Abstract
Mannan polysaccharides are widespread among plants, where they serve as structural elements in cell walls, as carbohydrate reserves, and potentially perform other important functions. Previous work has demonstrated that members of the cellulose synthase-like A (CslA) family of glycosyltransferases from Arabidopsis (Arabidopsis thaliana), guar (Cyamopsis tetragonolobus), and Populus trichocarpa catalyze β-1,4-mannan and glucomannan synthase reactions in vitro. Mannan polysaccharides and homologs of CslA genes appear to be present in all lineages of land plants analyzed to date. In many plants, the CslA genes are members of extended multigene families; however, it is not known whether all CslA proteins are glucomannan synthases. CslA proteins from diverse land plant species, including representatives of the mono- and dicotyledonous angiosperms, gymnosperms, and bryophytes, were produced in insect cells, and each CslA protein catalyzed mannan and glucomannan synthase reactions in vitro. Microarray mining and quantitative real-time reverse transcription-polymerase chain reaction analysis demonstrated that transcripts of Arabidopsis and loblolly pine (Pinus taeda) CslA genes display tissue-specific expression patterns in vegetative and floral tissues. Glycan microarray analysis of Arabidopsis indicated that mannans are present throughout the plant and are especially abundant in flowers, siliques, and stems. Mannans are also present in chloronemal and caulonemal filaments of Physcomitrella patens, where they are prevalent at cell junctions and in buds. Taken together, these results demonstrate that members of the CslA gene family from diverse plant species encode glucomannan synthases and support the hypothesis that mannans function in metabolic networks devoted to other cellular processes in addition to cell wall structure and carbohydrate storage.
Plant cells are enveloped by an extracellular matrix consisting of a highly organized and complex arrangement of carbohydrates, proteins, and often lignin. Among the many functions of plant cell walls, they define plant cell and organ shape, act as a barrier against plant pathogens, provide signals that direct growth and development, and supply strength and flexibility that enable plants to grow and respond to variable environmental conditions (Freshour et al., 2003; Somerville et al., 2004). Cellulose, pectins, and cross-linking glycans, including xyloglucans, xylans, mixed-linkage β-glucans, and mannans are the main constituents of plant cell walls. Variations in cell wall composition and architecture impart unique forms and functions to the variety of specialized cell types found in plants. Human uses of plant cell wall constituents are significant and wide ranging; plant cell walls are used as food, fuel, textiles and building materials.
Whereas cell wall composition has been examined in a variety of plant species, only a small proportion of the hundreds of proteins predicted to be involved in cell wall biosynthesis and metabolism have been identified and characterized. Glycan synthases and glycosyltransferases that biosynthesize noncellulosic cell wall polysaccharides have been especially difficult to identify because these integral membrane proteins are often present in low quantities and many have proven to be labile during purification procedures (Keegstra and Raikhel, 2001). Nonetheless, purification efforts have led to the identification of a number of cell wall biosynthetic enzymes, including xyloglucan fucosyltransferase, galactomannan galactosyltransferase, and homogalacturonan galacturonosyltransferase (Edwards et al., 1999; Perrin et al., 1999; Sterling et al., 2006). All of these enzymes are predicted to be type II integral membrane proteins, with globular catalytic domains facing the Golgi lumen. This topology may explain why these proteins maintain detectable enzymatic activity in the presence of detergent. Many other plant cell wall biosynthetic enzymes are thought to be type III integral membrane proteins, whose polytopic nature may make them less amenable to purification techniques involving solubilization.
Molecular genetic and functional genomic strategies have provided a powerful alternative suite of tools for identifying genes encoding glycosyltransferases and glycan synthases involved in plant cell wall biosynthesis (Lerouxel et al., 2006; Liepman et al., 2007). Many of the enzymes involved in cell wall metabolism are encoded by members of sizeable gene families (Yokoyama and Nishitani, 2004). For example, the cellulose synthase family of genes includes the authentic cellulose synthase (CesA) genes and at least eight additional groups, designated cellulose synthase-like (Csl) genes because of sequence similarity with the CesA genes. Not all plants contain the same Csl subgroups. The 30 Csl proteins of Arabidopsis (Arabidopsis thaliana) group into six Csl subfamilies: CslA, CslB, CslC, CslD, CslE, and CslG (Richmond and Somerville, 2000). The 33 Csl proteins of rice (Oryza sativa) also group into six families: CslA, CslC, CslD, CslE, CslF, and CslH (Keegstra and Walton, 2006). However, only the CslA, CslC, CslD, and CslE proteins are shared by Arabidopsis and rice. The Csl genes are considered the best candidates to encode enzymes that polymerize the backbones of β-linked noncellulosic polysaccharides (Richmond and Somerville, 2001; Hazen et al., 2002). Experimental evidence to support this hypothesis for the CslA family came first from Dhugga et al. (2004), who demonstrated that a CslA gene from guar (Cyamopsis tetragonolobus) encodes a β-mannan synthase. More recently, Liepman et al. (2005) expressed three Arabidopsis CslA genes in Drosophila Schneider 2 (S2) cells and demonstrated that the resulting CslA proteins were capable of producing β-mannans when supplied with GDP-Man and glucomannans when provided with a mixture of GDP-Man and GDP-Glc. Similar observations were made with CslA proteins from poplar (Populus spp.; Suzuki et al., 2006).
Mannan polysaccharides are widespread among land plants and are also present in many algal species, some of which completely lack cellulose in their cell walls (Frei and Preston, 1968; Preston, 1968). Several varieties of mannan polysaccharides have been characterized, including pure mannans, galactomannans, glucomannans, and galactoglucomannans. Structurally, each of these polysaccharides contains a β-1,4-linked backbone composed of Man or a combination of Glc and Man residues; this backbone may be substituted with α-1,6-linked Gal side chains. Two different types of enzymes involved in the biosynthesis of mannan polysaccharides have been identified: a β-1,4-(gluco) mannan synthase (GlcManS) that polymerizes the polysaccharide backbone, and an α-1,6-galactomannan galactosyltransferase that adds Gal residue side chains to the backbone (Edwards et al., 1999). Mannan polysaccharides appear to serve at least two functions: (1) structural, as hemicelluloses that bind cellulose and as crystalline fibrils in algae that lack cellulose-based cell walls; and (2) storage, as nonstarch carbohydrate reserves in endosperm walls and vacuoles of seeds and in vacuoles in vegetative tissues (Preston, 1968; Meier and Reid, 1982; Brennan et al., 1996). In the wood of gymnosperms, this polysaccharide is the most abundant cross-linking glycan (Maeda et al., 2000). In Arabidopsis, mannans have been immunolocalized to the thickened secondary cell walls of xylem and fiber cells and appear to be present in primary walls and possibly in the vacuoles of other cells (Handford et al., 2003). Man constitutes a significant percentage of total monosaccharides in Arabidopsis inflorescence stems, which contain vascular cells and fibers with thickened secondary cell walls (Zhong et al., 2001; Brown et al., 2005; Zhong et al., 2005; Harholt et al., 2006). In other tissues consisting mostly of cells lacking secondary walls, such as leaves, very little mannan is present (Zablackis et al., 1995). Crystalline mannans appear to take the place of cellulose as the primary structural polysaccharide in certain algal species (Frei and Preston, 1968; Preston, 1968).The seeds of certain legume species have rich galactomannan deposits in the cell walls and in vacuoles of endosperm cells, where they serve as storage polysaccharides. Pure mannans serve a similar function in the fruits of some species of palm (e.g. ivory nut palm [Metroxylon salomonense]). Vacuolar mannans and glucomannans have been observed in vegetative tissues of a variety of monocot species, including members of the Liliaceae, Orchidaceae, and Araceae (Meier and Reid, 1982).
Although it is hypothesized that many or even most of the Csl genes encode enzymes involved in plant cell wall biosynthesis, the biochemical functions of very few of these gene products have actually been demonstrated. It is tempting to speculate that biochemical activity is conserved within each Csl family. However, there is limited experimental evidence to support this hypothesis. The CslA family provides an excellent case study to test this hypothesis. Among the Csl gene families present in Arabidopsis and rice, the CslA family contains the most members, with nine present in each genome (Richmond and Somerville, 2001; Hazen et al., 2002). CslA gene sequences are unique to plants and have been observed in over 100 plant species, including representatives of the mosses (Ceratodon purpureus, Physcomitrella patens), ferns (Adiantum capillus-veneris), and gymnosperms (Picea sp., Pinus sp.), as well as mono- and dicotyledonous angiosperms (http://cellwall.stanford.edu/php/summary.php). Previous studies using heterologous expression revealed that CslA gene products from Arabidopsis, guar, and poplar catalyze β-mannan and glucomannan synthase reactions in vitro (Dhugga et al., 2004; Liepman et al., 2005; Suzuki et al., 2006). To determine whether these activities are conserved, CslA proteins from diverse land plant lineages were expressed in insect cells. Recombinant CslA proteins from representative mono- and dicotyledonous angiosperms, as well as a gymnosperm and a bryophyte each catalyzed mannan and glucomannan synthase reactions in vitro. Analysis of the expression patterns of CslA genes and deposition patterns of mannan polysaccharides indicate that the physiological roles of these polysaccharides may extend beyond structure and storage and that mannans are involved in other growth and developmental processes.
