaction mixture containing 50 mM Tris⋅HCl (pH 7.4), 1 mM DTT, 0.5 mM MnCl2, 10 μM [32P]phosphorylase a, and 0.5 μg/ml PP1. The reaction was initiated by the addition of 1 μl of PP1 to 20 μl of assay mixture containing the rest of the assay components. After 20 min at 30°C the reaction was terminated by adding 10 μl of 50% trichloroacetic acid to the assay mixture. The assay mixture was then cooled on ice and centrifuged. A 20-μl aliquot from the supernatant was spotted onto filter paper and placed in a scintillation counter to determine the amount of released [32P]Pi. [32P]Phosphorylase a used for PP1 assays was prepared at 30°C for 30 min as described (10). [32P]Phosphorylase a was dialyzed in 50 mM Tris⋅HCl, pH 7.4/1 mM EDTA/1 mM DTT and stored frozen at −80°C until used. Dephosphorylation of phosphorylated I-1 by PP1 was monitored as above. The concentration of 32P-I-1 in the assay was 0.1 mg/ml.
Phosphopeptide Purification and Peptide Sequencing.
GST-I-1 (0.6 mg) was phosphorylated by NCLK for 12 h as above. Phosphorylated GST-I-1 was desalted on a Sephadex G-25 column, lyophilized, redissolved in 0.2 ml of 50 mM NH4HCO3 (pH 8.0) containing 50 μg/ml trypsin, and incubated at 37°C for 12 h. The incubated sample was subjected to C18 reverse-phase HPLC (10). Radioactive HPLC fractions were combined, concentrated to ≈0.5 ml, and chromatographed through a Sephadex G-25 column equilibrated and eluted with 0.1% trifluoroacetic acid (TFA). Fractions (0.5 ml each) were collected and those containing radioactivity were combined, concentrated to 0.2 ml, and reinjected into an HPLC column as described above. The column was eluted with a 0–40% (vol/vol) acetonitrile gradient in 0.1% TFA in 60 min. Fractions containing radioactive peptide were subjected to amino acid sequencing at the University of Victoria Department of Biochemistry and Microbiology.
Partial Purification of GST-I-1(T35A) Kinase.
All procedures were carried out at 4°C. Fresh bovine brain (≈1 kg) was homogenized for 1 min in 2 liters of buffer A (20 mM Mops, pH 7.4/50 mM β-glycerophosphate/10 mM NaF/1 mM EDTA/1 mM DTT/15 mM MgCl2) containing 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and 5 mg/ml benzamidine. The homogenate was centrifuged at 104 × g for 30 min. The supernatant was then centrifuged at 105 × g for 40 min. The resulting clear supernatant was loaded onto a DEAE-Sephacel (Sigma) column (2.5 × 45 cm) previously equilibrated with buffer A. The column was eluted with 0.5 M NaCl in buffer A. The flow-through fraction containing GST-I-1(T35A) kinase activity was loaded onto a SP-Sepharose (Pharmacia) column (1 × 60 cm). The column was washed with buffer A and eluted with 600 ml of a linear gradient of 0–0.5 M NaCl in buffer A. Fractions containing kinase activity were combined and loaded onto a hydroxylapatite column (1.5 × 25 cm) preequilibrated in buffer A. The column was washed with buffer A and then eluted with a 250-ml linear gradient of K2HPO4 (0–0.4 M) in buffer A. Effluent fractions containing kinase activity were combined (≈50 ml), concentrated to ≈5 ml, and subjected to gel filtration chromatography on an FPLC Superose 12 gel filtration column (Pharmacia; 2 × 70 cm, and equilibrated and eluted in buffer A containing 200 mM NaCl).
Immunoprecipitation and Immunoblotting.
The gel filtration fraction containing kinase activity from Fig. 1C (≈1 ml) was cleared with 50 μl of staphylococcal protein A-agarose beads (Sigma) and divided into halves (450 μl each). To each half, 50 μl of either preimmune serum or anti-NCLK serum was added. Both halves were shaken end-over-end at 4°C. After 1 h of shaking, 50 μl of protein A-agarose beads preequilibrated in buffer A was added to each half, and shaking was continued for another hour. Protein A-agarose beads were collected by centrifugation, washed five times with ice-cold buffer A, and subjected to NCLK activity assay as described above except the trichloroacetic acid precipitation step was excluded. The assay mixture was centrifuged and the supernatant was spotted onto filter paper to determine amount of radioactivity incorporated into the peptide substrate. Western immunoblotting was carried out as described previously (9).
