Abstract
Pectin, one of the main components of plant cell wall, is secreted in a highly methylesterified form and is demethylesterified in muro by pectin methylesterase (PME). The action of PME is important in plant development and defense and makes pectin susceptible to hydrolysis by enzymes such as endopolygalacturonases. Regulation of PME activity by specific protein inhibitors (PMEIs) can, therefore, play a role in plant development as well as in defense by influencing the susceptibility of the wall to microbial endopolygalacturonases. To test this hypothesis, we have constitutively expressed the genes AtPMEI-1 and AtPMEI-2 in Arabidopsis (Arabidopsis thaliana) and targeted the proteins into the apoplast. The overexpression of the inhibitors resulted in a decrease of PME activity in transgenic plants, and two PME isoforms were identified that interacted with both inhibitors. While the content of uronic acids in transformed plants was not significantly different from that of wild type, the degree of pectin methylesterification was increased by about 16%. Moreover, differences in the fine structure of pectins of transformed plants were observed by enzymatic fingerprinting. Transformed plants showed a slight but significant increase in root length and were more resistant to the necrotrophic fungus Botrytis cinerea. The reduced symptoms caused by the fungus on transgenic plants were related to its impaired ability to grow on methylesterified pectins.
Pectin is a structurally complex polysaccharide that accounts for nearly 35% of the dicot and nongraminaceous monocot primary cell wall. A main component of pectin is homogalacturonan (HGA) consisting of a backbone of 1,4-linked α-d-GalUA units, with variable amounts of methylester in the C6 position. Pectins are secreted into the cell wall in a highly methylesterified form and, soon thereafter, are deesterified in muro by pectin methylesterase (PME; Brummell and Harpster, 2001; Willats et al., 2001). Demethylesterification produces free carboxyl groups and modifies the pH and charge of the wall, allowing the aggregation of polyuronides into a calcium-linked gel structure and increasing the wall firmness (Willats et al., 2001). In addition, the action of PMEs makes HGA susceptible to degradation by hydrolases such as endopolygalacturonases (endoPGs), contributing to the softening of the cell wall (Brummell and Harpster, 2001; Wakabayashi et al., 2003).
Plant PMEs are involved in important physiological processes such as microsporogenesis, pollen growth, pollen separation, seed germination, root development, polarity of leaf growth, stem elongation, fruit ripening, and loss of tissue integrity (Tieman and Handa, 1994; Wen et al., 1999; Micheli et al., 2000; Pilling et al., 2000, 2004; Micheli, 2001; Bosch et al., 2005; Jiang et al., 2005; Bosch and Hepler, 2006; Francis et al., 2006; Tian et al., 2006). They may also have a role in resistance to fungal and bacterial pathogens (McMillan et al., 1993; Boudart et al., 1998; Wietholter et al., 2003) and are required for the systemic spread of Tobacco mosaic virus through the plant (Dorokhov et al., 1999; Chen et al., 2000; Chen and Citovsky, 2003). Spatial and temporal regulation of PME activity during plant development is complex, due to the presence of a large family of isoforms (Markovic and Janecek, 2004). In the Arabidopsis (Arabidopsis thaliana) genome, 67 genes encode putative PMEs (Arabidopsis Genome Initiative, 2000; Markovic and Janecek, 2004). Nearly all the Arabidopsis PME genes are predicted to encode pre-pro-proteins (Micheli, 2001). The pre-region is required for protein targeting to the endoplasmic reticulum (Micheli, 2001; Dorokhov et al., 2006) and the pro-peptide is thought to play an autoinhibitory role of the enzyme during its secretion to the apoplast, where only the mature part of the protein is found (Giovane et al., 2004). In addition, a mechanism of regulation of PME activity is played by individually expressed specific protein inhibitors discovered in kiwi fruits (Actinidia chinensis; Balestrieri et al., 1990; Giovane et al., 1995) and recently was found in Arabidopsis (Wolf et al., 2003; Raiola et al., 2004). PME inhibitors named PMEI are encoded in Arabidopsis by a small gene family of two members, AtPMEI-1 and AtPMEI-2, located in different chromosomes. AtPMEI-1 and AtPMEI-2, which share about 47% identity at the amino acid level, consist of 151 amino acids (molecular mass of 16,266 D, predicted pI = 7.7) and 148 amino acids (molecular mass 15,615 D, predicted pI = 9.0), respectively (Wolf et al., 2003; Raiola et al., 2004). PMEIs from Arabidopsis and kiwi exhibit an α-helix up-and-down four-helical bundle fold and five strictly conserved Cys residues, four of which engaged in disulfide bridges for the maintenance of the protein structure (Hothorn et al., 2004; Di Matteo et al., 2005). PMEIs inhibit PMEs of plant origin by forming a noncovalent stoichiometric 1:1 complex and typically do not inhibit PMEs produced by plant pathogenic microorganisms (Mattei et al., 2002; D'Avino et al., 2003; Giovane et al., 2004; Di Matteo et al., 2005).
The specificity of PMEI toward plant PME suggests a physiological role in the modulation of endogenous PME activity during development and growth. However, given the effect of pectin methylesterification on the physicochemical properties of the walls, they may also have a role in defense against pathogens by influencing the susceptibility of the wall to cell wall-degrading enzymes, which in some cases have a major role in pathogenesis (Clark and Lorbeer, 1976; Collmer and Keen, 1986; Cole et al., 1998; De Lorenzo et al., 2001; D'Ovidio et al., 2004). Pectin hydrolysis often occurs in the diseases caused by soft rot bacteria and fungi and appears to be the prerequisite for degradation of other cell wall components (D'Ovidio et al., 2004). For example, Botrytis cinerea, a ubiquitous necrotrophic fungus, possesses an efficient pectinolytic machinery including a variety of endoPGs, some of which are important virulence factors (ten Have et al., 1998; Kars et al., 2005). Possibly for this reason, the host preference of B. cinerea is generally considered to be restricted to plants with relatively high pectin content (van Kan, 2006).
To investigate the role of pectin methylesterification in plant growth and in plant-pathogen interactions, we have constitutively expressed the genes AtPMEI-1 and AtPMEI-2 (Raiola et al., 2004) in Arabidopsis plants. We report here that the transgenic expression of the two inhibitors decreases the plant PME activity and alters the level of pectin methylesterification of the wall. The transformed plants show an altered growth response and a lower susceptibility to B. cinerea.
