Abstract
Various abilities to synthesize and accumulate glycine betaine (GB) are crucial for angiosperms to develop salt and drought tolerances. In higher plants, GB is synthesized by a two-step oxidation of choline via an intermediate form of betaine aldehyde, and catalyzed by choline monooxygenase and betaine aldehyde dehydrogenase (BADH). In this study, numerous truncated and/or recombinant transcripts of two BADH homologs resulting from an unusual posttranscriptional processing were detected in rice (Oryza sativa) and other cereal crops, including maize (Zea mays), wheat (Triticum aestivum), and barley (Hordeum vulgare). The observed events took place at the 5′ exonic region, and led to the insertion of exogenous gene sequences and a variety of deletions that resulted in the removal of translation initiation codon, loss of functional domain, and frame-shifts with premature termination by introducing stop codon. By contrast, the BADH transcripts from dicotyledonous species, such as spinach (Spinacia oleracea), Arabidopsis (Arabidopsis thaliana), and tomato (Solanum lycopersicum), had correctly processed mRNA. This suggests the differentiation of posttranscriptional processing in BADH genes potentially contributes to the variation of GB-synthesizing capacities among various plant species. In addition, comprehensive sequence analyses demonstrated that extensive sequence similarities (named as short, direct repeats) are of paired presence surrounding the junctions of both the deletion and/or insertion sites in the unusual BADH transcripts. The site selection for the deletion/insertion was altered in response to the stress conditions. This indicates that the sequence elements of short, direct repeats are probably required for the recognition of the deletion/insertion sites.
The higher plants, as sessile organisms, are generally characterized by a high degree of homeostatic plasticity in response to environmental fluctuations, thereby optimizing their growth and development in a way that maximizes their opportunities for survival and reproduction. Osmotic stresses, such as salinity and drought, signify the most severe environmental pressures (abiotic stresses) that significantly limit the growth and productivity of plant species (Boyer, 1982). The higher plants, as well as other organisms, have evolved a number of adaptive strategies to overcome such abiotic stresses (Tester and Davenport, 2003; Bartels and Sunkar, 2005). At the cellular level, the most common type of osmotic adaptation involves the accumulation of compatible solutes in cytoplasm, including amino acids, ammonium compounds, and polyols/sugars. Such solutes can lower the osmotic potential for cells without interfering with the metabolic processes or protein structuring and functioning and, consequently, maintain the water content of the cells under stresses (Yancey et al., 1982). Among the few classes of organic compounds that are commonly employed as osmoprotectants, the quaternary ammonium compounds (e.g. Gly betaine [GB]) are widely found in bacteria, cyanobacteria, algae, higher plants, and animals. These compounds are frequently detected being accumulated in those plant species that are exposed to drought or high salinity environments (Rhodes and Hanson, 1993). In principle, GB is known to provide a tolerance to cells under stresses by stabilizing the quaternary structure of the complex proteins and adjusting the osmotic potential in cytoplasm for maintaining water content. During photosynthesis, it stabilizes both PSII complexes and Rubisco under salinity and low temperatures (Incharoensakdi et al., 1986; Holmström et al., 2000). Exogenous supply of GB has been demonstrated to protect plants from stresses (Harinasut et al., 1996; Chen et al., 2000) and has also been suggested to have a cryoprotective effect on cell membranes (Sakamoto et al., 2000).
In the known biological systems, GB is synthesized via two distinct pathways from two distinct substrates: choline and Gly, respectively (Sakamoto and Murata, 2002). The conversion of choline into GB has been studied in a number of organisms, and its pathway involves one or two enzymes, depending on the mode of oxidation of choline. The two-enzyme pathway is commonly found across a wide range of plant and animal species and microorganisms, in which GB is formed as the result of a two-step oxidation of choline via the toxic intermediate betaine aldehyde. Different enzymes from plants, animals, and bacteria are involved in the first step of the pathway (Wilken et al., 1970; Ikuta et al., 1977; Lamark et al., 1991; Burnet et al., 1995). In plants, the first step is catalyzed by a novel Rieske-type iron-sulfur enzyme choline monooxygenase (CMO; Burnet et al., 1995; Rathinasabapathi et al., 1997). By contrast, the enzyme involved in the second step of the pathway is supposedly the same in plants, animals, and bacteria: it is NAD-dependent betaine aldehyde dehydrogenase (BADH; Weretilnyk and Hanson, 1989; Falkenberg and Strom, 1990; Chern and Pietruszko, 1995). BADH is a soluble, nonspecific aldehyde dehydrogenase, and its substrates include betaine aldehyde, aminoaldehydes, and dimethylsulfoniopropionaldehyde (Trossat et al., 1997; Livingstone et al., 2003). Plant BADH proteins are dimers (Weretilnyk and Hanson, 1989; Arakawa et al., 1990; Valenzuela-Soto and Munoz-Clares, 1994). The presence of two BADH homologs has been widely reported in cereal crops (Wood et al., 1996; Nakamura et al., 2001; Bradbury et al., 2005).
In many plant species under investigation, genes encoding for the two-step enzymatic process have been identified (Weretilnyk and Hanson, 1990; McCue and Hanson, 1992; Burnet et al., 1995; Ishitani et al., 1995; Wood et al., 1996; Nakamura et al., 1997; Russell et al., 1998). However, the abilities to accumulate GB are considerably different. In comparison with Chenopodiaceae species such as spinach (Spinacia oleracea) and sugar beet (Beta vulgaris) that usually accumulate abundant GB in response to water deficit or salt stress (Pan et al., 1981; Hanson and Wyse, 1982), much less GB (0–20 μmol/g fresh weight) occurs in Poaceous species such as maize (Zea mays), wheat (Triticum aestivum), sorghum (Sorghum bicolor), and barley (Hordeum vulgare; Hanson and Scott, 1980; Lerma et al., 1991; Ishitani et al., 1993; Jagendorf and Takabe, 2001; Yang et al., 2003). Moreover, studies showed that GB accumulation in rice (Oryza sativa) plants was undetectable (Flowers and Yeo, 1981; Ishitani et al., 1993; Rathinasabapathi et al., 1993; Shirasawa et al., 2006), although a recent genome sequence of rice revealed that it contains a CMO (accession no. AJ578494) and two conserved BADH homologs (accession nos. AK103582 and AK071221, named as OsBADH1 and OsBADH2, respectively) that are required for the two-step catalytic reactions. This prompts us to inspect the possible mechanism and molecular processes that underlie the large variation of GB accumulation among various species.
In this study, we extensively examined the expression of the BADH genes in rice and their response to stress conditions by including different degrees of ion concentrations and drought. We also examined differential transcriptional products derived from different tissues and treatments that had deletion(s) of 5′ exon material or insertion of exogenous gene sequences, probably resulting from an unusual posttranscriptional processing. Similar experiments were also conducted extending the examination of BADH homologs to other cereal crops, including maize, wheat, and barley, as well as dicotyledonous species, including spinach, Arabidopsis (Arabidopsis thaliana), and tomato (Solanum lycopersicum). In addition, we showed that extensive sequence similarities (named as short, direct repeats [SDRs]) are of paired presence surrounding the junctions at both deletions and insertions sites, and SDRs are probably required for recognition of the deletion/insertion sites in response to stress conditions. These data together with transgenic experiments by other groups (Sakamoto et al., 1998; Takabe et al., 1998; Kishitani et al., 2000) suggest that the lack of precise BADH gene products resulting from incorrectly processed BADH transcripts may contribute, at least in part, to the large variation in GB accumulation among various species.
RESULTS
Incorrectly Processed Transcripts Derived from Two Rice BADH Homologs
To determine the expression pattern of OsBADH1, we isolated the total RNAs from various rice tissues grown in different conditions (Table I) and analyzed the transcription by reverse transcription (RT)-PCR. The results showed that drought and salt stresses increased the transcription level of OsBADH1 (Fig. 1A). Using seedlings of rice japonica cv Nipponbare as an example, we found that the expression of OsBADH1 was very low under conditions without salt stress and detected the increased transcripts when stresses were added. However, a high ion concentration (0.5 m NaCl) inhibited transcription of this gene. Similar results were obtained when different rice varieties were used (Fig. 2, A–D).
Table I.