Figure 1.
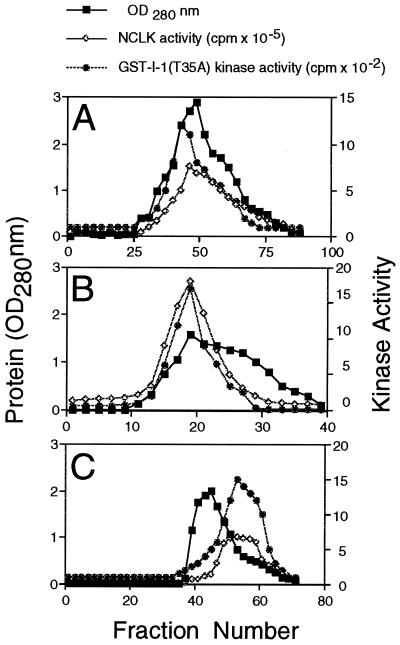
Coelution of GST-I-1(T35A) kinase and NCLK from various chromatographic columns. Bovine brain extract was chromatographed through a DEAE-Sephacel column. The flow-through fraction containing GST-I-1(T35A) kinase activity was subjected to SP-Sepharose (A), hydroxylapatite (B), and Superose 12 gel filtration (C) chromatographies sequentially. Aliquots from indicated fractions were withdrawn and assayed for GST-I-1(T35A) kinase and NCLK activities. Gel filtration was carried out with the Pharmacia FPLC system at a flow rate of 1 ml/min. The size of each fraction in A, B, and C was 5, 5, and 1 ml, respectively.
Results
GST-I-1(T35A) Kinase in Brain Extract.
We found a high level of GST-I-1(T35A) kinase activity in bovine brain extract. This activity was recovered in flow-through fractions when subjected to DEAE-Sephacel chromatography (data not shown). When the DEAE flow-through fraction was chromatographed through an SP-Sepharose column, GST-I-1(T35A) kinase activity eluted as a single symmetric peak, suggesting that there may be only one GST-I-1(T35A) kinase in the brain extract (Fig. 1A). We set out to determine the identity of this kinase.
To determine whether GST-I-1(T35A) kinase is one of the MAP kinases, 20 μl from various fractions in Fig. 1A was immunoblotted, using polyclonal antibody against MAP kinase. Our antibody that can detect nanograms of p43erk1 and p42erk1 showed no immunoreactivity (data not shown). Similar immunoblot analyses using polyclonal antibodies against casein kinase 2 and casein kinase 1 determined that both these kinases were absent from Fig. 1A fractions (data not shown). Fig. 1A fractions also did not have any detectable kemptide kinase (A kinase) activity (data not shown). On the basis of these observations we concluded that MAP kinases p43erk1 and p43erk2, casein kinase 1, casein kinase 2, and A kinase were not present in various column fractions in Fig. 1A. These kinases most likely bound to the DEAE column and were absent from DEAE flow-through fractions used to generate Fig. 1A. When 20 mM LiCl, a glycogen synthase kinase 3 inhibitor (12), was included in the GST-I-1(T35A) kinase assay mixture and various fractions in Fig. 1A were assayed, no effect was observed (data not shown). Similarly, 2 mM EGTA, a specific Ca2+ chelator, also did not inhibit GST-I-1(T35A) kinase activity in Fig. 1A. These observations indicated that GST-I-1(T35A) kinase in Fig. 1 could not be glycogen synthase kinase 3 or any Ca2+-dependent kinase (protein kinase C, calmodulin-dependent protein kinases, or phosphorylase kinase).
NCLK is a brain proline-directed protein kinase (9, 13). Because there is a proline at the carboxyl side of I-1 Ser67 (7), we assayed NCLK activity in various Fig. 1A fractions. A robust kinase activity that coeluted with GST-I-1(T35A) kinase was detected (Fig. 1A). To determine whether NCLK and GST-I-1(T35A) kinase were identical or different, various fractions containing kinase activity in Fig. 1A were subjected to hydroxylapatite and gel filtration chromatographies sequentially. As shown in Fig. 1 B and C, NCLK and GST-I-1(T35A) kinase activity coeluted from both columns. Using anti-NCLK antibody, we then immunoprecipitated NCLK from Fig. 1C peak column fractions. The resulting immune complex was subjected to NCLK activity assay. Anti-NCLK immune complex displayed intense GST-I-1(T35A) kinase activity (data not shown).