RESULTS
AtPMEI Reduces PME Activity in Transgenic Arabidopsis
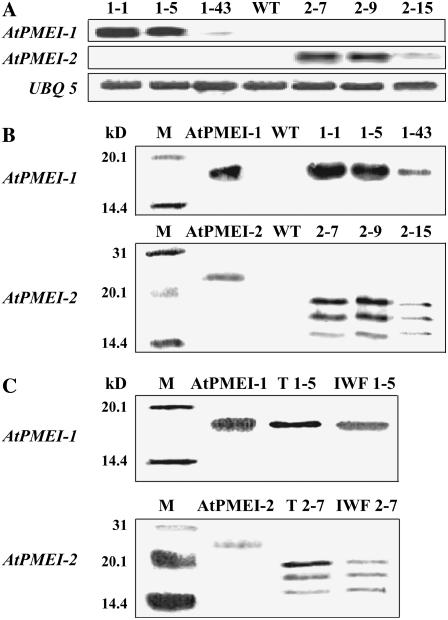
To explore the physiological role of AtPMEIs in Arabidopsis, transgenic plants constitutively expressing AtPMEI-1 or AtPMEI-2 genes were generated. To target the inhibitors into the apoplast, the sequence encoding the predicted N-terminal signal peptide for secretion was included in the coding sequence of each gene and both genes were placed under the control of the cauliflower mosaic virus 35S promoter. Strand-specific probes were used to analyze the accumulation of AtPMEI-1 or AtPMEI-2 mRNA in transformed rosette leaves, where the genes are expressed at low levels in untransformed plants (Wolf et al., 2003; Raiola et al., 2004). Thirty T2 transgenic plants exhibited a wide range of AtPMEI-1 or AtPMEI-2 mRNA levels (data not shown). Three transgenic lines transformed with AtPMEI-1 (1-1, 1-5, and 1-43) and three lines transformed with AtPMEI-2 (2-7, 2-9, and 2-15) were selected for analysis (Fig. 1A). Segregation analysis of the marker gene bar for resistance to the herbicide Basta in the self-crossed T2 progeny showed either a 3:1 Mendelian segregation ratio (1-1, 2-7, 2-9, and 2-15 lines) or a 15:1 segregation ratio (1-5 and 1-43 lines). These ratios correspond to one and two independent insertions of the transgene, respectively, and suggest a normal fitness of transformed pollen grains during fertilization (Bosch and Hepler, 2006). Lines 1-1 and 1-5 and lines 2-7 and 2-9 expressed levels of AtPMEI-1 or AtPMEI-2 transcripts about 90 and 200 times higher, respectively, than wild-type plants. These lines exhibited high levels of proteins as shown by immunoblotting analysis using polyclonal antibodies raised against AtPMEI-1 or AtPMEI-2 expressed in Pichia pastoris (Raiola et al., 2004; Fig. 1B). AtPMEI-1 was produced as a single band with an apparent molecular mass of 17 kD. AtPMEI-2 showed three bands with apparent molecular masses of 20, 18, and 16 kD. Upon treatment with N-glycosidase A, the two bands with the higher molecular mass disappeared with a concomitant increase of the 16-kD band, i.e. the band corresponding to the protein with the predicted molecular mass (15,615 D) of the nonglycosylated form. Glycosylation of AtPMEI-2 was also observed in the recombinant protein expressed in P. pastoris. Lines expressing AtPMEI-1 did not show any band when the immunoassay was performed with the antibodies raised against AtPMEI-2 and, vice versa, lines expressing AtPMEI-2 did not show any band when AtPMEI-1 antibodies were used (data not shown). Lines 1-43 and 2-15 expressed levels of AtPMEI-1 or AtPMEI-2 transcripts 5 and 20 times higher than wild-type plants and exhibited low levels of transgenic proteins, whereas no AtPMEI-1 or AtPMEI-2 proteins were detected in untransformed plants (Fig. 1B). The accumulation of the inhibitors was detected in transformed seedlings grown either under light and in the dark (data not shown). In lines 1-1 and 2-9, the proteins were detected in extracellular fluids isolated from mature leaves, indicating that both inhibitors have been correctly targeted to the apoplast (Fig. 1C).
Figure 1.
Expression of AtPMEI-1 and AtPMEI-2 in Arabidopsis transgenic plants. Numbers indicate the different AtPMEI-1 or AtPMEI-2 transformed lines; WT, untransformed wild-type plants. A, RNA gel-blot of T2 independent lines; UBQ 5, ubiquitin. B, Immunodetection of AtPMEI-1 and AtPMEI-2 in leaf extracts of transgenic lines using polyclonal antibodies generated against recombinant AtPMEI-1 or AtPMEI-2. M, Protein markers with relative molecular mass indicated at left; as standard, 30 ng of purified AtPMEI-1 or 20 ng of AtPMEI-2 expressed in P. pastoris was used (Raiola et al., 2004). C, Immunodetection of AtPMEI-1 and AtPMEI-2 in IWFs from rosette leaves. T, Total proteins from rosette leaves (9 μg); IWF, protein from intercellular washing fluids (3 μg).
We assessed whether the overexpression of AtPMEI-1 or AtPMEI-2 resulted in a decrease of PME activity in transgenic Arabidopsis. Total protein extracts from rosette leaves were assayed using the quantitative PME activity radial gel diffusion assay (Downie et al., 1998). Lines 1-1 and 1-5 and lines 2-7 and 2-9 showed 45% to 62% of PME activity as compared with the wild-type plants, whereas PME activity of lines 1-43 and 2-15 was only slightly lower than that detected in wild-type plants (Table I).
Table I.
Quantification of PME activity in AtPMEI-1, AtPMEI-2, and wild-type plants with radial gel diffusion assay
PME activities (%) were determined from leaves of AtPMEI-1 and AtPMEI-2 transgenic lines relative to wild type. Data represent average ± se of five biological replicates. The PME activity of the wild-type leaves (0.013 units) was set to 100%.
| Line | PME Activity |
|---|---|
| % | |
| Wild type | 100 |
| 1-43 | 85.9 ± 0.7 |
| 1-1 | 61.6 ± 2.3 |
| 1-5 | 46.7 ± 1.1 |
| 2-15 | 83.8 ± 0.2 |
| 2-7 | 52.4 ± 0.3 |
| 2-9 | 44.7 ± 0.8 |
To detect whether the inhibitors interact with endogenous PME(s), total proteins extracted from mature leaves of AtPMEI transformed lines 1-5 and 2-7 were separated by gel-filtration chromatography (Supplemental Fig. S2, A and B) and subjected to SDS-PAGE followed by western-blot analysis using polyclonal antibodies against AtPMEI-1 or AtPMEI-2 (see “Materials and Methods”). AtPMEI-1 and AtPMEI-2, expressed in P. pastoris and purified to homogeneity, eluted in fractions with a molecular mass of about 15 to 25 kD (i.e. 25–27), consistently with their molecular mass, and PME inhibitory activity was associated with the presence of the immunodetected bands (Supplemental Fig. S2D). Instead, AtPMEI-1 and AtPMEI-2 from transformed leaf extracts eluted in the fractions 22 to 24, which contain proteins with an estimated molecular mass of about 50 to 60 kD (Supplemental Fig. S2, C and E); no PME inhibitory activity was associated to the presence of the immunodetected bands. Moreover, PME activity was also eluted in fractions 25 and 26, which contained proteins with a molecular mass of about 30 to 40 KD. The chromatographic behavior of AtPMEI-1 and AtPMEI-2 from transformed plants and the absence of detectable inhibitory activity indicate that both inhibitors are eluted as inactive complexes with endogenous PME(s). On the other hand, the reduced enzymatic activity in the transformed tissues and the elution of free PME activity from the gel-filtration chromatography indicate that the enzyme is present in excess as an unbound active form.
Proteins were analyzed by SDS-PAGE and after staining were reduced, alkylated, digested with trypsin, and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Two PMEs were recognized in both AtPMEI-1- and AtPMEI-2-containing fractions: the most abundant was identified as the isoform AtPME3 (SwissProt accession Q9LUL7; locus At3g14310) and the less abundant second one was identified as a putative PME (SwissProt accession Q9SKX2, locus At2g43050; Supplemental Table S1).
Overexpression of AtPMEI in Arabidopsis Affects the Degree of Pectin Methylesterification
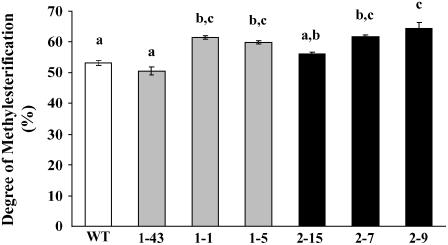
The degree of pectin methylesterification (DM) was determined in cell walls extracted from rosette leaves of transformed AtPMEI plants. As compared with the wild-type plants, a significant increase of DM of about 16% was detected in lines 1-1 and 1-5 and lines 2-7 and 2-9 expressing high levels of AtPMEI-1 and AtPMEI-2 (Fig. 2). Lines 1-43 and 2-15, both expressing a low level of inhibitor, showed no differences. The content of uronic acids in transformed plants was not significantly different from that of wild-type plants (Table II).
Figure 2.
Determinations of the DM of cell wall from transgenic plants transformed with AtPMEI-1 (lines 1-43, 1-1, 1-5), AtPMEI-2 (lines 2-15, 2-7, 2-9), and wild type. The different letters indicate data sets significantly different according to Tukey's Student range test (P < 0.01). Values represent the average ± se of three independent experiments with n = 4 plants each.