RESULTS
To extend our understanding of functional conservation within the CslA family, the enzymatic activities of CslA proteins from a bryophyte (P. patens; Roberts and Bushoven, 2007), a gymnosperm (loblolly pine [Pinus taeda]), a monocot (rice; Hazen et al., 2002), and a dicot (Arabidopsis; Richmond and Somerville, 2001) were examined using heterologous expression. CslA sequences from loblolly pine were identified by searching the Cell Wall database (http://www.cellwall.stanford.edu) and the Loblolly Pine expressed sequence tag database (http://pinetree.ccgb.umn.edu/) using Arabidopsis CslA protein sequences as queries. The sequences assembled into contigs that represented partial coding regions of two distinct CslA homologs. The sequences were verified by using gene-specific primers to amplify cDNA obtained from loblolly pine xylem. The corresponding full-length cDNA clones were obtained by screening a cDNA library from developing loblolly pine xylem and designated PtaCslA1 and PtaCslA2. Putative full-length polypeptide sequences were deduced from the nucleotide sequences of cDNA clones isolated for this study and obtained from stock centers (Supplemental Table S1; Seki et al., 2002; Kikuchi et al., 2003). The predicted proteins encoded by this collection of CslA genes are 55% to 96% identical to each other.
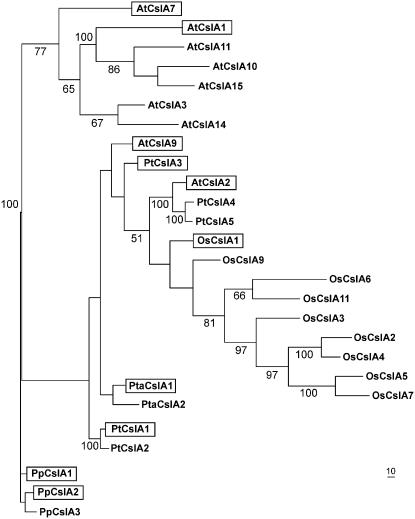
For phylogenetic analysis of CslA genes families, a parsimony phylogram was constructed using an alignment of full-length CslA polypeptide sequences from the complete genomes of Arabidopsis (Richmond and Somerville, 2001), rice (Hazen et al., 2002), Populus trichocarpa (Suzuki et al., 2006), and P. patens (Roberts and Bushoven, 2007) as well as the full-length CslA protein sequences from loblolly pine (Fig. 1). Although it was originally reported that the P. patens genome could contain as many as five CslA genes (Roberts and Bushoven, 2007), only three (PpCslA1, PpCslA2, and PpCslA3) were identified in the assembled genome (data not shown). The topology of the phylogram is characterized by several monospecific clades. These include a well-supported P. patens clade as has been shown previously (Roberts and Bushoven, 2007). In addition, a well-supported clade of four Arabidopsis sequences clusters with three other Arabidopsis sequences to form a moderately well-supported clade containing seven Arabidopsis sequences. Similarly, a well-supported clade of five rice sequences clusters with two additional rice sequences to form a moderately well supported clade of seven rice sequences. These rice and Arabidopsis clades are supported by a previous analysis (Suzuki et al., 2006). Pairing of P. trichocarpa sequences PtCslA1 with PtCslA2 and PtCslA4 with PtCslA5 is also consistent with a previous analysis of CslA protein sequences (Suzuki et al., 2006) and with evidence of a recent duplication of the P. trichocarpa genome (Tuskan et al., 2006). Strong support is also provided for a clade containing one Arabidopsis sequence and a pair of P. trichocarpa sequences. The placements of AtCslA9, PtCslA3, OsCslA1, OsCslA9, the loblolly pine sequences, and the PtCslA1/2 pair are not resolved.
Figure 1.
Phylogenetic analysis of CslA sequences. A parsimony phylogram that corresponds to the majority consensus of 1,000 bootstrap replicates (percent values above 50% shown) for full-length CslA polypeptide sequences deduced from the complete genomes of Arabidopsis (At), rice (Os), P. trichocarpa (Pt), and P. patens (Pp) and from cDNA sequences isolated from a loblolly pine (Pta) cDNA library is shown. Recombinant CslA proteins shown in boxes catalyze β-1,4-mannan and/or glucomannan synthase reactions in vitro. These activities were demonstrated in previous studies for AtCslA2, AtCslA7, and AtCslA9 by Liepman et al. (2005) and for PtCslA1 and PtCslA3 by Suzuki et al. (2006).
The hypothesis that mannan and/or glucomannan synthase activity is conserved among proteins encoded by CslA genes from Arabidopsis and other plant species was tested using a heterologous expression system. CslA proteins from Arabidopsis, rice, loblolly pine, and P. patens were expressed in Drosophila S2 cells, an expression system previously shown to produce enzymatically active CslA proteins, including AtCslA9, which was used in this study as a positive control. These cells are particularly well suited for this application, because background incorporation of the assay substrates GDP-Man and GDP-Glc into ethanol-insoluble products by endogenous enzymes present in microsomal membrane preparations is very low (Liepman et al., 2005). Transgenes encoding epitope-tagged CslA proteins under the control of the inducible metallothionein promoter were stably incorporated into S2 cells. Following induction of protein expression in transformed cell lines, cell lysates were fractionated by differential centrifugation and microsomal membrane fractions were collected. Five recombinant CslA proteins were detected by immunoblot analysis (AtCslA1, AtCslA9, OsCslA1, PpCslA1, and PtaCslA1; Supplemental Fig. S1), and another four were either not produced by the S2 cells or produced at levels below the detection limit of immunoblot analysis (AtCslA3, AtCslA14, OsCslA7, and PpCslA2; data not shown).
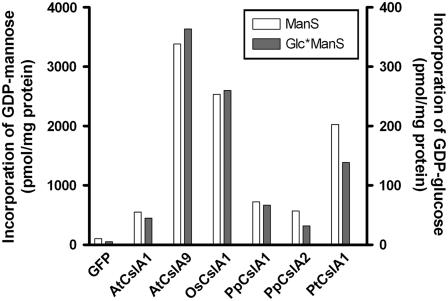
To determine whether recombinant CslA proteins catalyzed mannan and/or glucomannan synthase reactions, microsomal membrane fractions from S2 cells transformed with constructs containing CslA genes or a green fluorescent protein (GFP) control gene were assayed in vitro. Cell lines producing detectable quantities of CslA protein as measured by immunoblot analysis also contained significantly elevated mannan and glucomannan synthase activities compared with GFP control lines (Fig. 2). These activities were also elevated in membrane fractions from PpCslA2-expressing S2 cells, even though no detectable cross-reactive protein was detected. No mannan or glucomannan synthase activity or epitope-tagged Csl protein was detected in membrane fractions from S2 cells transformed with AtCslA3, AtCslA14, or OsCslA7 (data not shown).
Figure 2.
Glucomannan synthase activity assays of recombinant CslA proteins from diverse land plants produced in Drosophila S2 cells. Microsomal membranes of S2 cells producing either GFP or a CslA protein were assayed for ManS or GlcManS activity in vitro. The incorporation of GDP-Man or GDP-Glc into 70% ethanol-insoluble products by microsomal membrane fractions is graphed. The ManS product was labeled with GDP-[14C]Man, and the GlcManS product was labeled with GDP-[14C]Glc in the presence of nonradioactive GDP-Man.
