Abstract
The light-harvesting complexes (LHCs) of land plants and green algae have essential roles in light capture and photoprotection. Though the functional diversity of the individual LHC proteins are well described in many land plants, the extent of this family in the majority of green algal groups is unknown. To examine the evolution of the chlorophyll a/b antennae system and to infer its ancestral state, we initiated several expressed sequence tag projects from a taxonomically broad range of chlorophyll a/b-containing protists. This included representatives from the Ulvophyceae (Acetabularia acetabulum), the Mesostigmatophyceae (Mesostigma viride), and the Prasinophyceae (Micromonas sp.), as well as one representative from each of the Euglenozoa (Euglena gracilis) and Chlorarachniophyta (Bigelowiella natans), whose plastids evolved secondarily from a green alga. It is clear that the core antenna system was well developed prior to green algal diversification and likely consisted of the CP29 (Lhcb4) and CP26 (Lhcb5) proteins associated with photosystem II plus a photosystem I antenna composed of proteins encoded by at least Lhca3 and two green algal-specific proteins encoded by the Lhca2 and 9 genes. In organisms containing secondary plastids, we found no evidence for orthologs to the plant/algal antennae with the exception of CP29. We also identified PsbS homologs in the Ulvophyceae and the Prasinophyceae, indicating that this distinctive protein appeared prior to green algal diversification. This analysis provides a snapshot of the antenna systems in diverse green algae, and allows us to infer the changing complexity of the antenna system during green algal evolution.
Light-harvesting complexes (LHCs) are a superfamily of chlorophyll (Chl) and carotenoid-binding proteins present in photosynthetic eukaryotes that are responsible for the capture of light energy and its transfer to the photosynthetic reaction centers, where it is then used to drive oxygenic photosynthesis (Green and Durnford, 1996). The genes that encode the LHCs are nuclear encoded, translated in the cytosol, and their products are then posttranslationally directed to the chloroplasts where they associate with pigments and insert into the thylakoid membrane. LHCs are found in nearly all photosynthetic eukaryotes and can be divided into the Chl a/b-binding proteins of land plants and green algae, the Chl a-binding proteins of the red algae, and the Chl a/c-binding proteins of the chromalveolates. The presence of LHCs in the red and green algae has been cited as evidence that they share a common ancestor and that primary plastids are monophyletic (Wolfe et al., 1994; Durnford et al., 1999). To date, LHC relatives have not been detected in the glaucophytes but this group, like red algae, possesses phycobilisomes as the dominant antenna system. The Chl a-binding antennae of red algae are encoded by the Lhcr genes and associate specifically with PSI (Wolfe et al., 1994). Our knowledge about the LHC diversity in red algae is limited due to sampling but these LHCs seem to be confined to a red algal clade, which is becoming more complex with the discovery of red algal-like LHCs in various chromalveolates (Green, 2003).
The functional diversity and organization of the Chl a/b-binding protein family is best characterized in land plants; this provides a framework upon which one can examine and compare antenna complexity in other organisms. The antenna system is composed of distinct LHC proteins that are associated with PSI (LHCI) or PSII (LHCII). In Arabidopsis (Arabidopsis thaliana), there are six LHCI genes (Lhca1–Lhca6), three minor LHCII genes (Lhcb4 [CP29], Lhcb5 [CP26], and Lhcb6 [CP24]), and three classes of major LHCII genes (Lhcb1, Lhcb2, and Lhcb3; Jansson, 1999). The atomic structure of the major LHCII has been determined at different resolutions and each LHC monomer binds 14 Chls: eight Chl a and six Chl b (Kühlbrandt et al., 1994; Liu et al., 2004). The minor LHCII proteins CP26 and CP29 interact directly with the CP43 and D2 subunits of PSII (Yakushevska et al., 2003) and, aided by the third minor antenna protein CP24, are able to interact with one to three additional LHCII trimers (Horton and Ruban, 2005; Kouril et al., 2005). Interactions between adjacent trimers and PSII cores form a three-dimensional macrostructure in the stacked membranes of the grana (Horton and Ruban, 2005). The crystal structure of the plant PSI has been resolved to 4.4 Å (Ben-Shem et al., 2003) and LHCI was found to consist of two separate dimers (proteins encoded by Lhca1/Lhca4 and Lhca2/Lhca3) that were arranged in a half-moon-shaped belt (LHCI belt) located on the subunit F side of the reaction center. It has been proposed that the close proximity of the dimers and the presence of linker Chls allows for easy energy migration between the LHCI dimers and the reaction center (Ben-Shem et al., 2003).
