Abstract
Bread wheat (Triticum aestivum) has a greater ability to exclude Na+ from its leaves and is more salt tolerant than durum wheat (Triticum turgidum L. subsp. durum [Desf.]). A novel durum wheat, Line 149, was found to contain a major gene for Na+ exclusion, Nax2, which removes Na+ from the xylem in the roots and leads to a high K+-to-Na+ ratio in the leaves. Nax2 was mapped to the distal region on chromosome 5AL based on linkage to microsatellite markers. The Nax2 locus on 5AL coincides with the locus for a putative Na+ transporter, HKT1;5 (HKT8). The Nax2 region on 5AL is homoeologous to the region on chromosome 4DL containing the major Na+ exclusion locus in bread wheat, Kna1. A gene member of the HKT1;5 family colocates to the deletion bin containing Kna1 on chromosome 4DL. This work provides evidence that Nax2 and Kna1 are strongly associated with HKT1;5 genes.
Increase in salt tolerance of crops is needed to sustain agriculture in regions affected by natural or secondary salinity. Durum wheat (Triticum turgidum L. subsp. durum [Desf.]) is particularly sensitive to salinity and has a limited ability to exclude sodium, which is an important mechanism of salt tolerance in wheat (Tester and Davenport, 2003; Munns et al., 2006). Exclusion of Na+ from the leaves is due to low net Na+ uptake by cells in the root cortex and the tight control of net loading of the xylem by parenchyma cells in the stele (Tester and Davenport, 2003). To improve Na+ exclusion in durum wheat it is necessary to understand the molecular basis of these Na+ transport processes.
Modern durum cultivars do not exclude Na+ to the same extent as bread wheat (Triticum aestivum); however, a source of sodium exclusion in a novel durum wheat, Line 149, was described by Munns et al. (2000). Line 149 is derived from a cross between a Triticum monococcum (accession C68-101; AA) and a durum cultivar Marrocos (AABB; The, 1973). The T. monococcum is the donor of the sodium exclusion trait (James et al., 2006). The sodium exclusion in Line 149 is conferred by two major genes (Munns et al., 2003) named Nax1 and Nax2 (for Na+ exclusion), inherited from the T. monococcum. Line 149 was crossed with the durum cultivar Tamaroi and selected F2 lines were backcrossed into Tamaroi so that Nax1 and Nax2 were separated into two single gene BC5F2 families (James et al., 2006). Nax1 was mapped on chromosome 2AL (Lindsay et al., 2004). Nax1 is a putative Na+ transporter, a member of the HKT (high-affinity K+ transporter) family, T. monococcum HKT7-A2 (Huang et al., 2006). The Nax1 gene confers a reduced rate of transport of Na+ from root to shoot and retention of Na+ in the leaf sheath, thus giving a higher sheath-to-blade Na+ concentration ratio (James et al., 2006). The second gene, Nax2, also confers a lower rate of transport of Na+ from root to shoot and has a higher rate of K+ transport, resulting in enhanced K+ versus Na+ discrimination in the leaf (James et al., 2006).
HKT genes control Na+ transport in higher plants, as demonstrated in rice (Oryza sativa), barley (Hordeum vulgare), and Arabidopsis (Arabidopsis thaliana; Mäser et al., 2002a; Berthomieu et al., 2003; Rus et al., 2004; Haro et al., 2005; Ren et al., 2005; Sunarpi et al., 2005). HKT genes have been given a new nomenclature and separated into two groups based on amino acid sequence (Platten et al., 2006). A Gly/Ser residue in the first pore loop of the protein differs between group 1 and group 2 HKT genes.
Group 1 HKT genes have a Ser in the first pore loop; this may make them more selective for Na+ (Horie et al., 2001; Mäser et al., 2002b; Garciadeblas et al., 2003; Platten et al., 2006). Group 1 HKT genes, such as AtHKT1;1 (previous name AtHKT1) and OsHKT1;5 (previous name OsHKT8), transport Na+ only and may be involved in unloading of Na+ from the xylem (Uozumi et al., 2000; Garciadeblas et al., 2003; Ren et al., 2005; Sunarpi et al., 2005). In wheat, group 1 HKT genes are involved in Na+ transport and may confer a phenotype of low leaf Na+ concentration in leaves (Huang et al., 2006).
For the group 2 genes there is no consensus on the mechanism of action or whether the main function is to transport Na+ or K+ (Schachtman and Schroeder, 1994; Rubio et al., 1995; Walker et al., 1996; Wang et al., 1998; Horie et al., 2001; Golldack et al., 2002; Haro et al., 2005). Heterologous expression of TaHKT2;1 (previous name TaHKT1) in yeast (Saccharomyces cerevisiae) and Xenopus laevis oocytes indicated that this gene is likely to play a role in Na+ and K+ transport (Rubio et al., 1995; Mäser et al., 2002b). Rodriguez-Navarro and Rubio (2006) provide an explanation for the conflicting results for group 2 HKT genes. They suggest that in plants some HKT mRNA transcripts have alternative initiations of translation and in heterologous systems translation may not occur exactly as in plants, leading to expression of proteins with different kinetic properties. A physiologically relevant function for group 2 transporters might be to transport Na+ when there is a limited supply of K+ so that the plant may have a monovalent cation for use in osmotic adjustment in the vacuole (Rodriguez-Navarro and Rubio, 2006).
The Na+ transporter OsHKT1;5 localizes to the plasma membrane and is expressed in the xylem tissues (Ren et al., 2005). OsHKT1;5 is likely to function in transporting Na+ out of the xylem into the xylem parenchyma cells (Ren et al., 2005), where it may then be effluxed through the cortex to the epidermis and back into the soil. Evidence is increasing that a major mode of action of AtHKT1;1 is to increase Na+ retrieval from the xylem in mature roots (Sunarpi et al., 2005). Nax2 functions in a similar way, in transporting Na+ out of the xylem. A compartmental loading experiment where 22Na+ was fed only to the lower part of the roots showed that lines with Nax2 withdrew more of the total 22Na+ into the upper part of the root than lines without Nax2 (James et al., 2006). The rates of root Na+ uptake were identical in the lines with and without Nax2, indicating that differences in shoot uptake were due to the net rate of xylem loading in the root (James et al., 2006).
The major Na+ exclusion locus in bread wheat, Kna1, is located on the D genome, on the distal part of chromosome 4. The phenotype is Na+ exclusion from the leaves and discrimination of K+ over Na+ in leaves, but no difference in Na+ concentrations in roots (Gorham et al., 1990). As there is no difference in Na+ concentrations in the roots it is likely that Kna1 controls net xylem loading rather than net Na+ uptake (Gorham et al., 1990). Hence, an HKT-like gene is a good candidate for Kna1, as well as for Nax2.
Kna1 may be homoeologous to Nax2, the term homoeologous referring to a gene that used to be homologous in ancestral wheats before polyploidization of wheats and their related species. If so, one would expect that Nax2 would be located in the group 4 chromosomes. During the evolution of wheat the distal part of chromosome 4A that is homoeologous to the distal part of chromosome 4D was translocated with chromosome 5A (Nelson et al., 1995). Therefore, if Kna1 is located on the distal part of chromosome 4D and Nax2 is homoeologous to Kna1, then Nax2 would be physically located on the distal end of chromosome 5AL.