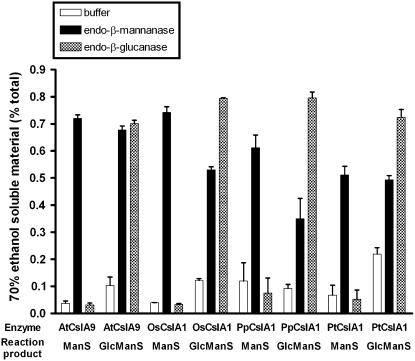
In vitro products synthesized by representative CslA proteins from a dicot (AtCslA9), a monocot (OsCslA1), a gymnosperm (PtaCslA1), and a bryophyte (PpCslA1) were further analyzed using hydrolytic enzymes that cleave specific carbohydrates. Each of the in vitro mannan synthase products was hydrolyzed by endo-β-mannanase but not by endo-β-glucanase. In contrast, products of glucomannan synthase reactions produced by enzymes from each species were susceptible to digestion by both endo-β-mannanase and endo-β-glucanase (Fig. 3). Consistent with previous studies using several CslA proteins from Arabidopsis and poplar (Liepman et al., 2005; Suzuki et al., 2006), these results indicate that representative CslA proteins from dicots, monocots, gymnosperms, and bryophytes all catalyze the biosynthesis of β-linked mannan and glucomannan polysaccharides.
Figure 3.
Characterization of in vitro mannan and glucomannan synthase products. Large-scale in vitro assay products of recombinant CslA proteins from P. patens (PpCslA1), loblolly pine (PtaCslA1), rice (OsCslA1), and Arabidopsis (AtCslA9) were treated with endo-β-glucanase, endo-β-mannanase, or buffer alone. The percentage of total radioactivity released from each product into the 70% ethanol-soluble fraction after each treatment is shown.
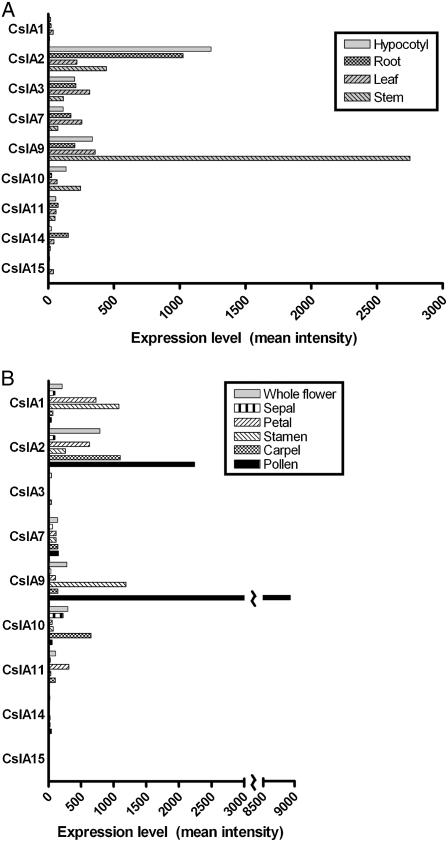
Previous analyses of the cell wall composition of various Arabidopsis tissues have revealed tissue-specific differences in Man and mannan abundance. While mannans were not detected by linkage analysis of Arabidopsis leaf carbohydrates (Zablackis et al., 1995), they were detected in this tissue using carbohydrate immunofluorescence (Handford et al., 2003). Man, potentially in the form of mannans, constitutes a significant percentage of the total monosaccharides in inflorescence stems, which contain vascular cells and fibers with thickened secondary cell walls (Zhong et al., 2001, 2005; Brown et al., 2005; Harholt et al., 2006). Indeed, mannans were detected in Arabidopsis inflorescence stems by carbohydrate immunofluorescence (Handford et al., 2003). RNA transcript abundance profiles of the nine Arabidopsis CslA genes were examined in various vegetative (Fig. 4A) and floral (Fig. 4B) tissues from the Arabidopsis AtGenExpress dataset (Schmid et al., 2005). As observed previously, Arabidopsis CslA genes displayed tissue-specific gene expression patterns (Hamann et al., 2004), and aside from the AtCslA15 gene, transcripts of each of the other AtCslA genes were detected in one or more tissues. Among vegetative tissues (Fig. 4A), transcripts of the AtCslA2 gene were the most abundant in hypocotyls and roots, whereas AtCslA9 transcripts were most abundant in leaves and inflorescence stems. Although mannans are present at very low levels in Arabidopsis leaves (Zablackis et al., 1995; Handford et al., 2003), transcripts of several AtCslA genes were present in this tissue. Transcripts of many AtCslA genes also displayed tissue-specific abundance patterns in various floral organs (Fig. 4B), where secondary cell walls are virtually absent. Transcripts of the AtCslA10 gene were most abundant in sepals, AtCslA1 transcripts in petals, AtCslA9 transcripts in stamens, and AtCslA2 transcripts in carpels. It is also noteworthy that in mature pollen, where AtCslA7 transcripts have been observed previously (Goubet et al., 2003; Honys and Twell, 2003), the level of AtCslA9 transcripts was very high.
Figure 4.
Microarray expression profiling of CslA genes in vegetative and reproductive tissues of Arabidopsis. A, Mean signal intensity of Arabidopsis CslA genes in various vegetative tissues from the AtGenExpress microarray dataset (Schmid et al., 2005) is plotted. Hypocotyl, root, and leaf tissues were from 7-d-old seedlings (ATGE_2, _3, _5, respectively), and stem tissue was from 21-d-old plants (ATGE_27). B, Mean signal intensity of Arabidopsis CslA genes in flowers, flower parts, and mature pollen. Whole stage 15 flowers as well as sepals, petals, stamens, and carpels from stage 15 flowers were from 21-d-old plants (ATGE_39, _41, _42, _43, respectively); mature pollen was from 42-d-old plants (ATGE_73). All tissues were harvested from plants grown under continuous light. Low expression levels of particular CslA genes in some tissues yielded signal intensity values below the level of significance (mean intensity < 100).
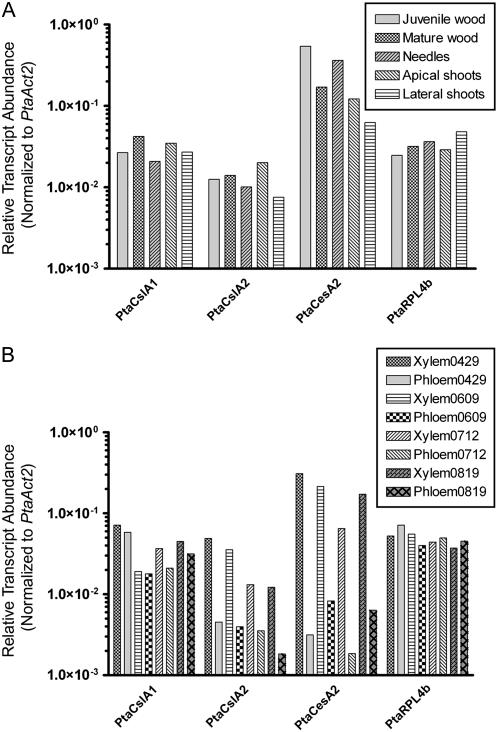
Quantitative real-time reverse transcription (RT)-PCR was used to examine the distribution of CslA transcripts in tissues of loblolly pine. Primer pairs suitable for quantitative real-time RT-PCR analysis of the PtaCslA1, PtaCslA2 genes, as well as several control genes (Actin 1, Actin 2, and Ribosomal protein L4b) were designed and their efficiencies tested (Supplemental Table S2). The PtaCesA2 gene was also included in this analysis because it encodes a cellulose synthase isoform thought to be involved in secondary cell wall synthesis (Nairn and Haselkorn, 2005). Because its expression level displayed the lowest sd across all of the samples, the PtaAct2 gene was selected for normalization of data (Supplemental Table S3). Transcripts of the PtaCslA1 and PtaCslA2 genes were detected in various vegetative tissues, including needles, juvenile wood, mature wood, and apical and lateral shoots (Fig. 5A).
Figure 5.
Quantitative real-time RT-PCR analysis of PtaCslA1 and PtaCslA2 genes in tissues of loblolly pine. A, The transcript abundance of two PtaCslA genes, a cellulose synthase gene (PtaCesA2), an actin gene (PtaAct1), and a ribosomal protein L4 gene (PtaRPL4b) normalized to an actin control gene (PtaAct2) was measured in juvenile wood, mature wood, needles, apical shoot tips, and lateral shoot tips. B, Gene expression levels in xylem and phloem tissues collected at four time points, gathered from April to August, 2005.