Table II.
Quantification of uronic acid content in AtPMEI-1 and AtPMEI-2 transformants and in wild-type cell walls
The same letter indicates not significant differences according to Tukey's Student range test (P < 0.05). Data represent average ± se of four biological replicates.
| Line | Uronic Acids |
|---|---|
| μmol mg−1 cell wall | |
| Wild type | 0.39 ± 0.06a |
| 1-43 | 0.49 ± 0.04a |
| 1-1 | 0.41 ± 0.04a |
| 1-5 | 0.45 ± 0.04a |
| 2-15 | 0.40 ± 0.1a |
| 2-7 | 0.42 ± 0.02a |
| 2-9 | 0.41 ± 0.03a |
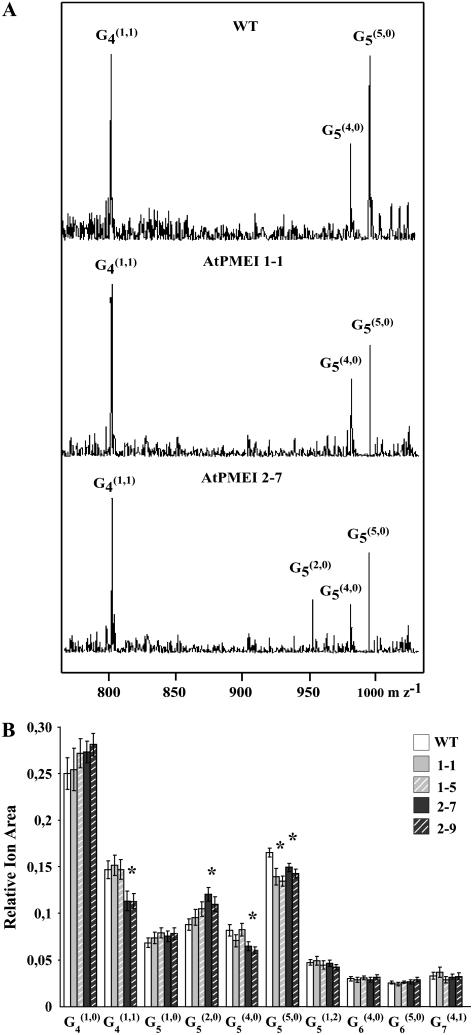
The fine structure of pectins of transformed plants was also analyzed by enzymatic fingerprinting (Lerouxel et al., 2002). Cell walls from wild-type and transformed seedlings were treated with a combination of fungal PME and endoPG, which releases methylesterified or O-acetylated HGA fragments (Obel et al., 2006). The water-soluble oligosaccharides were successively analyzed by matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF MS). The standard symbols reported by van Alebeek (van Alebeek et al., 2003), with some modifications, were used to define the pectic fragments. In particular, in the Gn(m,a) fragment, “Gn” denotes the number of GalUA residues in the backbone, and “m” and “a” indicate the number of methylesterified and of acetylated residues, respectively. The main ions detected by MALDI-TOF MS analysis were assigned to (M+Na+)+ adducts on the basis of their Mr and literature data (Daas et al., 1998; Table III). Differences in MALDI-TOF mass spectra were observed in both highly expressing AtPMEI-1 and AtPMEI-2 lines in comparison with wild type (Fig. 3A). The areas of ion signals were integrated, allowing the establishment of the relative abundance profile of pectic fragments in the cell wall digest. Figure 3B shows that G4(1,0), G4(1,1), and G5(5,0) were the main fragments released. Compared with the wild type, all the AtPMEI transformed plants analyzed showed a significantly reduced level of fully methylesterified G5(5,0) fragments. In addition, the decrease of G4(1,1) and G5(4,0) and the increase of G5(2,0) ions were detected in lines 2-7 and 2-9 with respect to the wild-type plants. The differences in oligosaccharide profiles observed among plants indicate alteration in pectic structure/accessibility possibly due to differences in degree and/or pattern of methylesterification. On the other hand, no significant differences among AtPMEI-1, AtPMEI-2, and wild-type seedlings were observed when xyloglucan structure was analyzed by enzymatic oligosaccharide profiling using a xyloglucan-specific endo β-(1→4)-d-glucanase (Lerouxel et al., 2002).
Table III.
Nominal masses, composition, and significance of ions generated by MALDI-TOF MS of the various AtPMEI transgenic lines
| Nominal Massa | Compositionb | 1-1c | 1-5c | 2-7c | 2-9c |
|---|---|---|---|---|---|
| 759 | G4(1,0) | ||||
| 801 | G4(1,1) | * | * | ||
| 935 | G5(1,0) | ||||
| 949 | G5(2,0) | * | * | ||
| 977 | G5(4,0) | * | * | ||
| 991 | G5(5,0) | * | * | * | * |
| 1033 | G5(1,2) | ||||
| 1153 | G6(4,0) | ||||
| 1167 | G6(5,0) | ||||
| 1386 | G7(4,1) |
Nominal mass (M+Na+)+ of pectic oligosaccharides released by endoPG/PME digestion of Arabidopsis etiolated seedling cell wall material.
Gn(m,a) fragment; “Gn” denotes the number of galacturonic acid residues in the backbone; and “m” and “a” the number of methylesterified and acetylated residues, respectively.
AtPMEI-1 or AtPMEI-2 transformed lines. * and bold, Ion areas significantly different compared with wild type by t test (P = 0.01; data were obtained from 30 independent samples for wild type and 20 independent samples for each transformed line; 10 seedlings were used for each sample).
Figure 3.
Comparison of the relative abundance of pectic oligosaccharides released by endoPG/PME digestion from transformed and wild-type seedlings. A, Positive-ion MALDI-TOF mass spectra of endoPG/PME-generated pectic fragments from cell wall of wild-type, AtPMEI-1, and AtPMEI-2 etiolated seedlings. B, The area of ion signals obtained from MALDI-TOF MS spectra of the different pectic fragments were integrated. Data represent the average ± sd of n = 30 independent samples for wild type and n = 20 independent samples for each transformed lines. Ten seedlings were used for each sample. Asterisks indicate the significant differences (P < 0.05).
Transgenic Arabidopsis Plants Exhibit Altered Growth and Reduced Susceptibility to B. cinerea
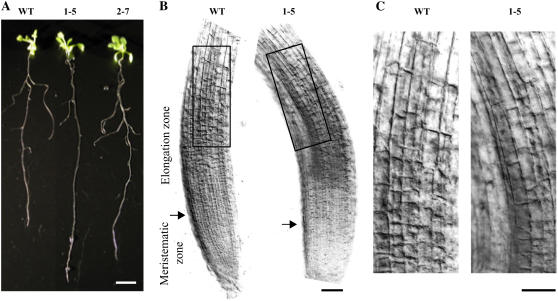
Transformed plants grown in greenhouse did not show any obvious differences in growth, development, and plant fertility in comparison with the untransformed ones. However, when seedlings were vertically grown on solid Murashige and Skoog medium, a significant 20% increase in root length was observed in lines 1-5 and 2-7 with respect to wild-type plants (Fig. 4A; Table IV). Root cells of the expanding zone, the region in which cell elongation mainly occurs, were elongated with respect to the controls (Fig. 4, B and C).
Figure 4.
Morphological analysis of vertically grown AtPMEI and wild-type plants. A, Wild type, AtPMEI-1 (line 1-5), and AtPMEI-2 (line 2–7) plants; bar = 10 mm. B, Optical microphotographs of epidermal root cells. The starting point of root elongation zone is indicated by the arrow. Corresponding root areas are delimited by black boxes. C, Magnification of the boxed areas. Bars = 50 μm. [See online article for color version on this figure.]
Table IV.