The incorrect processing of OsBADH1 transcripts in rice seedlings, callus, and mature leaves
| Rice Variety | Tissue and Growth Condition | No. of Clones | Position and Size of Deletion | SDR | Nearly Identical Sequences |
|---|---|---|---|---|---|
| Nipponbare | Seedling, unstressed | 1 | e1. 16-e2. 72 (172)a | CG | GCCGtCGgCG/GCCGCGaCG |
| Nipponbare | Seedling, drought, 24 h | 2 | e1. 10-e2. 72 (178) | GGCCGCG | GGCCGCGcCG/GGCCGCGaCG |
| Nipponbare | Seedling, 0.25 m KCl, 24 h | 2 | e1. 8-e2. 108 (216) | GGCCG | GGCCGcGC/GGCCGtGC |
| Nipponbare | Seedling, 0.1 m NaCl, 72 h | 3 | e1. 1-e2. 100 (215) | GCC | GCCaTGG/GCCTGG |
| Nipponbare | Seedling, 0.2 m NaCl, 72 h | 1 | e1. 11-e2. 97 (202) | CGCGC | GcCGCGCcG/GCGCGCG |
| Nipponbare | Seedling, 0.3 m NaCl, 72 h | 4 | e1. 16-e2. 72 (172) | CG | GCCGtCGgCG/GCCGCGaCG |
| Nipponbare | Seedling, 0.3 m NaCl, 72 h | 1 | e1. 7-e1. 99 (93) | GGCC | GGCCgCG/GGCCaCG |
| e2. 27-e2. 82 (56) | GGAG | GGAGgAC/GGAGAC | |||
| Nipponbare | Seedling, 0.5 m NaCl, 24 h | 1 | e1. 10-e2. 72 (178) | GGCCGCG | GGCCGCGcCG/GGCCGCGaCG |
| 93-11 | Seedling, unstressed | 1 | e1. 16-e2. 72 (172) | CG | GCCGtCGgCG/GCCGCGaCG |
| 93-11 | Seedling, drought, 24 h | 1 | e1. 8-e2. 108 (216) | GGCCG | GGCCGcGC/GGCCGtGC |
| 93-11 | Seedling, 0.25 m KCl, 24 h | 2 | e1. 10-e2. 72 (178) | GGCCGCG | GGCCGCGcCG/GGCCGCGaCG |
| 93-11 | Seedling, 0.1 m NaCl, 72 h | 1 | e1. 10-e2. 72 (178) | GGCCGCG | GGCCGCGcCG/GGCCGCGaCG |
| 93-11 | Seedling, 0.2 m NaCl, 72 h | 1 | e1. 16-e2. 72 (172) | CG | GCCGtCGgCG/GCCGCGaCG |
| 93-11 | Seedling, 0.3 m NaCl, 72 h | 1 | e1. 20-e2. 73(169) | GCGA | CgGCGAtC/CcGCGAC |
| 93-11 | Seedling, 0.5 m NaCl, 24 h | 1 | e1. 16-e2. 72 (172) | CG | GCCGtCGgCG/GCCGCGaCG |
| Suhui 527 | Seedling, 0.1 m NaCl, 72 h | 1 | e1. 7-e1. 34 (28) | GGCC | GGCCG/GGCCtG |
| e1. 44-e2. 72 (144) | CG | CGgCGGcGGG/CGaCGGtGGG | |||
| Chuanxiang 29B | Seedling, 0.1 m NaCl, 72 h | 1 | e1. 1-e2. 100 (215) | GCC | GCCaTGG/GCCTGG |
| Zhonghua 9 | Seedling, 0.1 m NaCl, 72 h | 1 | e1. 16-e2. 72 (172) | CG | GCCGtCGgCG/GCCGCGaCG |
| Yuanlixiagjing | Seedling, 0.1 m NaCl, 72 h | 2 | e1. 10-e2. 72 (178) | GGCCGCG | GGCCGCGcCG/GGCCGCGaCG |
| Newbonnet | Seedling, 0.1 m NaCl, 72 h | 2 | e1. 10-e2. 72 (178) | GGCCGCG | GGCCGCGcCG/GGCCGCGaCG |
| Tebonnet | Seedling, 0.1 m NaCl, 72 h | 2 | e1. 7-e1. 34 (28) | GGCC | GGCCG/GGCCtG |
| e1. 44-e2. 72 (144) | CG | CGgCGGcGGG/CGaCGGtGGG | |||
| Nipponbare | Callus, unstressed | 1 | e1. 16-e2. 72 (172) | CG | GCCGtCGgCG/GCCGCGaCG |
| Nipponbare | Callus, 0.5 m NaCl, 24 h | 1 | e1. 10-e2. 72 (178) | GGCCGCG | GGCCGCGcCG/GGCCGCGaCG |
| Nipponbare | Callus, 0.5 m NaCl, 24 h | 1 | e1. 13-e2. 72 (175) | CG | CGcCGtCGG/CGCGaCGG |
| 93-11 | Callus, 0.5 m NaCl, 24 h | 1 | e1. 13-e2. 72 (175) | CG | CGcCGtCGG/CGCGaCGG |
| Nipponbare | Mature leaf, field | 1 | e1. 7-e2. 116 (225) | GGCC | |
| 93-11 | Mature leaf, field | 1 | e1. 8-e2. 72 (180) | CG | GcCGCG/GCGaCG |
| Basmati | Mature leaf, field | 2 | e1. 14-e2. 109 (211) | GCCGT | GCCGTCGG/GCCGTgCGG |
| Della | Mature leaf, field | 1 | e1. 7-e1. 34 (28) | GGCC | GGCCG/GGCCtG |
| e1. 44-e2. 72 (144) | CG | CGgCGGcGGG/CGaCGGtGGG |
e1. 16-e2. 72 (172) denotes that exon materials of OsBADH1 from the 16th base of exon 1 to the 72nd of exon 2 were excised; 172 nucleotides in total were deleted.
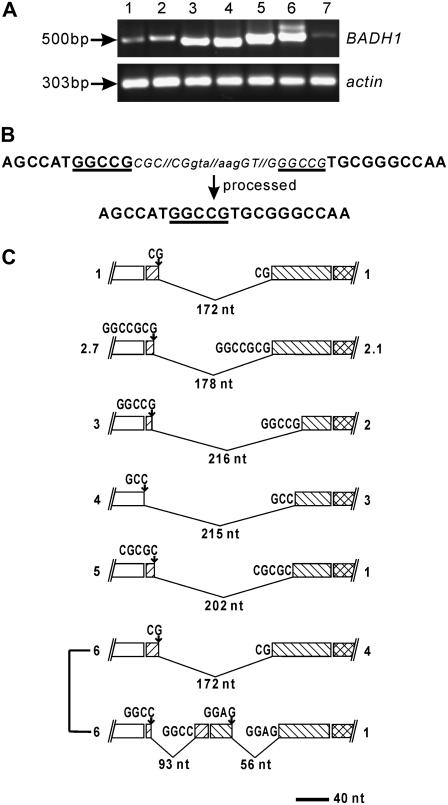
Figure 1.
Expression and structure of OsBADH1 RNAs derived from seedlings of japonica cv Nipponbare. A, Agarose gel electrophoresis analysis of RT-PCR amplification products. Comparison of expression abundance under conditions of no stress (lane 1), drought (lane 2), 0.25 m KCl (lane 3), and 0.1, 0.2, 0.3, and 0.5 m NaCl (lanes 4, 5, 6, and 7, respectively) is shown. The position of Mr marker is indicated. Actin was amplified as internal positive control. B, Incorrect processing in transcripts from lane 3 of A leading to a deletion comprising intron 1 and partial exons 1 and 2. Introns and exons are marked by lowercase and uppercase letters, respectively. The excised sequence is indicated by the smaller italic letters, while the sequence remaining in the resultant transcript is shown in bold letters. SDRs at deletion junctions are underlined. Notice that one copy of SDRs is excised and the other copy is retained in the resultant transcript. Interrupted lines represent the excised sequence not shown here. C, Structure of incorrectly processed cDNAs derived from OsBADH1. Clone numbers are indicated on the right. Numbers on the left correspond to the lane numbers in A. Exons are indicated as boxes, and excised exon materials are shown as folded lines, under which the size is indicated. The positions and sequences of 5′ UTR, exon 1, exon 2, and exon 3 are indicated as white boxes and boxes with slashes, backslashes, and cross-hatching, respectively. Short nucleotide sequences at the 5′ deletion site represent SDRs kept in transcripts, while the nucleotide sequences at the 3′ deletion site are shown as excised SDRs. Scale bar represents 40 nucleotides.
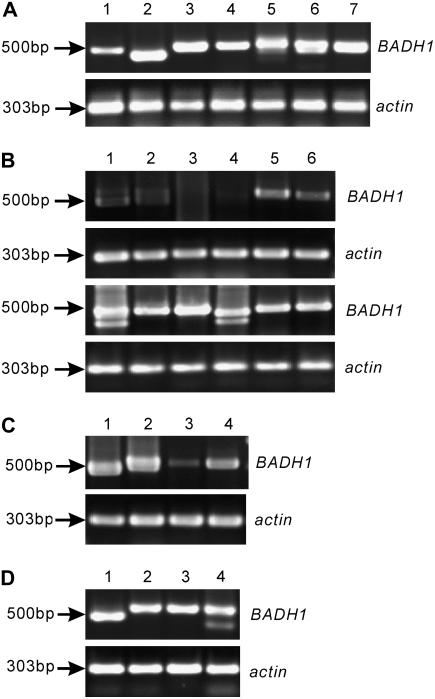
Figure 2.
Expression of OsBADH1 in different rice lines. Agarose gel electrophoresis analysis of RT-PCR amplification products is shown. Positions of Mr markers are indicated. Actin was amplified as internal positive control. A, Expressed products derived from seedlings of indica cv 93-11 under different growth conditions (lane 1, unstressed; lane 2, drought; lane 3, 0.25 m KCl; lane 4, 0.1 m NaCl; lane 5, 0.2 m NaCl; lane 6, 0.3 m NaCl; lane 7, 0.5 m NaCl). B, Comparison of expression abundance in seedlings between unstressed (0 m NaCl, top) and stressed (0.1 m NaCl, bottom) conditions. Lanes 1 to 6 represent Zhonghua 9, Yuanlixiangjing, Suhui 527, Chuanxiang 29B, Newbonnet, and Tebonnet, respectively. C, Comparison of expression abundance in callus between the unstressed (0 m NaCl) and stressed (0.5 m NaCl) conditions. Lanes 1 and 2 show transcripts derived from the untreated and treated callus of Nipponbare, respectively, and lanes 3 and 4 are indicated as the transcripts derived from untreated and treated callus of 93-11, respectively. D, Expression in mature leaf. Lanes 1 to 4 represent transcripts derived from Nipponbare, 93-11, Della, and Basmati, respectively.
To examine whether the expressed products were OsBADH1 gene, the RT-PCR-amplified fragments were cloned and sequenced. We used primers derived from 5′ and 3′ untranslated regions (UTRs) to isolate the full length of OsBADH1 cDNA clones (Table II). The resultant sequencing analysis revealed that the OsBADH1 cDNAs were truncated at the 5′ exonic region. Accordingly, we focused the analysis on a region comprising exons 1 to 6 using primers specific to the 5′ region. As shown in Figures 1A and 2, the observed expressed products were shorter than the expected size of 695 bp of the 5′ exonic region. To examine the structure of OsBADH1 transcripts, a total of 41 OsBADH1 cDNA clones were studied in detail. Sequence comparison of the cDNAs revealed a considerable variation in their structural compositions (Table I). All of the cDNAs contained a deletion of the 5′ coding sequence within the OsBADH1 gene. The deleted exon material ranged from 28 to 225 nucleotides in size. The start-point of the deletions in four cDNAs began with the first nucleotide of the coding sequence, which gave rise to the loss of translation initiation codon. Thirty-two cDNAs encoded derivatives with frame-shifts in the open reading frame (ORF), introducing various stop codons at different positions. Only five cDNA clones showed the potential to encode partial BADH1 proteins with deletions that coded for a part of the putative NAD+-binding domain. Most of the missing sequences from the truncated transcripts indicated above involved two different exons, and in a few cases the truncation took place within a single exon. In addition, two independent deletions of exon materials within a single cDNA clone were observed in five clones. Therefore, no cDNA was found to have the capacity to encode the full length of the OsBADH1 protein, indicating that correctly processed transcripts represented a very small proportion of the total cytoplasmic mature OsBADH1 RNA population and consequently that the majority of the OsBADH1 mRNAs were unlikely to encode functional proteins.
Table II.