Phosphorylation of I-1 by NCLK.
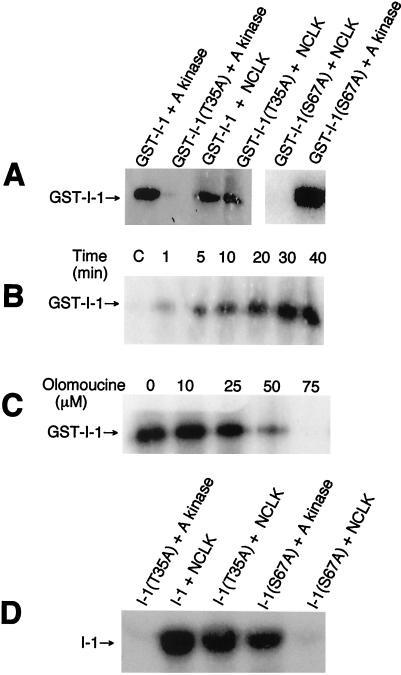
Our preceding data indicated the interesting possibilities that either NCLK is GST-I-1(T35A) kinase or GST-I-1(T35A) kinase is physically bound to NCLK in the brain extract. To discriminate between these possibilities, we examined the phosphorylation of various I-1 species by purified NCLK. As shown in Fig. 2A, NCLK phosphorylated GST-I-1(T35A) and GST-I-1 but failed to phosphorylate GST-I-1(S67A). A kinase, as expected, phosphorylated GST-I-1 and GST-I-1(S67A) but did not phosphorylate GST-I-1(T35A). Fig. 2B shows the time course of GST-I-1 phosphorylation by NCLK. In 4 h NCLK incorporated 0.5 mole of phosphate per mole of GST-I-1. Since NCLK used in these experiments was purified from the brain extract, we performed experiments to determine that observed GST-I-1 phosphorylation was caused by the action of NCLK and not by any contaminant kinase in our NCLK preparations.
Figure 2.
Phosphorylation of I-1 and inhibition of phosphorylation by olomoucine. Phosphorylation was carried out with 400 units/ml NCLK and 0.2 mg/ml I-1 species. The concentration of A kinase, when used, was 25 μg/ml. After the indicated times, 20 μl was withdrawn from each mixture and electrophoresed, and phosphorylation was analyzed by autoradiography of gels. (A) Phosphorylation of GST-I-1, GST-I-1(T35A), and GST-I-1(S67A) by NCLK and A kinase. The phosphorylation was carried out for 30 min. (B) Time course of GST-I-1 phosphorylation by NCLK. Control lane, C, contained GST-I-1 alone incubated with the rest of the assay components for 40 min. NCLK incubated alone with the rest of the assay components did not show any radioactive band with the size of GST-I-1 on the gel (data not shown). (C) Inhibition of GST-I-1 phosphorylation by olomoucine. GST-I-1 phosphorylation by NCLK was carried out for 30 min in the presence of olomoucine (Sigma) at the indicated concentrations. (D) Phosphorylation of I-1, I-1(T35A), and I-1(S67A) by NCLK and A kinase. Indicated I-1 species were phosphorylated by the indicated kinase for 30 min.
A kinase-inhibitory peptide PKI (50 μM) completely suppressed the I-1 phosphorylation by A kinase but displayed no effect on I-1 phosphorylation by NCLK (data not shown). Similarly, GST-I-1 phosphorylation by NCLK was unaffected by 20 mM LiCl or 2 mM EGTA (data not shown). These observations indicated that GST-I-1 phosphorylation in Fig. 2B was not caused by any contaminant A kinase, glycogen synthase kinase 3, or any Ca2+-dependent kinases. Immunoblot analyses using polyclonal antibodies against casein kinase 1, MAP kinase, and casein kinase 2 failed to show any cross-reactivity with all of our NCLK preparations. These results indicated that GST-I-1 phosphorylation by NCLK preparations was not caused by any contaminant MAP kinases, casein kinase 1, or casein kinase 2. Finally, we included olomoucine, a specific NCLK inhibitor (14), in the phosphorylation mixture and monitored GST-I-1 phosphorylation. As shown in Fig. 2C, olomoucine completely suppressed GST-I-1 phosphorylation. On the basis of these results along with our observations that all of the NCLK preparations used in this study phosphorylated GST-I-1 in a similar manner, we concluded that NCLK indeed phosphorylates GST-I-1.