Average lengths of AtPMEI-1, AtPMEI-2, and wild-type roots
Root lengths were measured in seedlings after 12 d of vertical growth. Data represent average ± se of three independent experiments with n = 10 plants each. Significant differences by Student's t test for P < 0.001 are indicated by asterisks.
| Line | Root Length |
|---|---|
| mm | |
| Wild type | 83.7 ± 1.2 |
| 1-43 | 88.2 ± 3.6 |
| 1-5 | 107.7 ± 1.5* |
| 2-15 | 88.1 ± 4 |
| 2-7 | 98.1 ± 2.6* |
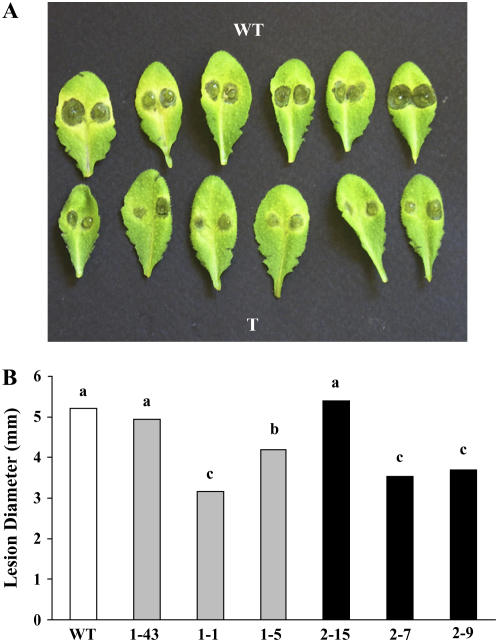
Resistance of transgenic plants to the necrotrophic fungus B. cinerea was assessed. Leaves of wild-type and transgenic plants were inoculated with fungal conidia and the lesion size was determined 3 d postinoculation (Fig. 5A). No differences in the ratio between the number of expanding lesions and the number of inoculated spots were observed among transformed lines and wild-type plants (data not shown). Instead, lines 1-5, 1-1, 2-7, and 2-9 displayed a reduced radial lesion size (P < 0.01) compared with wild type and lines 1-43 and 2-15 (Fig. 5B). No inhibitory activity of both purified AtPMEI-1 and AtPMEI-2 was observed against PME activity of B. cinerea, excluding a direct mechanism of these inhibitors in limiting fungal growth.
Figure 5.
Reduction of B. cinerea symptoms in Arabidopsis plants overexpressing AtPMEI-1 and AtPMEI-2. A, Detached leaves from wild type and AtPMEI transformed lines (T) were inoculated with B. cinerea conidia and the lesion size was determined 3 d postinoculation. B, The average diameter of the expanding lesions is shown, and different letters indicate data sets significantly different at P < 0.01 (lsd 0.01). The experiment was repeated three times with different plant batches, and statistical analysis of the results was performed by randomized-blocks ANOVA. [See online article for color version on this figure.]
To ascertain if the reduced symptoms produced by B. cinerea on the AtPMEI lines are related to an impaired ability of this fungus to grow on methylesterified pectins, we examined the fungal growth on liquid synthetic media containing polygalacturonic acid (PGA) or 81% methylesterified pectin (E81) as carbon source. B. cinerea grew significantly better on PGA (average milligram of mycelium dry weight ± sd, 30.2 ± 2.7; n = 6) than on E81 (12.6 ± 1.2; n = 6), indicating that the fungus prefers unesterified pectins as carbon source.
To determine if the cell wall composition of transformed plants affects B. cinerea growth, we measured the fungal growth on liquid medium containing cell walls isolated from wild type or from AtPMEI lines as a carbon source. A reduction of 14.3% and 28.5%, respectively, was observed on cell walls of lines 1-1 and 2-7 in comparison with the untransformed plants.
DISCUSSION
In this study we have generated Arabidopsis plants constitutively expressing the PME inhibitors AtPMEI-1 and AtPMEI-2 (Wolf et al., 2003; Raiola et al., 2004). Among the transformed lines obtained, the lines 1-1, 1-5, 2-7, and 2-9 showed the highest level of AtPMEI transcripts and proteins and a concomitant lower level of PME activity. The reduction of PME activity was likely due to the interaction between the overexpressed inhibitor(s) and endogenous enzyme(s). In gel-filtration experiments, the lack of detectable PMEI activity and the shift of the PMEI elution volume toward a higher Mr suggested that both PMEIs were engaged in the formation of a complex with endogenous PME. All three glycoforms of AtPMEI-2 present in the transformed plants may form a complex with endogenous PME, consistent with the previous observation that the fully glycosylated recombinant AtPMEI-2, expressed in P. pastoris, inhibits plant PMEs (Raiola et al., 2004). It is relevant that the two N-glycosylation sites are spatially located outside the contact interface identified in the PME/PMEI complex (Di Matteo et al., 2005).
PMEs interacting with AtPMEI-1 and AtPMEI-2 were identified as AtPME3 (At3g14310), shown to be mainly expressed in Arabidopsis hypocotyls, leaves, and roots (Micheli et al., 1998; Pina et al., 2005; Louvet et al., 2006) and At2g43050 mainly expressed in siliques and stems (Micheli et al., 1998; Pina et al., 2005; Louvet et al., 2006; see also the gene expression database: http://csbdb.mpimp-golm.mpg.de/). The finding that both PMEs were associated with both inhibitors is consistent with the large spectrum of recognition played by the inhibitors (Mattei et al., 2002; Ciardiello et al., 2004; Raiola et al., 2004; Di Matteo et al., 2005).
The DM in transformed plants was significantly higher than in wild-type plants. On the other hand, the total content of uronic acids of AtPMEI-1 and AtPMEI-2 plants was not significantly different from that of wild-type plants, suggesting that the susceptibility of pectin to endogenous hydrolases is not affected. However, qualitative difference in the fine structure of pectins could be detected by enzymatic fingerprinting (Lerouxel et al., 2002), and the pattern of alteration was highly reproducible within the same class of independent transformants (i.e. AtPMEI-1 or AtPMEI-2). Differences between AtPMEI-1 and AtPMEI-2 transformants suggest that the two inhibitors may differently influence PMEs acting on various pectic subdomains.
It is known that pectin methylesterification plays a crucial role in plant growth: it is maximal during the cell expansion phase and decreases as cell elongation ceases (Goldberg, 1984; McCann and Roberts, 1994). Transgenic plants whose specific PME activities were inhibited, using antisense mRNA, exhibited predictable effects on pectin chemistry but little or no alterations on plant growth (Tieman et al., 1992; Tieman and Handa, 1994), likely because the silenced PMEs are not involved in growth. Our transgenic AtPMEI-1 or AtPMEI-2 plants showed longer roots mainly due to an increase in cell elongation when seedlings were grown on agar in vertical position. The analysis of roots is considered an ideal system to study cell growth (Scheres et al., 2002), and our results are consistent with the notion that an increased DM promotes cell expansion and positively affects growth.
AtPMEI-1 and AtPMEI-2 are expressed in flowers and pollen, while AtPMEI-1, which shows a considerably higher expression level than AtPMEI-2, is also expressed in roots, as well as in seedlings, stems, and mature leaves (Wolf et al., 2003; Raiola et al., 2004). Due to the large number of PME members in Arabidopsis and to the specificity of their expression in different tissues and at different developmental stages, it is difficult to define the contribution of these inhibitors in the regulation of PME activity. However, the tissue distribution of PMEs in Arabidopsis (Supplemental Fig. S1) indicates a lower activity in flowers with respect to the other tissues, and this well correlates with the high expression level of the inhibitors in flower (Wolf et al., 2003; Raiola et al., 2004).