Oligonucleotide primers for genes of BADH, ubiquitin, and actin
| Gene (Accession No.) | Forward Primer (5′→3′) | Reverse Primer (5′→3′) | Expected Size |
|---|---|---|---|
| bp | |||
| Rice BADH1 (AK103582) | GCCGCCCCCCAACCGGAAGC | TTCTGTCCGTCCGTTCTG | 1,624 |
| GCCGCCCCCCAACCGGAAGC | GCACCAGCTTCAGTGCCCAG | 695 | |
| Rice BADH2 (AK071221) | CAGCTCCAGCTCCTCCTCGA | GGGCGTGTCATGCGTATGCGCA | 1,581 |
| CAGCTCCAGCTCCTCCTCGA | CCAACTACACCGATAGGCTCT | 507 | |
| Maize BADH1 (DV031390) | CAGAGTTCAGACCACCGCATC | GTGATAGCAGCAAGCAACTTTACC | 1,678 |
| CAGAGTTCAGACCACCGCATC | AAACAGGCTTAACCATTTGGGC | 802 | |
| Maize BADH2 (AY587278) | ACTAGCGTCCGCAACAGCAG | AACTCAACTCTCGCCTTTATCCTGC | 1,831 |
| ACTAGCGTCCGCAACAGCAG | CAAAGCACCCAGCAACATCA | 543 | |
| Wheat BADH1 (BJ259181) | CCCATACTCCTACTCACCTCCT | AAGTCCCAGCCAGGATAGAAC | 1,670 |
| CCCATACTCCTACTCACCTCCT | TAGTTCCAGGGAGTGATGAGTCC | 563 | |
| Wheat BADH2 (AY050316) | ATGGTCGTCACGGCGAAGATCC | CTCATGCCATTTATTTCGCAAG | 1,644 |
| ATGGTCGTCACGGCGAAGATCC | CCTTTTGTCCAAGGCTTCAG | 399 | |
| Barley BADH1 (AB063179) | CGCCCTCATAATACTCCTGC | TGGACGCCTCGTCTCAACTC | 1,630 |
| CGCCCTCATAATACTCCTGC | AGGATAGTTCCAGGGAGTGATG | 578 | |
| Barley BADH2 (AB063178) | ACCACCACGCAAGCTCACTC | GAACAGATACAGGTCCTCGAAAC | 1,582 |
| ACCACCACGCAAGCTCACTC | GCTCTAAGCAAGTCACGGAAG | 625 | |
| Spinach BADH (M31480) | ACCAAGAATGGCGTTCCCAA | CTGCATCAACCAACCCTTTATTC | 1,634 |
| Arabidopsis BADH (AY087395) | CACCACGAATCCACGATCCA | GATTTGAACAGACAGCATTTGGT | 1,632 |
| Arabidopsis BADH (AF370333) | GAGTATGGCGATTACGGTGC | CTCAGAGCTTGGAAGGAGGT | 1,517 |
| Tomato BADH (BI935476) | GCATACACTTTGACAAAATA | GCATGTAACGTGCAATTAGT | 1,798 |
| Rice BADH-like (AK068462) | GGGTTGGCGCCCCAGTCTCCG | CCGATGGGTACATCATAAGAGC | 1,879 |
| Rice actin (X16280) | AAGATCCTGACGGAGCGTGGTTAC | CTTCCTAATATCCACGTCGCACTTC | 303 |
| Maize ubiquitin (U29162) | CCACTTGGTGCTGCGTCTTAG | CCTTCTGAATGTTGTAATCCGCA | 218 |
| Wheat actin (AF326781) | GGTGATGAGGCGCAGTCCAAG | CGACCAGCGAGATCCAAACGA | 386 |
| Barley actin (AY145451) | CTGGTCGGGATCTCACGGAC | ATCGCTGGGCCAGACTCGTC | 565 |
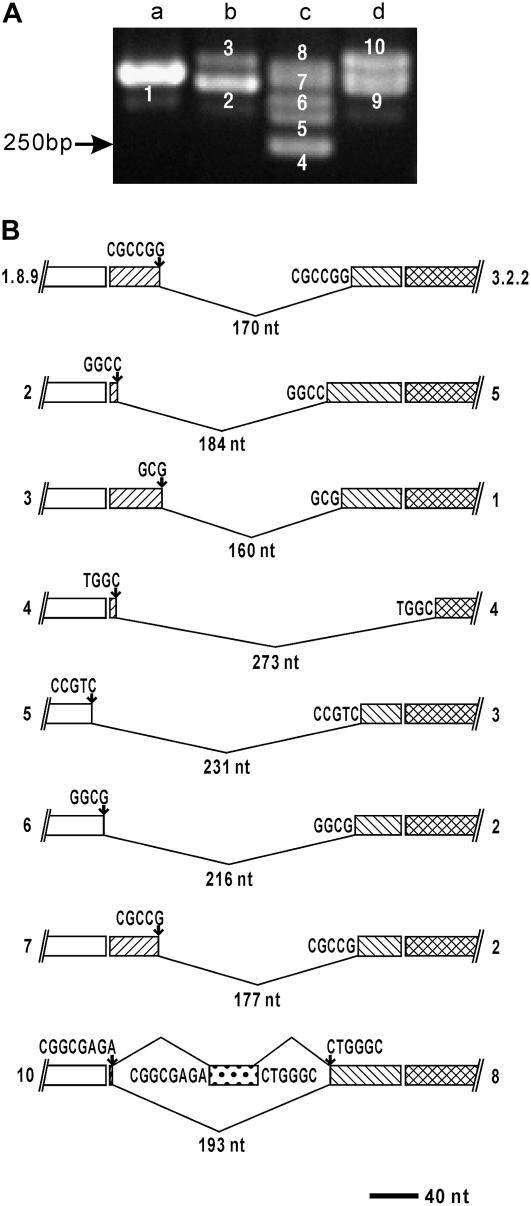
For OsBADH2, preliminary experiments based on RT-PCR showed that the mRNA was expressed constitutively and multiple transcription products were detected (Fig. 3A). Primers specific to the 5′ region were used in these experiments (Table II). To analyze the transcripts derived from the OsBADH2 gene, seedlings from different varieties under different growth conditions were harvested for the total RNA isolation. As a result, all the 59 cDNA clones sequenced also had deletions at the 5′ exonic region (Table III). Similar to that in the OsBADH1 gene, various unusual events in the OsBADH2 locus generated a number of truncated transcripts. The size of the deleted sequences from 5′ UTR and exon(s) ranged from 112 to 523 nucleotides. The start-points of deletions in the 15 cDNA clones varied from −12 to −2 bases of 5′ UTR, resulting in the failure of translation initiation. Twenty-seven cDNA clones encoded derivatives with frame-shifts in the ORF. Eight cDNA clones were able to encode the partial OsBADH2 proteins without frame-shifts. In addition, nine OsBADH2 cDNAs involved the intermolecular recombination of two different RNA molecules, and the exogenous gene fragments derived either from the putative acyl-CoA oxidase or peptidylprolyl isomerase (accession nos. XM_476282 and XM_463914, respectively) were incorporated into OsBADH2 transcripts. These results implicated that the abundance of precisely processed OsBADH2 transcripts was very low.
Figure 3.
Expression and structure of OsBADH2 RNAs derived from seedlings of Nipponbare. A, Agarose gel electrophoresis of RT-PCR amplification products. Product fragments are marked by numbers. Position of Mr marker is indicated. Lanes (a–d) are shown as transcripts under four different growth conditions (unstressed, drought, 0.25 m KCl, and 0.5 m NaCl, respectively). B, Structure of incorrectly processed cDNAs derived from OsBADH2. Clone numbers are indicated on the right. Numbers on the left correspond to the fragment numbers in A. Exons are indicated as boxes, and excised exon materials are shown as folded lines, under which the size is indicated. The positions and sequences of 5′ UTR, exon 1, exon 2, and exon 3 are indicated as white boxes and boxes with slashes, backslashes, and cross-hatching, respectively. Short nucleotide sequences at the 5′ deletion site represent SDRs kept in transcripts, while the nucleotide sequences at the 3′ deletion site are shown as excised SDRs. Exogenous gene fragment other than OsBADH2 is indicated as a stippled box. Scale bar represents 40 nucleotides.
Table III.