The completion of the Chlamydomonas reinhardtii and Ostreococcus tauri genomes has increased our understanding of the LHC family in green algae. In C. reinhardtii, the LHCI and LHCII proteins are each encoded by a multigene family, while the minor PSII light-harvesting polypeptides (CP26 and CP29) are each encoded by one gene (Teramoto et al., 2002; Elrad and Grossman, 2004). Though there are no true orthologs for the plant Lhcb1-3 genes, a study that modeled the C. reinhardtii LHCII-PSII supercomplex in C. reinhardtii indicated it is very similar in both structure and biochemical properties to the LHCII-PSII supercomplexes found in land plants, with approximately 200 Chl molecules associated with each PSII dimer (Nield et al., 2000). Major differences between the C. reinhardtii and plant LHCII-PSII supercomplexes include the fact that C. reinhardtii lacks a homolog to the minor antennae, CP24 (Elrad and Grossman, 2004), and that compared to Arabidopsis, C. reinhardtii has a greater LHCI gene diversity with roughly twice the number of distinct Lhca genes. Kargul et al. (2003) modeled the structure of the C. reinhardtii LHCI-PSI supercomplex and predicted that there were 11 LHCI proteins rather than four as found in land plants, and that these proteins were arranged in a crescent docked with each PSI complex. Eight of these LHCI proteins are predicted to form dimers with the residual three proteins remaining as monomers (Kargul et al., 2003). Other LHC relatives such as PsbS (Elrad and Grossman, 2004) and LI818 (Savard et al., 1996) are also present in C. reinhardtii, though their functions have not yet been determined. A novel Lhc gene, Lhcq, has also been identified in the Chlamydomonas genome project, though little is known about its encoded protein (Elrad and Grossman, 2004). A number of additional LHC-related proteins, such as the early light-inducible proteins have also been described (Teramoto et al., 2002; Elrad and Grossman, 2004).
Six et al. (2005) described the family of antennae proteins from the prasinophyte O. tauri that comprised an unexpected level of antennae complexity. Previously, it was believed that prasinophytes primarily used a prasinophyte-specific (Lhcp) antennae system that was shared between PSI and PSII (Rhiel and Mörschel, 1993). Genes encoding five separate LHCI proteins, five Lhcp proteins, and one each of CP26, CP29, LI818, and LHCQ proteins were reported.
Both the euglenophytes and chlorarachniophytes contain Chl a/b-binding proteins and obtained their plastids from eukaryotes that likely resembled green algae (Keeling, 2004). In Euglena gracilis (euglenophyte), the main Lhc genes consist of multiple Lhc coding regions concatenated together, producing very long mRNA molecules that are translated into a large, single polyprotein (Muchhal and Schwartzbach, 1992). Once inside the chloroplast, the polyproteins are cleaved at conserved decapeptide linkers to form individual LHC units (Sulli and Schwartzbach, 1996). Polyproteins for both LHCI (Houlné and Schantz, 1988) and LHCII (Muchhal and Schwartzbach, 1992) have been reported, and it has been suggested that E. gracilis photosystems use a common light-harvesting system composed of both LHCI and LHCII proteins (Doege et al., 2000). For the chlorarachniophytes, however, there are only a few known LHCII-like sequences and they are clearly related to the Chl a/b family (Durnford et al., 1999; Deane et al., 2000; Archibald et al., 2003). There is very little known about the antennae complexity and diversity within either the euglenophytes or the chlorarachniophytes.