Methylesterification of pectin may correlate with a lesser accessibility to pectin-degrading enzymes and therefore with an increased resistance to pathogens (McMillan et al., 1993; Marty et al., 1997; Boudart et al., 1998). Arabidopsis plants expressing high level of AtPMEI-1 or AtPMEI-2 showed a reduced spreading of disease symptoms after inoculation with B. cinerea as compared with inoculated wild-type plants. Since endoPG prefer PGA rather than methylesterified pectin (Kars et al., 2005), it is likely that the higher DM of pectins of AtPMEI plants hampers the activity of endoPGs produced by this fungus, thus delaying its colonization of the plant tissue. EndoPGs are the main cell wall-deconstructing enzymes secreted by B. cinerea, and two isoenzymes (BcPG1 and BcPG2) are required for its virulence on several hosts (van Kan, 2006). On the other hand, fungal PME activity, which is not inhibited by AtPMEIs, is thought to facilitate the activity of PG during plant infection. However, growth data in liquid cultures indicate that the fungal biomass on methylesterified pectin is lower than on PGA, suggesting that B. cinerea PME does not perform an efficient pectin demethylesterification. The possible induction of plant PMEs during fungal infection (Boudjeko et al., 2005; Abuqamar et al., 2006) can also be considered. In this case, the increase of PMEI content in transformed plants would be effective to impede the de-esterification of the pectic polymer by plant PME and, consequently, the accessibility of the fungal PG to its substrate.
A role of the DM of the cell wall pectins in plant disease resistance has been reported in several pathosystems. For example, highly methylesterified pectin has been related to resistance of potato (Solanum tuberosum) cultivars to Erwinia soft rot (McMillan et al., 1993; Marty et al., 1997). On the other hand, the DM and the distribution of methylesters along the pectic polymer can also influence the type of oligogalacturonides (OGs) released from the host cell wall. These, in turn, may affect the plant resistance to disease because of their elicitor activity. For example, pectic fragments with higher DM and superior elicitor activity have been reported to accumulate in bean (Phaseolus vulgaris) cultivars resistant to Colletotrichum lindemuthianum (Boudart et al., 1998). Also, a different methylester distribution was found in HGA of near-isogenic wheat (Triticum aestivum) lines resistant and susceptible to the stem rust fungus Puccinia graminis f. sp. tritici, and was considered to affect the type of OG elicitors released during the plant-fungus interaction (Wietholter et al., 2003). The possibility that AtPMEI affects the pattern of distribution of methyl esters within GalA polymer and, consequently, the type of OGs released and their elicitor activity remains to be investigated.
MATERIALS AND METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana) accession Columbia-0 was obtained from G. Redei and A.R. Kranz (Arabidopsis Information Service, Frankfurt). Arabidopsis plants were grown in a controlled environmental chamber maintained at 22°C, 70% relative humidity, with a 16-h photoperiod (100 μmol m−2 s−1 fluorescent light). For vertical growth, seeds were sterilized and germinated in vertical position for 12 d on agar plates containing Murashige and Skoog medium (Sigma-Aldrich) containing 0.8% agar and 1% Suc with 16-h photoperiod (100 μmol m−2 s−1 fluorescent light). Root elongation zones were microscopically examined, in vertically grown seedlings, using an Axiophot microscope (Carl Zeiss), and photomicrographs were taken using a Canon Powershot G3 photocamera. For MALDI-TOF MS analysis, seedlings were grown for 4 d in the dark to minimize environmental influences (Santoni et al., 1994; Lerouxel et al., 2002).
Pathogen Growth and Infection
Botrytis cinerea strain SF1 obtained from Dr. S. Ferrari (University of Padua, Italy) was grown for 15 d on malt extract agar at 24°C and 12-h photoperiod before spore collection. Conidia at the density of 5 × 105 conidia mL−1 were germinated in 12 g L−1 potato dextrose broth (Difco) at room temperature for 3 h. Fully developed leaves of 6-week-old Arabidopsis plants were detached and placed in petri dishes with petioles embedded in 0.7% agar. Two droplets of spore suspension (5 μL each) were placed on the surface of each leaf and incubated at 23°C and 12-h photoperiod and lesion diameter was measured after 3 d. The experiment was repeated three times with different plant batches, and statistical analysis of the results was performed by randomized-blocks ANOVA.
Fungal Growth on Different Pectic Substrates
B. cinerea was cultured on Czapeck Dox medium amended as sole carbon source, either with 0.5% (w/v) PGA (Sigma) or 0.5% (w/v) lime pectin with 81% degree of methylesterification (E81; DANISCO) or cell walls (40 mg of dried material dispersed in 50 mL of medium) isolated from transformed and wild-type leaves. The flasks were inoculated with 1 mL of conidia (4 × 105 conidia mL−1) and incubated on a rotary shaker at 100 rpm at 23°C for 2 d. Mycelium was harvested by filtration through tared crucibles and oven-dried at 65°C to a constant weight. Three flasks were collected for each medium.
Plant Transformation
For transgenic expression in Arabidopsis, the coding sequences of AtPMEI-1 and AtPMEI-2 were amplified from genomic DNA isolated from Arabidopsis seedlings with the NucleoSpin Plant kit (Macherey-Nagel) using Pfu DNA polymerase (Promega). The primers pairs used were AtPMEI-1/Fw (5′-TTGTCCATGGCTGCGAATCTAAG-3′) and AtPMEI-1/Rv (5′-GCGGAGCTCTTAATTACGTGGTAACAT-3′) for AtPMEI-1, and AtPMEI-2/Fw (5′-TTGTCCATGGCAGCATACCTGAC-3′) and AtPMEI-2/Rv (5′-GCGGAGCTCTCACATCATGTTTGAGATGAC-3′) for AtPMEI-2. The amplicons were subcloned between the NcoI and SacI sites (underlined in the above primer sequences) in the pJD301 plasmid (Luehrsen and Walbot, 1991) and sequenced. The cassettes, comprising the 35S promoter of Cauliflower mosaic virus, the Ω leader of Tobacco mosaic virus, the AtPMEI open reading frame, and the nopaline synthase 3′ sequence, were inserted into the XbaI site of the pCAMBIA 3300 vector (Cambia). Transgenic Arabidopsis plants were generated by Agrobacterium tumefaciens-mediated transformation as described previously (Clough and Bent, 1998). Following transformation, transgenic lines were selected in T1 generation on agarized Murashige and Skoog medium containing phosphinothricin (8 mg L−1). T2 lines resistant to phosphinothricin were selected for subsequent analysis.
RNA Gel-Blot Analysis
Total RNA was extracted from rosette leaves of 6-week-old plants, using the Trizol reagent (Life Technologies) and following manufacturer's instructions. Samples of 12 μg of total RNA were separated on a 1.2% agarose gel containing 1.2% formaldehyde and transferred onto Hybond-N+ membrane (Amersham). Prehybridization and hybridization reactions were performed at 45°C in the presence of DIG Easy Hyb solution (Roche Diagnostics). Filters were hybridized with a digoxigenin-labeled probe, incubated in CSPD (Roche Diagnostics), and exposed to x-ray film (X-Omat AR; Kodak). The probe used to detect the AtPMEI-1 and AtPMEI-2 mRNA was a cDNA corresponding to coding region of each AtPMEI gene labeled with digoxigenin-11-dUTP by PCR, using the following primers: Fw 5′-ATGGCTGCGAATCTAAG-3′ and Rv 5′-TTAATTACGTGGTAACAT-3′ for AtPMEI-1; and Fw 5′-ATGGCAGCATACCTGAC-3′ and Rv 5′-TCACATCATGTTTGAGATGAC-3′ for AtPMEI-2. Specific UBQ5 probe was prepared as described (Rogers and Ausubel, 1997).
Protein Purification and Analysis
Total protein extracts were obtained by homogenizing leaves of 6-week-old Arabidopsis plants in the presence of 1 m NaCl, 12.5 mm citric acid, 50 mm Na2HPO4[0], pH 6.5 (1 mL per g of tissue). The homogenate was then shaken for 1 h at 4°C, centrifuged at 15,000g for 15 min, and the supernatant collected. Protein concentration was determined according to Bradford (1976) using Bio-Rad reagents and bovine serum albumin as standard. SDS-PAGE was performed as described by Laemmli (1970); for each sample, 9 μg of protein extract was loaded into the gel. Immunoblot analysis was performed as described previously (Desiderio et al., 1997) using rabbit-specific antibodies raised against recombinant AtPMEI-1 or AtPMEI-2 purified to homogeneity (Raiola et al., 2004).