The incorrect processing of OsBADH2 transcripts in rice seedling and mature leaf
| Line | Tissue and Growth Condition | No. of Clones | Position and Size of Deletion | SDR | Nearly Identical Sequences |
|---|---|---|---|---|---|
| Nipponbare | Seedling, unstressed | 3 | e1. 42-e2. 102 (170)a | CGCCGG | CGtCGCCGGCG/CGCGCCGGgCG |
| Nipponbare | Seedling, drought | 5 | e1. 7-e2. 81 (184) | GGCC | GGCCaCG/GGCCgCG |
| Nipponbare | Seedling, drought | 1 | e1. 44-e2. 94 (160 bp) | GCG | GGCGaG/GcGCGcG |
| Nipponbare | Seedling, 0.25 m KCl, 24 h | 4 | e1. 6-e3. 26 (273 bp) | TGGC | GAGaTGGC/GAGcTGGC |
| Nipponbare | Seedling, 0.25 m KCl, 24 h | 3 | 5′ UTR −12-e2. 110 (231) | CCGTC | CCGTCgCG/CCGTCCG |
| Nipponbare | Seedling, 0.25 m KCl, 24 h | 2 | 5′ UTR −2-e2. 105 (216) | GGCG | CGGCG/CgGGCG |
| Nipponbare | Seedling, 0.25 m KCl, 24 h | 2 | e1. 41-e2. 108 (177) | CGCCG | GtCGCCGgC/GCGCCGtC |
| Nipponbare | Seedling, 0.25 m KCl, 24 h | 2 | e1. 42-e2. 102 (170) | CGCCGG | CGtCGCCGGCG/CGCGCCGGgCG |
| Nipponbare | Seedling, 0.5 m NaCl, 24 h | 2 | e1. 42-e2. 102 (170) | CGCCGG | CGtCGCCGGCG/CGCGCCGGgCG |
| Nipponbare | Seedling, 0.5 m NaCl, 24 h | 8 | e1. 2-e2. 85 (193)b 40-bp insertion from putative acyl-CoA oxidase | CGGCGAGA CTGGGC | CGGCGAGAtGGCCAC/CGGCGAGAgGGCCAC GaCTGGGCG/GcCTGGGCtG |
| 93-11 | Seedling, unstressed | 3 | e1. 53-e2. 55 (112) | GGCGCG | GtGGCGCG/GcGGCGCG |
| 93-11 | Seedling, 0.1 m NaCl, 72 h | 1 | e1. 55-e2. 123 (178) | CC | CCcCCGCGC/CCtCCGCGC |
| 2 | e2 (143 bp)c | ||||
| Minghui 63 | Seedling, unstressed | 1 | e1. 42-e2. 102 (170) | CGCCGG | CGtCGCCGGCG/CGCGCCGGgCG |
| Minghui 63 | Seedling, 0.1 m NaCl, 72 h | 1 | e1. 42-e2. 102 (170) | CGCCGG | CGtCGCCGGCG/CGCGCCGGgCG |
| Suhui 527 | Seedling, unstressed | 1 | e1. 21-e2. 96 (185)d 59-bp insertion from putative peptidylprolyl isomerase | GCA C | GCAgCGGCAG/GCAcCGGCgAG CgCCGgG/CaCCGcG |
| Suhui 527 | Seedling, 0.1 m NaCl, 72 h | 1 | 5′ UTR −11-e2. 112 (232) | CG | CCGTCG/CCGTcCG |
| Chuanxiang 29B | Seedling, unstressed | 1 | e1. 41-e2. 108 (177) | CGCCG | GtCGCCGgC/GCGCCGtC |
| Chuanxiang 29B | Seedling, 0.1 m NaCl, 72 h | 1 | e1. 23-e5. 63 (523) | CAGC | CAGCgG/CAGCtG |
| Zhonghua 9 | Seedling, unstressed | 1 | e1. 21-e2. 131 (220) | CGCA | TCCCGCA/TCCgCGCA |
| Zhonghua 9 | Seedling, 0.1 m NaCl, 72 h | 1 | 5′ UTR −2-e2. 101 (212) | CG | CGgCGaG/CGcCGgG |
| Yuanlixiangjing | Seedling, unstressed | 1 | e1. 52-e2. 106 (164) | GGCGC | GtGGCGCG/GGGCGCcG |
| Yuanlixiangjing | Seedling, 0.1 m NaCl, 72 h | 1 | 5′ UTR −8-e2. 85 (202) | CGCGA | GtCGCGAtC/GcCGCGAC |
| Leab | Seedling, unstressed | 1 | e1. 55-e2. 107 (162) | GCGCC | GGcGCGCC/GGGCGCC |
| Leab | Seedling, 0.1 m NaCl, 72 h | 1 | 5′ UTR −8-e2. 85 (202) | CGCGA | GtCGCGAtC/GcCGCGAC |
| Newbonnet | Seedling, unstressed | 1 | 5′ UTR −10-e2. 106 (225) | CGC | GtCGCG/GCGCcG |
| Newbonnet | Seedling, 0.1 m NaCl, 72 h | 1 | e1. 18-e2. 140 (232) | CC | |
| Tebonnet | Seedling, unstressed | 1 | e1. 24-e2. 98 (184) | GCG | CGCaGCG/CGCGCG |
| Tebonnet | Seedling, 0.1 m NaCl, 72 h | 1 | e1. 24-e2. 84 (170) | GCG | GCaGCGgC/GCcGCGaC |
| Nipponbare | Mature leaf, field | 2 | 5′ UTR −3-e2. 115 (227) | GGC | CGGCgA/CgGGCcA |
| 93-11 | Mature leaf, field | 1 | 5′ UTR −10-e2. 130 (249) | CGC | CCGtCGCgATCG/CCGCGCaATCG |
| Chuanxiang 29B | Mature leaf, field | 2 | 5′ UTR −3-e2. 99 (211) | GC | CGGCG/CGcGCcG |
e1. 42-e2. 102 (170) denotes that exon materials of OsBADH2 from the 42nd base of exon 1 to the 102nd of exon 2 were excised; 170 nucleotides in total were deleted.
In these cDNAs, the deletions were replaced by a fragment derived from putative acyl-CoA oxidase (accession no. XM_476282).
Selective deletion of entire exon 2.
In this cDNA, the deletion was replaced by a fragment derived from putative peptidylprolyl isomerase (accession no. XM_463914).
Paired Presence of SDRs at Deletion/Insertion Junctions in Rice BADH Transcripts
A total of 100 cDNA clones derived from rice BADH homologs were sequenced to characterize the sequence structure at deletion or insertion boundaries, and, interestingly, the site selection of deletion/insertion was altered in association with the change of stress conditions (Tables I and III; Figs. 1–3). Moreover, the sequence comparison unraveled extensive sequence similarities between the surrounding sequences at the 5′ and 3′ truncating and/or recombination sites in individual events (Figs. 1, B and C, and 3B). As shown in Figure 1B, deletion occurring in the transcripts in lane 3 of Figure 1A resulted in the excision of the first intron and 216-nucleotide exon materials. In this case, the 5′ and 3′ truncating sites involved exon 1 and exon 2, respectively, and the sequence of GGCCG at the 3′ end was identical to that of the immediate upstream 5′ end of the deleted sequence. We denominated the identical sequences as SDRs. Diagrams described in Figure 1C demonstrated that seven types of different SDRs (CG, GGCCGCG, GGCCG, GCC, CGCGC, GGCC, and GGAG) were identified in the deletion events from the OsBADH1 gene expressed in japonica rice cv Nipponbare. Actually, the length of almost all the SDRs can be extended to larger, nearly identical sequences (Table I). For example, two copies of dinucleotide SDR of CG that are separately located at 5′ and 3′ deletion sites, respectively, can be extended to larger, nearly identical sequences of GCCGtCGgCG and GCCGCGaCG (in which the lowercase letters indicate the differences). Similarly, two copies of trinucleotide SDRs of GCC at 5′ and 3′ deletion sites can be replaced by larger, nearly identical sequences of GCCaTGG and GCCTGG, respectively. It is noteworthy that each type of SDR was somewhat unique depending on the position of the truncating sites and contained GC-rich nucleotides. In many cases, only two copies of individual SDRs were present in a proximal genomic region, and each copy located separately at the 5′ or 3′ truncating boundaries. During posttranscriptional processing, one copy of the SDR was excised and the other one retained in the resultant transcripts. Nevertheless, in the case of multiple copies of the SDRs that occurred in pre-mRNA, only two of the copies were recruited by individual deletion events. For instance, four copies of GGCC were present at the seven, 34, and 99 bases of exon 1 and 116 bases of exon 2, respectively, and arranged three combinations of truncating-site choices, in which the copy GGCC at seven bases of exon 1 was most commonly selected for the 5′ excision site, while one of the other three copies was alternatively used for the downstream selection of the 3′ excision site (Table I). Moreover, as indicated in the last cDNA clone in Figure 1C, two independent deletion events were found within a single cDNA clone, and one excision event took place within a single exon, suggesting that the intron might not be required during the processing.
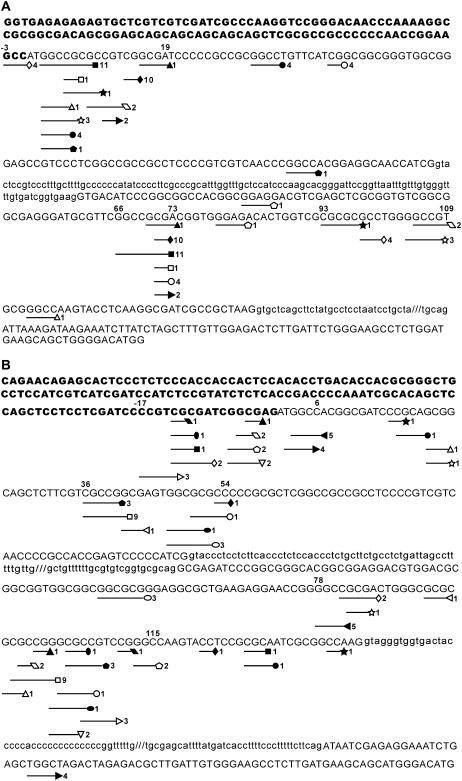
Similarly, as shown in Figure 3B, nine types of SDRs (CGCCGG, GGCC, GCG, TGGC, CCGTC, GGCG, CGCCG, CGGCGAGA, and CTGGGC) were found in transcripts of OsBADH2 induced in japonica rice cv Nipponbare under different stress conditions. These types of SDRs can be largely substituted by longer, nearly identical sequences (Table III). Moreover, in the last OsBADH2 cDNA shown in Figure 3B, the excised sequence was replaced by a 40-bp exogenous gene sequence of acyl-CoA oxidase (accession no. XM_476282). More intriguingly, two types of SDRs (CGGCGAGA and CTGGGC, respectively) were present at the junctions, but in this case only one copy of SDR derived from OsBADH2 and another copy from the acyl-CoA oxidase. A similar phenomenon was observed from another exogenous gene fragment (Table III). Based on the comparison of sequence composition in SDRs, we found a commonly used SDR (GGCC) by both of the rice BADH homologs for the selection of excision sites. A total of 14 and 23 SDRs were identified in OsBADH1 and OsBADH2 for their deletion events, respectively, and their positions in the genomic sequences are shown in Figure 4, A and B. For OsBADH1, SDRs at the 5′ deletion site were mainly distributed in a 22-bp region from −3 to 19 bases of exon 1, and SDRs at the 3′ excision site distributed in a relatively large region from 66 to 109 bases of exon 2. For OsBADH2, SDRs at the 5′ excision site spanned a region from −17 to 54 bases of exon 1, whereas SDRs at the 3′ excision site primarily covered a 38-bp region from 78 to 115 bases of exon 2.
Figure 4.
Distribution of SDRs in genomic sequences of rice BADH homologs. Exons are indicated as uppercase letters, and intron sequences are indicated as lowercase letters. 5′ UTRs are shown as bold letters. Positions of exon sequences are indicated by numbers. SDRs are underlined. Symbols following underlines indicate specificity and position of SDRs. Clone numbers for individual events are indicated on the right. Notice that those sequences between individual paired SDRs will be excised during the unusual posttranscriptional processing. Distribution of SDRs in OsBADH1 (A) and OsBADH2 (B) are shown. Those intron sequences not shown are indicated by interrupted lines.