To further evaluate the phosphorylation of I-1 by NCLK, various GST-I-1 species were treated with thrombin to remove GST tags. The resulting I-1 species were phosphorylated by NCLK and A kinase. As shown in Fig. 2D, NCLK phosphorylated I-1 and I-1(T35A) and did not phosphorylate I-1(S67A). Similarly A kinase phosphorylated I-1(S67A) but failed to phosphorylate I-1(T35A).
We compared GST-I-1 as the substrate of NCLK and A kinase (Table 1). The Km values indicate that GST-I-1 binds to NCLK better than to A kinase. However, the turnover rate (kcat) of A kinase phosphorylation is ≈13 times higher than NCLK phosphorylation. Overall, kcat/Km values indicate that GST-I-1 is ≈2 times better substrate of A kinase than of NCLK.
Table 1.
Kinetic parameters for phosphorylation of GST-I-1 by NCLK and A kinase
| Kinase | Km, μM | kcat, min−1 | kcat/Km, min−1⋅μM−1 |
|---|---|---|---|
| A kinase | 8.1 ± 0.8 | 318 ± 23 | 39.2 |
| NCLK | 1.2 ± 0.1 | 24 ± 4.8 | 20.0 |
Phosphorylation Site Determination.
NCLK-phosphorylated GST-I-1 was digested with trypsin and subjected to HPLC analysis. Only one radioactive peak with retention time ≈32 min eluted from the column (data not shown). Radioactive fractions were pooled and the phosphopeptide was purified. One phosphopeptide was isolated. This purified phosphopeptide was subjected to amino acid sequencing in a gas-phase amino acid sequencer (Table 2). Ser, Thr, Leu, Ala, Met, Pro, and Arg were identified as the first, second, third, fourth, fifth, seventh, and eighth residues, respectively, of this peptide. The sixth residue could not be identified. Since phenylthiohydantoin derivatives of phosphorylated amino acids are not identified by Edman degradation, the unidentified sixth residue must be a phosphorylated amino acid. On the basis of these observations, inability of NCLK to phosphorylate GST-I-1(S67A) and I-1(S67A) (Fig. 2 A and D), and the amino acid sequence of human I-1 (7), we concluded that the phosphopeptide extends from residue 62 to residue 69 and Ser67 is the phosphorylation site. To confirm that NCLK phosphorylates I-1 on Ser67, GST-I-1 and I-1 were phosphorylated by NCLK as above and digested with trypsin. Both tryptic digests were subjected to HPLC phosphopeptide mapping under identical conditions. Both maps looked identical and each had only one radioactive peak with retention time ≈32 min (data not shown). These observations indicated that NCLK phosphorylates I-1 on Ser67.
Table 2.
Sequence determination of 32P-labeled tryptic phosphopeptide
| Cycle | Amino acid | Yield, pmol |
|---|---|---|
| 1 | Ser | 18.0 |
| 2 | Thr | 8.8 |
| 3 | Leu | 5.4 |
| 4 | Ala | 2.6 |
| 5 | Met | 1.7 |
| 6 | Xaa | |
| 7 | Pro | 1.8 |
| 8 | Arg | 1.4 |
Effect of NCLK Phosphorylation on I-1 Function.
Previous studies have determined that I-1 inhibits PP1 only upon Thr35 phosphorylation by A kinase (4–7). Since we found that NCLK phosphorylates I-1 on Ser67, we examined the effect of NCLK phosphorylation on I-1 inhibitory activity. As shown in Fig. 3, NCLK-phosphorylated I-1 and I-1(T35A) inhibited PP1 in a dose-dependent manner. Table 3 compares the IC50 (concentration required for half-maximal inhibition) values of various I-1 species. A kinase-phosphorylated I-1 inhibited PP1 with IC50 value ≈7.5 nM. IC50 values for the inhibition of PP1 by NCLK-phosphorylated I-1 and I-1(T35A) are ≈9.5 nM and ≈13.8 nM, respectively. The NCLK/A kinase-phosphorylated I-1 inhibited PP1 with IC50 value 4.8 nM. These data indicate that A kinase-phosphorylated I-1 is ≈1.3 times more inhibitory than NCLK-phosphorylated I-1 and NCLK/A kinase-phosphorylated I-1 is ≈1.5 and ≈2 times more inhibitory than A kinase-phosphorylated I-1 and NCLK-phosphorylated I-1, respectively.