To partially purify the transgenic inhibitors, total proteins were precipitated with ammonium sulfate up to 80% saturation. The precipitate was suspended in 10 mm Tris-HCl, pH 7.5, 150 mm NaCl, loaded onto a Superdex75 (HR10/30), eluted with the same buffer at a flow rate of 0.5 mL min−1, and fractions of 0.5 mL were collected. For comparison, AtPMEI-1 and AtPMEI-2 expressed in Pichia pastoris and purified to homogeneity were run under the same conditions. SDS-PAGE and immunoblot analysis were performed using rabbit-specific antibodies raised against recombinant AtPMEI-1 and AtPMEI-2. To detect the presence of AtPMEI-2 glycoforms, Superdex fractions containing the AtPMEI-2 were boiled for 15 min and digested with 0.4 munits of N-glycosidase A (Roche Applied Science). The sample was run on SDS-PAGE and protein bands were detected by immunoblot analysis.
For LC-MS/MS analysis, 3 mg of total proteins isolated from AtPMEI-1 and AtPMEI-2 leaves was run on a Superdex75 preparative column (HR26/60) in the same buffer as above at a flow rate of 2 mL min−1, and fractions of 3 mL were collected and analyzed by immunoblot to select the fraction containing the inhibitor. The selected fractions were analyzed by SDS-PAGE and stained with Coomassie Brilliant Blue. After staining the protein bands were excised from the gel, reduced, alkylated, and digested with trypsin according to Hellman et al. (1995). Tryptic peptides were analyzed by LC-MS/MS (LC-ESI-quadrupole iontrap MS 1100 Series; Agilent Technologies) equipped with a column Zorbax SB-C18 5-μm 150 × 0.5 mm. Protein identification was performed by searching the Mass Spectrometry Protein Sequence DataBase (Imperial College London) using the Mascot search engine, which uses a probability-based scoring system. The following parameters were used for database searches: peptide and MS/MS mass tolerance of 1.5 D; peptide charge of +1, +2, or +3; trypsin as digesting enzyme with one missed cleavage allowed; and carbamidomethylation of Cys as a fixed modification.
Intercellular Washing Fluid Isolation
Intercellular washing fluids (IWFs) were collected from Arabidopsis leaves by centrifugation as described previously (Salvi et al., 1990) and according to Lohaus et al. (2001). Rosette leaves were excised from 6-week-old AtPMEI-1 and AtPMEI-2 plants. Leaves were stacked on a 105-μm nylon mesh in the bottom of a 20-mL plastic syringe. The packed leaves were washed with 12.5 mm citric acid, 50 mm Na2HPO4, pH 6.5, for 5 min. Afterward, they were vacuum-infiltrated for 5 min with the same buffer. IWF was recovered by centrifuging the vacuum-infiltrated stems at 500g for 5 min at 4°C. The amount of IWF obtained from 1 g of tissue (fresh weight) was 0.2 to 0.3 mL. Contamination of IWFs by cytoplasmic components was ruled out by measuring Glc-6-P dehydrogenase activity according to Takahama (1993), which accounted for less than 1% of total extractable activity.
Determination of PME Activity
PME activity was quantified by the radial gel diffusion assay as described by Downie et al. (1998) with some modifications. A gel was prepared with 0.1% (w/v) of 81% methylesterified lime pectin (E81; Danisco A/S), 1% (w/v) agarose, 12.5 mm citric acid, and 50 mm Na2HPO4, pH 7.0. The gel was cast into agar plates (13 mL per plate) and allowed to polymerize at room temperature. Wells with a diameter of 4 mm were made and the protein samples (2 μg in 20 μL) were loaded in each well. Plates were incubated at 30°C for 16 h. The gels were stained with 0.05% (w/v) ruthenium red for 45 min, destained with water, and the diameter of the red-stained areas, resulting from the hydrolysis of esterified pectin in the gel, was measured. PME activities were determined in protein extracts from rosette leaves carefully chosen at the same stage of development isolated by 6-week-old AtPMEI-1, AtPMEI-2, and wild-type plants. Each value was obtained from five independent biological replicates. PME activity of B. cinerea was determined by agar diffusion assay by testing aliquots of culture filtrate after 3 d of fungal growth in liquid Czapeck Dox medium containing 0.5% (w/v) commercial apple pectin (Sigma-Aldrich) as the sole carbon source. A standard curve was prepared using commercial orange peel PME (Sigma-Aldrich) showing a log-linear relationship between the stained zone diameter and the activity of PME applied to the well within a PME activity range going from 0.001 units (staining zone diameter = 5 mm) to 5 units (staining zone diameter = 25 mm). PME activity was calculated based on this standard curve.
Isolation of Cell Walls and Determination of the Degree of Methylesterification
Leaves were frozen in liquid nitrogen and homogenized using a Retschmill machine (model MM200; Retsch) at 25 Hz for 1 min. The ground tissue was washed twice in 70% ethanol, vortexed, and pelleted by centrifugation at 10,000g for 10 min. The pellet was suspended with a chloroform:methanol mixture (1:1, v/v). After centrifugation and evaporation of the solvent, 1 mg of sample was saponified and suspended in 0.25 m NaOH. The solution was incubated at room temperature for 1 h and afterward neutralized with HCl. After centrifugation at 10,000g, aliquots of the supernatant (20 μL) were loaded in microtiter plate (96-well cod.9018 from Costar) filling up with water to a total volume of 50 μL. Alcohol oxidase (50 μL) was added to each well (0.03 units in 0.1 m sodium phosphate, pH 7.5; Sigma), and this mixture was incubated at room temperature for 15 min on shaker. Thereafter, 100 μL of a mixture containing 0.02 m 2,4-pentanedione in 2 m ammonium acetate and 0.05 m acetic acid was added. After 10 min of incubation at 68°C, samples were cooled on ice and absorbance was measured at 412 nm in microplate reader (ETI-System Reader; Sorin Biomedica Cardio S.p.A.). The methanol content was estimated as the amount of formaldehyde produced from methanol by alcohol oxidase, according to Klavons and Bennett (1986), by comparison with a standard calibration curve.
For the determination of uronic acid content, the saponified samples were treated with 200 μL of 2 m TFA for 1 h at 121°C in screw-cap tube properly closed. After washing three times with 2-propanol, uronic acids were quantified by colorimetry using the automated sulfamate/m-hydroxy diphenyl assay (Filisetti-Cozzi and Carpita, 1991). Uronic acid concentration was estimated by comparison with a standard calibration curve using d-GalUA. The degree of methylesterification was expressed as methanol to uronic acid molar ratio (%).
Oligosaccharide Mass Profiling MS Analysis
Cell walls were isolated from etiolated seedlings as above described. Thirty or 20 independent samples, each originating from 10 individual seedlings, were used for wild type or each transformed line, respectively. Fifty millimolar ammonium formate, pH 4.5, containing 0.02 units of xyloglucan-specific endoglucanase (Pauly et al., 1999) or a combination of 0.05 units of PME from Aspergillus aculeatus and 0.15 units of endoPG from Aspergillus niger (Megazyme), was added to cell walls and incubated for 17 h at 37°C. The combination of these pectic enzymes released methylesterified or O-acetylated HGA fragments from cell walls (Obel et al., 2006). After digestion, the suspension was centrifuged and the entire supernatant, containing the solubilized oligosaccharides, was dried in a speed-vac concentrator.
