Abstract
Plant defense against insect herbivores is mediated in part by enzymes that impair digestive processes in the insect gut. Little is known about the evolutionary origins of these enzymes, their distribution in the plant kingdom, or the mechanisms by which they act in the protease-rich environment of the animal digestive tract. One example of such an enzyme is threonine (Thr) deaminase (TD), which in tomato (Solanum lycopersicum) serves a dual role in isoleucine (Ile) biosynthesis in planta and Thr degradation in the insect midgut. Here, we report that tomato uses different TD isozymes to perform these functions. Whereas the constitutively expressed TD1 has a housekeeping role in Ile biosynthesis, expression of TD2 in leaves is activated by the jasmonate signaling pathway in response to herbivore attack. Ingestion of tomato foliage by specialist (Manduca sexta) and generalist (Trichoplusia ni) insect herbivores triggered proteolytic removal of TD2's C-terminal regulatory domain, resulting in an enzyme that degrades Thr without being inhibited through feedback by Ile. This processed form (pTD2) of TD2 accumulated to high levels in the insect midgut and feces (frass). Purified pTD2 exhibited biochemical properties that are consistent with a postingestive role in defense. Shotgun proteomic analysis of frass from tomato-reared M. sexta identified pTD2 as one of the most abundant proteins in the excrement. Among the other tomato proteins identified were several jasmonate-inducible proteins that have a known or proposed role in anti-insect defense. Subtilisin-like proteases and other pathogenesis-related proteins, as well as proteins of unknown function, were also cataloged. We conclude that proteomic analysis of frass from insect herbivores provides a robust experimental approach to identify hyperstable plant proteins that serve important roles in defense.
The optimal growth of leaf-eating insects depends on their ability to acquire essential amino acids from dietary protein. The low protein content of plant tissue, however, poses a major nutritional challenge to phytophagous insects; protein is both the major macronutrient and the most commonly limiting nutrient for insect growth (Mattson, 1980; Bernays and Chapman, 1994). In addition to factors affecting protein quantity, evidence indicates that dietary protein quality also has a significant impact on plant-insect relations (Broadway and Duffey, 1988; Felton, 1996). Insect diets containing nutritionally unbalanced proteins pose a serious impediment to herbivory and may also influence patterns of host plant utilization among insect herbivores (Moran and Hamilton, 1980; Karowe and Martin, 1989; Haukioja et al., 1991; Berenbaum, 1995). The idea that variation in protein quality has evolved as a plant defense is supported by studies showing that certain classes of allelochemicals, such as tannins and phenolic resins, impair herbivore performance by interfering with the digestibility of dietary protein (Feeny, 1976; Rhoades and Cates, 1976).
Plants also produce defensive proteins that disrupt nutrient acquisition and other aspects of insect digestive physiology. Proteinase inhibitors (PIs) that impair the activity of digestive proteases are perhaps the best example of this type of postingestive defense (Green and Ryan, 1972; Ryan, 1990). Because PIs are not catalytic, their capacity to slow herbivore growth is dependent on accumulation to relatively high concentrations inside the gut lumen. Enzymes have the potential to exert defensive effects at much lower concentrations, but this hypothesis has received relatively little attention until recently (Duffey and Stout, 1996; Felton, 1996; Chen et al., 2005; Felton, 2005). Research on midgut-active plant enzymes has focused mainly on polyphenol oxidase and other oxidative enzymes that covalently modify dietary protein, thus reducing the digestibility of plant food (Constabel et al., 1995; Duffey and Stout, 1996; Felton, 1996; Wang and Constabel, 2004). Other defensive proteins directly target structural components of the insect digestive apparatus. Members of the Cys protease family of enzymes, for example, are thought to disrupt the integrity of the peritrophic membrane that protects the gut epithelium (Pechan et al., 2002; Konno et al., 2004; Mohan et al., 2006). These collective studies indicate that enzymes play a pivotal role in host plant defense and thus broaden the traditional view that secondary metabolites are the major determinants of host plant utilization and specialization (Fraenkel, 1959; Berenbaum, 1995).
Many plant anti-insect proteins are synthesized in response to wounding and herbivore attack. Induced expression of the vast majority of these proteins is regulated by the jasmonate signaling pathway (Walling, 2000; Gatehouse, 2002; Kessler and Baldwin, 2002; Howe, 2004; Schilmiller and Howe, 2005). Examples of jasmonate-inducible proteins (JIPs) that have a confirmed or proposed role in postingestive defense include polyphenol oxidase, arginase, Leu amino peptidase A (LAP-A), lipoxygenase, and a battery of PIs (Duffey and Felton, 1991; Felton et al., 1994; Constabel et al., 1995; Felton, 1996; Chen et al., 2005; Walling, 2006; Lison et al., 2006). A jasmonic acid (JA)-inducible acid phosphatase (VSP2) in Arabidopsis (Arabidopsis thaliana) was recently shown to exert insecticidal activity against coleopteran and dipteran insects (Liu et al., 2005). These observations indicate that a primary function of the jasmonate signaling pathway is to promote the expression of proteins that act postingestively to impair the growth and development of insect herbivores (Chen et al., 2005).
Biosynthetic Thr deaminase (TD) is a pyridoxal phosphate-dependent enzyme that converts l-Thr to α-ketobutyrate and ammonia. Plant TDs function in the chloroplast to catalyze the committed step in the biosynthesis of Ile. The enzyme contains an N-terminal catalytic domain and a C-terminal regulatory domain and is subject to negative feedback regulation by Ile (Gallagher et al., 1998). The physiological importance of TD in plant growth and development was demonstrated by studies of TD-deficient mutants of Nicotiana plumbaginifolia and, more recently, Nicotiana attenuata (Sidorov et al., 1981; Colau et al., 1987; Kang et al., 2006). TD expression in leaves of several solanaceous plants is massively induced by the jasmonate signaling pathway in response to wounding and herbivory (Hildmann et al., 1992; Samach et al., 1995; Hermsmeier et al., 2001; Li et al., 2004). In contrast to this expression pattern, TD is constitutively expressed to high levels in reproductive organs (Hildmann et al., 1992; Kang and Baldwin, 2006). TD is reported to be the most abundant protein in tomato (Solanum lycopersicum) flowers (Samach et al., 1991). The high level of TD expression in reproductive tissues is similar to the expression pattern of PIs and other JIPs that impair insect growth.
Direct evidence for the hypothesis that TD has a role in anti-insect defense came initially from studies showing that the enzyme accumulates in the midgut of tomato-reared Manduca sexta larvae (Chen et al., 2005). TD activity in the midgut was correlated with reduced levels of free Thr, which is a dietary requirement for phytophagous insects. A jasmonate-insensitive mutant (jai1) of tomato that fails to express TD is more susceptible to attack by M. sexta larvae. Because this mutant is defective in all jasmonate-signaled processes, however, decreased resistance of jai1 plants could not be linked directly to loss of TD function (Li et al., 2004; Chen et al., 2005). A recent study by Kang et al. (2006) showed that mutants of N. attenuata engineered specifically for TD deficiency are compromised in resistance to M. sexta larvae. Supplementation of N. attenuata leaves with Thr led to increased larval performance, indicating that Thr availability in the leaf diet is limiting for larval growth. The Ile deficiency in TD-silenced N. attenuata plants also resulted in decreased production of jasmonoyl-Ile (JA-Ile), which is an important signal for induced defense responses to pathogens (Staswick et al., 1998) and insects (Kang et al., 2006). Thus, TD's defensive function in N. attenuata was attributed both to its involvement in JA-Ile synthesis and its role in postingestive defense (Kang et al., 2006).
TD's dual function in primary metabolism and defense makes it an attractive subject for research aimed at understanding the evolutionary origins of plant enzymes that exert toxic or antinutritional effects on insect herbivores. Here, we show that tomato has two TD genes (designated TD1 and TD2) whose differential expression pattern is consistent with functional divergence of the two isoforms. Second, we show that ingestion of tomato foliage by specialist and generalist herbivores triggers proteolytic removal of the TD2 regulatory domain, resulting in an enzyme (processed TD2 [pTD2]) that effectively degrades Thr in the lepidopteran gut. Third, we show that the biochemical properties of pTD2 are consistent with a postingestive role in defense. Finally, we employed a shotgun proteomic approach to demonstrate that pTD2 is one of the most abundant proteins in frass from tomato-reared M. sexta larvae. Nineteen additional tomato proteins were cataloged in M. sexta frass. Among these were JIPs that have a known role in defense against insect herbivores, pathogenesis-related (PR) proteins, and proteins of unknown function. These findings provide insight into the evolution of plant anti-insect proteins and establish a robust experimental approach to identify hyperstable proteins that serve important roles in plant protection against biotic stress.
RESULTS
Tomato Has Two TD Genes That Are Differentially Expressed
Previous studies of TD-encoding genes in tomato and potato (Solanum tuberosum) have focused on a single orthologous gene whose expression in leaves is induced by various stress conditions, including wounding and jasmonate treatment (Samach et al., 1991, 1995; Hildmann et al., 1992; Schaller et al., 1995; Strassner et al., 2002; Li et al., 2004). We previously reported that the jai1 mutant of tomato, which is defective in all known jasmonate responses as a consequence of a null mutation in Coi1, fails to express this TD gene, but nevertheless does not exhibit symptoms (e.g. stunted growth) of Ile deficiency (Li et al., 2004). This observation led us to test the hypothesis that tomato uses a different TD isozyme for Ile biosynthesis. Indeed, searches of the tomato expressed sequence tag (EST) database provided evidence for a second expressed TD gene. The corresponding full-length cDNA is predicted to encode a 606-amino acid protein with a molecular mass of 66,182 D (Fig. 1A). The recombinant protein expressed in Escherichia coli converted Thr to α-ketobutyrate in a manner that was inhibited by exogenous Ile (data not shown), indicating that the enzyme is an authentic TD. For reasons explained below, we henceforth refer to this previously uncharacterized gene as SlTD1 and refer to the JA-inducible gene initially described by Samach et al. (1991) as SlTD2.
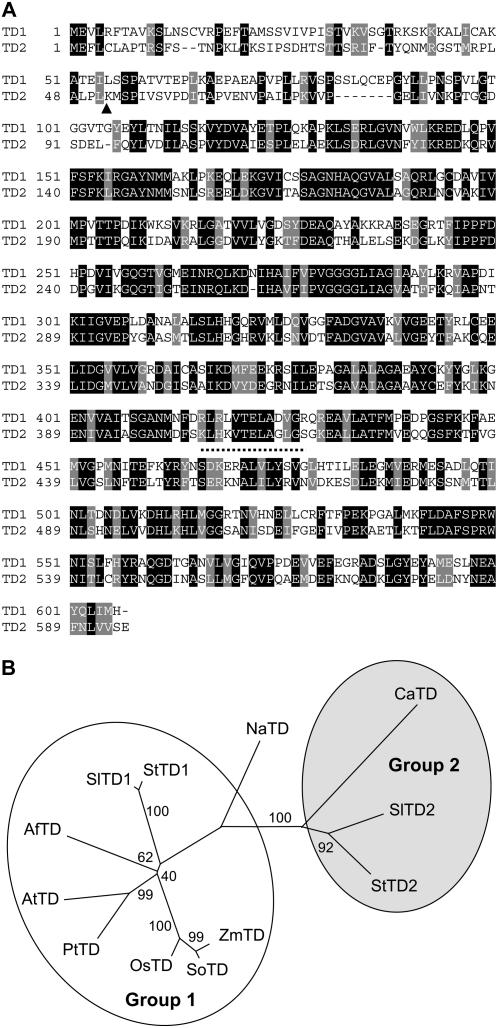
Figure 1.