In the 19 years since the Kna1 locus was first described (Gorham et al., 1987), Kna1 has been transferred from bread wheat to durum wheat (from the D genome to the B genome) by homoeologous recombination (Dvořák and Gorham, 1992). Kna1 has been mapped to the distal portion of the long arm of chromosome 4D and five markers have been identified that are linked to Kna1, within 2.2 cM (Dubcovsky et al., 1996; Luo et al., 1996), yet despite the extensive mapping work toward isolating Kna1 no gene has been identified as a candidate for Kna1.
In this study we test whether the two Na+ exclusion genes, Nax2 and Kna1, may be homoeologous, and we suggest that a putative Na+ transporter gene in wheat, HKT1;5, may correspond to both Nax2 and Kna1.
RESULTS
Nax2 Is a Single Major Gene
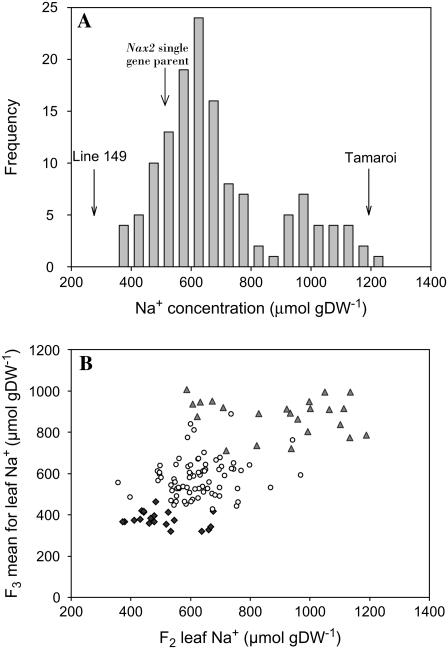
Line 149 is from a cross between T. monococcum (C68-101; AA) and Marrocos (AABB; The, 1973; James et al., 2006). A single gene family segregating for Nax2 was developed from Line 149, which contains the genes for Na+ exclusion Nax1 and Nax2, via backcrossing into the cultivar Tamaroi that lacks the two genes (James et al., 2006). Nax2 is responsible for a greater than 2-fold difference in leaf Na+ concentration. The mean leaf Na+ concentration of the BC4F2 Nax2 single gene parent was 473 ± 72 μmol g dry weight−1 compared to Tamaroi, which had a mean leaf Na+ concentration of 1,193 ± 48 μmol g dry weight−1. The frequency distribution of leaf Na+ concentration from 137 BC5F2 plants indicated that Nax2 was dominant, segregating in a 3:1 (low:high leaf blade Na+) ratio (Fig. 1A).
Figure 1.
A, Frequency distribution for leaf 3 Na+ concentrations of the BC5F2 lines in the single gene Nax2 mapping population. Arrows indicate parental means (n = 6); Line 149: 278 ± 37, Nax2 single gene parent: 473 ± 72, Tamaroi: 1,193 ± 48. Adapted from James et al. (2006). B, Relationship between the F2 and F2:3 progeny means for Na+ concentration of leaf 3. Plants were grown at 150 mm NaCl for 10 d. ▴, Homozygous lacking Nax2; ○, heterozygous for Nax2; ♦, homozygous for Nax2.
In this study, the 137 BC5F2 lines were progeny tested and the segregation of the Na+ exclusion trait was confirmed in the F2:3 families (773 individuals). The F2:3 families fitted the expected ratio for a single major gene (expected 94:31; observed 96:29; χ2 = 0.171, P ≥ 0.05; Fig. 1B). The mean leaf Na+ of the Nax2 single gene parent was 462 ± 23 μmol g dry weight−1 in the F2:3 generation, compared to 473 ± 72 μmol g dry weight−1 in the F2 generation.
HKT1;5 Cosegregates with Nax2
Publicly available cation transporter sequences from rice were used to screen the GenBank database of wheat expressed sequence tags (ESTs) to identify putative cation transporters in wheat. As part of this work, the protein sequence of OsHKT1;5 was used to search the wheat EST database. The search identified a single closely related partial wheat EST sequence (CK193616). This partial sequence (TaHKT1;5) shared 86% identity at the nucleotide level and contained parts of the corresponding sequences of exon 2 and exon 3 of OsHKT1;5 (Ren et al., 2005).
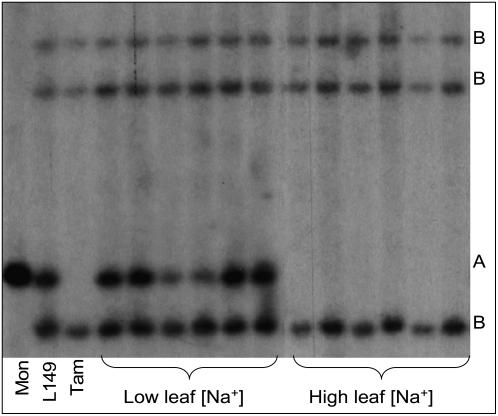
A probe designed from the partial HKT1;5 sequence (named HKT1;5 probe) identified RFLP between parental lines. Polymorphism found between the parents, Line 149 and Tamaroi, was due to the presence of an additional fragment in Line 149 when compared to Tamaroi (Fig. 2). The additional fragment in Line 149 cosegregated with the low Na+ accumulation phenotype in all of the 137 BC5F2 families tested. All of the lines in the mapping family verified as homozygous and heterozygous for Nax2 had the additional fragment and those lines that were homozygous for the high leaf Na+ phenotype lacked the fragment (a selection is shown in Fig. 2).
Figure 2.
Cosegregation of the A genome HKT1;5 fragment with Nax2. Southern-blot hybridization with the HKT1;5 probe after HindIII digestion of DNA from T. monococcum (Mon), Line 149 (L149), Tamaroi (Tam), and plants from the Nax2 BC5F3 mapping population with low leaf 3 Na+ concentrations and high leaf 3 Na+ concentrations. The genome origin of each band is shown on the right. In the Nax2 mapping family the A genome fragment (inherited from the monococcum) cosegregates with Nax2 in all of the 137 families.
Wheat HKT1;5 Gene Homoeologs
The HKT1;5 probe hybridized to a single restriction fragment in DNA from the diploid T. monococcum (C68-101), the putative donor of salt tolerance in Line 149. A fragment of the same size as the T. monococcum fragment was present in Line 149 but absent in Tamaroi. Three other fragments were present in both Line 149 and Tamaroi (Fig. 2). The monococcum fragment in Line 149 cosegregated with Nax2 (Fig. 2). Six restriction enzymes were tested and the HKT1;5 probe always hybridized to at least two fragments in Tamaroi and three fragments in Line 149 (data not shown). HindIII produced an additional fragment in both parents (Fig. 2). In summary, Tamaroi contained at least two HKT1;5-like genes, while Line 149 had the same two fragments plus one additional gene member that was inherited from T. monococcum. These results were confirmed with three addition probes (named HKT1;5 probe 2, HKT1;5 probe 3, and HKT1;5 probe 4) designed on different parts of the open reading frame (ORF) spanning from exon 1 to the 3′ untranslated region.