MALDI-TOF MS of the pectic oligosaccharides released after enzymatic digestions were recorded on a Voyager DE-Pro MALDI-TOF MS (Applied Biosystems) in positive mode with an acceleration voltage of 20,000 V and a delay time of 350 ns. Mass spectra were obtained in the reflectron mode using 2,5-dihydroxybenzoic acid (10 mg mL−1) as matrix mixed with the solubilized sugars 1:1 (v/v). The spectra were manually recorded and output-data files were analyzed, compared, and statistically evaluated by a Student's t test using the PERL program (Lerouxel et al., 2002). Pectic fragments were not detected in cell wall preparation without enzymes, and no detectable pectic fragments were observed by replacing the two pectic enzymes with endoPG alone (data not shown). In addition, pectic fragments generated from cell walls of Arabidopsis plant transformed with pCAMBIA 3300 plasmid alone showed no significant differences with respect to the wild type. Ions having 42 additional mass units were assigned to fragments containing O-acetyl substitutions on the Gal residues.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Quantification of PME activity in different tissues from wild-type plants with radial gel diffusion assay.
Supplemental Figure S2. Immunodetection of AtPMEI-1 and AtPMEI-2 in transformed leaf extracts after gel filtration.
Supplemental Table S1. Sequences of tryptic fragments obtained by LC-MS/MS analysis.
Supplementary Material
Acknowledgments
We thank Dr. Kirk Schnorr, Novozymes, for the gift of Aspergillus aculeatus PME and Dr. Ida Barbara Reca for generating transformed plants.
This work was supported by the Institute Pasteur-Fondazione Cenci Bolognetti and the Commission of European Communities (project no. QLK1–2000–00811 Gemini). V.L. was recipient of a short-term fellowship (ASTF 165.00–05) from EMBO.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Daniela Bellincampi (daniela.bellincampi@uniroma1.it).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Abuqamar S, Chen X, Dhawan R, Bluhm B, Salmeron J, Lam S, Dietrich RA, Mengiste T (2006) Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J 48 28–44 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Balestrieri C, Castaldo D, Giovane A, Quagliuolo L, Servillo L (1990) A glycoprotein inhibitor of pectin methylesterase in kiwi fruit (Actinidia chinensis). Eur J Biochem 193 183–187 [DOI] [PubMed] [Google Scholar]
- Bosch M, Cheung AY, Hepler PK (2005) Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol 138 1334–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Hepler PK (2006) Silencing of the tobacco pollen pectin methylesterase NtPPME1 results in retarded in vivo pollen tube growth. Planta 223 736–745 [DOI] [PubMed] [Google Scholar]
- Boudart G, Lafitte C, Barthe JP, Frasez D, Esquerré-Tugayé MT (1998) Differential elicitation of defense responses by pectic fragments in bean seedlings. Planta 206 86–94 [Google Scholar]
- Boudjeko T, Omokolo NA, Driouich A, Balangé AP (2005) Peroxidase and pectin methylesterase activities in cocoyam (Xanthosoma sagittifolium L. Schott) roots upon Phytium myriotylum inoculation. J Phytopathol 153 409–416 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH (2001) Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Mol Biol 47 311–340 [PubMed] [Google Scholar]
- Chen MH, Sheng J, Hind G, Handa AK, Citovsky V (2000) Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J 19 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M-H, Citovsky V (2003) Systemic movement of a tobamovirus requires host cell pectin methylesterase. Plant J 35 386–392 [DOI] [PubMed] [Google Scholar]
- Ciardiello MA, Tamburrini M, Tuppo L, Carratore V, Giovane A, Mattei B, Camardella L (2004) Pectin methylesterase from kiwi and kaki fruits: purification, characterization, and role of pH in the enzyme regulation and interaction with the kiwi proteinaceous inhibitor. J Agric Food Chem 52 7700–7703 [DOI] [PubMed] [Google Scholar]
- Clark CA, Lorbeer JW (1976) Comparative histopathology of Botrytis squamosa and B. cinerea on onion leaves. Phytopathology 66 1279–1289 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cole L, Dewey FM, Hawes CR (1998) Immunocytochemical studies of the infection mechanisms of Botrytis fabae. New Phytol 139 611–622 [Google Scholar]
- Collmer A, Keen NT (1986) The role of pectic enzymes in plant pathogenesis. Annu Rev Phytopathol 24 383–409 [Google Scholar]
- D'Avino R, Camardella L, Christensen TM, Giovane A, Servillo L (2003) Tomato pectin methylesterase: modeling, fluorescence, and inhibitor interaction studies-comparison with the bacterial (Erwinia chrysanthemi) enzyme. Proteins 53 830–839 [DOI] [PubMed] [Google Scholar]
- D'Ovidio R, Mattei B, Roberti S, Bellincampi D (2004) Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant-pathogen interactions. Biochim Biophys Acta 1696 237–244 [DOI] [PubMed] [Google Scholar]
- Daas PJH, Arisz PW, Schols HA, De Ruiter GA, Voragen AGJ (1998) Analysis of partially methyl-esterified galacturonic acid oligomers by high-performance anion-exchange chromatography and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Biochem 257 195–202 [DOI] [PubMed] [Google Scholar]
- De Lorenzo G, D'Ovidio R, Cervone F (2001) The role of polygalacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu Rev Phytopathol 39 313–335 [DOI] [PubMed] [Google Scholar]
- Desiderio A, Aracri B, Leckie F, Mattei B, Salvi G, Tigelaar H, Van Roekel JS, Baulcombe DC, Melchers LS, De Lorenzo G, et al (1997) Polygalacturonase-inhibiting proteins (PGIPs) with different specificities are expressed in Phaseolus vulgaris. Mol Plant Microbe Interact 10 852–860 [DOI] [PubMed] [Google Scholar]
- Di Matteo A, Giovane A, Raiola A, Camardella L, Bonivento D, De Lorenzo G, Cervone F, Bellincampi D, Tsernoglou D (2005) Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 17 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorokhov YL, Makinen K, Frolova OY, Merits A, Saarinen J, Kalkkinen N, Atabekov JG, Saarma M (1999) A novel function for a ubiquitous plant enzyme pectin methylesterase: the host-cell receptor for the tobacco mosaic virus movement protein. FEBS Lett 461 223–228 [DOI] [PubMed] [Google Scholar]
- Dorokhov YL, Skurat EV, Frolova OY, Gasanova TV, Ivanov PA, Ravin NV, Skryabin KG, Makinen KM, Klimyuk VI, Gleba YY, et al (2006) Role of the leader sequence in tobacco pectin methylesterase secretion. FEBS Lett 580 3329–3334 [DOI] [PubMed] [Google Scholar]
- Downie B, Dirk LM, Hadfield KA, Wilkins TA, Bennett AB, Bradford KJ (1998) A gel diffusion assay for quantification of pectin methylesterase activity. Anal Biochem 264 149–157 [DOI] [PubMed] [Google Scholar]
- Filisetti-Cozzi TMCC, Carpita NC (1991) Measurement of uronic acids without interference from neutral sugars. Anal Biochem 197 157–162 [DOI] [PubMed] [Google Scholar]
- Francis KE, Lam SY, Copenhaver GP (2006) Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol 142 1004–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovane A, Balestrieri C, Quagliuolo L, Castaldo D, Servillo L (1995) A glycoprotein inhibitor of pectin methylesterase in kiwi fruit: purification by affinity chromatography and evidence of a ripening-related precursor. Eur J Biochem 233 926–929 [DOI] [PubMed] [Google Scholar]
- Giovane A, Servillo L, Balestrieri C, Raiola A, D'Avino R, Tamburrini M, Ciardiello MA, Camardella L (2004) Pectin methylesterase inhibitor. Biochim Biophys Acta 1696 245–252 [DOI] [PubMed] [Google Scholar]
- Goldberg R (1984) Changes in the properties of cell wall pectin methylesterase along the Vigna radiata hypocotyl. Physiol Plant 61 58–63 [Google Scholar]
- Hellman U, Wernstedt C, Gonez J, Heldin CH (1995) Improvement of an “In-Gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal Biochem 224 451–455 [DOI] [PubMed] [Google Scholar]