Tomato has two distinct TD isoforms. A, Comparison of the deduced amino acid sequence of SlTD1 (TD1) and SlTD2 (TD2). Amino acids that are either identical (black) or similar (gray) between the two sequences are indicated. The inverted triangle denotes the site of cleavage (between Leu-51 and Lys-52) of the plastid-targeting peptide on TD2 (Samach et al., 1991). The dotted line denotes the neck region that connects the N-terminal catalytic and C-terminal regulatory domains, based on homology modeling with E. coli TD (Gallagher et al., 1998). B, Phylogenetic relationship of SlTD1 and SlTD2 to TDs from other plants. Shown is an unrooted neighbor-joining tree constructed with MEGA 3.1 (Kumar et al., 2004) from the following sequences: maize (Zea mays; ZmTD; CO446428); sugarcane (Saccharum officinarum; SoTD; CA208490); rice (OsTD; NP_001051069); poplar (PtTD; estExt_fgenesh4_pg.C_280257); Arabidopsis (AtTD; NP_187616); Aquilegia formosa × pubescens (AfTD; DT735861); tomato (SlTD1; ABK20067) (SlTD2; P25306); potato (StTD1; BI436101) (StTD2; X67846); N. attenuata (NaTD; AAX22214); chickpea (CaTD; Q39469). GenBank accession numbers are given in parentheses. The indicated bootstrap values (% of 1,000 repeated tree reconstructions) show the reliability of each branch of the inferred tree. The two major subgroups of the tree are designated group 1 and group 2.
SlTD1 and SlTD2 share 48% amino acid sequence identity (Fig. 1A). Both proteins contain a predicted chloroplast-targeting sequence, as well as canonical catalytic and regulatory domains found in other plant and bacterial TDs. Phylogenetic analysis showed that plant TD sequences cluster into two major groups (groups 1 and 2; Fig. 1B). SlTD1 was more similar to TDs from Arabidopsis (66% identity), poplar (Populus spp.; 68% identity), and rice (Oryza sativa; 69% identity) than it was to SlTD2. Because Arabidopsis, poplar, and rice each contain a single TD gene, this finding supports the idea that SlTD1 performs a housekeeping role in Ile biosynthesis. JA-inducible isozymes from tomato (SlTD2) and potato (StTD2) comprised a distinct subgroup of proteins that, interestingly, were closely related to a TD sequence from chickpea (Cicer arietinum). The JA-inducible TD from N. attenuata, which has a dual role in Ile biosynthesis and postingestive defense (Kang et al., 2006), occupied an intermediate position in the phylogeny and thus was not assigned to either group.
We used RNA-blot analysis to compare the developmental and stress-induced expression patterns of SlTD1 and SlTD2. SlTD1 was constitutively expressed in all tissues examined (Fig. 2A). In contrast, SlTD2 transcripts accumulated to very high levels in immature buds and unopened flowers, but were not detected in unstressed leaves and other vegetative tissues. Expression of SlTD2 in leaves was massively induced in response to methyl jasmonate (MeJA) application, as previously reported (Hildmann et al., 1992; Samach et al., 1995; Li et al., 2004; Fig. 2B). MeJA had little or no effect on SlTD1 transcript levels. SlTD2 expression was induced locally and systemically in response to mechanical wounding, whereas SlTD1 mRNA levels were not affected by wounding (Fig. 2C). These findings support the hypothesis that SlTD1 and SlTD2 serve distinct physiological roles.
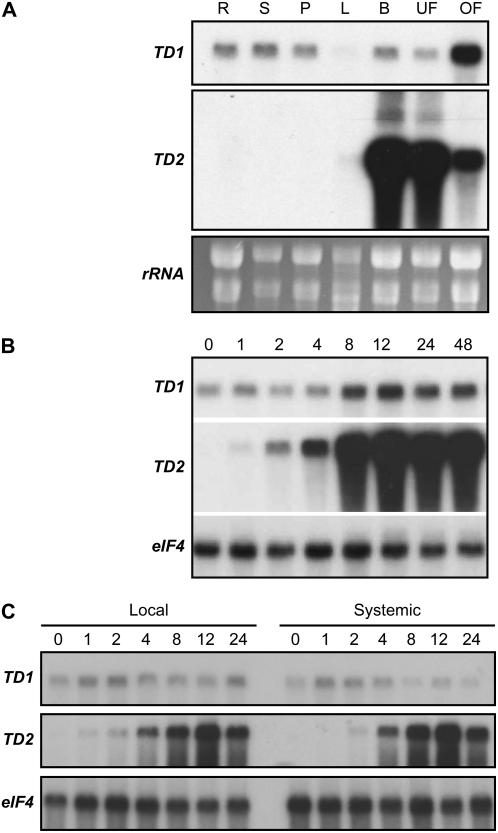
Figure 2.
Differential expression of SlTD1 and SlTD2 in tomato. A, Five micrograms of total RNA from root (R), stem (S), petiole (P), leaf (L), immature flower bud (B), unopened flower (UF), and opened flower (OF) were immobilized to a membrane and hybridized to full-length cDNA probes for SlTD1 and SlTD2. A duplicate blot (bottom) was stained with ethidium bromide to visualize rRNA as a means to determine the quality and quantity of the loaded RNA. B, Expression of SlTD1 and SlTD2 in response to treatment with MeJA. Four-week-old plants were exposed to MeJA vapor for the indicated length of time (h) in an enclosed box, after which leaves were harvested for RNA extraction. RNA isolated from untreated plants (0-h time point) was analyzed as a control. RNA gel blots were hybridized to cDNA probes for SlTD1 and SlTD2. Blots were also hybridized to an eIF4A probe as a loading control. C, Expression of SlTD1 and SlTD2 in response to mechanical wounding. Leaflets on the second and third fully expanded leaves (counted from the oldest leaf) of 4-week-old plants were wounded three times with a hemostat, perpendicular to the main vein. Total RNA was isolated separately from the lower wounded (local) and the upper unwounded (systemic) leaves at various times (h) after wounding. RNA was also isolated from unwounded plants (0 h time point) as a control. RNA gel blots were hybridized to cDNA probes for SlTD1 and SlTD2. Blots were also hybridized to an eIF4A probe as a loading control.
Digestion of Bulk Tomato Leaf Protein in the Gut Lumen of M. sexta Larvae
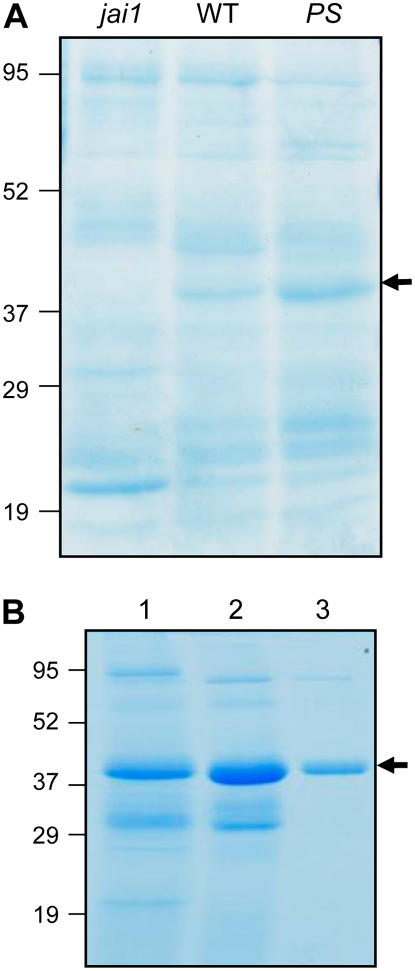
As a prelude to studying the fate of TD2 in the M. sexta digestive system, we used SDS-PAGE to qualitatively assess changes in bulk tomato leaf protein during passage through the insect. These studies were facilitated by analysis of caterpillars raised either on wild-type plants or on mutants that are affected in the JA signaling pathway. These mutants included a transgenic line (35S∷PS) that constitutively expresses high levels of JIPs (McGurl et al., 1994; Bergey et al., 1996), and the jai1 mutant that fails to express TD2 and other JIPs (Li et al., 2004). A phenol-based protein extraction procedure was used to isolate total protein from three sources: (1) insect-damaged leaves; (2) midgut content from actively feeding larvae; and (3) larval feces (i.e. frass). The polypeptide profile of total leaf protein was much different from that of protein isolated from midgut content or frass (Fig. 3A). One conspicuous difference was the large subunit (rbcL) of Rubisco. As the most abundant soluble protein in tomato leaves, rbcL is a major source of amino acids for phytophagous insects and a convenient marker for bulk leaf protein (Sheen, 1991; Bernays and Chapman, 1994; Felton, 1996). In contrast to the high level of rbcL in herbivore-damaged leaves, midgut and frass contained very little intact rbcL. Efficient digestion of chloroplast proteins within the midgut was confirmed by western-blot analysis with antibodies against the chloroplast outer envelope protein Toc75 (Fig. 3B; Reumann et al., 2005). We also found that a peroxisomal matrix protein, acyl-CoA oxidase1A (Acx1A), accumulated in leaves, but not in the midgut content or frass (Fig. 3C). These results demonstrate that bulk tomato leaf protein is efficiently degraded during passage through the M. sexta digestive system.
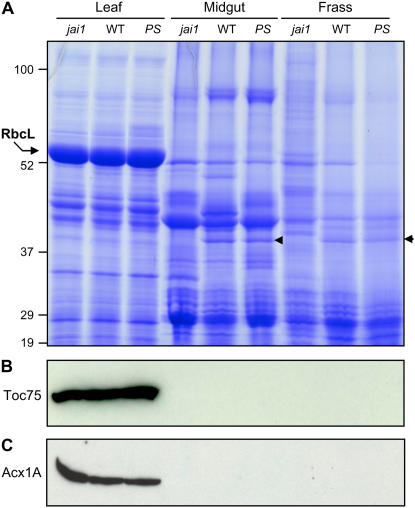
Figure 3.
Digestion of tomato foliar protein during passage through the M. sexta digestive tract. A, M. sexta larvae were reared to the fourth instar on jai1, wild-type (WT), or 35S∷PS (PS) tomato plants. Total protein was extracted from three different sources of material: tomato leaves that were heavily damaged by the herbivore (leaf); the midgut content of actively feeding, fourth instar larvae (midgut); and fecal droppings from actively feeding fourth instar larvae (frass). Forty micrograms of protein from each sample were analyzed on a 10% to 18% polyacrylamide gradient gel, which was stained with Coomassie Blue. The position of protein standards (kD) is shown on the left, as is the major polypeptide corresponding to rbcL. The arrowhead denotes a 40-kD polypeptide observed in midgut and frass of larvae reared on wild-type and 35S∷PS plants. B and C, Western-blot analysis of the protein samples shown in A with polyclonal antibodies raised against the chloroplast outer envelope protein Toc75 (B) or the peroxisomal matrix protein Acx1A (C). [See online article for color version of this figure.]