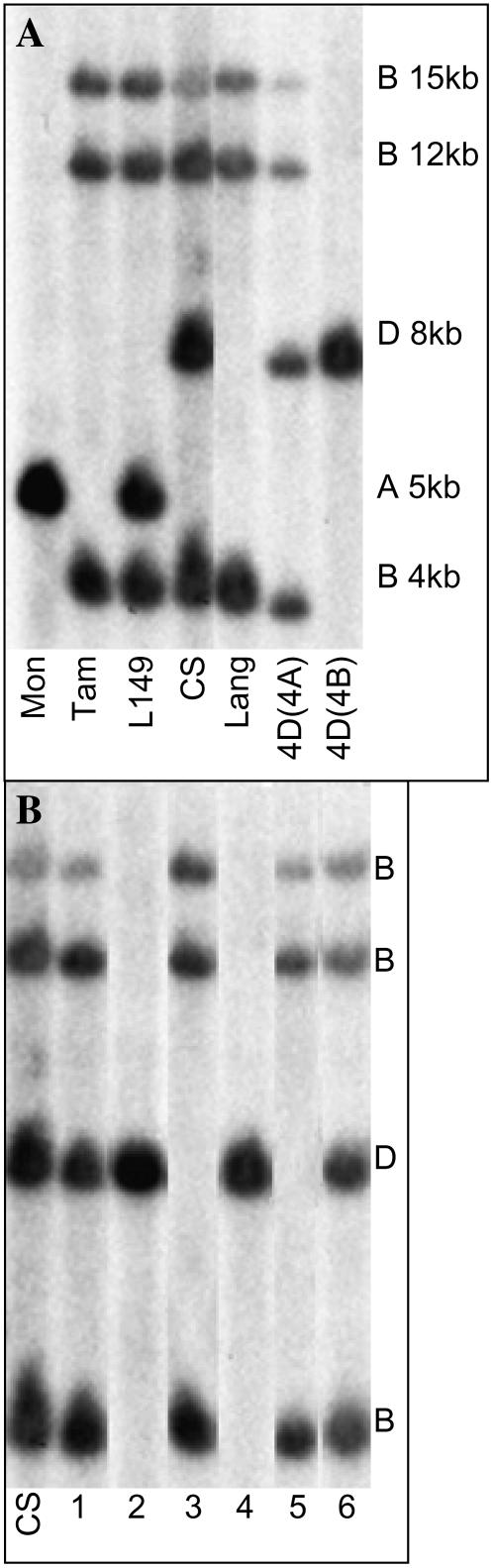
To identify the HKT1;5 gene family on the A, B, and D genomes of wheat, we analyzed DNA from the durum cultivar Langdon (AABB) carrying individual chromosome substitutions from the hexaploid Chinese Spring (CS; AABBDD; Joppa 1987). The HKT1;5 probe hybridized to at least two fragments in Langdon DNA, producing an identical pattern to Tamaroi. Hybridization of the partial HKT1;5 probe to CS DNA produced a pattern identical to that of Langdon and Tamaroi except for one additional chromosome 4D fragment. A HindIII digest of Langdon with chromosome 4A substituted for chromosome 4D of CS retained all three fragments and gained an additional fragment from chromosome 4D of CS. The Langdon substitution line with chromosome 4B replaced by chromosome 4D of CS had lost all three Langdon fragments but retained the 4D fragment from CS (Fig. 3A). These results indicate that all HKT1;5 hybridizing fragments in Langdon and Tamaroi are located on chromosome 4B and that Line 149 inherited an additional A genome member from T. monococcum.
Figure 3.
Chromosomal location of the HKT1;5 fragments. Southern-blot hybridization with the HKT1;5 probe after HindIII restriction digest. A, DNA from T. monococcum (Mon), Tamaroi (Tam), Line 149 (L149), CS, Langdon (Lang), and Langdon substitution lines 4D(4A) and 4D(4B). The genome locations of the HKT1;5 gene members and the approximate sizes (kilo bp) of the fragments are shown on the right. B, DNA from CS; CS chromosome arm deletion lines for chromosome 4B: N4AT4B (1), m4BT4A (2), N4DT4B (3); and CS ditelomeric lines Dt4BS (4), Dt4DS (5), and Dt4DL (6); n = nullitetrasomic (no copies); T = tetrasomic (four copies); m = monosomic (one copy); Dt = ditelomeric (for Dt4BS the long arm of 4B is missing and the short arm of 4B is present). The genome locations of the HKT1;5 gene members are shown on the right.
Analysis of nullitetrasomic CS lines of homoeologous group 4 (refer to “Materials and Methods”) confirmed that HKT1;5 fragments were located on either chromosome 4B or 4D, but not 4A (Fig. 3B). DNA hybridization of the partial HKT1;5 probe to ditelosomic lines of CS, where individual group 4B or 4D chromosome arms have been deleted, positioned these genes on the long arm of chromosome 4B (at least three members), and one member on the long arm of chromosome 4D, where the major Na+ exclusion locus in bread wheat, Kna1, is located (Fig. 3B).
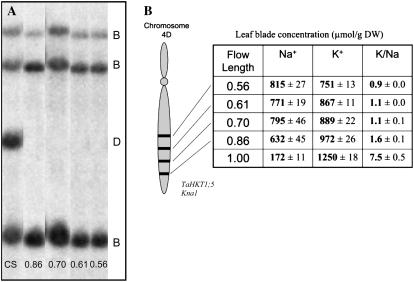
To study the relationship of Kna1 to the bread wheat HKT1;5-D gene member, we probed DNA from CS and a series of telomeric deletion lines generated from chromosome 4DL in CS (Endo and Gill, 1996). In the telomeric deletion lines the 4DL chromosome has been shortened to different lengths. The HKT1;5 gene member on chromosome 4DL was absent in the terminal deletion line flow length (Fl) 0.86 with approximately 14% of the chromosome arm deleted (Fig. 4A). The 4DL fragment was also missing in other lines with progressively larger terminal deletions (Fig. 4A). The loss of the chromosomal fragment containing the TaHKT1;5 gene member on the D genome (TaHKT1;5-D) corresponded to an increase in average Na+ concentrations in the leaf blade of the deletion lines compared with the euploid salt-tolerant CS, and a decrease in the K+-to-Na+ ratio from 7.5 to 1.2 (Fig. 4B).
Figure 4.
A, Southern-blot hybridization with HKT1;5 probe after HindIII restriction digest of DNA from CS and CS chromosome deletion lines 0.86, 0.70, 0.61, and 0.56. The TaHKT1;5-D gene member maps to the most distal deletion bin. The genome location of the TaHKT1;5 gene members are shown on the right. B, CS and CS chromosome deletion lines were grown in salt tanks (see “Materials and Methods”). Fl describes remaining fraction of chromosome 4DL. Kna1 maps to the same location (Dubcovsky et al., 1996) as TaHKT1;5-D. Leaf blade Na+ and K+ concentrations were recorded after leaf 3 had grown for 10 d in 50 mm NaCl.
Genetic Map Location of Nax2
To position Nax2 in the durum wheat genome, 34 microsatellite markers previously mapped to chromosome 4A were screened for polymorphism between parental lines and tested for linkage to Nax2 in the segregating family. None of the markers from chromosome 4A were linked to Nax2 (C.S. Byrt, unpublished data).
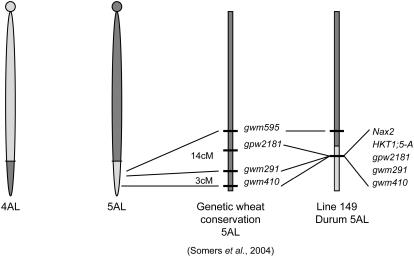
In an ancestor of modern wheats, chromosomes 4AL and 5AL exchanged short terminal segments (Liu et al., 1992). Considering this, we tested microsatellite markers that mapped to the distal segment of chromosome 5AL for linkage to Nax2. Microsatellite markers gwm291, gwm410, and gpw2181, previously mapped to the distal region of chromosome 5AL (Roder et al., 1998), cosegregated with Nax2 in the segregating family of 137 F3 lines. Therefore, Nax2 maps to the location that we might expect to find a homoeolog of Kna1.