In the wood of gymnosperms, glucomannans are the most abundant hemicellulose and may contribute up to 18% of the mass of the cell walls (Maeda et al., 2000). Quantitative real-time RT-PCR analysis revealed that both PtaCslA1 and PtaCslA2 are expressed in xylem and phloem tissues of loblolly pine (Fig. 5B). Transcripts of the PtaCslA1 gene were present at approximately equivalent levels in xylem and phloem tissues. In contrast, PtaCslA2 is expressed at approximately 4- to 11-fold higher levels in xylem tissue than in phloem tissue. The pattern of PtaCslA2 expression closely resembled that of the secondary cell wall-specific PtaCesA2 gene. The expression levels of each of the control genes were similar in xylem and phloem.
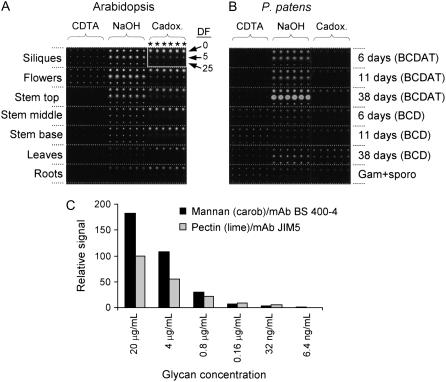
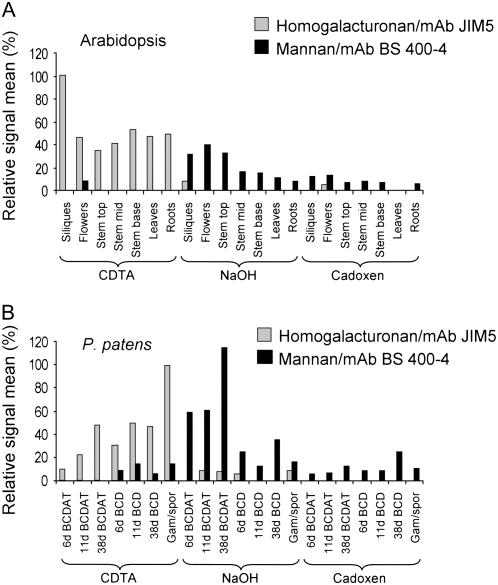
Although mannans have been detected in various tissues of Arabidopsis using carbohydrate immunofluorescence (Handford et al., 2003), it is not clear whether the high level of Man observed in certain Arabidopsis tissues (Brown et al., 2005; Zhong et al., 2005; Harholt et al., 2006) originates from mannan polysaccharides. A novel technique using glycan microarrays (comprehensive microarray polymer profiling [CoMPP]) was used to investigate the differences in mannan content between Arabidopsis organs. Polysaccharides were sequentially extracted from various tissues of Arabidopsis using cyclohexane diamine tetraacetic acid (CDTA), NaOH, and 1,2-diaminoethane and cadmium oxide (cadoxen), spotted onto membranes, and probed with monoclonal antibodies (mAbs) with specificity for mannan polysaccharides (Pettolino et al., 2001) and, for comparison, homogalacturonan (HG; Clausen et al., 2003). An example of an Arabidopsis CoMPP array is shown (Fig. 6A). To determine if the level of spotted glycan was proportional to CoMPP spot signals, defined samples of mannan (from carob [Ceratonia siliqua]) and pectin (containing HG domains, from lime [Citrus aurantifolia]) were spotted as dilution series on arrays in the range 20 μg/mL to 6.4 ng/mL. These arrays were probed with the antimannan and anti-HG mAbs used for the CoMPP analysis. As shown in Figure 6C, signal values reflected the amount of spotted glycans. In this study, mannans were extracted predominantly by NaOH from all Arabidopsis organs examined (Fig. 7A). The highest levels of mannans were detected in flowers, siliques, and the upper regions of inflorescence stems. As expected, HG was detected primarily in the CDTA-soluble fraction.
Figure 6.
Microarrays of cell wall glycan polymers. A and B, Examples of microarrays generated from polysaccharides extracted from Arabidopsis (A) and P. patens (B) probed with mannan-specific mAb (BS 400–4). Polysaccharides were sequentially extracted from the tissues/organs indicated using CDTA, followed by NaOH, then cadoxen. In the case of P. patens, cultures were grown on either BCD or BCDAT mediums as indicated. Three dilution factors (DF) of the extractions (0, 5, and 25) were printed in sextuplet (*) such that each organ/tissue is represented by 18 spots on the arrays (as indicted by the white box in A). C, Arrays were created in which samples of mannan and lime pectin were spotted at the dilutions shown and probed with antimannan or anti-HG (JIM5) mAbs. Spot signals were quantified using ImaGene 6.0 and the maximal signal for JIM5 set at 100%. Note that these values should not be used to calibrate glycan levels in Figure 7.
Figure 7.
CoMPP analysis of mannan polysaccharides in Arabidopsis and P. patens. A and B, CoMPP data indicating the relative levels of HG (gray bars) and mannan (black bars) in Arabidopsis (A) and P. patens (B) organs/tissues. Mean spot signals were quantified using ImaGene 6.0 and for each species, the maximal signal obtained for HG in each plant was set at 100%. Note that signals can be used to infer difference in the relative glycan levels within, but not between, the two species.
Man is also present at moderate to high levels in primary cell walls of a variety of bryophytes (mosses) at levels typically exceeding those found in primary cell walls of vascular plants (Popper and Fry, 2003). To determine whether mannans are also found in the cell walls of P. patens, tissues of this moss cultured for various durations on BCD or BCDAT medium were also examined using CoMPP analysis. An example of a P. patens CoMPP array is shown in Figure 6B. Tissue cultured on BCDAT included only chloronemal filaments after 6 d, chloronemal filaments with some rapidly growing caulonemal filaments and occasional buds after 11 d, and mixed filaments with some leafy gametophores after 38 d. Tissue cultured on BCD included chloronemal and caulonemal filaments with buds after 6 d, mixed filaments with leafy gametophores after 11 d, and leafy gametophores with a few caulonemal filaments after 38 d. Figure 7B shows the relative levels of HG and mannan polysaccharides in P. patens tissues. Mannans were detected in the NaOH-soluble fraction of P. patens at all growth stages examined, but the abundance varied with medium and culture duration. Mannan abundance was higher in P. patens plants cultured on BCDAT compared to those cultured on BCD. On both culture media, the highest levels of mannans were detected in older cultures. CoMPP analysis provided information about the relative levels of mannan and HG but is not a quantitative technique for the accurate measurement of absolute levels of glycans, and polymer abundance cannot be directly compared between Arabidopsis and P. patens. One reason this is true is because only extractable glycans can be analyzed by CoMPP, and while the solvents used are generally effective for releasing many cell wall components, it is possible that the extractions do not work equally well in all sample types (Fry, 2000). Nevertheless, CoMPP analysis clearly indicates that mannan-containing polysaccharides are a component of the cell walls of both Arabidopsis and P. patens. In both plants, these polysaccharides are extracted most effectively with NaOH. Because the antimannan mAb recognizes a variety of mannan, galactomannan, and, to a lesser extent, glucomannan polysaccharides, we were unable to determine the relative level of each type of polymer in these plants.
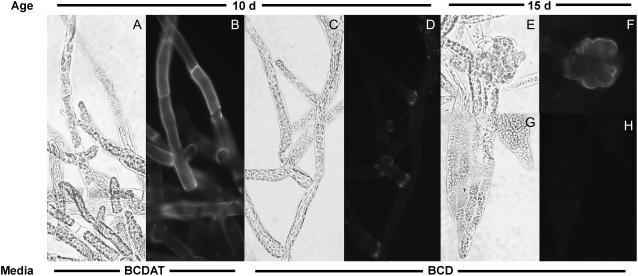
Mannan-specific mAbs were used to examine the distribution of these polysaccharides in cells of P. patens plants using immunofluorescence microscopy (Fig. 8). Mannan epitopes were strongly labeled on chloronemal and caulonemal filaments from P. patens cultures grown on BCDAT, whereas both types of filaments from cultures grown on BCD were only weakly labeled. Mannans were most abundant in BCD-cultured cells at the bases of branch cells. Buds were labeled strongly with the antimannan antibodies; however, labeling of leafy gametophytes was very weak. No labeling was detected in control samples lacking primary antibody or using primary antibody preadsorbed with mannan (data not shown).
Figure 8.
Immunolocalization of mannans in P. patens. Tissue from P. patens cultured on BCDAT medium (A and B) or BCD medium (C–H) for 10 d (A–D) or 15 d (E–H), labeled with monoclonal antimannan, and imaged with brightfield (A, C, E, and G) and epifluorescence optics (B, D, F, and H). A and B, Chloronemal filaments. C and D, Chloronemal and caulonemal filaments. E and F, Bud. G and H, Gametophore leaf. No signal was observed in control samples lacking primary antibodies or incubated with antibodies that had been preadsorbed with carob galactomannan.