- Hothorn M, Wolf S, Aloy P, Greiner S, Scheffzek K (2004) Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins. Plant Cell 16 3437–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Yang SL, Xie LF, Puah CS, Zhang XQ, Yang WC, Sundaresan V, Ye D (2005) VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kars I, Krooshof GH, Wagemakers L, Joosten R, Benen JA, van Kan JA (2005) Necrotizing activity of five Botrytis cinerea endopolygalacturonases produced in Pichia pastoris. Plant J 43 213–225 [DOI] [PubMed] [Google Scholar]
- Klavons JA, Bennett RD (1986) Determination of methanol using alcohol oxidase and its application to methyl ester content of pectins. J Agric Food Chem 34 597–599 [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lerouxel O, Choo TS, Seveno M, Usadel B, Faye L, Lerouge P, Pauly M (2002) Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiol 130 1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohaus G, Pennewiss K, Sattelmacher B, Hussmann M, Hermann MK (2001) Is the infiltration-centrifugation technique appropriate for the isolation of apoplastic fluid? A critical evaluation with different plant species. Physiol Plant 111 457–465 [DOI] [PubMed] [Google Scholar]
- Louvet R, Cavel E, Gutierrez L, Guenin S, Roger D, Gillet F, Guerineau F, Pelloux J (2006) Comprehensive expression profiling of the pectin methylesterase gene family during silique development in Arabidopsis thaliana. Planta 224 782–791 [DOI] [PubMed] [Google Scholar]
- Luehrsen KR, Walbot V (1991) Intron enhancement of gene expression and the splicing efficiency of introns in maize cells. Mol Gen Genet 225 81–93 [DOI] [PubMed] [Google Scholar]
- Markovic O, Janecek S (2004) Pectin methylesterases: sequence-structural features and phylogenetic relationships. Carbohydr Res 339 2281–2295 [DOI] [PubMed] [Google Scholar]
- Marty P, Jouan B, Bertheau Y, Vian B, Goldberg R (1997) Charge density in stem cell walls of solanum tuberosum genotypes and susceptibility to blackleg. Phytochemistry 44 1435–1441 [Google Scholar]
- Mattei B, Raiola A, Caprari C, Federici L, Bellincampi D, De Lorenzo G, Cervone F, Giovane A, Camardella L (2002) Studies on plant inhibitors of pectin modifying enzymes: polygalacturonase-inhibiting protein (PGIP) and pectin methylesterase inhibitor (PMEI). In TT Teeri, B Svensson, HJ Gilbert, T Feizi, eds, Carbohydrate Bioengineering: Interdisciplinary Approaches. Royal Society of Chemistry, Cambridge, UK, pp 160–167
- McCann MC, Roberts K (1994) Changes in cell wall architecture during cell elongation. J Exp Bot 45 1683–1691 [Google Scholar]
- McMillan GP, Hedley D, Fyffe L, Pérombelon MCM (1993) Potato resistance to soft-rot erwinias is related to cell wall pectin esterification. Physiol Mol Plant Pathol 42 279–289 [Google Scholar]
- Micheli F (2001) Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci 6 414–419 [DOI] [PubMed] [Google Scholar]
- Micheli F, Holliger C, Goldberg R, Richard L (1998) Characterization of the pectin methylesterase-like gene AtPME3: a new member of a gene family comprising at least 12 genes in Arabidopsis thaliana. Gene 220 13–20 [DOI] [PubMed] [Google Scholar]
- Micheli F, Sundberg B, Goldberg R, Richard L (2000) Radial distribution pattern of pectin methylesterases across the cambial region of hybrid aspen at activity and dormancy. Plant Physiol 124 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obel N, Erben V, Pauly M (2006) Functional wall glycomics through oligosaccharide mass profiling. In T Hayashi, ed, The Science and Lore of the Plant Cell Wall. Brown Walker Press, Boca Raton, FL, pp 258–266
- Pauly M, Albersheim P, Darvill A, York WS (1999) Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant J 20 629–639 [DOI] [PubMed] [Google Scholar]
- Pilling J, Willmitzer L, Bucking H, Fisahn J (2004) Inhibition of a ubiquitously expressed pectin methyl esterase in Solanum tuberosum L. affects plant growth, leaf growth polarity, and ion partitioning. Planta 219 32–40 [DOI] [PubMed] [Google Scholar]
- Pilling J, Willmitzer L, Fisahn J (2000) Expression of a Petunia inflata pectin methyl esterase in Solanum tuberosum L. enhances stem elongation and modifies cation distribution. Planta 210 391–399 [DOI] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijo JA, Becker JD (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiola A, Camardella L, Giovane A, Mattei B, De Lorenzo G, Cervone F, Bellincampi D (2004) Two Arabidopsis thaliana genes encode functional pectin methylesterase inhibitors. FEBS Lett 557 199–203 [DOI] [PubMed] [Google Scholar]
- Rogers EE, Ausubel FM (1997) Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi G, Giarrizzo F, De Lorenzo G, Cervone F (1990) A polygalacturonase-inhibiting protein in the flowers of Phaseolus vulgaris L. J Plant Physiol 136 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni V, Bellini C, Caboche M (1994) Use of two dimensional protein pattern analysis for the characterization of Arabidopsis thaliana mutants. Planta 192 557–566 [Google Scholar]
- Scheres B, Benfey P, Dolan L (2002) Root development. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0101, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Takahama U (1993) Redox state of ascorbic acid in the apoplast of stems of Kalanchoë daigremontiana. Physiol Plant 89 791–798 [Google Scholar]
- ten Have A, Mulder W, Visser J, van Kan JA (1998) The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol Plant Microbe Interact 11 1009–1016 [DOI] [PubMed] [Google Scholar]
- Tian GW, Chen MH, Zaltsman A, Citovsky V (2006) Pollen-specific pectin methylesterase involved in pollen tube growth. Dev Biol 294 83–91 [DOI] [PubMed] [Google Scholar]
- Tieman DM, Handa AK (1994) Reduction in pectin methylesterase activity modifies tissue integrity and cation levels in ripening tomato (Lycopersicon esculentum Mill.) fruits. Plant Physiol 106 429–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Harriman RW, Ramamohan G, Handa AK (1992) An antisense pectin methylesterase gene alters pectin chemistry and soluble solids in tomato fruit. Plant Cell 4 667–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Alebeek GJ, van Scherpenzeel K, Beldman G, Schols HA, Voragen AG (2003) Partially esterified oligogalacturonides are the preferred substrates for pectin methylesterase of Aspergillus niger. Biochem J 372 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kan JAL (2006) Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci 11 247–253 [DOI] [PubMed] [Google Scholar]
- Wakabayashi K, Hoson T, Huber DJ (2003) Methyl de-esterification as a major factor regulating the extent of pectin depolymerization during fruit ripening: a comparison of the action of avocado (Persea americana) and tomato (Lycopersicon esculentum) polygalacturonases. J Plant Physiol 160 667–673 [DOI] [PubMed] [Google Scholar]
- Wen FS, Zhu YM, Hawes MC (1999) Effect of pectin methylesterase gene expression on pea root development. Plant Cell 11 1129–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wietholter N, Graessner B, Mierau M, Mort AJ, Moerschbacher BM (2003) Differences in the methyl ester distribution of homogalacturonans from near-isogenic wheat lines resistant and susceptible to the wheat stem rust fungus. Mol Plant Microbe Interact 16 945–952 [DOI] [PubMed] [Google Scholar]
- Willats WG, McCartney L, Mackie W, Knox JP (2001) Pectin: cell biology and prospects for functional analysis. Plant Mol Biol 47 9–27 [PubMed] [Google Scholar]
- Wolf S, Grsic-Rausch S, Rausch T, Greiner S (2003) Identification of pollen-expressed pectin methylesterase inhibitors in Arabidopsis. FEBS Lett 555 551–555 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.