Numerous discretely sized polypeptides exhibiting a wide range of molecular masses were present in frass extracts (Fig. 3A). The polypeptide profile of frass from larvae reared on the various genotypes exhibited several reproducible differences. For example, frass extracts from larvae grown on jai1 plants contained more discretely sized, higher molecular mass polypeptides in comparison to the wild type and 35S∷PS frass samples (Fig. 3A). A second host genotype-specific difference was a protein of approximately 40 kD that accumulated in wild-type and 35S∷PS frass, but not in frass from jai1-reared larvae (Fig. 3A, arrow). Differential accumulation of this protein was also observed in the midgut content, but not in herbivore-damaged leaves. This pattern of accumulation suggests that the 40-kD polypeptide is a JIP that is stable in the M. sexta gut.
Proteolytic Processing of TD2 in the Lepidopteran Gut
We previously reported mass spectrometry (MS) evidence indicating that a form of TD2 lacking the enzyme's regulatory domain accumulates in M. sexta midgut content and frass (Chen et al., 2005). The predicted size of this TD2 variant was consistent with its identity as the above-mentioned 40-kD protein (Fig. 3A). To test this hypothesis, a gel slice containing the 40-kD protein was digested with trypsin and the resulting peptides were sequenced by liquid chromatography (LC)-MS/MS. Among 18 unique peptides that were confidently identified (P < 0.05), all showed an exact match to the catalytic domain of TD2 (Fig. 4A). The predicted molecular mass of the protein defined by LC-MS/MS (Met-52 to Lys-418) was 38,890 D, which was in good agreement with the size observed by SDS-PAGE.
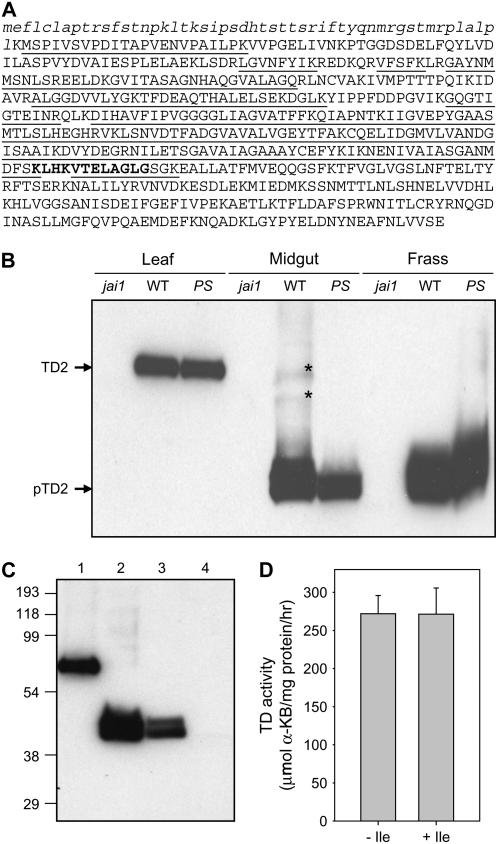
Figure 4.
Proteolytic processing of TD2 in the digestive tract of M. sexta and T. ni larvae. A, MS-based identification of a truncated form (pTD2) of TD2 in M. sexta frass. Underlined letters denote the amino acid sequence of TD2 that was identified by LC-MS/MS analysis of a gel slice containing the 40-kD protein. The chloroplast-targeting peptide of TD2 is denoted by lowercase italicized letters. The neck region that connects the N-terminal catalytic and C-terminal regulatory domains is indicated by bold letters. B, Proteolytic processing of TD2 in M. sexta. Protein (10 μg/lane) isolated from tomato leaf, M. sexta midgut content, and M. sexta frass was separated by SDS-PAGE. The gel was subjected to western-blot analysis with an anti-TD2 antibody. See Figure 3A for a description of the samples. The cross-reacting polypeptide labeled TD2 corresponds to the 55-kD mature form of the protein that accumulates in herbivore-damaged leaves. The polypeptide labeled pTD2 is the proteolytically processed form of TD2 that lacks the C-terminal regulatory domain. Asterisks denote faint bands corresponding to incompletely processed TD2. C, Proteolytic processing of TD2 in the digestive tract of T. ni larvae. Protein was analyzed by western-blot analysis as described in B. Protein was isolated from the following material: lane 1, herbivore-damaged wild-type leaves; lane 2, frass from M. sexta larvae reared on wild-type plants; lane 3, frass from T. ni larvae reared on wild-type plants; lane 4, frass from T. ni larvae reared on jai1 plants. D, TD activity in frass from T. ni larvae reared on wild-type tomato plants. Frass extracts were assayed for TD activity in the absence or presence of 10 mm Ile. Data indicate the mean and sd of measurements from four different pools of frass.
Mature TD2 isolated from tomato tissues has an apparent molecular mass of 55 kD (Samach et al., 1991, 1995). We used western-blot analysis to determine whether there is a product-precursor relationship between the truncated TD2 variant, designated pTD2, and the 55-kD protein. Anti-TD2 antibodies cross-reacted with an approximately 55-kD protein in herbivore-damaged wild-type and 35S∷PS leaves (Fig. 4B). The absence of this polypeptide in jai1 leaves, in which TD2 is not expressed, confirmed the specificity of the antibody for TD2. In contrast to leaf tissue, pTD2 was the predominant form of the protein in midgut and frass extracts. These results indicate that TD2 is proteolytically processed to pTD2 following ingestion of foliage by M. sexta. A small amount of unprocessed TD2 in midgut extracts was observed upon prolonged development of western blots (Fig. 4B, asterisks). Shorter exposure times showed that pTD2 migrates as a doublet, suggesting heterogeneity in the size of the processed protein.
M. sexta is highly specialized for feeding on tomato and other solanaceous plants. To determine whether proteolytic processing of TD2 occurs in the gut lumen of a generalist herbivore, we analyzed the TD2 content in frass from Trichoplusia ni (cabbage looper) caterpillars that were raised on tomato foliage. Western-blot analysis showed that T. ni frass contained a form of TD2 that comigrated with pTD2 from M. sexta frass (Fig. 4C). The absence of this polypeptide in frass from jai1-reared T. ni larvae confirmed that the cross-reacting protein is derived from TD2. TD activity was detected in frass from T. ni larvae grown on wild-type plants (Fig. 4D). Consistent with a processing event that removes the regulatory domain, this activity was insensitive to feedback inhibition by 10 mm Ile. We conclude that ingestion of tomato foliage by both specialist (M. sexta) and generalist (T. ni) insect herbivores results in proteolytic removal of the regulatory domain of TD2.
Biochemical Properties of pTD2
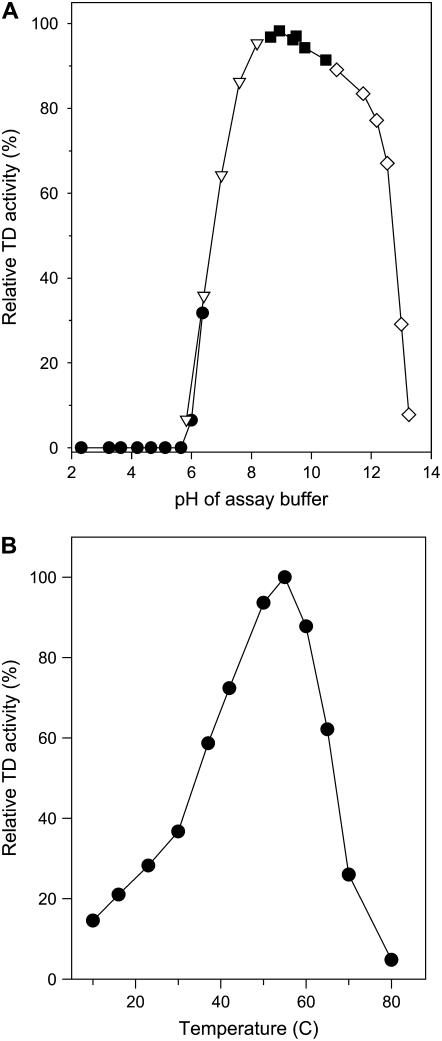
To investigate the biochemical properties of pTD2 in more detail, we purified the enzyme from M. sexta frass. An aqueous buffer system effectively extracted active pTD2 from frass (Fig. 5A). The 40-kD polypeptide copurified with Ile-insensitive TD activity during subsequent purification steps (data not shown). Following the final stage of purification by gel filtration chromatography, we estimated that pTD2 was at least 90% pure as determined by SDS-PAGE (Fig. 5B). The purified enzyme was active against l-Thr and l-Ser. Kinetic analysis showed that the apparent Km of l-Thr and l-Ser was 2.3 and 3.0 mm, respectively. The Vmax for l-Thr was approximately 5,000 μmol mg protein−1 h−1, which was about 1.5 times higher than the Vmax for l-Ser. The enzyme was highly active in an alkaline pH range that matches that of the lepidopteran midgut; little or no activity was observed at pH values below 6.0 (Fig. 6A). pTD2 was also active over a wide range of temperatures. Optimal enzyme activity against l-Thr was observed at 58°C (Fig. 6B).
Figure 5.
Purification of pTD2 from M. sexta frass. A, Frass pellets collected from M. sexta larvae grown on the indicated host plant genotype were extracted with an aqueous buffer to maintain TD activity. The resulting protein (approximately 60 μg) was separated by SDS-PAGE, and the gel was stained with Coomassie Blue. The arrow indicates pTD2, which accumulates in frass from larvae grown on wild-type and 35S∷PS plants, but not in frass from jai1-reared larvae. Migration position of molecular mass markers (kD) are shown on the left. B, Frass from M. sexta larvae grown on wild-type tomato foliage was used as the starting material for purification of pTD2. The Coomassie-stained gel shows pTD2 at various steps of the purification procedure: lane 1, 65% (NH4)2SO4 cut; lane 2, pooled TD-containing fractions from DEAE-cellulose chromatography; lane 3, pooled TD-containing fractions from Superose-12 gel filtration chromatography. Migration position of molecular mass markers (kD) are shown on the left. The arrow indicates the polypeptide corresponding to pTD2. [See online article for color version of this figure.]
Figure 6.
Biochemical features of purified pTD2. A, pH optimum of pTD2 activity assayed against l-Thr in the following buffer systems: sodium citrate (black circles); NaPO4 (inverted triangles); Gly (black squares); and KH2PO4 (white diamonds). Data are expressed relative to the activity at pH 9.0. B, Effect of temperature on pTD2 activity. Enzyme activity was assayed against l-Thr at the indicated temperature (°C). Data are expressed relative to the activity at 58°C.