The lack of recombination between the HKT1;5 gene on the A genome (HKT1;5-A) and gwm291, gwm410, and gpw218 raised the question whether the introgressed segment carrying Nax2 from T. monococcum was able to recombine with the homologous region in Tamaroi. To investigate this, we tested other microsatellite markers that were previously positioned on the distal end of chromosome 5AL. Four microsatellite markers, gwm595, gwm179, gwm126 (Roder et al., 1998), and barc232 (http://wheat.pw.usda.gov) were polymorphic between Line 149 and Tamaroi but were not retained in the BC4 parent and therefore failed to segregate in the BC-derived family (C.S. Byrt, unpublished data). These results show that recombination did occur between the Nax2 gene and gwm595 and the chromosomal region on 5AL proximal to gpw2181 was replaced by Tamaroi during the process of five backcrossing steps of transferring Nax2 into Tamaroi (Fig. 5).
Figure 5.
Part of chromosome 5AL originated from part of chromosome 4AL due to an ancient reciprocal translocation between the distal ends of chromosomes 4AL and 5AL. Durum 5AL represents the fragment containing Nax2 that introgressed from Line 149 into the Tamaroi background in the Nax2 mapping population. Proximal to the introgression is gwm595. Nax2 is linked to gwm291, gwm410, and gpw2181. (See Somers et al. [2004].)
In summary, Nax2 is located on the ancestral segment of chromosome 4AL that is attached to the distal end of chromosome 5AL. Hence, the tightly linked TmHKT1;5-A gene member from T. monococcum was also located on chromosome 5AL, consistent with the map locations of other TaHKT1;5 members on homoeologous chromosomes 4BL and 4DL.
Isolation of Full-Length HKT1;5 Gene Members
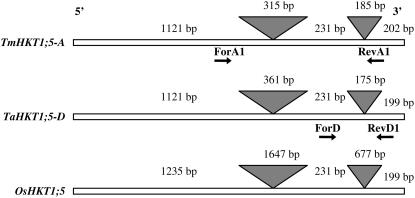
The partial HKT1;5 probe was used to screen a cDNA library from root tissue of CS. Several positive phagemid clones were isolated and when sequenced revealed identical DNA sequences with insert size varying between 812 and 1,741 bp. The cDNA sequences were identical to genomic sequence derived from a bacterial artificial chromosome clone that was previously isolated from a bacterial artificial chromosome library made from the diploid D genome progenitor species Aegilops tauschii Coss (E.S. Lagudah, unpublished data), suggesting that the cDNA sequence isolated from CS was derived from the D genome (TaHKT1;5-D). The cDNA was predicted to encode a full-length gene based on the comparison of its predicted amino acid sequence to OsHKT1;5 (SKC1) in rice (C.S. Byrt, unpublished data). Reverse-transcriptase PCR with primers designed from the 5′ and 3′ untranslated regions of TaHKT1;5-D amplified the corresponding A gene member, TmHKT1;5-A, from T. monococcum and Line 149. The predicted ORF of TmHKT1;5-A is 1,554 bp and the predicted ORF of TaHKT1;5-D is 1,551 bp. OsHKT1;5, TmHKT1;5-A, and TaHKT1;5-D each have two introns. The predicted amino acid sequence of TmHKT1;5-A and TaHKT1;5-D shared 94% identity and were closely related to the rice Na+ transporter OsHKT1;5 (66% identity). The intron and exon structure of the TmHKT1;5-A, TaHKT1;5-D, and OsHKT1;5 genes are shown in Figure 6.
Figure 6.
Gene structures of TmHKT1;5-A, TaHKT1;5-D, and OsHKT1;5. The gray triangles represent the intron regions of the gene. The arrows indicate the primers designed for gene expression analysis (Fig. 7).
Expression of HKT1;5 Gene Members
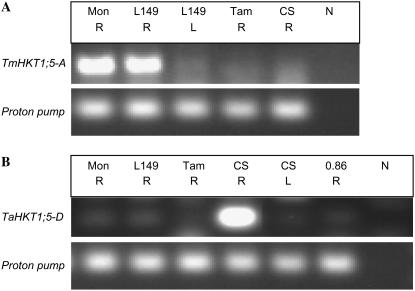
Reverse-transcriptase PCR with specific primers for A and D genome members was used to analyze the expression of the HKT1;5 A and D gene members in T. monococcum, Line 149, Tamaroi, and CS. TmHKT1;5-A was expressed in the roots of T. monococcum and Line 149, but not in the shoots (Fig. 7A). HKT1;5-A was not expressed in Tamaroi or CS. The TaHKT1;5-D gene member was expressed in CS roots but not shoots (Fig. 7B). The TaHKT1;5-D gene member was not expressed in the CS deletion line missing the distal 14% of chromosome 4DL (0.86; Fig. 7B). The expression results for the HKT1;5-D gene member are consistent with the mapping results, indicating that HKT1;5-D is missing from the CS deletion line 0.86 and is therefore positioned in the same region as Kna1.
Figure 7.
Expression of HKT1;5 in wheat analyzed using reverse-transcriptase PCR. A, Expression of TmHKT1;5-A. A product of the expected size (442 bp) was observed for the T. monococcum and Line 149 root samples. B, Expression of TaHKT1;5-D. A product of the expected size (147 bp) was observed for the CS root sample. Mon = T. monococcum; L149 = Line 149; Tam = Tamaroi; 0.86 = CS deletion line missing the distal 14% of chromosome 4DL; n = no template control; R = root tissue; L = leaf tissue. Plants were grown in hydroponic solution for 2 weeks before addition of 50 mm NaCl, tissue was harvested 48 h after addition of NaCl. Proton pump was used as a control.
DISCUSSION
Mapping of Nax2
Nax2 is a single dominant gene, as Nax2 segregated in a 3:1 ratio of low:high leaf blade Na+ concentration in the BC5F2 plants (Fig. 1). Nax2 is located on the distal part of chromosome 5AL. The HKT1;5 gene member on the A genome mapped to the same region as Nax2 and cosegregated with Nax2.
HKT genes were considered to be the best candidates for Nax2 based on the role of other HKT transporters in higher plants. HKT7 cosegregates with Nax1, which confers Na+ exclusion from leaves in wheat (Huang et al., 2006), AtHKT1;1 confers Na+ exclusion from leaves in Arabidopsis (Uozumi et al., 2000; Sunarpi et al., 2005), and OsHKT1;5 (SKC1) confers Na+ exclusion from leaves in rice (Ren et al., 2005).
There are up to five HKT1;5 genes in wheat, a partial wheat HKT1;5 probe detected one gene member on the D genome, two or three on the B genome, and one gene member on the A genome derived from T. monococcum (C68-101). The predicted amino acid identity between the wheat HKT1;5-A and HKT1;5-D gene members is 94%. The most closely related gene in rice, OsHKT1;5, shares 66% identity (75% positives) with the predicted wheat HKT1;5 sequences.