The Conservation of an Unusual Posttranscriptional Processing Pattern in BADH Homologs in Cereal Crop Species
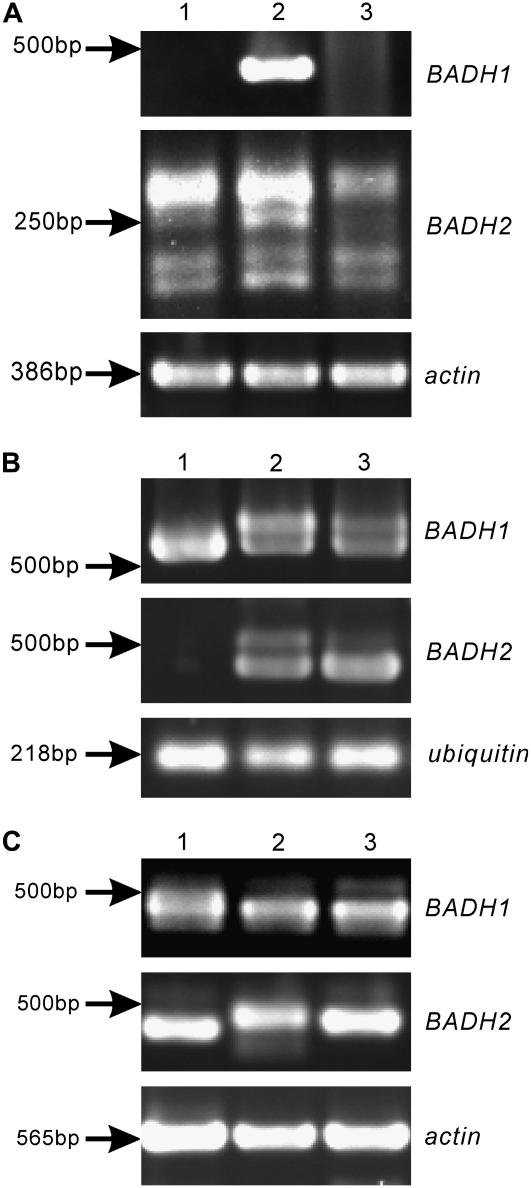
To determine whether the unusual events occurring in the BADH transcripts were specific only to the rice genome, we carried out RT-PCR experiments using the total RNA extracted from seedlings of other cereal crop species, i.e. maize, wheat, and barley (Fig. 5, A–C). These experiments used primers either to amplify the full length of mRNA or exclusively the 5′ region of BADH homologs corresponding to those in rice (Table II). The sequencing data from a total of 52 cDNA clones (four clones for wheat BADH1, 22 for wheat BADH2, six for maize BADH1, nine for maize BADH2, six for barley BADH1, and five for barley BADH2) demonstrated that all the tested cDNA clones had deletion(s) of the 5′ exonic sequences resulting from the unusual posttranscriptional processing. SDRs and/or their extended, nearly identical sequences at the truncating junctions were identified with no exception for every sequence deletion event (Table IV). Multiple deletion events were also observed within a single cDNA clone. These results suggested a highly conserved posttranscriptional processing pattern in BADH homologs that distinctly evolved in cereal crop species.
Figure 5.
Expression of BADH homologs in related cereal crop species. The position of the Mr marker is indicated. Actin or ubiquitin was amplified as internal positive control. Lanes 1 to 3 represent three different growth conditions (unstressed, 0.1, and 0.5 m NaCl, respectively). Agarose gel electrophoresis analysis of RT-PCR amplification products derived from wheat (A), maize (B), and barley (C) BADH homologs are shown.
Table IV.
The incorrectly processed transcripts of BADH homologs in seedlings of wheat cv Chuanmai 43, maize cv 095, and barley cv Chuannongda 3
| Gene | Growth Condition | No. of Clones | Position and Size of Deletion | SDR | Nearly Identical Sequences |
|---|---|---|---|---|---|
| Wheat BADH1 | 0.1 m NaCl, 72 h | 4 | 5′ UTR −13-CDS No. 157 (170)a | CTCGCCG | AcCTCGCCG/AgCTCGCCG |
| Wheat BADH2 | Unstressed | 4 | CDS Nos. 44–250 (207) | ATCG | CATCGaCG/CgATCGcCG |
| Wheat BADH2 | Unstressed | 1 | CDS Nos. 55–204 (150) | GCGACTGG | CgGCGACTGGcGC/CGCGACTGGGC |
| Wheat BADH2 | Unstressed | 2 | CDS Nos. 49–176 (128) | CGGC | GaCGGC/GCGGC |
| Wheat BADH2 | 0.1 m NaCl, 72 h | 3 | CDS Nos. 44–250 (207) | ATCG | CATCGaCG/CgATCGcCG |
| Wheat BADH2 | 0.1 m NaCl, 72 h | 1 | CDS Nos. 30–253 (224) | CG | GCGgCA/GcCGcCA |
| Wheat BADH2 | 0.1 m NaCl, 72 h | 3 | CDS Nos. 55–204 (150) | GCGACTGG | CgGCGACTGGcGC/CGCGACTGGGC |
| Wheat BADH2 | 0.1 m NaCl, 72 h | 1 | CDS Nos. 49–176 (128) | CGGC | GaCGGC/GCGGC |
| Wheat BADH2 | 0.5 m NaCl, 24 h | 3 | CDS Nos. 44–250 (207) | ATCG | CATCGaCG/CgATCGcCG |
| Wheat BADH2 | 0.5 m NaCl, 24 h | 1 | CDS Nos. 30–253 (224) | CG | GCGgCA/GcCGcCA |
| Wheat BADH2 | 0.5 m NaCl, 24 h | 1 | CDS Nos. 55–204 (150) | GCGACTGG | CgGCGACTGGcGC/CGCGACTGGGC |
| Wheat BADH2 | 0.5 m NaCl, 24 h | 1 | CDS Nos. 49–176 (128) | CGGC | GaCGGC/GCGGC |
| Wheat BADH2 | 0.5 m NaCl, 24 h | 1 | CDS Nos. 50–174 (125) | GGCG | CGGCGaC/CgGGCGgC |
| Maize BADH1 | Unstressed | 1 | CDS Nos. 22–249 (228) | CGATC | CGATCcCC/CGATCgCC |
| Maize BADH1 | 0.1 m NaCl, 72 h | 1 | CDS Nos. 22–249 (228) | CGATC | CGATCcCC/CGATCgCC |
| Maize BADH1 | 0.1 m NaCl, 72 h | 1 | CDS Nos. 49–244 (196) | CGGG | CGGGG/CGGGcG |
| Maize BADH1 | 0.5 m NaCl, 24 h | 1 | 5′ UTR −1-CDS No. 252 (252) | ATCGCC | CGcATCGCC/CGATCGCC |
| Maize BADH1 | 0.5 m NaCl, 24 h | 1 | CDS Nos. 47–246 (200) | GGCG | |
| Maize BADH1 | 0.5 m NaCl, 24 h | 1 | CDS Nos. 31–75 (45) | GGCGC | GGCGCgGCCT/GGCGCcGCCT |
| CDS Nos. 120–247 (128) | GGCGA | CGGCGAC/CgGGCGAtC | |||
| Maize BADH2 | 0.1 m NaCl, 72 h | 5 | CDS Nos. 117–259 (143) | GG | GGcGA/GGtGA |
| Maize BADH2 | 0.1 m NaCl, 72 h | 1 | CDS Nos. 50–168 (119) | CGGCG | GtCGaCGGCG/GCGgCGGCG |
| Maize BADH2 | 0.5 m NaCl, 24 h | 3 | CDS Nos. 117–259 (143) | GG | GGcGA/GGtGA |
| Barley BADH1 | Unstressed | 1 | 5′ UTR −24-CDS No. 210 (234) | GCGC | GGCGCC/GcGCGCgC |
| Barley BADH1 | Unstressed | 1 | 5′ UTR −24-CDS No. 208 (232) | GGCGC | GGCGCC/GGCGCgC |
| Barley BADH1 | Unstressed | 2 | 5′ UTR −5-CDS No. 197 (202) | GAG | |
| Barley BADH1 | 0.1 m NaCl, 72 h | 1 | 5′ UTR −24-CDS No. 208 (232) | GGCGC | GGCGCC/GGCGCgC |
| Barley BADH1 | 0.5 m NaCl, 24 h | 1 | 5′ UTR −24-CDS No. 208 (232) | GGCGC | GGCGCC/GGCGCgC |
| Barley BADH2 | Unstressed | 2 | 5′ UTR −7-CDS No. 178 (184) | CCGCCC | CCGCCCgC/CCGCCCtC |
| Barley BADH2 | 0.1 m NaCl, 72 h | 2 | CDS Nos. 64–214 (151) | CGCGCCC | CgCGCGCCCgCG/CCGCGCCCcCG |
| Barley BADH2 | 0.5 m NaCl, 24 h | 1 | CDS Nos. 58–221 (164) | GGCGC | GGCGCgCG/GGCGCCG |
5′ UTR −13-CDS No. 157 (170) means the exon materials from the −13 of 5′ UTR to the 157th base of coding sequence were excised; 170 nucleotides in total were deleted. CDS, Coding sequence.
To compare the posttranscriptional processing patterns of the BADH homologs between cereal crop species and more distantly related dicotyledonous species, we conducted RT-PCR experiments using total RNA extracted from seedlings of spinach, Arabidopsis, and tomato. Primers designed to amplify the full length of mRNA of BADH homologs were used (Table II). As anticipated, the RT-PCR products of BADH homologs from spinach (accession no. M31480), Arabidopsis (accession nos. AY087395 and AF370333), and tomato (accession no. BI935476) were of expected size for correctly processed transcripts. Sequencing analysis of 11 cDNA clones (four from spinach, four from Arabidopsis, and three from tomato) confirmed the correct processing. We also sequenced two cDNA clones derived from a distantly related BADH-like gene in rice (accession no. AK068462), and no aberrant transcripts were observed.
Based on the sequence comparison of the 5′ exonic region including exons 1 and 2, we did not find, in the distantly related BADH homologs, such extensive sequence similarity (SDRs) with paired presence at the deletion/insertion junctions in the tested cereal crop species. This consistently suggests that SDRs may be required for the recognition of deletion/insertion sites.