Identification of Plant Defensive Proteins by Shotgun Proteomic Analysis of Insect Frass
Excretion of pTD2 as an active enzyme from M. sexta and T. ni led us to hypothesize that insect frass may be a useful source of material in which to identify other defense-related proteins. To test this idea, we used a shotgun proteomic approach to catalog and quantify tomato proteins in frass from M. sexta caterpillars reared on tomato foliage. The total protein content of frass was digested with trypsin and the resulting peptide mixture was subjected to LC-MS/MS. Protein identifications were considered positive if at least two peptides derived from the same protein were confidently detected in searches of the MS/MS data against the tomato EST database. These stringent criteria resulted in identification of 20 distinct tomato proteins with probability scores of P < 10−4 (Table I; Supplemental Tables S1 and S2).
Table I.
Host plant proteins identified in frass of tomato-reared M. sexta larvae
| Protein Identity or Best BLAST Hita | Accession No. | Unigeneb | No. Peptides | Localc |
|---|---|---|---|---|
| JIP and Wound-Inducible Proteins | ||||
| pTD2 | AAA34171 | U312574 | 25 | CP |
| LAP-A | AAC49456 | U312377 | 23 | CP |
| Trypsin inhibitor-like protein | AAA80497 | U313384 | 4 | SP |
| CDI/chymotrypsin inhibitor 21 | CAC00536 | U312623 | 4 | SP |
| YjgF family proteind | BT013249 | U313029 | 5 | CP |
| Stress-induced LH2 domain protein | BI209796 | U315202 | 3 | SP |
| Aspartic protease inhibitor | BI929912 | U312622 | 2 | SP |
| GLP | CN384576 | U318102 | 2 | SP |
| PR Proteins | ||||
| P69B (PR-7) | CAA71234 | U313775 | 20 | SP |
| P69A (PR-7) | CAA64566 | U313772 | 7 | SP |
| Lignin-forming peroxidase (PR-9) | CAA50597 | U321126 | 10 | SP |
| β-1,3-Glucanase (PR-2) | CAA52872 | U314382 | 7 | SP |
| Endoglucanase inhibitor protein | AAN87262 | U314071 | 3 | SP |
| PR protein P2 (PR-4) | CAA41439 | U316008 | 2 | SP |
| Other Proteins | ||||
| Plastocyanin | CAA32121 | U312690 | 16 | CP |
| Malate dehydrogenase | AAU29198 | U313128 | 7 | MT |
| Ferredoxin | BI931178 | U312380 | 3 | CP |
| Superoxide dismutase | AAQ09007 | U315384 | 2 | CP |
| Carbonic anhydrase | AW093720 | U319550 | 2 | CP |
| Chlorophyll a/b-binding protein | CAA84525 | U312438 | 2 | CP |
In cases where the BLAST hit did not perfectly match a known tomato protein, the best BLAST hit is listed.
Unigene indicates the tomato gene nomenclature provided by the SOL Genomics Network at http://www.sgn.cornell.edu/index.pl.
Local denotes the predicted protein location. The presence of an N-terminal signal peptide (SP) for protein secretion was analyzed with SignalP software (http://www.cbs.dtu.dk/services/SignalP). Proteins having a signal peptide probability score P > 0.75 are indicated. CP, Chloroplast-targeted protein; MT, mitochondrial-targeted protein.
Annotated in the tomato EST database (http://compbio.dfci.harvard.edu/tgi/plant.html) as a protein translation inhibitor (Li et al., 2004).
Wound-inducible proteins and JIPs comprised the largest group of tomato proteins in M. sexta frass (Table I). Among the proteins previously implicated in defense against lepidopteran insects were TD2, LAP-A (Gu et al., 1999; Chen et al., 2005), cathepsin D inhibitor (CDI; Lison et al., 2006), and a germin-like protein (GLP) similar to a GLP isozyme from N. attenuata (Lou and Baldwin, 2006). Two stress-inducible proteins of unknown function were also identified. One of these is a member of a plant-specific group of stress-related proteins that contain a lipoxygenase homology (LH) domain (Coker et al., 2005). The second uncharacterized protein is a chloroplast-targeted member of the highly conserved YjgF family of proteins (Leitner-Dagan et al., 2006). We previously showed that the gene encoding this protein, which is annotated in the tomato EST database as a protein translation inhibitor, is regulated by the jasmonate signaling pathway (Li et al., 2004).
Proteins implicated in plant defense against pathogens were also identified in M. sexta frass (Table I). Among the PR proteins identified were the P69A and B members of the subtilisin-like family of endoproteases (PR-7; Tornero et al., 1996, 1997), β-1,3-glucanase (PR-2; Domingo et al., 1994), lignin-forming peroxidase (PR-9; Vera et al., 1993), and a hevein-like protein P2 (PR-4; Linthorst et al., 1991). A xyloglucan-specific fungal endoglucanase inhibitor protein previously reported from tomato, potato, and tobacco (Naqvi et al., 2005) was also identified. All of these proteins contain an N-terminal signal peptide for targeting to the secretory pathway (Table I) and most have been shown to be expressed in response to pathogen infection or wounding (Supplemental Table S1).
All other tomato proteins identified in M. sexta frass, with the exception of mitochondrial malate dehydrogenase, were chloroplastic metalloproteins (Table I). These included plastocyanin, ferredoxin, superoxide dismutase, and carbonic anhydrase. Given that TD2, LAP-A, and the YjgF-related protein (see above) are also plastid localized, it would appear that chloroplast proteins are highly represented in the frass. Failure to identify peptides corresponding to Rubisco in this experiment argues against the possibility that this phenomenon results from passage of the intact plastids through the insect digestive tract.
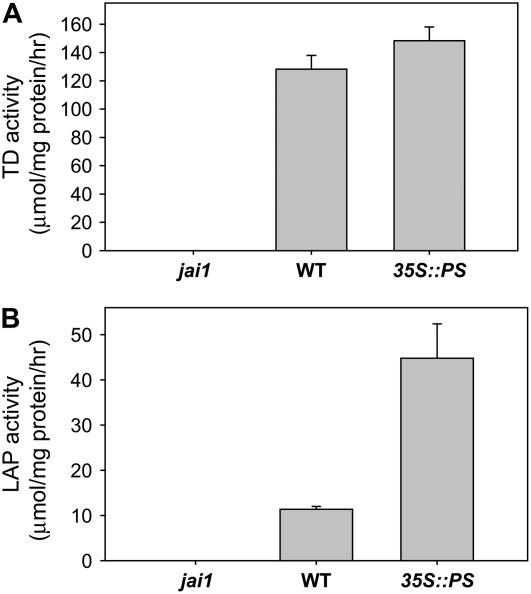
The number of mass spectral counts obtained for a given protein by LC-MS/MS provides a quantitative measure of the protein's abundance in the extract (Old et al., 2005; Gilchrist et al., 2006). Based on this information, pTD2 and LAP-A were among the most abundant tomato proteins in the frass (Table I). LAP activity assays were performed to determine whether LAP-A, like pTD2, is excreted as an active enzyme. Both LAP and TD activity was detected in frass from larvae grown on wild-type plants (Fig. 7). The lack of activity in frass from jai1-reared larvae indicated that the activity was specific for the JA-inducible isozymes LAP-A and pTD2. LAP activity in frass from insects reared on 35S∷PS plants was significantly greater than that in the wild-type frass, which is consistent with the fact that LAP-A expression in tomato foliage is up-regulated by systemin (Chao et al., 1999). These findings indicate that LAP-A, like TD2, is excreted from M. sexta as an active enzyme.
Figure 7.
LAP-A is excreted from M. sexta as an active enzyme. A, TD activity was measured in frass collected from M. sexta larvae grown on the indicated host plant genotype. Data represent the mean and sd of three independent measurements. B, LAP activity in the same extracts used in A. Data represent the mean and sd of three independent measurements.
DISCUSSION
Functional Diversification of Two TD Isoforms in Tomato
The role of TD in producing Ile for protein synthesis is essential for all aspects of plant growth and development. Ile is also required for the synthesis of JA-Ile, which is an important signal for activation of jasmonate-based defenses (Staswick et al., 1998; Kang et al., 2006). The broad distribution of jasmonates in the plant kingdom indicates that TD's participation in JA-Ile synthesis is likely conserved in all plants. In contrast, TD's function as a postingestive defense against insect herbivores appears to be restricted to certain plant lineages. Here, we provide evidence that tomato employs different TD isozymes to fulfill distinct roles in Ile biosynthesis in planta and postingestive defense. This contrasts with the situation in native tobacco, which uses a single TD isoform to perform both functions (Kang et al., 2006).
Several observations lead us to conclude that SlTD1 performs a role in Ile biosynthesis. First, the deduced amino acid sequence of SlTD1 is more similar to TDs in plants such as Arabidopsis, rice, and poplar, which all harbor a single housekeeping form of the enzyme, than it is to SlTD2. Second, constitutive expression of TD1 in all tissues is consistent with a general role in amino acid biosynthesis. Third, recombinant TD1 expressed in E. coli exhibits Ile-sensitive TD activity. Finally, the jai1-1 mutant, which lacks detectable TD2 expression in leaves (Fig. 4B), does not exhibit chlorosis or other signs of Ile deficiency (Li et al., 2004). This finding provides functional evidence that TD1 can produce Ile pools that are utilized for normal growth and development in the absence of TD2.
A specialized role for TD2 in postingestive defense is supported by the fact that this isozyme accumulates in the midgut and frass of tomato-reared caterpillars (Figs. 3 and 4). The gut-accumulating form of the enzyme (i.e. pTD2) has biochemical features that presumably facilitate its action in the midgut environment. These features include protease resistance, an alkaline pH optimum, and the capacity to degrade Thr in the presence of high concentrations of Ile. The high temperature optimum of pTD2 indicates that the enzyme would be active at elevated body temperatures, which for M. sexta caterpillars in natural field conditions can easily exceed 35°C (Casey, 1976). The expression pattern of TD2 also supports a role in anti-insect defense. TD2 is coordinately induced with other defensive genes in response to wounding and JA treatment (Hildmann et al., 1992; Samach et al., 1995; Li et al., 2004). In reproductive tissues, TD2 is expressed constitutively at extraordinarily high levels (Samach et al., 1991, 1995). Many other JA-regulated defensive proteins, including PIs, arginase, LAP-A, and AtVSP2, exhibit a similar expression pattern (Hildmann et al., 1992; Utsugi et al., 1998; Chao et al., 1999; Chen et al., 2004). These observations support the idea that accumulation of TD2 and other JIPs in floral tissues protects reproductive structures from insect herbivores. Induction of TD2 expression by diverse types of biotic and abiotic stress (Hildmann et al., 1992; Zhao et al., 2003) raises the possibility that the enzyme performs other physiological roles in planta. For example, it is possible that a proteolytically processed form of TD2 accounts for the biodegrative TD activity observed in senescing tomato leaves (Szamosi et al., 1993).