Dephosphorylation of NCLK-phosphorylated I-1 by PP1.
Thr35-phosphorylated I-1 is dephosphorylated by PP1 in the presence of Mn2+ (15). To examine dephosphorylation of Ser67-phosphorylated I-1 by PP1, A kinase-phosphorylated and NCLK-phosphorylated I-1 species were prepared. Dephosphorylation of these species by PP1 was monitored. Both A kinase- and NCLK-phosphorylated I-1 species were dephosphorylated by PP1. A kinase-phosphorylated I-1 was dephosphorylated 3 times faster than NCLK-phosphorylated I-1 (data not shown). These data indicate that PP1 dephosphorylates I-1 phosphorylated on both Thr35 and Ser67 in the presence of Mn2+.
Discussion
Nonphosphorylated I-1 does not inhibit PP1. Upon Thr35 phosphorylation by A kinase, I-1 becomes a potent PP1 inhibitor (6, 7). Thr35 phosphorylation was suggested to be essential for I-1 inhibitory activity (3–7). However, we found that Ser67 phosphorylated I-1 by NCLK inhibits PP1, and NCLK-phosphorylated I-1(T35A) is an excellent PP1 inhibitor (Fig. 3). These observations indicate that Ser67-phosphorylated I-1 inhibits PP1, and I-1 is activated independently by phosphorylation on two different sites, Thr35 and Ser67.
Aitken et al. (6) found that I-1 is phosphorylated in vivo on Ser67 in addition to Thr35. These authors did not directly examine the effect of Ser67 phosphorylation. However, on the basis of their observation that among several cyanogen bromide-cleaved I-1 peptides only CB1, which contained I-1 residues 2–66, was fully active upon A kinase phosphorylation, they suggested that Ser67 phosphorylation does not appear to regulate I-1 function (6).
In this study using bacterially expressed recombinant I-1, we have determined that Ser67 phosphorylation activates I-1 in a manner similar to Thr35 phosphorylation, contradicting the suggestion of Aitken et al. (6). Consistent with our data, Huang and Glinsmann (16), who originally described I-1, showed that I-1 exists as phosphorylated active and nonphosphorylated inactive forms and only the nonphosphorylated inactive form requires A kinase phosphorylation to become active. Because Aitken et al. determined that I-1 when isolated from tissues is completely dephosphorylated on Thr35 (6) and ≈50% phosphorylated on Ser67 (1, 6), the phosphorylated active form described by Huang and Glinsmann (16) is likely to be Ser67-phosphorylated I-1. Furthermore, the observation of Aitken et al. (6) that only CB1 peptide inhibits PP1 is not surprising and does not rule out the possibility that Ser67 phosphorylation activates I-1. Now it is well established that in addition to Thr35, the N-terminal region of I-1 residues 8–12 containing the PP1 binding motif RKIQF is essential for I-1 function (refs. 7 and 17; see below). A cyanogen bromide cleavage of I-1 will yield several peptides that will include so-called CB1 containing The35 and an octamer, residues 67–74, containing Ser67. Only CB1 contains the RKIQF sequence and therefore is likely to show PP1-inhibitory activity.
The PP1 active site consists of a catalytic cavity with a substrate-binding acidic groove and a hydrophobic groove (17). The phosphothreonine/serine of the substrate occupy the PP1 catalytic cleft. The basic and hydrophobic residues that flank the phosphothreonine/serine of the substrate interact with the PP1 acidic and hydrophobic grooves (17). It was suggested that Thr35-phosphorylated I-1 is an active site inhibitor and binds to PP1 in a manner similar to a substrate blocking PP1–substrate interaction (17). In this work we found that Ser67-phosphorylated I-1 also inhibits PP1. More studies will be required to elucidate the biochemical mechanism of PP1 inhibition by Ser67-phosphorylated I-1. However, I-1 contains four basic residues, Arg69, Arg71, Lys72, and Lys73, located at the carboxyl region of Ser67 (6, 7). Similarly, Ser67 is preceded by two hydrophobic residues, Leu64 and Ala65. Ser67 has all of the structural requirements to be a PP1 active site inhibitor upon phosphorylation.