DISCUSSION
Defective BADH Transcripts Resulted from Unusual Posttranscriptional Processing in Cereal Crop Species
Extensive sequence analysis from this study revealed that a number of aberrant BADH transcripts were derived from an unusual posttranscriptional processing in cereal crop species. The observed events took place at the 5′ exonic region, leading to the removal of translation initiation codon, deletion of functional domain, insertion of exogenous gene fragments, and frame-shifts with premature termination by introducing stop codon. We found that the majority of the incorrectly processed BADH transcripts were prematurely terminated by incorporation of stop codon. These transcripts are apparent targets of nonsense-mediated mRNA decay, a surveillance mechanism that selectively degrades nonsense mRNAs (Stamm et al., 2005). This finding suggested that very few functional BADH transcripts were made during RNA processing. The phenomenon of abnormally processed transcripts in plant genes has been described previously in maize and wheat (Burr et al., 1996; McKibbin et al., 2002). Most of the transcripts in maize In1 transcription-factor gene were spliced incorrectly, and missplicing occurred before the helix-loop-helix DNA-binding domain, indicating that truncated protein would not contain specific DNA-binding activity (Burr et al., 1996). Extensive missplicing of transcripts has also been observed for wheat Vp-1 transcription factor gene. Analysis of Vp-1 transcript structure showed many combinations of intron insertion/exon deletions, resulting in termination of the ORF before the B3 domain (McKibbin et al., 2002). The highly conserved B3 domain has been shown in maize to bind DNA in a sequence-specific, highly cooperative manner, activating transcription through the Sph cis-element present in the C1 promoter (Suzuki et al., 1997).
Our analysis of the cDNA sequence structure from various BADH genes revealed that numerous deletion events came about within about 200 bp in the 5′ exonic region that encodes a part of the putative NAD+-binding domain (Incharoensakdi et al., 2000). A variety of partial deletions involving exons 1 and 2 directly led to the termination of ORF before the catalytic domain. A substantial portion of the transcripts had deletions without the original translation initiation codon. These transcripts still contain a shorter ORF with the catalytic and oligomerization domains (Incharoensakdi et al., 2000). Consequently, from all the BADH cDNA clones sequenced in this study, no single cDNA clone was found to be processed correctly. However, this type of processing event may not be well represented in the RT-PCR products, as described in the missplicing of Vp-1 gene (McKibbin et al., 2002). Indeed, correctly spliced BADH mRNAs are found in published cDNA sequences in rice (accession nos. AK103582 and AK071221) as well as other cereal crop species (accession nos. AY587278, AY050316, AB063179, and AB063178). As a result, a residual BADH protein activity likely exists in these cereal crops, even though the precise mRNAs were decreased considerably by the unusual posttranscriptional processing.
Nevertheless, GB level accumulated in plants could be altered by any limiting factor in the complex biosynthetic pathway. Either BADH or CMO protein activity is crucial for the ultimate GB-synthesizing ability. Indeed, rice is the only cereal crop that does not accumulate GB (Rathinasabapathi et al., 1993). Although the rice genome contains both CMO and BADH homologs required for GB synthesis, their activities of coding proteins have not yet been characterized experimentally. In addition to the extensively defective RNA processing of rice BADH homologs found in this study, recent transgenic experiments suggest a functional defect in both CMO and BADH homologs in rice. Kishitani et al. (2000) reported the transgenic rice plants constitutively expressing a precise barley BADH1 converted high levels of exogenously applied betaine aldehyde to GB more efficiently than did wild-type plants. The lower conversion efficiency in the wild-type plants probably results from the limitation of precise native BADH proteins found in this study. More recently, Shirasawa et al. (2006) demonstrated transgenic rice plants harboring a single copy of expressed spinach CMO accumulated detectable GB and had enhanced tolerance to salt stress and temperature stress, strongly suggesting a severe deficit of native CMO protein activity in rice. Because CMO alone only converts choline into betaine aldehyde, these transgenic plants still need native functional BADH proteins for conversion of betaine aldehyde into GB, suggesting an active BADH protein present in rice. In addition, analyses of other transgenic studies have shown that stress tolerance levels as measured by increased GB accumulation could be manipulated via expression of the BADH orthologs (Sakamoto et al., 1998; Takabe et al., 1998), implying that the functional transgene is capable of complementing the functional defect in rice BADH genes. It is noteworthy that rice may take other defense strategies instead of accumulating GB against osmotic stresses. Ren et al. (2005) reported recently a major quantitative trait locus encoding a sodium transporter confers salt tolerance in rice.
By contrast, other cereal crops have been known to synthesize GB and tolerate various degrees of osmotic stresses. Their precisely spliced BADH mRNAs exist in vivo, as evidenced by the published cDNA sequences, and, particularly, the activity of barley BADH protein has been demonstrated in the transgenic experiment (Kishitani et al., 2000). Obviously, barley plants synthesize GB through catalytic reaction of the functional BADH protein, even though a large number of incorrectly processed BADH transcripts observed in this study may considerably reduce the precise gene products.
Interestingly, tremendous variation in GB accumulation among dicotyledonous species exists. Spinach has been documented to accumulate far more GB than do cereal crops in response to osmotic stresses (Hanson and Scott, 1980; Pan et al., 1981). We may associate these features with the demonstrated enzyme activities of spinach CMO and BADH proteins that have been well documented in a number of biochemical and genetic engineering experiments (Weretilnyk and Hanson, 1989; Rathinasabapathi et al., 1994; Burnet et al., 1995; Shirasawa et al., 2006). We did not find any abnormal events in splicing of spinach BADH homolog after intensive studies. However, the case in Arabidopsis is completely different. Arabidopsis contains one CMO-like (accession no. NM_119135) and two BADH homologs (accession nos. AY087395 and AF370333) but does not accumulate this compound of GB (Sakamoto and Murata, 2002). Although our study showed the existence of potential BADH function from correct splicing, another experiment demonstrated that the protein coding for CMO-like gene was absent in Arabidopsis plants and there was no enzyme activity in the CMO-like protein expressed in Escherichia coli (Hibino et al., 2002). This is sufficient to abrogate the biosynthetic ability of GB in Arabidopsis.
Because no abnormal BADH transcripts were detected in the studied dicotyledonous species, it appears that the RNA processing pattern of BADH homologs was considerably differentiated between monocotyledons and dicotyledons. But why maintain over such a long evolutionary span a stress-responsive gene and then reduce its activity by incorrect processing in monocotyledons? One possible reason is the differential tolerance to the deleterious effect of the accumulated GB among different plant species. The toxicity was indicated by a destabilizing effect of exogenous supplementation of GB in plant tissues during viability tests of membrane stability (Gibon et al., 1997). Cereal crops may suffer from overaccumulation of GB, and the appropriate level of GB accumulation may be regulated at both the transcriptional and posttranscriptional levels; namely, the transcription is induced abundantly in response to the osmotic stresses, while the proper amount of precise gene products is balanced by posttranscriptional processing. In this view, keeping the appropriate active protein level by the unusual posttranscriptional processing is subject to a favorable evolutionary selection. However, more detailed studies need to be conducted to confirm this hypothesis.
The Selection of Deletion/Insertion Sites Probably Regulated by SDRs
Posttranscriptional splicing accomplishes the excision of introns and the joining of exons into the mature sequences of RNA. Introns are removed from the nuclear pre-mRNA of higher eukaryotes by a system that recognizes only short consensus sequences at exon-intron boundaries and within the intron. In the classic U2-type of spliceosome-mediated splicing, the consensus sequences of the 5′ and 3′ splices sites, AG/GTAAGT and TGCAG/G (the slash denotes the exon-intron boundary), are highly conserved in higher plants and are similar to those in vertebrates (Liu and Filipowicz, 1996; Lorkovic et al., 2000). In this way, intron starts with dinucleotide GT and ends with the dinucleotide AG. Intron border sequences are required for the selection of splice sites by splice machinery. A minor class of nuclear pre-mRNA introns, referred to as U12-type, has recently been described (Tam and Steitz, 1997; Russell et al., 2006). U12-type introns frequently start with AT and terminate with AC. Moreover, primary transcripts of many genes are alternatively spliced by selection of different splice sites, producing different mRNA forms that encode proteins with functional differences (Lorkovic et al., 2000). In plants, the biological relevance of alternative exon inclusion and retention of an unspliced intron regulated by SR (Ser/Arg-rich) proteins has been documented (Isshiki et al., 2006).
In this study, the posttranscriptional processing pattern found in BADH homologs in the cereal crops does not conform to either the major GT-AG or the minor AT-AC roles. From a wide variety of unusual deletion/insertion events involving exon materials, we were not able to identify such short consensus at the ends of deleted or inserted sequences. Instead, we did find extensive sequence similarities surrounding the deletion/insertion junctions, at which the putative reactions of excision and joining during the processing were likely directed by the identical sequences called SDRs. SDRs are characterized by a short size (2–7 GC-rich nucleotides), paired recruitment, and direct orientation. More careful inspection of sequence data revealed that di- or trinucleotide SDRs could be extended to relatively larger, nearly identical sequences that might be required for the recognition as signature sequences (Tables I, III, and IV). For each unusual processing event, there are two copies of the same SDRs present in the pre-mRNA, and only one is always excised precisely. There are several sorts of deletion/insertion events that have consistently shown the selection of the deletion/insertion sites relies specifically on the presence and positioning of SDRs in pre-mRNA, although we cannot completely rule out other possibilities. First, the majority of deletion events involve two different exons that are composed of the same SDRs. This type of deletion also occurred in a published maize BADH1 cDNA (accession no. DW475114) with two copies of the sequence CGCC located separately at exon 1 and exon 2, respectively. Second, deletion detected within exons is probably the result of the presence of two copies of SDRs in the same exon, suggesting that the presence of an intron is not required. Third, in the case of multiple copies of an SDR present in the proximity of the genomic sequence, only two of them were mutually recruited in a manner that is quite similar to alternative splicing for the selection of splice site (Black, 2003). Fourth, in the form of intermolecular recombination (exogenous sequence insertion) identified in this experiment, two copies of the SDRs are of paired presence between different RNA molecules. Probably due to the sequence differentiation and absence of paired SDRs or nearly identical sequences, we did not observe any abnormal processing event in BADH homologs from spinach, tomato, and Arabidopsis, and in a distantly related BADH-like gene from rice. This implicates a sequence-dependent manner for the determination of the processing patterns.