Functional divergence of two TD isozymes in tomato raises interesting questions about the evolutionary origins of plant TDs that participate in postingestive defense. It is reasonable to assume that SlTD2 arose from a gene duplication event and that selective pressure imposed by insect herbivores led to the evolution of this isoform as a defensive enzyme. A key feature acquired by both SlTD2 and N. attenuata TD during evolution was regulation via the jasmonate signaling pathway. Whether or not these enzymes evolved novel biochemical or structural features that enhance their ability to impair insect digestive physiology is unclear. Future studies aimed at comparing the structure, stability, and activity of SlTD1 and SlTD2 promise to provide insight into this question. The dual role of N. attenuata TD in Ile synthesis and postingestive defense (Kang et al., 2006) is consistent with the intermediate position of this protein in the TD phylogenetic tree (Fig. 1B). The evolution of N. attenuata TD as a midgut-active enzyme may be constrained, however, by its essential role in Ile biosynthesis. Tomato TD2 is presumably not subjected to such constraint and thus may be better adapted to function in the lepidopteran gut.
Proteolytic Processing of TD2
Our results confirm and extend previous evidence (Chen et al., 2005) indicating that TD2 is proteolytically processed following ingestion of foliage by the herbivore. LC-MS/MS and other biochemical data demonstrated that the midgut-active form of the enzyme (pTD2) contains the entire catalytic domain, but lacks the C-terminal regulatory domain. That very little unprocessed TD2 was observed in midgut content suggests that the processing reaction occurs rapidly upon maceration of leaf tissue by the caterpillar. Based on the structural organization of TD2 into distinct catalytic and regulatory domains, we propose that processing involves an endoprotease that cleaves the neck region between the two domains. Additional work is needed to test this hypothesis and to determine whether the processing enzyme is of plant or insect origin. The finding that TD2 is processed to an Ile-insensitive enzyme in the digestive tract of the generalist caterpillar T. ni indicates that the processing phenomenon likely occurs in a broad range of tomato-insect interactions.
An important consequence of proteolytic removal of the regulatory domain is loss of feedback inhibition by Ile. The midgut content of M. sexta larvae reared on tomato plants contains levels of Ile (approximately 2.5 mm) that are sufficient to inhibit TD2 activity (H. Chen and G.A. Howe, unpublished data). Thus, proteolytic cleavage of TD2 is required to activate the enzyme's ability to degrade Thr in the amino acid-rich environment of the midgut. This interpretation is consistent with the fact that Thr content in the midgut of M. sexta larvae reared on TD2-containing tomato foliage is much less than that in the gut of insects grown on TD2-deficient foliage (Chen et al., 2005). Given that dietary Thr is limiting for M. sexta growth on native tobacco (Kang et al., 2006), these results strongly support the notion that postingestive processing of TD2 has evolved as a plant defense to deplete Thr levels in the midgut. It is also possible that the defensive function of pTD2 is related to its ability to produce ammonia, which at alkaline pH is highly toxic to biological systems (Visek, 1984).
Herbivore-induced processing of TD2 provides support for the more general concept that proteolysis of dietary protein is part of the plant's overall defense response against insect attack. Other examples of plant defensive proteins that are activated by digestive proteases include polyphenol oxidase (Wang and Constabel, 2004) and urease (Ferreira-DaSilva et al., 2000). Schmelz and coworkers (2006) identified a peptide elicitor from the oral secretion of insect herbivores that promotes the expression of plant defense responses. Interestingly, this elicitor is a proteolytic fragment of the γ-subunit of chloroplastic ATP synthase. It was thus proposed that proteolysis of dietary proteins by insect digestive proteases can generate peptide signals that are introduced to the host plant via insect oral secretions. The accumulation of tomato LAP-A and P69 proteins in the M. sexta gut leads us to suggest that plant proteases, in addition to insect proteases, play a role in the digestion of dietary protein.
Shotgun Proteomic Analysis of Insect Frass
Anal droppings of insect herbivores are a rich repository of biological information (Weiss, 2006). It is well established, for example, that frass is an important source of compounds involved in host selection by insect parasitoids (Vinson, 1976). The proteomic analysis reported herein shows that insect frass is enriched in defense-related plant proteins. Many of these proteins were previously shown to accumulate in the M. sexta midgut (Chen et al., 2005) and, significantly, have an established role in anti-insect defense. CDI is a chymotrypsin inhibitor belonging to the family of Ser PIs (rather than Asp PIs) that exerts potent growth-inhibiting effects on lepidopteran larvae (Lison et al., 2006). The frass-accumulating GLP is closely related to a MeJA-inducible GLP from N. attenuata that has a role in resistance to M. sexta attack (Lou and Baldwin, 2006). Detection of this protein in frass raises the possibility that GLPs exert defensive effects (e.g. H2O2 production) in the herbivore gut. A role for LAP-A in postingestive defense is supported by the stability of the protein in the lepidopteran digestive tract (Chen et al., 2005; this study), the enzyme's high pH optimum (Gu et al., 1999), coexpression with other midgut-active defensive proteins (Li et al., 2004; Chen et al., 2005), and increased susceptibility of LapA-silenced plants to herbivory (Walling, 2006). There is also evidence indicating that LAP-A performs a signaling role in jasmonate-induced expression of defensive proteins (Walling, 2006).
Shotgun proteomic analysis also identified proteins that had not previously been implicated in plant defense. These included stress-inducible isoforms of an LH2 domain protein that may participate in lipid metabolism (Coker et al., 2005) and a member of the YjgF family of proteins that is conserved in bacteria, yeast, animals, and plants. In the context of TD function, it is noteworthy that YjgF and related proteins have been implicated in the regulation of Ile biosynthesis and Thr degradation (Datta et al., 1987; Kim et al., 2001; Parsons et al., 2003). A recent study (Leitner-Dagan et al., 2006) showed that the YjgF-related tomato protein accumulates in chloroplasts of stressed leaves and is required for optimal photosynthetic function. Additional work is needed to test the hypothesis that this protein has a role in defense against insects.
The expression of many tomato proteins identified in M. sexta frass is promoted by the jasmonate signaling pathway. Genes encoding these JIPs tend to be among the most highly induced following wounding or jasmonate treatment. For example, a DNA microarray study identified TD2 and LapA as the most highly expressed JA-responsive genes among all elements on the array (Li et al., 2004), whereas proteomic analysis showed that TD2 and LAP-A are two of the most abundant tomato proteins in the midgut and frass of tomato-reared M. sexta larvae (Chen et al., 2005; this study). Thus, there is a strong correlation between the level of induced mRNA accumulation in leaves and protein accumulation in the insect gut. Similar correlations hold for the YjgF-related protein, CDI, and other PIs. We suggest that jasmonate-induced accumulation of defensive proteins in leaves, together with the stability of these proteins in the gut lumen, provide complementary mechanisms to maximize the effectiveness of postingestive plant defense. The correlation between gene and protein expression suggests that microarray data can be used as a starting point to identify novel anti-insect proteins.
Several tomato PR proteins were excreted in M. sexta frass. Nearly all of these proteins have been shown to be highly expressed in response to pathogen infection or wounding (Supplemental Table S1) and secreted into the extracellular space where they presumably interact directly with invading pathogens (van Loon et al., 2006). The biological significance of PR protein accumulation in frass is unclear. It is possible that these proteins accumulate in frass simply because they are highly resistant to proteolysis. This idea is supported by studies showing that PR proteins are extremely stable (Ferreira et al., 2001; Flamini and De Rosso, 2006; van Loon et al., 2006). It is also possible that PR proteins perform a physiological role in the digestive system of insect herbivores. The alkaline pH optimum of subtilisin-like P69 proteases (Vera and Conejero, 1988), which appear to be the most abundant PR proteins in M. sexta frass, indicates that these extracellular proteases may be activated upon entry of macerated leaf tissue into the lepidopteran gut. Studies of Cys proteases establish a precedent for the role of plant proteases in postingestive defense against insect herbivores (Pechan et al., 2002; Konno et al., 2004).
An important conclusion from this and previous (Chen et al., 2005) work is that foliar proteins have a wide range of stability in the gut lumen of phytophagous insects. Whereas bulk dietary protein (e.g. Rubisco) is efficiently degraded in the M. sexta midgut, other plastidic proteins, such as TD2 and LAP-A, remain active following passage through the gut. The most straightforward interpretation of these results is that midgut-active defensive proteins are highly resistant to digestive proteases and, as a consequence, are selectively enriched during passage of the food bolus through the animal. The biophysical properties that allow pTD2 and LAP-A to accumulate and function in the extreme environment of the lepidopteran gut remain to be determined. In this context, it is worth noting that hyperstable (as well as alkaliphilic) enzymes are of significant commercial interest for their use as industrial biocatalysts (Hough and Danson, 1999). Although research in this area has focused mainly on extremophilic bacteria and Archaea, our results suggest that frass-accumulating plant proteins can be exploited as a new source of hyperstable enzymes.
Seminal work by Green and Ryan (1972) introduced the idea that wound-inducible plant proteins act directly in the insect gut as a defense. The recent discovery of TD, arginase, VSP, and proteases as anti-insect proteins extends this concept to include plant enzymes that impair digestive physiology (Pechan et al., 2002; Chen et al., 2005; Liu et al., 2005; Kang et al., 2006). Proteomic-based technologies provide a powerful tool to address the question of how variation in the quantity and quality of dietary protein influences plant-insect relations. Only recently have these approaches been used to assess the effects of herbivory on large-scale changes in plant protein content (Francis et al., 2006; Giri et al., 2006; Lippert et al., 2007). Our results demonstrate that proteomic analysis of midgut content (Chen et al., 2005) and frass (this study) can be used to track the fate of the plant proteome during passage through the insect digestive tract, thus providing insight into how the plant proteome interacts with components of the insect gut. Additional work is needed to determine the limitations of this approach for identifying midgut-active plant defense proteins. For example, it is conceivable that some defensive proteins are degraded by microbial flora in the insect gut or frass, or that covalent modification of dietary polypeptides in the insect gut (Felton, 1996) prevents protein identification by MS. These limitations notwithstanding, we conclude that proteomic analysis of frass has general utility for large-scale identification of plant defensive proteins in virtually any plant-insect interaction for which appropriate sequence databases are available.
MATERIALS AND METHODS
Biological Material and Growth Conditions
Tomato (Solanum lycopersicum) cv Castlemart was used as the wild type for all experiments, except where otherwise noted. 35S∷PS and jai1 mutant lines and conditions for plant growth were previously described (Chen et al., 2005). Manduca sexta eggs were obtained from the Department of Entomology, North Carolina State University. Newly hatched larvae were transferred directly to 3-week-old tomato plants. Larval midguts were dissected from cold-anesthetized larvae (fourth to fifth instar) that were actively feeding at the time of collection. Total midgut content was isolated by removing the food bolus from the dissected midgut. Care was taken to avoid mixing the midgut content with insect tissue. Midgut content from three to five larvae was pooled and frozen at −20°C until further use for protein extraction. For collection of M. sexta frass, third to fourth instar larvae were transferred to a Tupperware box containing cut leaves from approximately 6-week-old tomato plants that were heavily damaged by M. sexta feeding. The petiole of the cut leaf was inserted through the closed cap (in which a hole was punctured) of a 1.5-mL plastic microcentrifuge tube containing water. Cut leaves were replaced on a daily basis. Frass was collected at least once daily and stored at −20°C until needed for protein extraction. The use of host genotypes that are noninducible (jai1) or constitutively induced (35S∷PS) helped to control for possible effects of wounds that were generated by leaf cutting. Care was taken to avoid contamination of frass with intact leaf tissue.