The location of the wheat HKT1;5 genes and the location of the rice OsHKT1;5 genes are not syntenic. OsHKT1;5 is located on chromosome 1S of rice (Lin et al., 2004; Ren et al., 2005). This region is syntenous to chromosome 3 in wheat (Sorrells et al., 2003), however, TmHKT1;5-A is located on chromosome 5AL and TaHKT1;5-D is located on chromosome 4DL.
There may be a second gene in the Nax2 region having an effect on leaf Na+, but it would have to be closely linked, as every time that the HKT1;5-A gene member is lost, leaf Na+ increases significantly. This has been demonstrated in the Nax2 mapping population (Fig. 2) and when HKT1;5-A was transferred into other genetic backgrounds including bread wheat. In moderately saline field conditions (100 mm) Tamaroi had an average leaf Na+ concentration of 125 μmol g dry weight−1 whereas BC4F2-derived lines with HKT1;5-A had an average leaf Na+ concentration of 25 μmol g dry weight−1, a 5-fold difference (R. Hare and R. Munns, unpublished data). In a completely different field environment with the same lines HKT1;5 conferred a 2-fold reduction in leaf Na+ concentration (A. Rathjen and R. Munns, unpublished data).
Overall, in field and glasshouse experiments, all the lines with the HKT1;5-A gene had at least 2-fold lower leaf Na+ than all those without the HKT1;5-A gene. When transferred into the bread wheat Westonia, which already contains the Na+ exclusion locus Kna1 on the D genome, HKT1;5-A reduced leaf Na+ by a further 25% (R. Munns and R. James, unpublished data).
Relationship of Nax2 to the Major Salt-Tolerant Gene in Hexaploid Wheat, Kna1
The HKT1;5-A genome member, which is the candidate for Nax2, is physically located on the distal part of chromosome 5AL, which ancestrally corresponds to the distal part of chromosome 4AL (Fig. 7). Kna1 maps to the distal region of chromosome 4DL of wheat (Dubcovsky et al., 1996), raising the possibility that Nax2 and Kna1 are homoeologous genes located on ancestral group 4 chromosomes. The TaHKT1;5-D genome member maps to the distal 14% of chromosome 4DL in hexaploid bread wheat (Fig. 4A), coinciding with the map location of Kna1 (Dubcovsky et al., 1996). The locus for Kna1 on chromosome 4DL is homoeologous to the location for Nax2 on chromosome 5AL and the TaHKT1;5-D genome member may be Kna1.
The loss of the region containing the TaHKT1;5-D gene member from CS deletion lines corresponded to an increase in average Na+ concentrations in the leaf blade and a 6-fold decrease in the K+-to-Na+ ratio from 7.5 to 1.2 (Fig. 4B), when plants were grown at 50 mm NaCl. These results are consistent with our hypothesis that the HKT1;5 probe detects not only a candidate gene for Nax2 in durum wheat, but also a candidate gene for Kna1 in hexaploid bread wheat and that both genes are located in homoeologous regions of the wheat genome. These results are also consistent with other data on the effect of Kna1 on leaf K+ and Na+ concentrations. When the Kna1 region was transferred from the bread wheat, CS, into the durum wheat cultivar, Langdon, lines with Kna1 had a greater leaf K+-to-Na+ ratio (Dvořák and Gorham, 1992). Lines of Langdon containing the Kna1 region had a leaf K+-to-Na+ ratio approximately 8 times higher than those without Kna1 when plants were grown at 50 mm, and 6 times higher when grown at 150 mm NaCl (Gorham et al., 1987, 1990).
Similarity of Phenotype between Sodium Excluding Genes in Rice and Wheat
There are five phenotypic characteristics in common between Nax2, Kna1, and SKC1: (1) low Na+ concentration in the leaves; (2) enhanced discrimination of K+ over Na+ in transport from the roots to the shoots; (3) regulation of the K+-to-Na+ ratio in the leaves; (4) no effect on root Na+ concentration; and (5) no effect on the sheath-to-blade Na+ ratio (Gorham et al., 1990; Davenport et al., 2005; Ren et al., 2005; James et al., 2006). Nax1, which cosegregates with T. monococcum HKT7, has a distinctive mechanism that differs from Nax2, Kna1, and SKC1. The characteristic of Nax1 is a high sheath-to-blade Na+ concentration ratio. Nax1 confers low leaf blade Na+ and high leaf blade K+-to-Na+ ratio by unloading Na+ in the leaf sheath and displacing K+ in the leaf blade (James et al., 2006). In the Tamaroi background, BC5F2 lines with Nax1 (HKT7-A2) have a 4-fold lower leaf Na+ concentration than lines without Nax1 (Huang et al., 2006).
The mechanism behind the common phenotype for the Nax2, SKC1, and Kna1 genes may be unloading of Na+ from the xylem. We know that Nax2 unloads Na+ from the xylem as experiments with 22Na+ showed that the rate of unloading of Na+ from the xylem was double that in lines with Nax2 than in those without (James et al., 2006). SKC1 in rice also unloads Na+ from the xylem (Ren et al., 2005). Under salt stress the shoots and the xylem sap of near-isogenic lines with SKC1 contained more K+ and less Na+ than those without SKC1 (Ren et al., 2005). There is also indirect evidence that Kna1 may unload Na+ from the xylem. One is the observation that there is no difference in the Na+ concentration in the roots of lines with and without Kna1 (Gorham et al., 1990), and the other is that the membrane potential-dependent uptake of Na+ was no different in vesicles from hexaploid wheat (cv Troy), which has Kna1, and tetraploid wheat (cv Langdon), which does not have Kna1 (Allen et al., 1995). Gorham et al. (1997) suggested that this observation of Allen et al. (1995) would be expected if Kna1 acts specifically on xylem loading.
A single HKT gene may be sufficient to explain the Nax2 or Kna1 phenotypes. The Nax1 gene in wheat and the SKC1 gene in rice are both HKT genes and they both have a strong affect on the leaf K+-to-Na+ ratio and Na+ concentration. Wheat lines with Nax1 (TmHKT7-A2) have a 4 times greater leaf K+-to-Na+ ratio, and 4 times less leaf Na+ than lines without Nax1 (Huang et al., 2006; James et al., 2006). Transformed rice lines with SKC1 (OsHKT1;5) have a 2.5 times greater leaf K+-to-Na+ ratio and 2 times less leaf Na+ than transformed lines without SKC1 (Ren et al., 2005). Wheat lines with Nax2 have a 3.6 times greater leaf K+-to-Na+ ratio and approximately 2.5 times less leaf Na+ than lines without Nax2. One HKT1;5-A gene could account for these differences. Likewise, for Kna1, one HKT1;5-D gene may account for the 6 times greater leaf K+-to-Na+ ratio and the 4 times less leaf Na+ in CS compared to the CS deletion lines lacking Kna1.
Allelic Variation in HKT Genes
In rice and Arabidopsis, allelic variation for OsHKT1;5 and AtHKT1;1, respectively, has been linked to variation in function (Ren et al., 2005; Rus et al., 2006). In rice, six nucleotide substitutions in the coding region of OsHKT1;5 differ between the tolerant allele from Nona Bokra and the sensitive allele from Koshihikari, leading to four amino acid residues differing (Ren et al., 2005). Under salt stress Koshihikari had almost double the leaf Na+ concentration of near isogenic lines with the Nona Bokra SKC1 (OsHKT1;5) allele. Changes in the coding region, rather than differences in the promoter, are likely to account for the functional variation in the two alleles (Ren et al., 2005). The data that support this include a genetic complementation test, where the Nona Bokra promoter and ORF was transferred into a sensitive line and recovered the SKC1 phenotype, and an observation that there were no differences in the expression patterns for the different OsHKT1;5 alleles (Ren et al., 2005).