DISCUSSION
The Enzymatic Function of CslA Proteins Is Conserved across Major Land Plant Lineages
Genes encoding plant cell wall biosynthetic enzymes are often members of extended gene families (Yokoyama and Nishitani, 2004), as exemplified by the CesA and Csl genes. Representatives of these groups have been implicated in the biosynthesis of cellulose (CesA), β-mannans (CslA), and mixed-linkage β-glucans (CslF; Pear et al., 1996; Dhugga et al., 2004; Burton et al., 2006). The remaining Csl genes are considered excellent candidates to encode hemicellulosic glycan synthases. Whether all members of a particular Csl subgroup encode enzymes with the same catalytic function remains an open question, and in no case have functions been ascribed to more than a few members of a family. The CslA family provides an excellent opportunity to address this question because: (1) CslA genes have been identified in the major groups of land plants, including bryophytes, ferns, gymnosperms, and angiosperms; (2) many plants, including Arabidopsis, have multiple CslA genes that encode proteins with relatively high sequence identity; and (3) a heterologous expression system that yields enzymatically active CslA proteins has been developed.
Heterologous expression of representative CslA proteins from a bryophyte (P. patens), a gymnosperm (loblolly pine), a monocot (rice), and three dicots including guar (Dhugga et al., 2004), Arabidopsis (Liepman et al., 2005), and P. trichocarpa (Suzuki et al., 2006) has demonstrated that each catalyzes β-1,4-mannan and glucomannan synthase reactions. An exception was PtCslA5, for which no activity was observed in vitro (Suzuki et al., 2006). Multiple explanations are possible for the lack of ManS activity observed in membrane preparations containing recombinant PtCslA5. One interpretation is that PtCslA5 does not catalyze ManS and/or GlcManS reactions (Suzuki et al., 2006). Another interpretation is that PtCslA5 catalyzes these reactions, but issues relating to the expression or assay of this particular sequence prevented the detection of these enzymatic activities. Further analysis is needed to distinguish between these possibilities. Biochemical characterization of CslA proteins from a range of species, including two or more paralogs from Arabidopsis, P. trichocarpa, and P. patens demonstrates conserved function among many members of the CslA subfamily in land plants. Additionally, as other Csl families exhibit levels of sequence identity similar to that found among members of the CslA family, it seems reasonable to hypothesize that each Csl family contains members that catalyze a single enzymatic reaction.
The Majority of CslA Diversification Occurred after the Divergence of Monocots and Eudicots
The CslA proteins from four complete genomes (i.e. Arabidopsis, rice, P. trichocarpa, and P. patens) were subjected to phylogenetic analysis along with two CslA proteins from loblolly pine. The analysis clearly demonstrates independent diversification within the lineages that gave rise to Arabidopsis and rice. In contrast, CslA diversification appears to have been less extensive in P. trichocarpa, although pairing of four of the five P. trichocarpa CslA proteins is consistent with diversification resulting from a recent genome duplication (Tuskan et al., 2006). The well-supported clustering of AtCslA2 with the PtCslA4/5 pair provides evidence that at least some CslA diversification occurred prior to the divergence of the Arabidopsis and P. trichocarpa lineages. However, we are unable to infer additional orthologous groups due to uncertain placement of several Arabidopsis, rice, and P. trichocarpa sequences. A previous analysis using the neighbor-joining utility in ClustalX inferred orthology of AtCslA9 with PtCslA1/2 and PtCslA3 (Suzuki et al., 2006). However, this was not supported by parsimony analysis.
The presence of CslA genes in P. patens is consistent with an ancient origin of the CslA family inferred from the similarity between vascular plant CslAs and bacterial cellulose synthase genes (Nobles and Brown, 2004). It has been shown previously that the CslA gene family (as well as the CesA, CslC, and CslD families) diversified independently in P. patens and vascular plants (Roberts and Bushoven, 2007). Thus, mosses and vascular plants may have inherited a single CslA gene from their common ancestor. The extent of CslA diversification prior to the divergence of the gymnosperm and angiosperm lineages is unknown. Although loblolly pine (a gymnosperm) has at least two CslA genes, the phylogenetic relationships of these genes to angiosperm counterparts were not resolved in our analysis. Identification and analysis of additional loblolly pine CslA sequences may clarify these relationships. Failure to resolve orthologous groups among the CslA sequences of Arabidopsis, rice, P. trichocarpa, and loblolly pine contrasts with analyses of the CesA sequences from these species, among which six orthologous groups can be clearly identified (e.g. Djerbi et al., 2005; Nairn and Haselkorn, 2005).
Biological Functions of Mannan and Glucomannan Polysaccharides
Whereas the roles of mannans as structural and storage polysaccharides in plants and algae are well documented (Frei and Preston, 1968; Meier and Reid, 1982), accumulating evidence suggests that these polysaccharides may serve additional roles during plant growth and development. Molecular genetic studies have revealed that two Arabidopsis mutants with lesions in CslA genes display morphological and developmental abnormalities. The SGT4425 (cslA7) mutant exhibits pleiotropic phenotypic abnormalities, including defects in pollen tube growth and lethal embryo development (Goubet et al., 2003). Cells of Agrobacterium tumefaciens bind less effectively to roots of rat4 (cslA9) mutants, rendering them resistant to Agrobacterium transformation. These plants also grew fewer lateral roots than wild-type plants (Zhu et al., 2003). In tomato (Lycopersicon esculentum), mannan metabolic enzymes, including endo-β-mannanase and mannan transglycosylase, are present in a variety of tissues. Endo-β-mannanase activity is observed in tomato fruits, anthers, and pollen grains (Filichkin et al., 2004). Mannan transglycosylase activity, potentially involved in reorganization of mannan polysaccharides, is present in tissues from a variety of plants (Schroder et al., 2004), and recently, a tomato endo-β-mannanase (LeMAN4) was shown to catalyze this reaction (Schroder et al., 2006). Galactoglucomannan-derived oligosaccharides also appear to elicit growth and developmental responses when applied to a variety of plant tissues and cell cultures (Auxtova et al., 1995; Liskova et al., 1995; Bilisics et al., 2004; Benova-Kakosova et al., 2006). Little is known about the roles mannans play in these processes or the mechanisms of their involvement. However, these studies suggest that mannans may act as signaling molecules.
Supporting the potentially diverse roles of mannan polysaccharides, CslA gene transcripts were present in many vegetative and reproductive tissues. In Arabidopsis and pine, transcripts of CslA genes showed distinct patterns of accumulation throughout the plant. CslA gene transcripts have also been observed in a variety of tissues of poplar (Geisler-Lee et al., 2006) and are particularly abundant in wood-forming tissues (Aspeborg et al., 2005). Certain CslA transcripts were surprisingly highly expressed in particular Arabidopsis floral organs and mature pollen grains.
Consistent with CslA gene expression profiles, mannan polysaccharides were detected in a range of Arabidopsis tissues and were most abundant in stems, flowers, and siliques. Because stems are rich in secondary cell walls, the mannans observed in this tissue may serve primarily structural roles. The significance of mannans in tissues with a lower proportion of secondary walls, such as flowers and siliques, is less clear. Mannans were also detected in the moss P. patens, where they were more abundant in cultures grown in the presence of ammonia compared to cultures not supplemented with this nitrogen source. Immunolocalization experiments confirmed the marked differences in mannan content between moss cultures grown in the presence or absence of ammonia and demonstrated that both chloronemal and caulonemal filaments of P. patens contain more mannans when cultured in the presence of ammonia. The primary effect of ammonia is inhibition of the development of caulonemal filaments and leafy gametophores. It is not clear why mannan is more abundant in cultures containing ammonia, because mannan does not appear to be restricted to chloronemal cells. Intense labeling of cell junctions and buds may indicate that mannan is deposited in the cell walls of recently divided cells. In tip-growing branch cells, the cell wall components deposited prior to cell expansion would remain at the cell base. In contrast, diffuse growth would dilute cell wall components deposited prior to cell expansion in gametophore leaf cells.