Trichoplusia ni eggs were obtained from Benzon Research and hatched at 30°C. Within 8 h of hatching, larvae were transferred to 3-week-old tomato plants. Frass pellets were collected daily from fourth to fifth instar larvae grown on cut tomato leaves. Leaves were replaced daily. Pellets were stored at −20°C until further use.
Protein Extraction and Enzyme Assays
A modified version of a phenol-based protein extraction method (Constabel et al., 1995) was used to isolate total protein for SDS-PAGE (Fig. 3A) and immunoblot analysis. Frozen leaf tissue, midgut content, and frass were ground in liquid nitrogen to a fine powder. One volume equivalent of the ground tissue was mixed with 2 volumes of protein extraction buffer (0.7 m Suc, 100 mm Tris-HCl [pH 6.8], 20 mm EDTA, 100 mm KCl, 2% [v/v] 2-mercaptoethanol, and 1 mm phenylmethylsulfonyl fluoride [PMSF]). Following the final protein precipitation step (Constabel et al., 1995), protein pellets were resuspended in solution containing 9.5 m urea, 2% (v/v) Nonidet detergent, and 5% 2-mercaptoethanol. Samples were heated to 65°C for 5 min prior to SDS-PAGE.
An aqueous buffer system (Chen et al., 2005) was used to prepare frass extracts for TD and LAP enzyme assays, purification of pTD2, and shotgun proteomic analysis. TD activity measurements in the presence or absence of Ile were as previously described (Chen et al., 2005). LAP activity was measured as previously described (Gu et al., 1999; Nampoothiri et al., 2005) with some modifications. Briefly, the reaction mixture contained 1 mL of a 2.5 mm solution of l-Leu-p-nitroanilide substrate in 100 mm NaOH-Gly buffer (pH 8.5), 1 mL 0.5 mm MnCl2 in 100 mm NaOH-Gly buffer (pH 8.5), and 0.5 mL water. Reactions were initiated by the addition of 5 μL of protein extract prepared from frass. Following 30-min incubation at 37°C, the reaction was stopped by the addition of 1 mL glacial acetic acid. The absorbance was measured at 405 nm against a mock reaction devoid of enzyme. A standard curve was prepared with p-nitroaniline. l-Leu-p-nitroanilide and p-nitroaniline were purchased from Sigma.
Purification of pTD2 from M. sexta Frass
Frass obtained from tomato-reared M. sexta larvae was ground in liquid nitrogen to a fine powder. Ten grams of powder were extracted with approximately 2 volumes of 100 mm Tris buffer (pH 7.5) containing 1 mm EDTA, 1% (v/v) 2-mercaptoethanol, and 0.1 mm PMSF. The mixture was centrifuged at 20,000g for 10 min and the resulting supernatant was filtered through a 0.45-μm filter (Millipore). Protein in the supernatant was brought to 30% (w/v) saturation with ammonium sulfate and stirred for 2 h at 4°C. Precipitated proteins were discarded following centrifugation at 20,000g for 15 min. The supernatant was adjusted to 65% saturation with ammonium sulfate and stirred for 4 h at 4°C. Following centrifugation at 20,000g for 15 min, the supernatant was discarded. The protein precipitate was dissolved in 15 mm Tris buffer (pH 7.5) and then desalted on a Sephadex G-25 column (Amersham-Pharmacia Biosciences) that was equilibrated with the same buffer. The desalted extract was applied to a Whatman DEAE-cellulose (DE52) column (30- × 1.5-cm i.d.) that was equilibrated with the same buffer. Proteins were eluted from the column with a linear gradient of 0 to 0.5 m NaCl in 15 mm Tris-HCl (pH 7.5). Fractions (1.5 mL) were collected with a Gilson fraction collector (model FC-203B). Fractions containing the bulk of TD activity (approximately two-thirds of the activity peak height) were pooled and concentrated with a 10-kD molecular mass cutoff Amicon centrifugal filter (Millipore). Concentrated enzyme preparation (0.2 mL) was loaded on a Superose-12 gel filtration column (Pharmacia) that was pre-equilibrated with 50 mm Tris-HCl (pH 7.5) containing 100 mm NaCl. Proteins were eluted with the same buffer at a flow rate of 0.6 mL/min on a Waters HPLC system equipped with a model 600 pump, a 996 photodiode array detector, and a 717-plus autosampler. Fractions (1.0 mL) were collected manually and assayed for TD activity. The specific activity of TD increased approximately 30-fold during the purification procedure. Protein concentrations were determined by the Bradford method, using bovine serum albumin as a standard. The relative purity of protein samples was assessed by SDS-PAGE and staining of gels with Coomassie Brilliant Blue R-250.
Cloning and Expression Analysis of SlTD1 and SlTD2
A search of the tomato EST database (version 11.0, released June 21, 2006) at The Gene Index Project (http://compbio.dfci.harvard.edu/tgi/plant.html) identified a tentative consensus sequence (TC176654) annotated as a TD. This sequence, which we designated as SlTD1, was distinct from the published tomato TD sequence (Samach et al., 1991). Three cDNA clones (cTOF22A12, cTOC4022, and cTOD22M4) corresponding to SlTD1 were obtained from the Boyce Thompson Institute and sequenced in their entirety. Overlapping regions between the clones showed that the three cDNAs corresponded to the same transcript. The assembled full-length cDNA sequence of SlTD1, deposited in GenBank (accession no. EF026088), has a 1,821-bp open reading frame, a 91-bp 5′-untranslated region upstream of the ATG initiation codon, and a 235-bp 3′-untranslated region excluding poly(A) residues.
Full-length SlTD2 cDNA was obtained by reverse transcription-PCR (DuraScript; Sigma) of total RNA isolated from leaves of tomato plants (cv Castlemart) that were treated with MeJA for 24 h. The PCR primers for the cDNA amplification step were TD5 (forward) 5′-ATGGAATTCCTTTGTTTAGCCCCA-3′ and TD3-2 (reverse) 5′-GCCATTACATTACATTGGATACAT-3′. The resulting PCR product was cloned into the pGEM-T Easy vector (Promega) to yield pGEM-SlTD2. The sequence of the cDNA insert perfectly matched that of the TD sequence reported by Samach et al. (1991).
RNA-blot experiments were performed with total RNA isolated from wild-type tomato plants (cv Micro-Tom), as previously described (Howe et al., 2000). Roots, stems, petioles, and leaves were collected from 3-week-old plants. Floral tissues were harvested from 6-week-old plants. Wound and MeJA treatments were performed according to published methods (Howe et al., 2000; Li and Howe, 2001). RNA blots were probed with 32P-labeled SlTD1 and SlTD2 cDNAs or with a cDNA for eIF4A as a loading control.
Antibody Production and Western-Blot Analysis
The pET30TD plasmid for expression of AtTD was kindly provided by Dr. Renaud Dumas (Wessel et al., 2000). This vector, which is derived from pET30a+, was digested with NdeI and SalI to release the AtTD cDNA. The resulting linearized vector was ligated to a PCR product containing a modified SlTD2 cDNA. This cDNA was prepared by PCR amplification of pGEM-SlTD2 with the following primer sets. The forward primer (5′-TGATTAATATGATGTCACCAATTGTTTCTGTG-3′) was designed with an AseI restriction site that is compatible with the NdeI site on the vector. The underlined ATG sequence in the forward primer represents the initiation codon in the resulting recombinant protein. This ATG codon replaces the first amino acid (Lys-52) of the mature protein, thus eliminating the 51-amino acid chloroplast-targeting sequence (Wessel et al., 2000). The reverse primer was designed with a TGA stop codon upstream of the SalI site (5′-ATGTCGACTCACTCACTTACTACAAGGAA-3′). The amplified PCR product was cloned into the pET30 vector (described above) to yield a plasmid called pET30-TD2. This expression vector produces a truncated form of SlTD2 that lacks 51 amino acids corresponding to the N-terminal chloroplast-targeting sequence.
TD2 expression and purification were performed as described previously (Wessel et al., 2000) with minor modifications. A 750-mL log-phase culture of Escherichia coli BL21 (DE3) cells containing the pET30-TD2 plasmid grown at 37°C was induced by the addition of isopropylthio-β-galactoside to a final concentration of 0.5 mm. The induced cells were incubated with agitation at 28°C for 15 h and then harvested by centrifugation at 20,000g for 15 min. The cell pellet was resuspended in buffer A (50 mm HEPES [pH 7.5] and 1 mm EDTA) containing 0.1 mm PMSF and 1 mm dithiothreitol. The cell suspension was treated with lysozyme (1 mg/mL) for 30 min at 30°C, followed by sonication. Cell debris was removed by centrifugation at 20,000g for 15 min and the supernatant (crude lysate) was saved. The crude lysate was brought to 30% (w/v) saturation with ammonium sulfate and stirred for 1 h at 4°C. The precipitated proteins were discarded and the solution was adjusted to 65% saturation with ammonium sulfate. The precipitate was collected by centrifugation at 20,000g for 15 min, resuspended in buffer A, applied to a Sephadex G-25 column, and eluted with buffer A. The eluate was collected and loaded onto a Whatman DEAE-cellulose DE52 column (30 × 1.5 cm) equilibrated with buffer A. The column was eluted with 600 mL of a 0 to 400 mm KCl gradient. Elution was monitored by A280 and enzyme activity. Fractions containing TD activity were pooled, concentrated with an Amicon Ultra-15 30-kD filter (Millipore), and subjected to further purification on a Pharmacia Biotech ÄKTA Basic FPLC system. Specifically, the concentrate was applied to a HiLoad26/60 Superdex 200 column (Amersham-Pharmacia Biotech) previously equilibrated with buffer A. The column was eluted with 1.5 column volumes of buffer A containing 150 mm KCl. Fractions containing TD activity were pooled and loaded directly into a HiPrep 16/10 column (Pharmacia Biotech) equilibrated in buffer A. The column was eluted with 20 column volumes of a 0 to 400 mm KCl gradient. The enzyme was concentrated with an Amicon Ultra-15 30-kD filter to a concentration of 4.5 mg/mL. The purified enzyme was determined to be >95% pure as determined by SDS-PAGE. Rabbit polyclonal antibodies against purified TD2 antigen were produced by a commercial vendor (Cocalico Biologicals) according to their standard protocol, using 0.5 mg of the purified protein as antigen.
Western-blot analysis was performed as previously described (Schilmiller et al., 2007), using anti-SlTD2 antibodies that were diluted 1:2,000 in Tris-buffered saline with 0.1% Tween 20 containing 1% nonfat milk. Blots were washed three times with Tris-buffered saline with 0.1% Tween 20 and then incubated with a peroxidase-conjugated anti-rabbit secondary antibody (1:10,000 dilution; Sigma). TD2 protein-antibody complexes were visualized with the SuperSignal West Pico chemiluminescent substrate (Pierce) according to the manufacturer's instructions.