In Arabidopsis, allelic variation in the promoter of AtHKT1;1, rather than the coding region, may account for functional differences between AtHKT1;1 from Columbia-0 and two natural variants, Tsu-1 and Ts-1 (Rus et al., 2006). The AtHKT1;1 alleles from Tsu-1 and Ts-1 are linked to higher leaf Na+. Sequence differences included 19 single nucleotide polymorphisms in the coding region, leading to seven different amino acid residues (Rus et al., 2006), but a deletion in a tandem repeat sequence in the promoter may be responsible for the high leaf Na+ as there was reduced expression of AtHKT1;1 in the roots (Rus et al., 2006). Curiously, the Tsu-1 allele linked to higher leaf Na+ was also linked to NaCl tolerance, whereas the loss of function mutant Athkt1;1 has higher leaf Na+ than wild type (Columia-0) but is more NaCl sensitive (Rus et al., 2004), although, as Rus et al. (2006) note, a second gene that influences NaCl tolerance may be segregating with AtHKT1;1 in the Tsu-1 population.
In contrast, no allelic variation for HKT1;5-A has been identified in wheat. The difference in leaf Na+ between lines with and without HKT1;5-A seems to be determined by the presence or absence of the gene, rather than allelic variation (Fig. 2). We found a single HKT1;5 gene member on the A genome of families containing Nax2 that was absent in those without Nax2. This gene member originated in T. monococcum (C68-101), was introgressed into Line 149 by crossing T. monococcum with the durum cultivar Marrocos (The, 1973), and inherited in all the near-isogenic lines containing Nax2 that were derived from the cross between Line 149 and cv Tamaroi (James et al., 2006). There is no homoeolog of HKT1;5 on the A genome of Marrocos or Tamaroi, or in the durum cv Langdon, or the bread wheat CS (Fig. 3A). It is absent in all the modern wheats that we have tested (data not shown). We suggest that the HKT1;5 gene was not present in the A genome diploid ancestor that gave rise to modern wheat. An extensive screen of diverse genetic material to look for allelic variation for HKT1;5-A in wheat ancestors and wild A genome species, including T. monococcum and Triticum urartu, is under way.
The HKT1;5-D allele isolated from CS has the same ORF as the HKT1;5-D gene members isolated from three Triticum tauschii accessions (data not shown). Screening for allelic variation in HKT1;5-D in a diverse range of genetic material is also under way.
Concluding Remarks
The data presented in this work supports the hypothesis that TmHKT1;5-A is a candidate for Nax2 and is a homoeolog of Kna1. To test whether Nax2 is TmHKT1;5-A and whether Kna1 is TaHKT1;5-D, functional assays of the HKT1;5-A and HKT1;5-D transporters are necessary. RNA interference constructs against TaHKT1;5-D will be introduced into hexaploid bread wheat to test if silencing TaHKT1;5-D results in the same phenotype that we have observed in the CS deletion lines lacking the Kna1 region. We will also express the wheat HKT1;5 A, B, and D gene members in yeast and X. laevis oocytes to characterize the transport properties of the proteins.
Incorporation of HKT1;5 into durum wheat breeding programs may provide a mechanism for Na+ exclusion for durum wheat of similar potency to that conferred by Kna1 in bread wheat.
MATERIALS AND METHODS
Plant Material
Parent material for the mapping population was durum wheat (Triticum turgidum subsp. durum [Desf.] Line 149; Munns et al., 2000) and the cultivar Tamaroi. Line 149 was derived from Triticum monococcum (C68-101; low leaf Na+) and the durum variety Marrocos (high leaf Na+; T. monococcum/3*Marrocos; The, 1973). T. monococcum C68-101 is the putative donor of Nax1 and Nax2 (James et al., 2006). Selected F2 lines from the cross between Line 149 and Tamaroi were backcrossed into Tamaroi four times. A selected BC4F2 individual that carried Nax2, but not Nax1, was backcrossed once more into Tamaroi to produce a BC5F2 family of 137 F2 individuals (James et al., 2006). This family containing the Nax2 gene but not the Nax1 gene formed the basis for this study.
Plant material for the CS deletion line experiment included lines with the following Fls: 0.09, 0.31, 0.38, 0.41, 0.46, 0.53, 0.56, 0.61, 0.70, 0.71, 0.86, and 1.00 (wild-type CS; Endo and Gill, 1996).
Plant material for DNA extraction for Southern-blot hybridization work included CS nullisomic-tetrasomic lines (Sears, 1954) and CS deletion lines (Endo and Gill, 1996) and Langdon substitution lines (Joppa, 1987). The Langdon substitution lines included 4D(4A) and 4D(4B), where chromosome 4D from CS had been substituted for chromosome 4A or 4B of Langdon, respectively.
Phenotyping
Plants were grown in half-strength Hoagland solution in supported hydroponics in a method adapted from Munns et al. (2000). Salt treatment commenced when leaf 2 was half emerged. The NaCl concentration of the hydroponic solution was increased by 25 mm twice daily over 3 d to reach a final concentration of 150 mm. Supplemental calcium [Ca(NO3)2] was added to achieve a Na+-to-Ca2+ ratio of 15:1. Leaf 3 was harvested after 10 d growing in salt and the Na+ was extracted with nitric acid (0.5 m). The Na+ concentration was measured using inductively coupled plasma analysis. Parental lines were replicated 10 times. Based on the score of F2 individuals, nine plants from every family predicted to be homozygous low were tested and five plants were tested for every family predicted to be homozygous high or heterozygous. These numbers were based upon recommended population sizes required in biparental populations to obtain at least one target homozygous genotype in later generations for segregating loci (Bonnett et al., 2005).
Genotyping
Plants grown in salt tanks for phenotyping were transplanted into soil and allowed to grow for approximately 4 weeks prior to DNA extraction. One or two plants were retained from each of the F2:3 families. For families with a homozygous low Na+ accumulation phenotype, the plant with the lowest leaf Na+ concentration was used. The plant with the highest leaf Na+ was used from families with a homozygous high Na+ accumulation phenotype. For half of those families with a heterozygous phenotype, the lowest of the low Na+ accumulating plants was used, and for the other half of the families with a heterozygous phenotype, the highest of the high Na+ accumulating plant was used. Leaf material from plants was harvested and DNA extracted as per Lagudah et al. (1991).
RFLP Probe Development
A search of the public database identified a wheat EST (CK193616) with strong homology, 86% nucleotide sequence identity, to the rice SKC1 candidate gene (DQ148410) subsequently named OsHKT1;5 (Platten et al., 2006). The wheat EST was named HKT1;5 as recommended by Platten et al. (2006). Primers designed internal to CK193616 were HKT1;5For01 (5′-CATCACCGTCGAGGTTATCAG-3′) and HKT1;5Rev01 (5′-TTGAGGTACTCGGCATA-3′. These primers were used to amplify a 332 bp product from T. monococcum genomic DNA. The product was cloned into pGEMT-easy vector (Promega Corporation). The PCR-amplified product was radiolabeled with 32P-CTP using the Megaprime DNA labeling system (Amersham) according to the manufacturer's instructions.