A proteolytic fragment containing I-1 residues 9–54 inhibited PP1 (18). It was suggested that the inhibitory region lies within the I-1 amino-terminal residues 9–54 (18). Studies using synthetic peptides have determined that within this I-1 amino-terminal region there are two PP1-interacting domains (7, 19–21). The first domain consists of residues 8–12 with sequence RKIQF found in many PP1-binding proteins (3) that bind to PP1 on sites remote from the catalytic center (22). The second domain, the Thr35 domain, occupies the PP1 catalytic center upon phosphorylation of Thr35 and blocks substrate binding (17). Both domains are suggested to be essential and to act in concert for PP1 inhibition (7, 19–21).
We do not know the mechanism of PP1 inhibition by Ser67-phosphorylated I-1. However, as discussed above, the amino acid sequence (7) suggests that Ser67-phosphorylated Ser67 domain may act as a PP1 active site inhibitor. Thus, the mechanisms of PP1 inhibition by the Thr35 domain and by the Ser67 domain may be similar. If so, like the Thr35 domain (7, 19–21), the Ser67 domain may also require a 8RKIQF12 domain for PP1 inhibition.
I-1 is activated by many cellular events that elevate cAMP (4, 23–25), and activation is thought to be through A kinase I-1 phosphorylation (1–4, 6, 7). In this study we found that NCLK phosphorylation also activates I-1. Therefore, the intracellular events that activate NCLK will also activate I-1. Thus, A kinase is not the sole I-1 regulator. Recent findings suggest that there are intracellular cAMP receptors other than A kinase, and these receptors mediate cell signaling in response to cAMP (26, 27). It will be of interest to find out whether NCLK participates in any of these events that activate I-1. After all, the catalytic cdk5 subunit of NCLK is widely expressed in various tissues and cell lines (28–30).
Acknowledgments
We express our sincere appreciation to Dr. Shirish Shenolikar of Duke University for providing I-1 and I-1(T35A) cDNA clones, Dr. Ernest Y. C. Lee of New York Medical College for the gift of purified recombinant PP1α, and Patrick W. Cafferty for helping in writing this manuscript. This work was supported by grants from the Medical Research Council of Canada and the American Health Assistance Foundation (U.S.A.). H.K.P. is the recipient of a Fraser Monat and McPherson Scholarship from McGill University.
Abbreviations
- A kinase
cAMP-dependent protein kinase
- GST
glutathione S-transferase
- I-1
inhibitor 1
- MAP kinase
mitogen-activated protein kinase
- NCLK
neuronal cdc2-like protein kinase
- PP1
protein phosphatase 1
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100460897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100460897
References
- 1.Cohen P. Annu Rev Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 2.Mumby M C, Walter G. Physiol Rev. 1993;73:673–699. doi: 10.1152/physrev.1993.73.4.673. [DOI] [PubMed] [Google Scholar]
- 3.Lee E Y C, Zhang L, Zhao S, Wei Q, Zhang J, Qi Z Q, Belmonte E R. Frontiers Biosci. 1999;4:270–285. doi: 10.2741/lee. [DOI] [PubMed] [Google Scholar]
- 4.Oliver C J, Shenolikar S. Frontiers Biosci. 1998;3:961–972. doi: 10.2741/a336. [DOI] [PubMed] [Google Scholar]
- 5.Shenolikar S. Annu Rev Cell Biol. 1994;10:55–86. doi: 10.1146/annurev.cb.10.110194.000415. [DOI] [PubMed] [Google Scholar]
- 6.Aitken A, Bilham T, Cohen P. Eur J Biochem. 1982;126:235–246. doi: 10.1111/j.1432-1033.1982.tb06771.x. [DOI] [PubMed] [Google Scholar]