We examined the evolution of the light-harvesting antennae in a very diverse cross section of Chl a/b-containing photosynthetic organisms consisting of land plants, green algae, euglenophytes, and chlorarachniophytes. The green algae/land plants are represented by two major evolutionary lineages, Chlorophyta and Streptophyta. The former contains the well-supported Ulvophyceae, Trebouxiophyceae, and Chorophyceae (UTC) clade plus the polyphyletic members of the Prasinophyceae that group at the base of the Chlorophyta (Lewis and McCourt, 2004). The Streptophytes contain the embryophytes (land plants), the charophytes, and the early diverging Mesostigmatophyceae (Karol et al., 2001; Lemieux et al., 2007). In this study we analyzed LHCs from Acetabularia acetabulum (Ulvophyceae), Micromonas sp. (a prasinophyte of the order Mamiellales), Mesostigma viride (Mesostigmatophyceae), plus E. gracilis (a euglenophyte) and Bigelowiella natans (a chlorarachniophyte) that acquired plastids secondarily from green algae but are otherwise unrelated. With this broader sampling from within the Chl a/b-containing organisms, our goal is to predict the composition of the antenna system during green algal diversification and trace the evolution of antenna complexity, which may have implications for functional capabilities.
RESULTS
Complexity of the LHC Superfamily
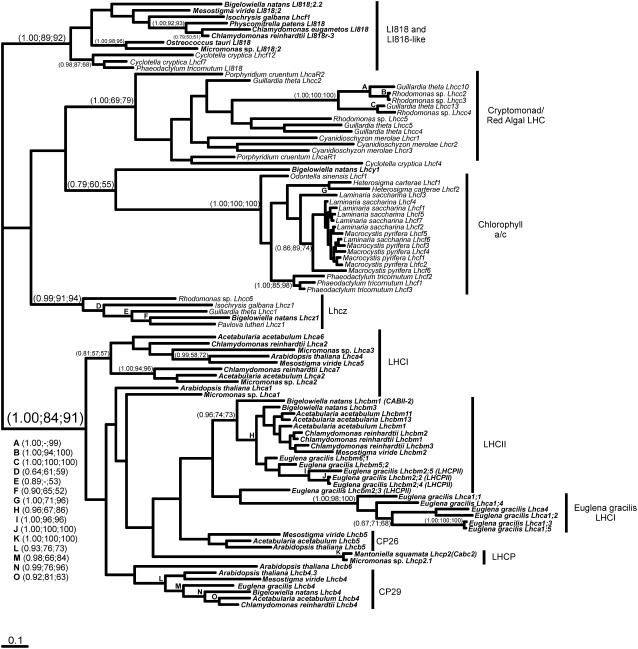
A phylogenetic tree containing the sequences of the Chl a/b- and Chl a/c-containing organisms is shown in Fig. 1. There were five major divisions within the tree, including the Chl a/b-binding proteins (Lhcb/Lhca genes), the LI818 and LI818-like proteins, the red algal/cryptomonad LHCs (Lhcr/Lhcc genes), the fucoxanthin-Chl a/c-binding proteins (Lhcf genes), and a new clade that we called Lhcz (Lhcz genes) that was composed of members from the cryptomonads, haptophytes, and chlorarachniophytes. Lhcz was used as there is no biochemical evidence hinting at functions or localizations, so z was selected as temporary designations to denote a unique class of genes. The most pronounced division (with support of 1.00, 84, 91) was between the genes that code for the Chl a/b-binding proteins (the green line) and the genes encoding the red algal/cryptomonad PSI-associated light-harvesting antenna proteins, the Chl a/c-binding proteins, and the LI818 proteins (the red line). The four clades within the red line (LI818, Lhcr/Lhcc, Lhcz, and Lhcf genes; Fig.1) were strongly supported though the relationships between them were not resolved. The LI818 branch is unique in that it contains homologs from many of the major groups of algae, including green algae (C. reinhardtii, Micromonas sp., M. viride, and O. tauri), chlorarachniophytes (B. natans), diatoms (Cyclotella cryptica and Phaeodactylum tricornutum), and haptophytes (Isochrysis galbana), agreeing with previous reports (Eppard et al., 2000; Richard et al., 2000). This clade even contained a LI818 homolog from a bryophyte (Physcomitrella patens) in contrast to the absence of this protein in angiosperms, suggesting it was lost at some point between bryophyte divergence and emergence of the angiosperms. The strongly supported (1.00, 100, 100) fucoxanthin-Chl a/c division comprised Lhcf genes of various stramenopiles.
Figure 1.