The HKT1;5 probe spans from exon 2, 1,692 bp into the ORF, to the 3′ untranslated region. Three additional probes were developed using the same method. The additional probes were named HKT1;5 probe 2, HKT1;5 probe 3, and HKT1;5 probe 4. They were used to confirm the mapping and cosegregation data. The sizes of HKT1;5 probe 2, HKT1;5 probe 3, and HKT1;5 probe 4 were 315, 324, and 321 bp in, respectively. Relative to the ORF they start at 935, 1,003, and 1,765 bp, respectively. Together, the four probes span from exon 1 through to the 3′ untranslated region.
DNA from the parental lines (Line 149 and Tamaroi) T. monococcum (AUS# 90382) and the BC5F2:3 progeny were digested individually with HindIII and/or EcoRV, EcoRI, NcoI, SacI, and/or XbaI. After gel electrophoresis, the gels were blotted onto a nitrocellulose membrane (Amersham Biosciences Hybond-N+) and hybridized with the HKT1;5 probe. DNA samples from wheat nullisomic-tetrasomic lines and ditelomeric lines in a CS background were also screened with the HKT1;5 probe as described above. In the nullisomic-tetrasomic lines each pair of homoeologous chromosomes, 4A, 4B, or 4D have been substituted by one of the other pairs, in the ditelomeric lines the short or long chromosome arms have been deleted.
Microsatellite Markers
Microsatellite markers were used to establish the chromosomal location of Nax2 in the durum Line 149. A group of 470 wheat microsatellite markers were used to screen DNA from the parental lines, Tamaroi and Line 149, for polymorphisms (Roder et al., 1998). Twenty five were found to be polymorphic between Line 149 and Tamaroi (C.S. Byrt, unpublished data). These were tested on bulked DNA samples from the BC5F2:3 lines, two bulks each containing pooled DNA from 10 low Na+ lines and two bulks each pooled from 10 high Na+ lines. Three wheat microsatellite markers cosegregated with Nax2, namely gwm291, gwm410, and gpw2181. These markers had been mapped previously to the distal end of chromosome 5AL (Roder et al., 1998).
To confirm cosegregation of these markers with Nax2, the genotype of 19 BC5F2:3 plants with a homozygous low Na+ phenotype were tested using the markers gwm291, gwm410, and gpw2181. All 19 had the same genotype as Line 149. In contrast, 11 BC5F2:3 plants that had a homozygous high Na+ phenotype all had the same genotypic pattern with gwm291, gwm410, and gpw2181 as Tamaroi.
Isolation of TaHKT1;5-D
The CS cDNA library was kindly supplied by Professor Timothy J. Close. The library was constructed from drought-stressed root tissue at full tillering. Approximately 120,000 clones of the mass excised phagemid library were plated and screened with the partial TmHKT1;5 probe according to standard protocols (Sambrook et al., 1989). Eleven colonies hybridized to the probe. These were grown at 37°C overnight in Luria broth (Sambrook et al., 1989) and plasmid DNA was isolated using a Qiaspin miniprep kit according to the manufacturer's instructions (Qiagen). Plasmid DNA was digested with EcoRI and XhoI to liberate the cloned inserts. The inserts were sequenced using T7 and SP6 universal primers (Invitrogen).
Isolation of RNA, Reverse-Transcriptase PCR, and Isolation of TmHKT1;5-A
Plants were grown in hydroponic solution described in “Plant Material.” After 2 weeks plants were exposed to 50 mm NaCl. After 48 h leaf and root tissues were harvested separately and snap frozen in liquid nitrogen. RNA was extracted using TRIzol Reagent (Invitrogen) as per the manufacturer's instructions. Reverse-transcriptase PCR to amplify TmHKT1;5 was undertaken using primers that were external to the coding sequence named 5primeUTRFor (5′-AGAAGTCTCTACACAACTTACAG-3′) and 3primeUTRRev (5′-GATCATTGAGAAATATGCAGTCC-3′) using a Qiagen OneStep RT-PCR kit as per the manufacturer's instructions. DNA fragments of the appropriate size were amplified from T. monococcum and Line 149. These fragments were cut out and purified using a Qiagen gel extraction kit according to the manufacturer's instructions. The fragments were ligated into the pGEM-T vector using the pGEM-T easy vector system 1 kit (Promega).
Reverse-transcriptase PCR to observe presence or absence of expression of the HKT1;5 A and D gene homoeologs was undertaken using A gene specific primers named ForA1 (5′-GAGTGGGGCTCCGACGGGCTGAA-3′) and RevA1 (5′-CGTCAGGCGTCACCTGCCGGCCG-3′) and D gene specific primers ForD1 (5′-GCTTGGCCATCTTCATCGCCGTG-3′) and RevD1 (5′-GGCCACAGCTGTACCCGGTGCTG-3′) using a Qiagen OneStep RT-PCR kit as per the manufacturer's instructions. Primer locations are shown in Figure 6. The PCR was conducted under standard conditions with the following cycling protocol: 50°C, 20 min; 94°C, 15 min; then 35 cycles of 94°C, 30 s; 58°C, 30 s; 68°C, 1 min; and finally 72°C, 2 min. The forward and reverse primers in each primer set were designed in different exons so as to include an intron in between them. Therefore, products that amplified from trace DNA in the RNA samples differed in size from the products amplified from coding DNA. The expected product size for the A gene specific primers was 942 bp from genomic DNA and 442 bp from coding DNA. The expected product size for the D gene specific primers was 322 bp from genomic DNA and 147 bp from coding DNA.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ646339 (TmHKT1;5-A) and DQ646342 (TaHKT1;5-D).
Acknowledgments
We thank Dr. Ray Hare for providing Tamaroi, Line 149, and T. monococcum lines, with information of the unusual pedigree of Line 149, Professor Timothy J. Close for the CS cDNA library, Lorraine Mason for Na+ and K+ analysis by inductively coupled plasma, Karen Glover and Kylie Groom for screening microsatellite markers, Marianne Bloemsma for expert technical assistance, and Dr. Shaobai Huang for scholarly and methodological advice.
This work was supported by the Commonwealth Scientific and Industrial Research Organization, Australian Centre for Plant Functional Genomics, and University of Adelaide (scholarship to C.S.B.); and by the New South Wales Agricultural Genomics Centre (J.D.P., E.S.D.); Australian Research Council Federation Fellowship (M.T.); and Grains Research and Development Corporation (R.A.J., M.T., R.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Caitlin S. Byrt (caitlin.byrt@csiro.au).
Open Access articles can be viewed online without a subscription.