Anti-Acx1A antibodies (Schilmiller et al., 2007) were used at a 1:1,000 dilution. Toc75 antibodies (Tranel et al., 1995) were used at a 1:2,000 dilution.
LC-MS/MS-Based Identification of Tomato Proteins in M. sexta Frass
Frass was collected from M. sexta larvae (third to fourth instar) that were grown on wild-type (cv Castlemart) tomato plants as described above. Frass was frozen in liquid nitrogen, ground to a fine powder, and extracted with 100 mm Tris buffer (pH 7.5) containing 1 mm EDTA, 1% (v/v) β-mercaptoethanol, and 0.1 mm PMSF. Extracts were centrifuged at 20,000g for 10 min at 4°C. Approximately 60 μg protein were electrophoresed through a 4% SDS-polyacrylamide stacking gel (1.5 cm) and approximately 1 cm into a 12% resolving gel. Gels were stained with Coomassie Blue and the protein-stained region of the gel was excised. Proteins within the gel piece were reduced and alkylated, followed by digestion with trypsin as previously described (Chen et al., 2005).
The extracted peptides were automatically injected by a Michrom Paradigm Endurance Bio-Cool Autosampler onto a Paradigm Platinum Peptide Nanotrap (C18; 0.15 × 50 mm) and washed for 5 min. The bound peptides were eluted onto a 10-cm × 75-μm New Objectives Picofrit column packed with Michrom Magic C18 AQ packing material. Peptides were eluted from this column over 90 min with a gradient of 5% B to 90% B, with constant 10% C in 76 min using a Michrom Paradigm MDLC (buffer A, 100% water; buffer B, 100% acetonitrile; buffer C, 1% formic acid), at a flow rate of 300 nL/min. Eluted peptides were analyzed with a ThermoElectron LTQ linear ion trap mass spectrometer (Thermo Electron). The top five ions in each survey scan were subjected to data-dependent zoom scans followed by low-energy collision-induced dissociation. The resulting MS/MS spectra were converted to peak lists using BioWorks Browser Version 3.2.
The X!-tandem algorithm (Craig and Beavis, 2003, 2004) was used to search MS/MS spectra against the tomato EST database from The Institute of Genomic Research (TIGR) Gene Indices (http://compbio.dfci.harvard.edu/tgi). Protein identifications were considered positive if two or more peptides from the same protein were identified, each with a probability score of P ≤ 0.01 (Eriksson and Fenyo, 2004). Of the 20 tomato proteins cataloged by this procedure, 18 proteins were identified on the basis of two or more unique peptides. A complete list of all peptides identified for each protein is provided in Supplemental Table S2.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EF026088 for SlTD1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Shotgun proteomic analysis of frass from M. sexta larvae reared on tomato foliage.
Supplemental Table S2. Unique peptides identified for tomato proteins excreted in M. sexta frass.
Supplementary Material
Acknowledgments
We thank Doug Whitten of the Michigan Proteome Consortium for assistance with proteomic experiments and bioinformatic analysis of LC-MS/MS data. Guanghui Liu and Rob Larkin (MSU) are acknowledged for assistance with the expression and purification of recombinant TD. John Froehlich (MSU) and Leron Katsir are acknowledged for the anti-Toc75 antibody and assistance with constructing SlTD homology models, respectively. Bonnie St. John provided helpful assistance with immunoblot analysis. We also thank R. Dumas for the pET30-TD expression vector for Arabidopsis TD. Tomato EST clones were obtained from the Boyce Thompson Institute at Cornell University.
This work was supported by the U.S. Department of Agriculture (grant no. 2007–35604–17791) and by the U.S. Department of Energy (grant no. DE–FG02–91ER20021 to G.A.H.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Gregg A. Howe (howeg@msu.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Berenbaum MR (1995) Turnabout is fair play—secondary roles for primary compounds. J Chem Ecol 21 925–940 [DOI] [PubMed] [Google Scholar]
- Bergey DR, Howe GA, Ryan CA (1996) Polypeptide signaling for plant defensive genes exhibits analogies to defense signaling in animals. Proc Natl Acad Sci USA 93 12053–12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernays EA, Chapman RF (1994) Host-Plant Selection by Phytophagous Insects. Chapman & Hall, New York
- Broadway RM, Duffey SS (1988) The effect of plant protein quality on insect digestive physiology and the toxicity of plant proteinase inhibitors. J Insect Physiol 34 1111–1117 [Google Scholar]
- Casey TM (1976) Activity patterns, body temperature and thermal ecology in two desert caterpillars (Lepidoptera-Sphingidae). Ecology 57 485–497 [Google Scholar]
- Chao WS, Gu YQ, Pautot VV, Bray EA, Walling LL (1999) Leucine aminopeptidase RNAs, proteins, and activities increase in response to water deficit, salinity, and the wound signals systemin, methyl jasmonate, and abscisic acid. Plant Physiol 120 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, McCaig BC, Melotto M, He SY, Howe GA (2004) Regulation of plant arginase by wounding, jasmonate, and the phytotoxin coronatine. J Biol Chem 279 45998–46007 [DOI] [PubMed] [Google Scholar]
- Chen H, Wilkerson CG, Kuchar JA, Phinney BS, Howe GA (2005) Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc Natl Acad Sci USA 102 19237–19242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker JS, Vian A, Davies E (2005) Identification, accumulation, and functional prediction of novel tomato transcripts systemically upregulated after fire damage. Physiol Plant 124 311–322 [Google Scholar]
- Colau D, Negrutiu I, Vanmontagu M, Hernalsteens JP (1987) Complementation of a threonine dehydratase-deficient Nicotiana plumbaginifolia mutant after Agrobacterium tumefaciens-mediated transfer of the Saccharomyces cerevisiae Ilv1 gene. Mol Cell Biol 7 2552–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constabel CP, Bergey DR, Ryan CA (1995) Systemin activates synthesis of wound-inducible tomato leaf polyphenol oxidase via the octadecanoid defense signaling pathway. Proc Natl Acad Sci USA 92 407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, Beavis RC (2003) A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun Mass Spectrom 17 2310–2316 [DOI] [PubMed] [Google Scholar]
- Craig R, Beavis RC (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20 1466–1467 [DOI] [PubMed] [Google Scholar]
- Datta P, Goss TJ, Omnaas JR, Patil RV (1987) Covalent structure of biodegradative threonine dehydratase of Escherichia coli—homology with other dehydratases. Proc Natl Acad Sci USA 84 393–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo C, Conejero V, Vera P (1994) Genes encoding acidic and basic class III β-1,3-glucanases are expressed in tomato plants upon viroid infection. Plant Mol Biol 24 725–732 [DOI] [PubMed] [Google Scholar]
- Duffey SS, Felton GW (1991) Enzymatic antinutritive defenses of the tomato plant against insects. ACS Symp Ser 449 166–197 [Google Scholar]
- Duffey SS, Stout MJ (1996) Antinutritive and toxic components of plant defense against insects. Arch Insect Biochem Physiol 32 3–37 [Google Scholar]
- Eriksson J, Fenyo D (2004) Probity: a protein identification algorithm with accurate assignment of the statistical significance of the results. J Proteome Res 3 32–36 [DOI] [PubMed] [Google Scholar]
- Feeny P (1976) Plant apparency in chemical defense. Recent Adv Phytochem 10 1–40 [Google Scholar]
- Felton GW (1996) Nutritive quality of plant protein: sources of variation and insect herbivore responses. Arch Insect Biochem Physiol 32 107–130 [Google Scholar]
- Felton GW (2005) Indigestion is a plant's best defense. Proc Natl Acad Sci USA 102 18771–18772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton GW, Bi JL, Summers CB, Mueller AJ, Duffey SS (1994) Potential role of lipoxygenases in defense against insect herbivory. J Chem Ecol 20 651–666 [DOI] [PubMed] [Google Scholar]
- Ferreira RB, Picarra-Pereira MA, Monteiro S, Loureiro VB, Teixeira AR (2001) The wine proteins. Trends Food Sci Technol 12 230–239 [DOI] [PubMed] [Google Scholar]
- Ferreira-DaSilva CT, Gombarovits MEC, Masuda H, Oliveira CM, Carlini CR (2000) Proteolytic activation of canatoxin, a plant toxic protein, by insect cathepsin-like enzymes. Arch Insect Biochem Physiol 44 162–171 [DOI] [PubMed] [Google Scholar]
- Flamini R, De Rosso M (2006) Mass spectrometry in the analysis of grape and wine proteins. Expert Rev Proteomics 3 321–331 [DOI] [PubMed] [Google Scholar]
- Fraenkel GS (1959) The raison d'etre of secondary plant substances; these odd chemicals arose as a means of protecting plants from insects and now guide insects to food. Science 129 1466–1470 [DOI] [PubMed] [Google Scholar]
- Francis F, Gerkens P, Harmel N, Mazzucchelli G, De Pauw E, Haubruge E (2006) Proteomics in Myzus persicae: effect of aphid host plant switch. Insect Biochem Mol Biol 36 219–227 [DOI] [PubMed] [Google Scholar]
- Gallagher DT, Gilliland GL, Xiao GY, Zondlo J, Fisher KE, Chinchilla D, Eisenstein E (1998) Structure and control of pyridoxal phosphate dependent allosteric threonine deaminase. Structure 6 465–475 [DOI] [PubMed] [Google Scholar]
- Gatehouse JA (2002) Plant resistance towards insect herbivores: a dynamic interaction. New Phytol 156 145–169 [DOI] [PubMed] [Google Scholar]
- Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, Nagaya H, Roy L, Gosline SJ, Hallett M, et al (2006) Quantitative proteomics analysis of the secretory pathway. Cell 127 1265–1281 [DOI] [PubMed] [Google Scholar]
- Giri AP, Wunsche H, Mitra S, Zavala JA, Muck A, Svatos A, Baldwin IT (2006) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. VII. Changes in the plant's proteome. Plant Physiol 142 1621–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TR, Ryan CA (1972) Wound-induced proteinase inhibitor in plant leaves. Possible defense mechanism against insects. Science 175 776–777 [DOI] [PubMed] [Google Scholar]
- Gu YQ, Holzer FM, Walling LL (1999) Overexpression, purification and biochemical characterization of the wound-induced leucine aminopeptidase of tomato. Eur J Biochem 263 726–735 [DOI] [PubMed] [Google Scholar]
- Haukioja E, Ruohomaki K, Suomela J, Vuorisalo T (1991) Nutritional quality as a defense against herbivores. For Ecol Manage 39 237–245 [Google Scholar]
- Hermsmeier D, Schittko U, Baldwin IT (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. I. Large-scale changes in the accumulation of growth- and defense-related plant mRNAs. Plant Physiol 125 683–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildmann T, Ebneth M, Pena-Cortes H, Sanchez-Serrano JJ, Willmitzer L, Prat S (1992) General roles of abscisic and jasmonic acids in gene activation as a result of mechanical wounding. Plant Cell 4 1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough DW, Danson MJ (1999) Extremozymes. Curr Opin Chem Biol 3 39–46 [DOI] [PubMed] [Google Scholar]