The CslA genes of loblolly pine are of particular interest because conifers contain higher mannan content in their wood relative to angiosperms. From the perspective of the pulp and paper industry, glucomannans are undesirable components of gymnosperm wood because they interfere with the pulping process, decreasing its efficiency. However, high levels of glucomannans might prove advantageous for biofuel production because of their potential contribution to elevated ethanol yields (Suzuki et al., 2006). Two pine CslA genes, PtaCslA1 and PtaCslA2, displayed different expression profiles. While transcripts of both genes were detected in a variety of tissues, marked differences between the expression of the PtaCslA1 and PtaCslA2 genes were observed in xylem and phloem tissues. The PtaCslA1 gene was expressed at a similar level in xylem and phloem, suggesting that the product of the PtaCslA1 gene functions during both primary and secondary wall synthesis. The pattern of PtaCslA2 expression closely resembled that of the PtaCesA2 gene, the product of which is a putative secondary cell wall-specific cellulose synthase isoform (Nairn and Haselkorn, 2005). Transcripts of both PtaCslA1 and PtaCesA2 were more abundant in xylem than phloem tissues; the similarity between the patterns of expression of the PtaCslA2 and PtaCesA2 genes suggests that the product of the PtaCslA2 gene is involved in a secondary cell wall biosynthesis. The identification of genes that regulate wood composition is an important objective in forest biotechnology; PtaCslA2 may be an excellent candidate for modification of wood properties.
CONCLUSION
Previous studies have demonstrated that members of the CslA gene family encode glucomannan synthase enzymes that polymerize the β-1,4-linked backbones of mannan polysaccharides (Dhugga et al., 2004; Liepman et al., 2005; Suzuki et al., 2006). This study extends previous analyses and demonstrates that CslA gene products from taxonomically diverse land plants catalyze β-1,4-mannan and glucomannan synthase reactions, indicating that this enzymatic function may be conserved among all members of the CslA family. The evidence presented here and elsewhere indicates that mannans are multifunctional cell wall polymers with widespread distribution in plant tissues. In addition to their roles as structural elements of cell walls, they function in storage and may serve additional important functions.
MATERIALS AND METHODS
Reagents and Enzymes
Unless otherwise noted, all chemicals and reagents were purchased from Sigma-Aldrich. GDP-[14C]Man (10.7 GBq/mmol) was obtained from GE Healthcare, and GDP-[14C]Glc (11.1 GBq/mmol) was obtained from American Radiolabeled Chemicals. Purified endo-1,4-β-mannanase from Bacillus sp. (catalog no. E-BMABS), endo-1,4-β-glucanase (cellulase; catalog no. E-CELTR) from Trichoderma sp., and carob (Ceratonia siliqua) galactomannan were from Megazyme International. Lime (Citrus aurantifolia) pectin was from Danisco. Oligonucleotide primers were synthesized by Integrated DNA Technologies. Magnetic beads were from Dynal Biotech and SYBR Green Supermix was from Bio-Rad. Gateway vectors, competent Escherichia coli cells, platinum pfx DNA polymerase, SuperScript III first-strand synthesis system, LR clonase, S2 cells, vectors, and culture media were from Invitrogen. Horseradish peroxidase-conjugated mouse mAbs against the T7 epitope tag were obtained from Novagen. Antimannan mAbs were from Biosupplies Australia, anti-HG antibodies were from PlantProbes. Alexa-fluor 488 conjugated goat antimouse antibodies and Slowfade glycerol mounting medium were from Molecular Probes. Complete, mini protease inhibitor tablets lacking EDTA were from Roche. The bicinchoninic acid protein assay kit and the Super Signal Pico West chemiluminescence system were from Pierce. Immobilon-P polyvinylidene difluoride membrane was from Millipore.
Isolation of CslA Genes from Loblolly Pine
CslA protein sequences from Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) were used to query the translated expressed sequence tag database for loblolly pine (Pinus taeda). Contigs were assembled using GCG software (Genetics Computer Group) and yielded two unigene sets representing putative CslA genes. Primers were designed and used to amplify partial cDNA clones from xylem cDNA. A full-length enriched cDNA library was screened using the partial CslA cDNA clones (Nairn and Haselkorn, 2005). Positive clones containing full-length coding regions were identified and sequenced by primer walking using the dideoxy chain termination method (Sanger et al., 1977) with BigDye Terminator v3.1 and analyzed using an ABI 3730XL (Applied Biosystems). Two full-length CslA cDNA clones were identified, and these were designated PtaCslA1 and PtaCslA2.
Phylogenetic Analysis
Full-length CslA polypeptide sequences from the complete genomes of Arabidopsis (The Arabidopsis Information Resource locus IDs: At4g16590, At5g22740, At1g23480, At2g35650, At5g03760, At1g24070, At5g16190, At3g56000, and At4g13410; Richmond and Somerville, 2001), rice (The Institute for Genomic Research locus ID: Os02g09930, Os10g26630, Os06g12460, Os03g07350, Os03g26044 Os02g51060, Os07g43710, Os06g42020, and Os08g33740; Keegstra and Walton, 2006), Populus trichocarpa (JGI PopulusDB [http://www.populus.db.umu.se/] protein ID: 686549, 687416, 589559, 594843, and 556940; Suzuki et al., 2006), and Physcomitrella patens (GenBank accession no. DQ417756-7 and JGI protein ID 207003; Roberts and Bushoven, 2007), and the full-length sequences from loblolly pine (GenBank accession no. DQ641986-7) were aligned using ClustalW (Thompson et al., 1994) with the Gonnet protein weight matrix, pairwise gap opening/extension penalties of 10/0.1, and multiple alignment gap opening/extension penalties of 10/0.2. The alignment was edited using BioEdit (Hall, 1999) to remove gaps and segments of uncertain homology (Baldauf, 2003). Phylograms were constructed from the aligned sequences using the heuristic search method in PAUP* (version 4.1b10, Sinauer Associates) with all characters given equal weight. The topology was tested with 1,000 bootstrap replicates using the parsimony method.
Preparation of Epitope-Tagged CslA Proteins and Heterologous Expression in Drosophila S2 Cells
Open reading frames were amplified using PCR as previously described (Liepman et al., 2005). To facilitate the detection of recombinant proteins expressed in S2 cells, a short oligonucleotide sequence encoding the 11-amino acid T7 epitope tag (MASMTGGQQMG) was engineered directly upstream and in frame with the start codon of each CslA gene. The sequences of forward and reverse primers used for various CslA constructs are included in the supplemental data (Supplemental Table S4). Heterologous expression of CslA proteins in S2 cells was carried out as previously described (Liepman et al., 2005).
Preparation of Microsomal Membranes from Drosophila S2 Cells and Immunoblot Analysis
Microsomal membrane fractions from S2 cells were prepared as previously described (Liepman et al., 2005). Protein content was measured using the enhanced bicinchoninic acid assay with bovine serum albumin as standard. Approximately 40 μg samples of S2 microsomal membrane fractions resolved on 10% SDS polyacrylamide gels were transferred to polyvinylidene difluoride membranes using standard methods (Harlow and Lane, 1988) and probed with peroxidase-conjugated antibodies that recognize the T7 epitope tag (Liepman et al., 2005).
Enzyme Assays and Characterization of Radiolabeled Polysaccharides
Small-scale mannan synthase and glucomannan synthase assays were conducted at room temperature for 30 min in a total reaction volume of 40 μL, as described by Liepman et al. (2005). For mannan synthase assays, 18 to 25 μm nonradioactive GDP-Man concentrations and 2.1 μm GDP-[14C]Man were included. For glucomannan synthase assays, 8 to 15 μm nonradioactive GDP-Glc, 10 to 25 μm nonradioactive GDP-Man, and 2.1 μm GDP-[14C]Glc were included. In vitro assays were scaled up 5- to 10-fold to make products for characterization; however, carrier carob galactomannan was omitted from the precipitation procedure. To characterize the structures of in vitro assay products, aliquots of large-scale reactions were treated with buffer alone or incubated with purified preparations of endo-β-mannanase or cellulase. Digestion reactions were carried out at optimal pH for 2 h at 55°C. Typical reactions were conducted in 150 μL total volume containing 10 to 20 mU of hydrolytic enzyme. Reactions were terminated, washed, and analyzed as described previously (Liepman et al., 2005).
RNA Isolation and cDNA Synthesis
Materials for analysis of gene expression in loblolly pine were harvested, immediately flash frozen in liquid nitrogen, transported from the field on dry ice, and stored at −80°C. Developing wood of a ≥40-year-old loblolly pine was collected as previously described from crown (juvenile wood) and trunk (mature wood; Lorenz and Dean, 2002). Apical shoot tips (above the first internode) were harvested from 3- to 4-year-old greenhouse-grown trees. Expanding young needles and emerging lateral shoots (candles) were harvested from field-grown plants in early spring. Xylem and phloem tissues were harvested from a single mature loblolly pine individual at four time points between April 29, 2005 and August 19, 2005. Total RNA from tissues of loblolly pine was isolated as described by Chang et al. (1993). Poly(A+) mRNA was purified from total RNA using oligo(dT) magnetic beads following the manufacturer's protocol. Template cDNA for real-time quantitative RT-PCR was synthesized using 100 ng poly(A+) enriched RNA in 20-μL reactions. Reactions were carried out using SuperScript III First Strand Synthesis system for RT-PCR using oligo(dT) and random hexamer oligonucleotide primers. Each 20-μL cDNA reaction was then diluted 20-fold with water.