However, the special mechanism that enforces the exclusive choice of deletion/insertion sites through recognition of SDRs is elusive. Because the proportion of unusual transcripts derived from BADH genes is so great, the event seems unlikely a missplicing that usually fails to recognize exon-intron junction by spliceosome. Instead, an unusual alternative splicing may be responsible for the high incidence of unusual BADH transcripts. This investigation indicates that the nature of the putatively SDR-directed deletion/insertion is similar to the intron splicing in conventional RNA processing, both seemingly involving reactions of excision and rejoining of RNA sequences. Similar identical trinucleotide AGG exists at the U2-type spliceosome-mediated splicing junctions (AG/GTAAGT and TGCAG/G at 5′ and 3′ splice sites, respectively) in the same manner of SDRs found in this experiment (Lorkovic et al., 2000). Hence, we argue that the conventional spliceosome or at least some components from it might be involved in the unusual, SDR-mediated posttranscriptional RNA processing. In fact, many components of the splicing machinery appear to be shared by both U2- and U12-type spliceosomes, even though distinct differences (GT-AG versus AT-AC) in signature sequences at U2- and U12-type intron borders exist (Tam and Steitz, 1997; Russell et al., 2006). Possibly, these components from conventional spliceosome could also be shared by the unusual alternative splicing. Additionally, SR proteins and other splicing factors and/or hnRNP proteins found in rice and other organisms play an important role in regulating splice-site selection by binding to sequence elements (enhancers or silencers) present in exons and introns (Lorkovic et al., 2000; Isshiki et al., 2006). Cereal crop genomes probably encode a special type of splicing factors to interact with the sequence elements of SDRs.
Alternatively, the unusual events result from intermolecular and/or intramolecular recombination occurring in the posttranscriptional processing mediated by SDRs without participation of splice machinery. This probably will need additional protein factors other than spliceosome to bring together the sequence elements of SDRs that are not complementary and separately located at putatively 5′ and 3′ recombination sites and to allow the reaction of recombination. Actually, a model of intramolecular recombination mediated by SDRs has been reported in the wheat chloroplast genome (Ogihara et al., 1988). This form of intramolecular recombination mediated by SDRs has also been observed within the ORF of rice chloroplast gene (Kanno et al., 1993). It seems reasonable to propose that numerous unusual BADH transcripts were derived from this form of recombination mediated by SDRs.
Interestingly, stressed conditions induced several intermolecular recombinant transcripts derived from different RNA molecules (Table III). Analogous examples of trans-splicing in which separate precursor transcripts contribute sequences to the mature mRNA through intermolecular reactions have been identified in plant chloroplasts and mitochondria (Wissinger et al., 1991). In this type of trans-splicing, discrete pre-mRNA molecules that are independently transcribed have sequences with features characteristic of group II introns. Also, the intermolecular recombination was documented in a rice calcium-dependent protein kinase gene encoded in nuclei (Kawasaki et al., 1999). In this case, the splicing process does not involve consensus splicing signals. Obviously, the currently observed unusual intermolecular recombination is different from above trans-splicing events. This process is possibly mediated by SDRs of paired presence with a new kind of mechanism. We should also consider the possibility that intermolecular recombination between two molecules from the same locus may have occurred.
MATERIALS AND METHODS
Plant Materials
Varieties or lines from seven plant species, rice (Oryza sativa), maize (Zea mays), wheat (Triticum aestivum), barley (Hordeum vulgare), spinach (Spinacia oleracea), tomato (Solanum lycopersicum), and Arabidopsis (Arabidopsis thaliana), used in this study are listed in Table V. Seeds of each variety or line were surface sterilized with 1% (v/v) sodium hypochlorite and germinated in a growth chamber (25°C, 14-h/10-h photoperiod at 200 μmol photons m−2 s−1). Ten days after germination, seedlings were subjected to different stresses of salt (0–0.5 m NaCl or 0.25 m KCl) or drought (by withdrawing water for 24 h) treatments. Seedling leaves were collected for RNA extraction and analysis. Mature rice leaves were obtained for analysis from plants grown in an experimental field at the Sichuan University. Rice callus was induced in the medium as described (Hiei, et al., 1994).
Table V.
Plant materials used for RNA isolation
| Species | Variety/Lines |
|---|---|
| Rice subsp. japonica | Nipponbare |
| Rice subsp. japonica | Zhonghua 9 |
| Rice subsp. japonica | Yuanlixiangjing |
| Rice subsp. japonica | Newbonnet |
| Rice subsp. japonica | Tebonnet |
| Rice subsp. japonica | Leab |
| Rice subsp. japonica | Della |
| Rice subsp. indica | 93-11 |
| Rice subsp. indica | Minghui 63 |
| Rice subsp. indica | Suhui 527 |
| Rice subsp. indica | Chuanxiang 29B |
| Rice subsp. indica | Basmati |
| Maize | 095 |
| Wheat | Chuanmai 43 |
| Barley | Chuannongda 3 |
| Spinach | Dayuanye |
| Spinach | Chunqiudayebo |
| Tomato | Ailsa Craig |
| Arabidopsis | Columbia |
RNA Isolation and RT-PCR
Total RNAs were extracted by using Trizol reagent following the protocol provided by the manufacturer (Invitrogen). First-strand cDNA was synthesized by using the First Strand cDNA Synthesis kit (Toyobo). The oligonucleotide primers used for amplifying genes of BADH, actin, ubiquitin, and rice BADH-like are summarized in Table II. PCR was carried out by using a Taq DNA Polymerase (Takara) in MJ MiniPCR (Bio-Rad) following the instruction given by the manufacturer. Actin and ubiquitin genes were employed as positive internal controls (Quaggiotti et al., 2003; Liu et al., 2005).
Sequencing and Data Analysis
RT-PCR products containing multiple bands were separated by 2% agarose gel and purified using the QIAquick Gel-Extraction kit (Qiagen). The isolated fragments were cloned into a pMD18-T vector (Takara). Cycle sequencing was performed with the ABI Prism BigDye Terminators v2.0 cycle sequencing reaction kit (Applied Biosystems). Sequences were determined with an ABI Prism 377 genetic analyzer (Applied Biosystems) and edited with the computer program BioEdit v4.7.8 (Hall, 1999). The sequence alignments were performed using the software ClustalX v1.83 (Thompson et al., 1997), and DNAMEN v5.0 (Lynnon Biosoft) was used to translate DNA sequences into protein sequences to aid the analysis of RNA processing events.
Sequence data from this study can be found in the GenBank data libraries under accession numbers AK103582 (OsBADH1), AK071221 (OsBADH2), DV031390 (maize BADH1), AY587278 (maize BADH2), BJ259181 (wheat BADH1), AY050316 (wheat BADH2), AB063179 (barley BADH1), AB063178 (barley BADH2), AK068462 (rice BADH-like), M31480 (spinach BADH), AY087395 (Arabidopsis BADH), AF370333 (Arabidopsis BADH), BI935476 (tomato BADH), X16280 (rice actin), U29162 (maize ubiquitin), AF326781 (wheat actin), and AY145451 (barley actin).
Acknowledgments
We thank Drs. Shigui Li (from Rice Research Institute, Sichuan Agricultural University), Zongyun Feng (from College of Agronomy, Sichuan Agricultural University), Wuyun Yang, and Duanping Yang (from Crop Research Institute, Sichuan Academy of Agricultural Sciences) for providing the seeds of rice, barley, wheat, and maize, respectively.
This work was supported by the Chinese Ministry of Science and Technology (973 Program, grant no. 2006CB100205), by the local government of Sichuan Province (application basis project, grant no. 2006Z–05–0039), and by Sichuan University (985 youth talent program, grant no. 0082204127106).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yongsheng Liu (liuyongsheng1122@yahoo.com.cn).
References
- Arakawa K, Katayama M, Takabe T (1990) Levels of betaine and betaine aldehyde dehydrogenase activity in the green leaves, and etiolated leaves and roots of barley. Plant Cell Physiol 31 797–803 [Google Scholar]
- Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. CRC Crit Rev Plant Sci 24 23–58 [Google Scholar]
- Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72 291–336 [DOI] [PubMed] [Google Scholar]
- Boyer JS (1982) Plant productivity and environment. Science 218 443–448 [DOI] [PubMed] [Google Scholar]
- Bradbury LMT, Fitzgerald TL, Henry RJ, Jin Q, Waters DLE (2005) The gene for fragrance in rice. Plant Biotechnol J 3 363–370 [DOI] [PubMed] [Google Scholar]
- Burnet M, Lafontaine PJ, Hanson AD (1995) Assay, purification, and partial characterization of choline monooxygenase from spinach. Plant Physiol 108 581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr FA, Burr B, Scheffler BE, Blewitt M, Wienand U, Matz EC (1996) The maize repressor-like gene intensifier1 shares homology with the r1/b1 multigene family of transcription factors and exhibits missplicing. Plant Cell 8 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WP, Li PH, Chen THH (2000) Glycinebetaine increases chilling tolerance and reduces chilling-induced lipid peroxidation in Zea mays L. Plant Cell Environ 23 609–618 [Google Scholar]
- Chern MK, Pietruszko R (1995) Human aldehyde dehydrogenase E3 isozyme is a betaine aldehyde dehydrogenase. Biochem Bioph Res Commun 213 561–568 [DOI] [PubMed] [Google Scholar]
- Falkenberg P, Strom AR (1990) Purification and characterization of osmoregulatory betaine aldehyde dehydrogenase of Escherichia coli. Biochim Biophys Acta 1034 253–259 [DOI] [PubMed] [Google Scholar]
- Flowers TJ, Yeo AR (1981) Variability in the resistance of sodium chloride salinity within rice varieties. New Phytol 88 363–373 [Google Scholar]
- Gibon Y, Bessieres MA, Larher F (1997) Is glycine betaine a non-compatible solute in higher plants that do not accumulate it? Plant Cell Environ 20 329–340 [Google Scholar]
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41 95–98 [Google Scholar]
- Hanson AD, Scott NA (1980) Betaine synthesis from radioactive precursors in attached, water-stressed barley leaves. Plant Physiol 66 342–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Wyse R (1982) Biosynthesis, translocation, and accumulation of betaine in sugar beet and its progenitors in relation to salinity. Plant Physiol 70 1191–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harinasut P, Tsutsui K, Takabe T, Nomura M, Takabe T, Kishitani S (1996) Exogenous glycinebetaine accumulation and increased salt-tolerance in rice seedlings. Biosci Biotechnol Biochem 60 366–368 [DOI] [PubMed] [Google Scholar]