- 7.Endo S, Zhou X, Connor J, Wang B, Shenolikar S. Biochemistry. 1996;35:5220–5228. doi: 10.1021/bi952940f. [DOI] [PubMed] [Google Scholar]
- 8.Sarker G, Sommer S S. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 9.Paudel H K, Lew J, Ali Z, Wang J H. J Biol Chem. 1993;268:23512–23518. [PubMed] [Google Scholar]
- 10.Paudel H K. J Biol Chem. 1997;272:1777–1785. [PubMed] [Google Scholar]
- 11.Roskoski R., Jr Methods Enzymol. 1983;99:3–6. doi: 10.1016/0076-6879(83)99034-1. [DOI] [PubMed] [Google Scholar]
- 12.Klein P S, Melton D A. Proc Natl Acad Sci USA. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lew J, Winkfein R J, Paudel H K, Wang J H. J Biol Chem. 1992;267:25922–25926. [PubMed] [Google Scholar]
- 14.Vesely J, Havlicek L, Strand M, Blow J J, Donella-Deana A, Pinna L, Letham D S, Kato J-Y, Detivaud L, Leclerc S, Meijer L. Eur J Biochem. 1994;224:771–786. doi: 10.1111/j.1432-1033.1994.00771.x. [DOI] [PubMed] [Google Scholar]
- 15.Foulkes J G, Strada S J, Henderson P J F, Cohen P. Eur J Biochem. 1983;132:309–313. doi: 10.1111/j.1432-1033.1983.tb07363.x. [DOI] [PubMed] [Google Scholar]
- 16.Huang F L, Glinsmann W H. Eur J Biochem. 1976;70:419–426. doi: 10.1111/j.1432-1033.1976.tb11032.x. [DOI] [PubMed] [Google Scholar]
- 17.Goldberg J, Huang H-b, Kwon Y-G, Greengard P, Nairn A C, Kuriyan J. Nature (London) 1995;376:745–753. doi: 10.1038/376745a0. [DOI] [PubMed] [Google Scholar]
- 18.Aitken A, Cohen P. FEBS Lett. 1984;147:54–58. doi: 10.1016/0014-5793(82)81010-7. [DOI] [PubMed] [Google Scholar]
- 19.Kwon Y-G, Huang H-B, Desdouits F, Girault J-A, Greengard P, Nairn A C. Proc Natl Acad Sci USA. 1997;94:3536–3541. doi: 10.1073/pnas.94.8.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hemmings H C, Jr, Nairn A C, Elliott J I, Greengard P. J Biol Chem. 1990;265:20369–20376. [PubMed] [Google Scholar]
- 21.Huang H-b, Horiuchi A, Watanabe T, Shih S-R, Tsay H-J, Li H-C, Greengard P, Nairn A C. J Biol Chem. 1999;274:7870–7878. doi: 10.1074/jbc.274.12.7870. [DOI] [PubMed] [Google Scholar]
- 22.Egloff M-P, Johnson D F, Moorhead G, Cohen P T W, Cohen P, Barford D. EMBO J. 1997;16:1876–1887. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann J, Gupta R C, Schmitz W, Scholz H, Nairn A C, Watanabe A M. Circ Res. 1991;69:1450–1457. doi: 10.1161/01.res.69.6.1450. [DOI] [PubMed] [Google Scholar]
- 24.Gupta R C, Neumann J, Watanbe A M, Lesch M, Sabbah H N. Am J Physiol. 1996;270:1159–1164. doi: 10.1152/ajpheart.1996.270.4.H1159. [DOI] [PubMed] [Google Scholar]
- 25.Mulkey R M, Endo S, Shenolikar S, Malenka R C. Nature. 1994;369:486–488. doi: 10.1038/369486a0. [DOI] [PubMed] [Google Scholar]
- 26.Kawasaki H, Springett G M, Mochizuki N, Toki S, Nakaya M, Matsuda M, Housman D E, Graybiel A M. Science. 1998;282:2275–2279. doi: 10.1126/science.282.5397.2275. [DOI] [PubMed] [Google Scholar]
- 27.de Rooij J, Zwartkruis F J T, Verheijen M H G, Cool R H, Nijman S M B, Wittinghofer A, Bos J L. Nature (London) 1998;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 28.Meyerson M, Enders G H, Wu C-L, Su L-K, Gorka C, Nelson C, Harlow E, Tsai L-H. EMBO J. 1992;11:2909–2917. doi: 10.1002/j.1460-2075.1992.tb05360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Philpott A, Porro E B, Kirschner M W, Tsai L-H. Genes Dev. 1997;11:1409–1421. doi: 10.1101/gad.11.11.1409. [DOI] [PubMed] [Google Scholar]
- 30.Lazaro J-B, Kitzmann M, Poul M-A, Vandromme M, Lamb N J C, Fernandez A. J Cell Sci. 1997;110:1251–1260. doi: 10.1242/jcs.110.10.1251. [DOI] [PubMed] [Google Scholar]