Primer Design and Validation for Real-Time Quantitative RT-PCR
Gene-specific primer pairs were designed for CslA, CesA, and potential control genes, including PtAct2, using the Primer3 program (Rozen and Skaletsky, 2000). Parameters were set for a primer length of 19 to 21 bp, primer melting temperature of 60.0°C ± 1.0°C, and amplimer length of 65 to 75 bp. Primers were validated using serial dilutions of purified plasmids containing the corresponding gene and/or genomic DNA. Standard curves were plotted and primers with efficiencies of 100% ± 10% selected for quantitative RT-PCR analysis of gene expression. The nucleotide sequences and characteristics of primers used for quantitative RT-PCR analysis are presented in the supplemental data (Supplemental Table S2).
Real-Time Quantitative RT-PCR
Gene expression profiling was conducted by real-time quantitative RT-PCR using triplicate reactions for each tissue sample and gene-specific primer pair. Reactions were assembled with 5 μL cDNA template, 5 μL primer mix containing 125 nm each of gene-specific forward and reverse primers, and 10 μL SYBR Green Supermix in 96-well plates sealed with optical film. Reactions were conducted using a MyiQ Single-Color Real-Time PCR Detection system (Bio-Rad). The amplification protocol was 95°C for 3 min, 40 cycles of 95°C for 30 s, 65°C for 45 s, 78°C for 20 s, then 95°C for 1 min, and 55°C for 1 min. Dissociation curves were obtained to confirm that single, specific products were produced in each reaction.
Calculation of Gene Transcript Levels
Threshold cycle (Ct) values from triplicate cDNA-primer samples were averaged and the sd was calculated. The logarithmic average Ct value for each gene and the control gene was converted to a linear value using the 2−Ct term (Livak and Schmittgen, 2001). The sd of Ct values was lowest for the PtAct2 gene among those examined for use as control genes and was selected for normalization of data. Converted values were normalized to the PtAct2 control gene by dividing average value for each gene by the average value of the control gene PtAct2.
Analysis of Cell Wall Polymers Using CoMPP
CoMPP analysis of cell wall polymers was performed on the various Arabidopsis organs and P. patens growth stages. Polymers were sequentially extracted using CDTA, NaOH, and cadoxen; supernatants from the extractions were spotted onto nitrocellulose membrane using a microarray robot (MicroGrid II, Applied Biosystems). Arrays were probed with mAbs specific for mannans (Pettolino et al., 2001) or pectic HG (Clausen et al., 2003). Spot signals were scanned and converted to 16-bit grayscale TIFFS, then quantified by means of microarray analysis software (ImaGene 4.0, BioDiscovery). Dilution series arrays were prepared using samples of carob galactomannan and lime pectin; these were probed as described above.
Culture of P. patens
Chloronemal tissue from P. patens that had been subcultured weekly on BCDAT medium overlain by cellophane (Knight et al., 2002) was blended, plated on media with (BCDAT) and without (BCD) ammonium tartrate, and incubated at 26°C with constant illumination at 45 μmol m−2 s−1. All media contained 1 mm CaCl2 added after autoclaving. For CoMPP analysis, tissue was harvested 6, 11, and 38 d after inoculation. Sporophyte tissue was obtained from cultures grown on peat pellets for 4 weeks at 26°C with constant illumination at 45 μmol m−2 s−1 followed by 12 weeks at 16°C with an 8-h photoperiod at 12 μmol m−2 s−1 (Knight et al., 2002). For immunofluorescence, tissue grown on BCDAT or BCD medium was harvested 10 and 15 d after inoculation.
Immunofluorescence Microscopy
Tissue from P. patens cultured on BCDAT or BCD medium was prepared for immunofluorescent localization of mannan as described previously (McCartney et al., 2003). Briefly, tissue was fixed in PEM buffer containing 4% formaldehyde and washed in phosphate-buffered saline (PBS). The washed tissue was incubated for 30 min in a blocking solution consisting of PBS with 3% nonfat dry milk, then for 1 h in blocking solution containing 4% antimannan hybridoma supernatant or without antibody as a control. After washing in blocking solution, tissue was incubated for 1 h in blocking solution containing 1% Alexa-fluor 488 conjugated goat antimouse antibodies. Finally, tissue was washed in PBS and mounted in SlowFade glycerol mounting medium. Images were recorded using a Spot RT slider digital camera (Diagnostic Instruments) mounted on an Olympus BHS microscope equipped with epifluorescence, 20× PlanApo UV objective, BP490 filter cube (Olympus), and KP560 barrier filter (Zeiss) for blocking chlorophyll autofluorescence. All epifluorescence images were recorded using identical optical conditions with 1-s manual exposure and were not processed digitally.
All of the results presented are from representative experiments that were performed at least twice with similar results.
Sequence data from this article are listed in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Immunoblot analysis of recombinant CslA proteins produced in Drosophila S2 cells.
Supplemental Table S1. Clones corresponding to CslA genes examined in this study.
Supplemental Table S2. Characteristics of primers used for quantitative real-time RT-PCR analysis.
Supplemental Table S3. Real-time quantitative RT-PCR analysis average Ct and converted values normalized to the endogenous control gene PtaAct2.
Supplemental Table S4. Oligonucleotide primer sequences used for heterologous expression studies.
Supplementary Material
Acknowledgments
We thank Linda Danhof for skillful culture and handling of Drosophila S2 cells, Katherine Krive for taxonomic analysis of the distribution of ManS genes, Alicia Wood-Jones for technical assistance in cloning and sequencing of the loblolly pine CslA genes, Jyl Venditti for assistance with immunofluorescence microscopy, all of the members of the Cell Wall Group at Michigan State University and the University of California at Riverside for helpful discussions and technical advice, and Karen Bird for editorial assistance.