- Howe GA (2004) Jasmonates as signals in the wound response. J Plant Growth Regul 23 223–237 [Google Scholar]
- Howe GA, Lee GI, Itoh A, Li L, DeRocher AE (2000) Cytochrome P450-dependent metabolism of oxylipins in tomato. Cloning and expression of allene oxide synthase and fatty acid hydroperoxide lyase. Plant Physiol 123 711–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Baldwin IT (2006) Isolation and characterization of the threonine deaminase promoter in Nicotiana attenuata. Plant Sci 171 435–440 [DOI] [PubMed] [Google Scholar]
- Kang JH, Wang L, Giri AP, Baldwin IT (2006) Silencing threonine deaminase and the JAR1 homologue in Nicotiana attenuata impairs JA-isoleucine-mediated defenses against the specialist herbivore, Manduca sexta. Plant Cell 18 3303–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karowe DN, Martin MM (1989) The effects of quantity and quality of diet nitrogen on the growth, efficiency of food utilization, nitrogen budget, and metabolic rate of 5th instar Spodoptera eridania larvae (Lepidoptera, Noctuidae). J Insect Physiol 35 699–708 [Google Scholar]
- Kessler A, Baldwin IT (2002) Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol 53 299–328 [DOI] [PubMed] [Google Scholar]
- Kim JM, Yoshikawa H, Shirahige K (2001) A member of the YER057c/yjgf/Uk114 family links isoleucine biosynthesis and intact mitochondria maintenance in Saccharomyces cerevisiae. Genes Cells 6 507–517 [DOI] [PubMed] [Google Scholar]
- Konno K, Hirayama C, Nakamura M, Tateishi K, Tamura Y, Hattori M, Kohno K (2004) Papain protects papaya trees from herbivorous insects: role of cysteine proteases in latex. Plant J 37 370–378 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5 150–163 [DOI] [PubMed] [Google Scholar]
- Leitner-Dagan Y, Ovadis M, Zuker A, Shklarman E, Ohad I, Tzfira T, Vainstein A (2006) CHRD, a plant member of the evolutionarily conserved YjgF family, influences photosynthesis and chromoplastogenesis. Planta 225 89–102 [DOI] [PubMed] [Google Scholar]
- Li L, Howe GA (2001) Alternative splicing of prosystemin pre-mRNA produces two isoforms that are active as signals in the wound response pathway. Plant Mol Biol 46 409–419 [DOI] [PubMed] [Google Scholar]
- Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA (2004) The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16 126–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linthorst HJM, Danhash N, Brederode FT, Vankan JAL, Dewit P, Bol JF (1991) Tobacco and tomato PR proteins homologous to Win and Pro-hevein lack the hevein domain. Mol Plant Microbe Interact 4 586–592 [DOI] [PubMed] [Google Scholar]
- Lippert D, Chowrira S, Ralph SG, Zhuang J, Aeschliman D, Ritland C, Ritland K, Bohlmann J (2007) Conifer defense against insects: proteome analysis of Sitka spruce (Picea sitchensis) bark induced by mechanical wounding or feeding by white pine weevils (Pissodes strobi). Proteomics 7 248–270 [DOI] [PubMed] [Google Scholar]
- Lison P, Rodrigo I, Conejero V (2006) A novel function for the cathepsin D inhibitor in tomato. Plant Physiol 142 1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Ahn JE, Datta S, Salzman RA, Moon J, Huyghues-Despointes B, Pittendrigh B, Murdock LL, Koiwa H, Zhu-Salzman K (2005) Arabidopsis vegetative storage protein is an anti-insect acid phosphatase. Plant Physiol 139 1545–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou YG, Baldwin IT (2006) Silencing of a germin-like gene in Nicotiana attenuata improves performance of native herbivores. Plant Physiol 140 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson WJ (1980) Herbivory in relation to plant nitrogen content. Annu Rev Ecol Syst 11 119–161 [Google Scholar]
- McGurl B, Orozco-Cardenas M, Pearce G, Ryan CA (1994) Overexpression of the prosystemin gene in transgenic tomato plants generates a systemic signal that constitutively induces proteinase inhibitor synthesis. Proc Natl Acad Sci USA 91 9799–9802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan S, Ma PWK, Pechan T, Bassford ER, Williams WP, Luthe DS (2006) Degradation of the S. frugiperda peritrophic matrix by an inducible maize cysteine protease. J Insect Physiol 52 21–28 [DOI] [PubMed] [Google Scholar]
- Moran N, Hamilton WD (1980) Low nutritive quality as defense against herbivores. J Theor Biol 86 247–254 [Google Scholar]
- Nampoothiri KM, Nagy V, Kovacs K, Szakacs G, Pandey A (2005) l-Leucine aminopeptidase production by filamentous Aspergillus fungi. Lett Appl Microbiol 41 498–504 [DOI] [PubMed] [Google Scholar]
- Naqvi SMS, Harper A, Carter C, Ren G, Guirgis A, York WS, Thornburg RW (2005) Nectarin IV, a potent endoglucanase inhibitor secreted into the nectar of ornamental tobacco plants. Isolation, cloning, and characterization. Plant Physiol 139 1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG (2005) Comparison of label free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics 4 1487–1502 [DOI] [PubMed] [Google Scholar]
- Parsons L, Bonander N, Eisenstein E, Gilson M, Kairys V, Orban J (2003) Solution structure and functional ligand screening of HI0719, a highly conserved protein from bacteria to humans in the YjgF/YER057c/UK114 family. Biochemistry 42 80–89 [DOI] [PubMed] [Google Scholar]
- Pechan T, Cohen A, Williams WP, Luthe DS (2002) Insect feeding mobilizes a unique plant defense protease that disrupts the peritrophic matrix of caterpillars. Proc Natl Acad Sci USA 99 13319–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Inoue K, Keegstra K (2005) Evolution of the general protein import pathway of plastids (review). Mol Membr Biol 22 73–86 [DOI] [PubMed] [Google Scholar]
- Rhoades DF, Cates RG (1976) Toward a general theory of plant antiherbivore chemistry. Rec Adv Phytochem 10 168–213 [Google Scholar]
- Ryan CA (1990) Protease inhibitors in plants. Genes for improving defenses against insects and pathogens. Annu Rev Phytopathol 28 425–449 [Google Scholar]
- Samach A, Broday L, Hareven D, Lifschitz E (1995) Expression of an amino acid biosynthesis gene in tomato flowers: developmental upregulation and MeJa response are parenchyma-specific and mutually compatible. Plant J 8 391–406 [DOI] [PubMed] [Google Scholar]
- Samach A, Hareven D, Gutfinger T, Ken-Dror S, Lifschitz E (1991) Biosynthetic threonine deaminase gene of tomato: isolation, structure, and upregulation in floral organs. Proc Natl Acad Sci USA 88 2678–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A, Bergey DR, Ryan CA (1995) Induction of wound response genes in tomato leaves by bestatin, an inhibitor of aminopeptidases. Plant Cell 7 1893–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Howe GA (2005) Systemic signaling in the wound response. Curr Opin Plant Biol 8 369–377 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Koo AJ, Howe GA (2007) Functional diversification of acyl-CoA oxidases in jasmonic acid biosynthesis and action. Plant Physiol 143 812–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Carroll MJ, LeClere S, Phipps SM, Meredith J, Chourey PS, Alborn HT, Teal PEA (2006) Fragments of ATP synthase mediate plant perception of insect attack. Proc Natl Acad Sci USA 103 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen SJ (1991) Comparison of chemical and functional properties of soluble leaf proteins from four plant species. J Agric Food Chem 39 681–685 [Google Scholar]
- Sidorov V, Menczel L, Maliga P (1981) Isoleucine requiring Nicotiana plant deficient in threonine deaminase. Nature 294 87–88 [Google Scholar]
- Staswick PE, Yuen GY, Lehman CC (1998) Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J 15 747–754 [DOI] [PubMed] [Google Scholar]
- Strassner J, Schaller F, Frick UB, Howe GA, Weiler EW, Amrhein N, Macheroux P, Schaller A (2002) Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J 32 585–601 [DOI] [PubMed] [Google Scholar]
- Szamosi I, Shaner DL, Singh BK (1993) Identification and characterization of a biodegradative form of threonine dehydratase in senescing tomato (Lycopersicon esculentum) leaf. Plant Physiol 101 999–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P, Conejero V, Vera P (1996) Primary structure and expression of a pathogen-induced protease (PR-P69) in tomato plants: similarity of functional domains to subtilisin-like endoproteases. Proc Natl Acad Sci USA 93 6332–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P, Conejero V, Vera P (1997) Identification of a new pathogen-induced member of the subtilisin-like processing protease family from plants. J Biol Chem 272 14412–14419 [DOI] [PubMed] [Google Scholar]
- Tranel PJ, Froehlich J, Goyal A, Keegstra K (1995) A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J 14 2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsugi S, Sakamoto W, Murata M, Motoyoshi F (1998) Arabidopsis thaliana vegetative storage protein (VSP) genes: gene organization and tissue-specific expression. Plant Mol Biol 38 565–576 [DOI] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CM (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44 135–162 [DOI] [PubMed] [Google Scholar]
- Vera P, Conejero V (1988) Pathogenesis related proteins of tomato. P-69 as an alkaline endoproteinase. Plant Physiol 87 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera P, Tornero P, Conejero V (1993) Cloning and expression analysis of a viroid-induced peroxidase from tomato plants. Mol Plant Microbe Interact 6 790–794 [PubMed] [Google Scholar]
- Vinson SB (1976) Host selection by insect parasitoids. Annu Rev Entomol 21 109–133 [Google Scholar]
- Visek WJ (1984) Ammonia: its effects on biological systems, metabolic hormones, and reproduction. J Dairy Sci 67 481–498 [DOI] [PubMed] [Google Scholar]
- Walling LL (2000) The myriad plant responses to herbivores. J Plant Growth Regul 19 195–216 [DOI] [PubMed] [Google Scholar]
- Walling LL (2006) Recycling or regulation? The role of amino-terminal modifying enzymes. Curr Opin Plant Biol 9 227–233 [DOI] [PubMed] [Google Scholar]
- Wang JH, Constabel CP (2004) Polyphenol oxidase overexpression in transgenic Populus enhances resistance to herbivory by forest tent caterpillar (Malacosoma disstria). Planta 220 87–96 [DOI] [PubMed] [Google Scholar]
- Weiss MR (2006) Defecation behavior and ecology of insects. Annu Rev Entomol 51 635–661 [DOI] [PubMed] [Google Scholar]
- Wessel PM, Graciet E, Douce R, Dumas R (2000) Evidence for two distinct effector-binding sites in threonine deaminase by site-directed mutagenesis, kinetic, and binding experiments. Biochemistry 39 15136–15143 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Thilmony R, Bender CL, Schaller A, He SY, Howe GA (2003) Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J 36 485–499 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.