Phylogenetic reconstruction of the LHC superfamily. The analysis includes sequences from several major light-harvesting divisions, including LI818 and LI818-like proteins, Chl a/c-binding proteins of the chromalveolates, the cryptomonad/red algal LHCs, the cryptophyte/haptophyte LHCs, and the Chl a/b-binding proteins. A MrBayes tree is shown (−lnL= 13,545.54, α = 1.419) with the support values for all analyses shown at specific nodes in the following order: MrBayes (posterior probabilities), NJD, PHYML. A total of 129 characters were included and the proportion of invariable sites was 0.041. The average sd of the split frequencies was 0.0126. A total of 100 sequences were included in the analyses and all sequences were either novel sequences generated in conjunction with the Protist EST Program (Table II), were present in GenBank, or were retrieved from individual genome projects (C. merolae). P. patens LI818 sequence compiled from individual ESTs available in dbEST (BJ958734, BJ957315, BJ955694, BJ954266, BJ958980, and BJ857081). Sequences from Chl a/b-containing organisms are in bold.
The positions of three chlorarachniophyte antenna proteins were hard to resolve in this analysis. B. natans LI818;2.2 groups within the LI818 clade (1.00, 89, 92), while the position of a more divergent LHC did not significantly associate with any of the five major clades, thus we called it Lhcy1. The third chlorarachniophyte Lhc (B. natans Lhcz1) is particularly interesting as it associates specifically with a diverse group of organisms composed of cryptomonads (Guillardia theta and Rhodomonas sp.) and haptophytes (Pavlova lutheri and I. galbana) with very good support (0.99, 91, 94). To our knowledge, this is the first reported Lhc clade that includes diverse organisms possessing only secondary/tertiary plastids.
While we were able to resolve a separation between the green and red line LHCs, resolution of distinct clusters within the Chl a/b clade was generally weak in this global analysis with the exception of the prasinophyte-type Lhcp (1.00, 100, 100).
Chl a/b LHC Superfamily
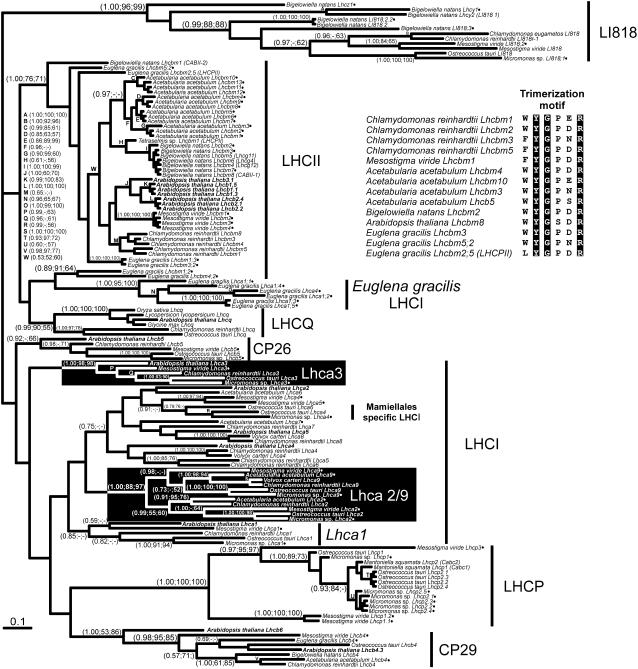
We further analyzed the LHC proteins in the Chl a/b clade to determine the presence of LHC homologs and to infer when they evolved. A phylogenetic tree of a larger number of diverse Chl a/b-binding LHC-like proteins with the LI818 proteins as the outgroup is shown in Figure 2.
Figure 2.
Phylogenetic reconstruction of the Chl a/b LHC superfamily. A MrBayes tree is shown with the posterior probabilities/support values for all three phylogeny programs shown at specific nodes (MrBayes, ProtDist, and PHYML). A total of 136 amino acid positions and 139 sequences were used with the proportion of invariable sites being 0.040. The γ shape distribution parameter (α) was 1.426 and the −lnL for the tree was 13,126.69. The average sd of the split frequencies was 0.0344. Novel sequences obtained from PEPdb have been differentiated from the other sequences by the addition of a black circle following the sequence names. The Arabidopsis LHC gene family is in bold for easy recognition of the plant LHCs. Solid black boxes surround specific Lhca clades (see text). The conserved trimerization motif for several LHCII sequences is shown, with the conserved residues highlighted.