This work was supported by the Energy Biosciences Program at the U.S. Department of Energy (to K.K.), by the Plant Genome Research Program at the National Science Foundation, and by the U.S. Department of Agriculture Forest Service and the Warnell School of Forestry and Natural Resources (to C.J.N.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Aaron H. Liepman (aliepman@emich.edu) and Kenneth Keegstra (keegstra@msu.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aspeborg H, Schrader J, Coutinho PM, Stam M, Kallas A, Djerbi S, Nilsson P, Denman S, Amini B, Sterky F, et al (2005) Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol 137 983–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auxtova O, Liskova D, Kakoniova D, Kubackova M, Karacsonyi S, Bilisics L (1995) Effect of galactoglucomannan-derived oligosaccharides on elongation growth of pea and spruce stem segments stimulated by auxin. Planta 196 420–424 [Google Scholar]
- Baldauf SL (2003) Phylogeny for the faint of heart: a tutorial. Trends Genet 19 345–351 [DOI] [PubMed] [Google Scholar]
- Benova-Kakosova A, Digonnet C, Goubet F, Ranocha P, Jauneau A, Pesquet E, Barbier O, Zhang Z, Capek P, Dupree P, et al (2006) Galactoglucomannans increase cell population density and alter the protoxylem/metaxylem tracheary element ratio in xylogenic cultures of zinnia. Plant Physiol 142 696–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilisics L, Vojtassak J, Capek P, Kollarova K, Liskova D (2004) Changes in glycosidase activities during galactoglucomannan oligosaccharide inhibition of auxin induced growth. Phytochemistry 65 1903–1909 [DOI] [PubMed] [Google Scholar]
- Brennan CS, Blake DE, Ellis PR, Schofield JD (1996) Effects of guar galactomannan on wheat bread microstructure and on the in vitro and in vivo digestibility of starch in bread. J Cereal Sci 24 151–160 [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB (2006) Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans. Science 311 1940–1942 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11 113–116 [Google Scholar]
- Clausen MH, Willats WG, Knox JP (2003) Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr Res 338 1797–1800 [DOI] [PubMed] [Google Scholar]
- Dhugga KS, Barreiro R, Whitten B, Stecca K, Hazebroek J, Randhawa GS, Dolan M, Kinney AJ, Tomes D, Nichols S, et al (2004) Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science 303 363–366 [DOI] [PubMed] [Google Scholar]
- Djerbi S, Lindskog M, Arvestad L, Sterky F, Teeri TT (2005) The genome sequence of black cottonwood (Populus trichocarpa) reveals 18 conserved cellulose synthase (CesA) genes. Planta 221 739–746 [DOI] [PubMed] [Google Scholar]
- Edwards ME, Dickson CA, Chengappa S, Sidebottom C, Gidley MJ, Reid JSG (1999) Molecular characterization of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J 19 691–697 [DOI] [PubMed] [Google Scholar]
- Filichkin SA, Leonard JM, Monteros A, Liu PP, Nonogaki H (2004) A novel endo-β-mannanase gene in tomato LeMAN5 is associated with anther and pollen development. Plant Physiol 134 1080–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei E, Preston RD (1968) Non-cellulosic structural polysaccharides in algal cell walls. III. Mannan in siphoneous green algae. Proc R Soc Lond B Biol Sci 169 127–145 [DOI] [PubMed] [Google Scholar]
- Freshour G, Bonin CP, Reiter WD, Albersheim P, Darvill AG, Hahn MG (2003) Distribution of fucose-containing xyloglucans in cell walls of the mur1 mutant of Arabidopsis. Plant Physiol 131 1602–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC (2000) The Growing Plant Cell Wall: Chemical and Metabolic Analysis. Blackburn Press, Caldwell, NJ
- Geisler-Lee J, Geisler M, Coutinho PM, Segerman B, Nishikubo N, Takahashi J, Aspeborg H, Djerbi S, Master E, Andersson-Gunneras S, et al (2006) Poplar carbohydrate-active enzymes: gene identification and expression analyses. Plant Physiol 140 946–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet F, Misrahi A, Park SK, Zhang Z, Twell D, Dupree P (2003) AtCslA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol 131 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41 95–98 [Google Scholar]
- Hamann T, Osborne E, Youngs H, Misson J, Nussaume L, Somerville C (2004) Global expression analysis of CESA and CSL genes in Arabidopsis. Cellulose 11 279–286 [Google Scholar]
- Handford MG, Baldwin TC, Goubet F, Prime TA, Miles J, Yu X, Dupree P (2003) Localisation and characterisation of cell wall mannan polysaccharides in Arabidopsis thaliana. Planta 218 27–36 [DOI] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Sorensen SO, Orfila C, Pauly M, Scheller HV (2006) ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol 140 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Hazen SP, Scott-Craig JS, Walton JD (2002) Cellulose synthase-like (CSL) genes of rice. Plant Physiol 128 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K, Raikhel NV (2001) Plant glycosyltransferases. Curr Opin Plant Biol 4 219–224 [DOI] [PubMed] [Google Scholar]
- Keegstra K, Walton J (2006) Plant science: beta-glucans—brewer's bane, dietician's delight. Science 311 1872–1873 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301 376–379 [DOI] [PubMed] [Google Scholar]
- Knight CD, Cove DJ, Cuming AC, Quatrano RS (2002) Moss gene technology. In PM Gilmartin, C Bowler, eds, Molecular Plant Biology: A Practical Approach. Oxford Press, Oxford, pp 285–301
- Lerouxel O, Cavalier DM, Liepman AH, Keegstra K (2006) Biosynthesis of plant cell wall polysaccharides: a complex process. Curr Opin Plant Biol 9 621–630 [DOI] [PubMed] [Google Scholar]
- Liepman AH, Cavalier DM, Lerouxel O, Keegstra K (2007) Cell wall structure, biosynthesis, and assembly. In J Roberts, Z Gonzalez-Carranza, eds, Plant Cell Separation and Adhesion. Blackwell Publishing, Oxford
- Liepman AH, Wilkerson CG, Keegstra K (2005) Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA 102 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskova D, Auxtova O, Kakoniova D, Kubackova M, Karacsonyi S, Bilisics L (1995) Biological activity of galactoglucomannan-derived oligosaccharides. Planta 196 425–429 [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Lorenz WW, Dean JFD (2002) SAGE profiling and demonstration of differential gene expression along the axial developmental gradient of lignifying xylem in loblolly pine (Pinus taeda). Tree Physiol 22 301–310 [DOI] [PubMed] [Google Scholar]
- Maeda Y, Awano T, Takabe K, Fujita M (2000) Immunolocalization of glucomannans in the cell wall of differentiating tracheids in Chamaecyparis obtusa. Protoplasma 213 148–156 [Google Scholar]
- McCartney L, Steele-King CG, Jordan E, Knox JP (2003) Cell wall pectic (1→4)-beta-d-galactan marks the acceleration of cell elongation in the Arabidopsis seedling root meristem. Plant J 33 447–454 [DOI] [PubMed] [Google Scholar]
- Meier H, Reid JSG (1982) Reserve polysaccharides other than starch in higher plants. In FA Loewus, W Tanner, eds, Encyclopedia of Plant Physiology, Vol 13A. Springer, Berlin, pp 418–471
- Nairn CJ, Haselkorn T (2005) Three loblolly pine CesA genes expressed in developing xylem are orthologous to secondary wall CesA genes of angiosperms. New Phytol 166 907–915 [DOI] [PubMed] [Google Scholar]
- Nobles DR, Brown RM (2004) The pivotal role of cyanobacteria in the evolution of cellulose synthases and cellulose synthase-like proteins. Cellulose 11 437–448 [Google Scholar]
- Pear JR, Kawagoe Y, Schrechengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93 12637–12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K (1999) Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284 1976–1979 [DOI] [PubMed] [Google Scholar]
- Pettolino FA, Hoogenraad NJ, Ferguson C, Bacic A, Johnson E, Stone BA (2001) A (1→4)-β-mannan-specific monoclonal antibody and its use in the immunocytochemical location of galactomannans. Planta 214 235–242 [DOI] [PubMed] [Google Scholar]
- Popper ZA, Fry SC (2003) Primary cell wall composition of bryophytes and charophytes. Ann Bot (Lond) 91 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston RD (1968) Plants without cellulose. Sci Am 216 102–108 [Google Scholar]
- Richmond T, Somerville CR (2001) Integrative approaches to determining Csl function. Plant Mol Biol 47 131–143 [PubMed] [Google Scholar]
- Richmond TA, Somerville CR (2000) The cellulose synthase superfamily. Plant Physiol 124 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AW, Bushoven JT (2007) The cellulose synthase (CESA) gene superfamily of the moss Physcomitrella patens. Plant Mol Biol 63 207–219 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In S Krawetz, S Misener, eds, Bioinformatics Methods and Protocols. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed]
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Schroder R, Wegrzyn TF, Bolitho KM, Redgwell RJ (2004) Mannan transglycosylase: a novel enzyme activity in cell walls of higher plants. Planta 219 590–600 [DOI] [PubMed] [Google Scholar]
- Schroder R, Wegrzyn TF, Sharma NN, Atkinson RG (2006) LeMAN4 endo-β-mannanase from ripe tomato fruit can act as a mannan transglycosylase or hydrolase. Planta 224 1091–1102 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296 141–145 [DOI] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Perrson S, Raab T, et al (2004) Toward a systems approach to understanding plant cell walls. Science 306 2206–2211 [DOI] [PubMed] [Google Scholar]
- Sterling JD, Atmodjo MA, Inwood SE, Kumar Kolli VS, Quigley HF, Hahn MG, Mohnen D (2006) Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc Natl Acad Sci USA 103 5236–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Li L, Sun YH, Chiang VL (2006) The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol 142 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K (2004) Genomic basis for cell-wall diversity in plants: a comparative approach to gene families in rice and Arabidopsis. Plant Cell Physiol 45 1111–1121 [DOI] [PubMed] [Google Scholar]
- Zablackis E, Huang J, Darvill AG, Albersheim P (1995) Characterization of the cell wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol 107 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Burk DH, Ye ZH (2001) Fibers: a model for studying cell differentiation, cell elongation, and cell wall biosynthesis. Plant Physiol 126 477–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Pena MJ, Zhou GK, Nairn CJ, Wood-Jones A, Richardson EA, Morrison WH III, Darvill AG, York WS, Ye ZH (2005) Arabidopsis fragile fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17 3390–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Nam J, Carpita N, Matthysse AG, Gelvin SB (2003) Agrobacterium-mediated root transformation is inhibited by mutation of an Arabidopsis cellulose synthase-like gene. Plant Physiol 133 1000–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.