References
- Allen GJ, Wyn Jones RG, Leigh RA (1995) Sodium transport measured in plasma membrane vesicles isolated from wheat genotypes with differing K+/Na+ discrimination traits. Plant Cell Environ 18 105–115 [Google Scholar]
- Berthomieu P, Conéjéro G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J 22 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnett DG, Rebetzke GJ, Spielmeyer W (2005) Strategies for efficient implementation of molecular markers in wheat breeding. Mol Breed 15 75–85 [Google Scholar]
- Davenport R, James RA, Zakrisson-Plogander A, Tester M, Munns R (2005) Control of sodium transport in durum wheat. Plant Physiol 137 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, María GS, Epstein E, Luo MC, Dvořák J (1996) Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theor Appl Genet 92 448–454 [DOI] [PubMed] [Google Scholar]
- Dvořák J, Gorham J (1992) Methodology of gene-transfer by homoeologous recombination into Triticum-turgidum—transfer of K+/Na+ discrimination from Triticum-aestivum. Genome 35 639–646 [Google Scholar]
- Endo T, Gill B (1996) The deletion stocks of common wheat. J Hered 87 295–307 [Google Scholar]
- Garciadeblas B, Senn ME, Banuelos MA, Rodriguez-Navarro A (2003) Sodium transport and HKT transporters: the rice model. Plant J 34 788–801 [DOI] [PubMed] [Google Scholar]
- Golldack D, Su H, Quigley F, Kamasani UR, Munoz-Garay C, Balderas E, Popova OV, Bennett J, Bohnert HJ, Pantoja O (2002) Characterization of a HKT-type transporter in rice as a general alkali cation transporter. Plant J 31 529–542 [DOI] [PubMed] [Google Scholar]
- Gorham J, Bridges J, Dubcovsky J, Dvorak J, Hollington PA, Luo M-C, Khan JA (1997) Genetic analysis and physiology of a trait for enhanced K+/Na+ discrimination in wheat. New Phytol 137 109–116 [Google Scholar]
- Gorham J, Hardy C, Wyn Jones RG, Joppa LR, Law CN (1987) Chromosomal location of a K/Na discrimination character in the D genome of wheat. Theor Appl Genet 74 584–588 [DOI] [PubMed] [Google Scholar]
- Gorham J, Wyn Jones RG, Bristol A (1990) Parial characterization of the trait for enhanced K+-Na+ discrimination in the D genome of wheat. Planta 180 590–597 [DOI] [PubMed] [Google Scholar]
- Haro R, Banuelos MA, Senn ME, Barrero-Gil J, Rodriguez-Navarro A (2005) HKT1 mediates sodium uniport in roots: pitfalls in the expression of HKT1 in yeast. Plant Physiol 139 1495–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A (2001) Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J 27 129–138 [DOI] [PubMed] [Google Scholar]
- Huang S, Spielmeyer W, Lagudah ES, James RA, Platten JD, Dennis ES, Munns R (2006) A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol 142 1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James RA, Davenport RJ, Munns R (2006) Physiological characterisation of two genes for Na+ exclusion in durum wheat: Nax1 and Nax2. Plant Physiol 142 1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joppa LR (1987) Aneuploid analysis in tetraploid wheat. In Heyne EG, ed, Wheat and Wheat Improvement, Monograph 13. Am Soc Agron, Madison, WI, pp 151–166
- Lagudah ES, Appels R, Brown A, McNeil D (1991) The molecular-genetic analysis of Triticum tauschii, the D-genome donor to hexaploid wheat. Genome 34 375–386 [Google Scholar]
- Lin HX, Zhu MZ, Yano M, Gao JP, Liang ZW, Su WA, Hu ZH, Ren ZH, Chao DY (2004) QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor Appl Genet 108 253–260 [DOI] [PubMed] [Google Scholar]
- Lindsay MP, Lagudah ES, Hare RA, Munns R (2004) A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct Plant Biol 31 1105–1114 [DOI] [PubMed] [Google Scholar]
- Liu CJ, Atkinson MD, Chinoy CN, Devos KM, Gale MD (1992) Nonhomoeologous translocations between group 4, 5 and 7 chromosomes within wheat and rye. Theor Appl Genet 83 305–312 [DOI] [PubMed] [Google Scholar]
- Luo MC, Dubcovsky J, Goyal S, Dvořák J (1996) Engineering of interstitial foreign chromosome segments containing the K+/Na+ selectivity gene Kna1 by sequential homoeologous recombination in durum wheat. Theor Appl Genet 93 1180–1184 [DOI] [PubMed] [Google Scholar]
- Mäser P, Gierth M, Schroeder JI (2002. a) Molecular mechanisms of potassium and sodium uptake in plants. Plant Soil 247 43–54 [Google Scholar]
- Mäser P, Hosoo Y, Goshima S, Horie T, Eckelman B, Yamada K, Yoshida K, Bakker EP, Shinmyo A, Oiki S, et al (2002. b) Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc Natl Acad Sci USA 99 6428–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, Hare RA, James RA, Rebetzke GJ (2000) Genetic variation for improving the salt tolerance of durum wheat. Aust J Agric Res 51 69–74 [Google Scholar]
- Munns R, James RA, Läuchli A (2006) Approaches to increasing the salt tolerance of wheat and other cereals. J Exp Bot 57 1025–1043 [DOI] [PubMed] [Google Scholar]
- Munns R, Rebetzke GJ, Husain S, James RA, Hare RA (2003) Genetic control of sodium exclusion in durum wheat. Aust J Agric Res 54 627–635 [Google Scholar]
- Nelson JC, Sorrells ME, Van-Deynze AE, Lu YH, Atkinson M, Bernard M, Leroy P, Faris JD, Anderson JA (1995) Molecular mapping of wheat: major genes and rearrangements in homoeologous groups 4, 5, and 7. Genetics 141 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten JD, Cotsaftis O, Berthomieu P, Bohnert H, Davenport RJ, Fairbairn DJ, Horie T, Leigh RA, Lin HX, Luan S (2006) Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci 11 372–374 [DOI] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37 1141–1146 [DOI] [PubMed] [Google Scholar]
- Roder MS, Korzun V, Wendehake K, Plaschke J, Tixier M-H, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149 2007–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro A, Rubio F (2006) High-affinity potassium and sodium transport systems in plants. J Exp Bot 57 1149–1160 [DOI] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI (1995) Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270 1660–1663 [DOI] [PubMed] [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE (2006) Natural variants of AtHKT1 enhance Na+ accumulation in two wild population of Arabidopsis. PLoS Genetics 2 1964–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Lee B-h, Munoz-Mayor A, Sharkhuu A, Miura K, Zhu J-K, Bressan RA, Hasegawa PM (2004) AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol 136 2500–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schachtman DP, Schroeder JI (1994) Structure and transport mechanism of a high-affinity potassium uptake transporter from higher-plants. Nature 370 655–658 [DOI] [PubMed] [Google Scholar]
- Sears ER (1954) The aneuploids of common wheat. University of Missouri-Columbia Agricultural Experiment Station Bulletin 572 1–59 [Google Scholar]
- Somers D, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109 1105–1114 [DOI] [PubMed] [Google Scholar]
- Sorrells ME, La Rota M, Bermudez-Kandianis CE, Greene RA, Kantety R, Munkvold JD, Miftahudin, Mahmoud A, Ma X, Gustafson PJ (2003) Comparative DNA sequence analysis of wheat and rice genomes. Genome Res 13 1818–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunarpi, Horie T, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan WY, Leung HY, Hattori K (2005) Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J 44 928–938 [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot (Lond) 91 503–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The TT (1973) Transfer of resistance to stem rust from Triticum monococcum L. to hexaploid wheat. PhD thesis. University of Sydney, Sydney
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI (2000) The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol 122 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker NA, Maathuis FJM, Rubio F, Gassmann W, Schroeder JI (1996) High-affinity potassium uptake in plants. Science 273 977–979 [DOI] [PubMed] [Google Scholar]
- Wang T-B, Gassmann W, Rubio F, Schroeder JI, Glass ADM (1998) Rapid up-regulation of HKT1, a high-affinity potassium transporter gene, in roots of barley and wheat following withdrawal of potassium. Plant Physiol 118 651–659 [DOI] [PMC free article] [PubMed] [Google Scholar]