- Hibino T, Waditee R, Araki E, Ishikawa H, Aoki K, Tanaka Y, Takabe T (2002) Functional characterization of choline monooxygenase, an enzyme for betaine synthesis in plants. J Biol Chem 277 41352–41360 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6 271–282 [DOI] [PubMed] [Google Scholar]
- Holmström KO, Somersalo S, Mandal A, Palva TE, Welin B (2000) Improved tolerance to salinity and low temperature in transgenic tobacco producing glycine betaine. J Exp Bot 51 177–185 [DOI] [PubMed] [Google Scholar]
- Ikuta S, Imamura S, Misaki H, Horiuti Y (1977) Purification and characterization of choline oxidase from Arthrobacter globiformis. J Biochem (Tokyo) 82 1741–1749 [DOI] [PubMed] [Google Scholar]
- Incharoensakdi A, Matsuda N, Hibino T, Meng YL, Ishikawa H, Hara A, Funaguma T, Takabe T, Takabe T (2000) Overproduction of spinach betaine aldehyde dehydrogenase in Escherichia coli. Eur J Biochem 267 7015–7023 [DOI] [PubMed] [Google Scholar]
- Incharoensakdi A, Takabe T, Akazawa T (1986) Effect of betaine on enzyme activity and subunit interaction of ribulose-1,5-bisphosphate carboxylase/oxygenase from Aphanothece halophytical. Plant Physiol 81 1044–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Arakawa K, Mizuno K, Kishitani S, Takabe T (1993) Betaine aldehyde in leaves of both betain-accumulating and non-accumulating cereal plants. Plant Cell Physiol 34 493–495 [Google Scholar]
- Ishitani M, Nakamura T, Han SY, Takabe T (1995) Expression of the betaine aldehyde dehydrogenase gene in barley in response to osmotic stress and abscisic acid. Plant Mol Biol 27 307–315 [DOI] [PubMed] [Google Scholar]
- Isshiki M, Tsumoto A, Shimamoto K (2006) The serine/arginine-rich protein family in rice plays important roles in constitutive and alternative splicing of Pre-mRNA. Plant Cell 18 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagendorf AT, Takabe T (2001) Inducers of glycinebetaine synthesis in barley. Plant Physiol 127 1827–1835 [PMC free article] [PubMed] [Google Scholar]
- Kanno A, Watanabe N, Nakamura I, Hirai A (1993) Variations in chloroplast DNA from rice (Oryza sativa): differences between deletions mediated by short direct-repeat sequences within a single species. Theor Appl Genet 86 579–584 [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Okumura S, Kishimoto N, Shimada H, Higo K, Ichikawa N (1999) RNA maturation of the rice SPK gene may involve trans-splicing. Plant J 18 625–632 [DOI] [PubMed] [Google Scholar]
- Kishitani S, Takanami T, Suzuki M, Oikawa M, Yokoi S, Ishitani M, Alvarez-Nakase AM, Takabe T, Takabe T (2000) Compatibility of glycinebetaine in rice plants: evaluation using transgenic rice plants with a gene for peroxisomal betaine aldehyde dehydrogenase from barley. Plant Cell Environ 23 107–114 [Google Scholar]
- Lamark T, Kaasen I, Eshoo MW, Falkenberg P, McDougall J, Strom AR (1991) DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol Microbiol 5 1049–1064 [DOI] [PubMed] [Google Scholar]
- Lerma C, Rich PJ, Ju GC, Yang WJ, Hanson AD, Rhodes D (1991) Betaine deficiency in maize. Plant Physiol 95 1113–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Filipowicz W (1996) Mapping of branchpoint nucleotides in mutant pre-mRNAs expressed in plant cells. Plant J 9 381–389 [DOI] [PubMed] [Google Scholar]
- Liu JG, Yao QH, Zhang Z, Peng RH, Xiong AS, Xu F, Zhu H (2005) Isolation and characterization of a cDNA encoding two novel heat-shock factor OsHSF6 and OsHSF12 in Oryza sativa L. J Biochem Mol Biol 38 602–608 [DOI] [PubMed] [Google Scholar]
- Livingstone JR, Maruo T, Yoshida I, Tarui Y, Hirooka K, Yamamoto Y, Tsutui N, Hirasawa E (2003) Purification and properties of betaine aldehyde dehydrogenase from Avena sativa. J Plant Res 116 133–140 [DOI] [PubMed] [Google Scholar]
- Lorkovic ZJ, Kirk DAW, Lambermon MHL, Filipowicz W (2000) Pre-mRNA splicing in higher plants. Trends Plant Sci 5 160–167 [DOI] [PubMed] [Google Scholar]
- McCue KF, Hanson AD (1992) Salt-inducible betaine aldehyde dehydrogenase from sugar beet: cDNA cloning and expression. Plant Mol Biol 18 1–11 [DOI] [PubMed] [Google Scholar]
- McKibbin RS, Wilkinson MD, Bailey PC, Flintham JE, Andrew LM, Lazzeri PA, Gale MD, Lenton JR, Holdsworth MJ (2002) Transcripts of Vp-1 homeologues are misspliced in modern wheat and ancestral species. Proc Natl Acad Sci USA 99 10203–10208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Nomura M, Mori H, Jagendorf AT, Ueda A, Takabe T (2001) An isozyme of betaine aldehyde dehydrogenase in barley. Plant Cell Physiol 42 1088–1092 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Yokota S, Muramoto Y, Tsutsui K, Oguri Y, Fukui K, Takabe T (1997) Expression of a betaine aldehyde dehydrogenase gene in rice, a glycinebetaine nonaccumulator, and possible localization of its protein in peroxisomes. Plant J 11 1115–1120 [DOI] [PubMed] [Google Scholar]
- Ogihara Y, Terachi T, Sasakuma T (1988) Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc Natl Acad Sci USA 85 8573–8577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan SM, Moreau RA, Yu C, Huang AHC (1981) Betaine accumulation and betaine-aldehyde dehydrogenase in spinach leaves. Plant Physiol 67 1105–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaggiotti S, Ruperti B, Borsa P, Destro T, Malagoli M (2003) Expression of a putative high-affinity NO3− transporter and of an H+-ATPase in relation to whole plant nitrate transport physiology in two maize genotypes differently responsive to low nitrogen availability. J Exp Bot 54 1023–1031 [DOI] [PubMed] [Google Scholar]
- Rathinasabapathi B, Burnet M, Russell BL, Gage DA, Liao PC, Nye GJ, Scott P, Golbeck JH, Hanson AD (1997) Choline monooxygenase, an unusual iron-sulfur enzyme catalyzing the first step of glycine betaine synthesis in plants: prosthetic group characterization and cDNA cloning. Proc Natl Acad Sci USA 94 3454–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinasabapathi B, Gage DA, Mackill DJ, Hanson AD (1993) Cultivated and wild rices do not accumulate glycinebetaine due to deficiencies in two biosynthetic steps. Crop Sci 33 534–538 [Google Scholar]
- Rathinasabapathi B, McCue KF, Gage DA, Hanson AD (1994) Metabolic engineering of glycine betaine synthesis: plant betaine aldehyde dehydrogenases lacking typical transit peptides are targeted to tobacco chloroplasts where they confer betaine aldehyde resistance. Planta 193 155–162 [DOI] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37 1141–1146 [DOI] [PubMed] [Google Scholar]
- Rhodes D, Hanson AD (1993) Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu Rev Plant Physiol 44 357–384 [Google Scholar]
- Russell AG, Charette JM, Spencer DF, Gray MW (2006) An early evolutionary origin for the minor spliceosome. Nature 443 863–866 [DOI] [PubMed] [Google Scholar]
- Russell BL, Rathinasabapathi B, Hanson AD (1998) Osmotic stress induces expression of choline monooxygenase in sugar beet and amaranth. Plant Physiol 116 859–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A, Alia, Murata N (1998) Metabolic engineering of rice leading to biosynthesis of glycinebetaine and tolerance to salt and cold. Plant Mol Biol 38 1011–1019 [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Murata N (2002) The role of glycine betaine in the protection of plants from stress: clues from transgenic plants. Plant Cell Environ 25 163–171 [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Valverde R, Alia, Chen THH, Murata N (2000) Transformation of Arabidopsis with the codA gene for choline oxidase enhances freezing tolerance of plants. Plant J 22 449–453 [DOI] [PubMed] [Google Scholar]
- Shirasawa K, Takabe T, Takabe T, Kishitani S (2006) Accumulation of glycinebetaine in rice plants that overexpress choline monooxygenase from spinach and evaluation of their tolerance to abiotic stress. Ann Bot (Lond) 98 565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H (2005) Function of alternative splicing. Gene 344 1–20 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR (1997) The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9 799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabe T, Hayashi Y, Tanaka A, Takabe T, Kishitani S (1998) Evaluation of glycinebetaine accumulation for stress tolerance in transgenic rice plants. In Proceedings of International Workshop on Breeding and Biotechnology for Environmental Stress in Rice. Hokkaido Agricultural Experiment Station, Sapporo, Japan, pp 63–68
- Tam WY, Steitz JA (1997) Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem Sci 22 132–137 [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot (Lond) 91 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trossat C, Rathinasabapathi B, Hanson AD (1997) Transgenically expressed betaine aldehyde dehydrogenase efficiently catalyzes oxidation of dimethylsulfoniopropionaldehyde and omega-aminoaldehydes. Plant Physiol 113 1457–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela-Soto EM, Munoz-Clares RA (1994) Purification and properties of betaine aldehyde dehydrogenase extracted from detached leaves of Amaranthus hypochondriacus L. subjected to water deficit. Plant Physiol 143 145–152 [Google Scholar]
- Weretilnyk EA, Hanson AD (1989) Betaine aldehyde dehydrogenase from spinach leaves: purification, in vitro translation of the mRNA, and regulation by salinity. Arch Biochem Biophys 271 56–63 [DOI] [PubMed] [Google Scholar]
- Weretilnyk EA, Hanson AD (1990) Molecular cloning of a plant betaine-aldehyde dehydrogenase, an enzyme implicated in adaptation to salinity and drought. Proc Natl Acad Sci USA 87 2745–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilken DR, McMacken ML, Rodriquez A (1970) Choline and betaine aldehyde oxidation by rat liver mitochondria. Biochim Biophys Acta 216 305–317 [DOI] [PubMed] [Google Scholar]
- Wissinger B, Schuster W, Brennicke A (1991) Trans splicing in Oenothera mitochondria: nadi mRNAs are edited in exon and trans-splicing group II intron sequences. Cell 65 473–482 [DOI] [PubMed] [Google Scholar]
- Wood AJ, Saneoka H, Rhodes D, Joly RJ, Goldsbrough PB (1996) Betaine aldehyde dehydrogenase in sorghum. Plant Physiol 110 1301–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN (1982) Living with water stress: evolution of osmolyte systems. Science 217 1214–1222 [DOI] [PubMed] [Google Scholar]
- Yang WJ, Rich PJ, Axtell JD, Wood KV, Bonham CC, Ejeta G, Mickelbart MV, Rhodes D (2003) Genotypic variation for glycinebetaine in sorghum. Crop Sci 43 162–169 [Google Scholar]