The minor PSII antenna protein, CP29 (Lhcb4), was identified in all the cDNA libraries we surveyed, with the exception of Micromonas sp., and support for the CP29 clade was very strong (Fig. 2; 0.98, 95, 85). Additionally, all CP29 homologs shared a unique insert prior to the first transmembrane helix, supporting this identification. Another minor PSII antenna protein, CP26 (Lhcb5), was also identified in the green algal lineages containing primary plastids, including Micromonas sp. and M. viride (Fig. 2). We also detected an Lhcb5 homolog in A. acetabulum, but because the sequence was not full length we excluded it from the final analysis. However, clear Lhcb5 orthologs were not detected in the protists with secondarily derived plastids E. gracilis and B. natans. The third minor PSII-associated antenna in land plants, CP24 (Lhcb6; Jansson, 1999), was not found in any of the expressed sequence tag (EST) projects, nor in O. tauri (Six et al., 2005) or C. reinhardtii genomes (Elrad and Grossman, 2004). Additionally, we screened all LHC sequences for Lhcb6-specific indels (including a deletion preceding and an insertion following the second transmembrane helix), but still failed to identify any candidates.
The major LHCII proteins of C. reinhardtii, A. acetabulum, Arabidopsis, B. natans, and M. viride grouped together, but lacked support. There are multiple paralogs for the LHCII proteins for each organism and these consistently formed taxon-specific clades, though support for these branches was often poor. The presence of taxon-specific LHCII groups was an indication that there were no orthologs to either the C. reinhardtii or the Arabidopsis LHCII gene complements (Table I). Many of the E. gracilis LHCII sequences were at the base of the main LHCII group (Fig. 2). The position of the novel LHCQ protein with an unknown function is unresolved. Previously, Lhcq homologs have been discovered in genome projects of C. reinhardtii (Elrad and Grossman, 2004) and O. tauri (Six et al., 2005), and we have detected Lhcq-like homologs from a variety of land plants in GenBank; however, we did not find any Lhcq homologs in any of our libraries.
Table I.
Putative Lhc gene and protein complements in A. acetabulum (Aa), O. tauri (Ot), M. viride (Mv), Micromonas sp. (Ms), E. gracilis (Eg), and B. natans (Bn) compared to complements in Arabidopsis and C. reinhardtii
A filled circle indicates that the presence of the ortholog is relatively certain, while an empty circle indicates that the presence of the ortholog is less certain.
| Arabidopsis Gene (Protein) | C. reinhardtii | Aa | Ot | Mv | Ms | Eg | Bn |
|---|---|---|---|---|---|---|---|
| Lhcb1-3 (LHCII types 1–3) | |||||||
| Lhcbm1-6, 8-9, 11 | |||||||
| Lhcb4 (CP29) | Lhcb4 | • | • | • | • | • | |
| Lhcb5 (CP26) | Lhcb5 | • | • | • | • | ||
| Lhcb6 (CP24) | |||||||
| Lhca1 (LHCI type 1) | Lhca1 | ○ | ○ | ○ | |||
| Lhca2 (LHCI type 2) | |||||||
| Lhca3 (LHCI type 3) | Lhca3 | • | • | • | • | ||
| Lhca4-6 (LHCI types 4–6) | |||||||
| Lhca2 | • | • | • | • | |||
| Lhca4-8 | |||||||
| Lhca9 | • | • | • | • | |||
| Lhcq (LHCQ) | Lhcq | • | |||||
| LI818 | • | • | • | ○ |
The primary antenna for prasinophytes has long been recognized as unusual, both in sequence divergence and in pigment-binding properties (Rhiel and Mörschel, 1993). The prasinophyte-specific LHCs (named Lhcp by Six et al., 2005) bind Chl a, b, and a Chl c-like pigment in addition to several carotenoids. As expected, a number of Micromonas sp. LHCs cluster within a prasinophyte-specific Lhcp clade that is strongly supported by all methods (Fig. 2; 1.00, 100, 100). Micromonas sp. possesses several types of Lhcp genes and, while M. viride was also found to possess two Lhcp genes, they did not appear to be orthologous to any specific Lhcp sequence. It is interesting to note that M. viride, unlike Mantoniella squamata and O. tauri, has the typical green algal LHCII proteins as well as the prasinophyte-specific complex. To our knowledge, this is the first report of a green alga possessing both classes of antenna proteins. Though the prasinophyte-specific Lhcp clade is strongly supported, its relationship with the other LHCs was impossible to resolve.
Our understanding of LHCI diversity and function is defined by work in land plants, but it is clear that in green algae there is a large diversity in LHCI-like genes (Teramoto et al., 2002; Elrad and Grossman, 2004; Six et al., 2005). In this study, the only clear land plant homolog in all the green algae examined was Lhca3 (Fig. 2). This clade was strongly supported (1.00, 98, 98) and representatives from M. viride, Micromonas sp., and A. acetabulum were discovered. While a number of Lhca1-like sequences consistently group together, including the strongly supported Mamiellales-specific Lhca1 group (1.00, 91, 94), there was no support for the monophyly of this clade. We also did not detect any homologs for the plant Lhca2 and 4 genes in any of the green algae examined. Six et al. (2005) found five separate Lhca genes, of which Lhca3 grouped strongly with its land plant homolog. Our data supports this conclusion though the C. reinhardtii Lhca6 sequence in our study does not group with the Lhca3 clade as they found.
If the green algal LHCI antennae are compared, it is clear that there are orthologs for the C. reinhardtii Lhca2 and Lhca9 genes in the green algae examined (Fig. 2). It also appears that Lhca2 and Lhca9 are close paralogs as they form a strongly supported group in all three analyses (1.00, 88, 97). Due to the high sequence identity between these paralogs, the separation between Lhca2 and Lhca9 is moderately supported (Fig. 2). We also found an Lhca2 sequence in the O. tauri genome (http://bioinformatics.psb.ugent.be/blast/public/?project=ostreococcus) not previously reported by Six et al. (2005), which grouped with the Micromonas sp. Lhca2 sequence with strong support (1.00, 100, 98). In terms of nomenclature this poses a problem as there is already an O. tauri sequence named Lhca2, which we have renamed Lhca6. We found that it is more related to the Micromonas sp. O. tauri Lhca4 sequences, which is likely part of a Mamiellales-specific LHCI clade. As more Lhc sequences become available, the nomenclature will have to be amended so that the names reflect the shared orthologs in all green algae.
It is interesting to note that we detected no LHCI homologs in either of the two species examined with secondarily derived plastids. E. gracilis possesses a cluster of proteins that are referred to LHCI simply due to their exclusion from the LHCII branch. The E. gracilis LHCI branch was monophyletic (1.00, 95, 100), but its position in relation to other LHCs is unresolved.
PsbS
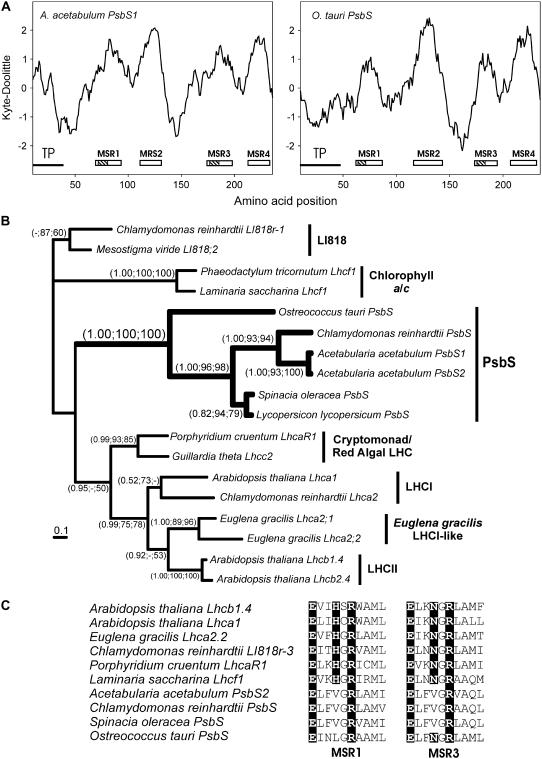
After an exhaustive search of all the EST databases, we only detected PsbS in A. acetabulum. Two different sequences were found in this library (PsbS1 and PsbS2), and both sequences have four predicted transmembrane helices, a characteristic of PsbS (Kim et al., 1992; Fig. 3A). Though we did not find a PsbS cDNA in M. viride or Micromonas sp., this is likely due to the depth of sampling and the conditions under which the libraries were made. We also mined the O. tauri genome and discovered a putative PsbS gene based on its hydropathy profile, which predicted four hydrophobic domains (Fig. 3A). To confirm that the A. acetabulum and O. tauri sequences were indeed related to known PsbS sequences, we generated an alignment possessing all major groups of LHC relatives. Due to the fact that PsbS proteins have four transmembrane helices rather than the three found in LHCs, a large amount of sequence data had to be trimmed. Nevertheless, clustering of the A. acetabulum and O. tauri proteins within the PsbS group was strongly supported by all three phylogenetic methods (1.00, 100, 100; Fig. 3B). However, the position of the PsbS branch within the LHC superfamily is not discernable due to the divergence of these proteins. The A. acetabulum and O. tauri PsbS proteins both lacked the conserved histidine in the Chl-binding motif of the first membrane-spanning region (MSR1) and instead had a valine or leucine, respectively, a notable characteristic of other PsbS proteins (Kim et al., 1992; Fig. 3C). However, while the A. acetabulum PsbS sequences had the characteristic valine rather than an asparagine in the third MSR, the O. tauri PsbS was more similar to the classic LHC and possessed the conserved asparagine (Fig. 3C).
Figure 3.
Analysis of green algal PsbS proteins. A, hydrophobicity plots of A. acetabulum PsbS1 and O. tauri PsbS sequences calculated with a window size of 19. Putative transmembrane regions are indicated by a white rectangle. Hatched areas identify the location of the putative Chl-binding motifs. TP denotes transit peptide location as determined with ChloroP. B, Phylogenetic analysis of PsbS sequences. A total of 120 amino acid positions were analyzed and the proportion of invariable sites was 0.113. The average sd of the split frequencies was 0.0015. A MrBayes tree is shown with the support values for all three phylogeny programs shown at specific nodes (MrBayes, ProtDist, and PHYML). The γ shape distribution parameter (α) was 4.123 and the −lnL value of the best PHYML tree was 3621.81. C, The conserved Chl-binding motifs for both MSR1 and MSR3 are shown for a variety of LHCs and organisms. The conserved residues in LHCs are highlighted.
DISCUSSION
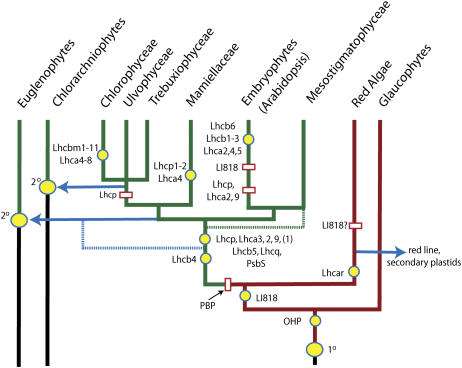
Examining diverse members of the green algae for the presence/absence of specific LHC homologs should allow us to assess the evolutionary changes in light harvesting and to predict the ancestral state of the antenna. Though the LHC antenna systems in the green and red lines evolved independently as they share few orthologous Lhc complexes, the LI818 clade is the notable exception as LI818-like sequences are present in a diverse group of photosynthetic organisms as shown here and elsewhere (Eppard et al., 2000; Richard et al., 2000; Green, 2003), indicating that the LI818 proteins were amongst the first eukaryotic LHCs (Fig. 4; Richard et al., 2000). However, the detection of a cryptomonad/haptophyte-like sequence in B. natans (Lhcz) would suggest that other Lhc types were present prior to red/green separation, but were subsequently lost (Fig. 4), though acquisition by lateral transfer is also a possibility. It was following the establishment of these first LHCs and the subsequent loss of phycobilisomes that the specialization into distinct PSI and PSII antennae likely occurred (Fig. 4).
Figure 4.
Scheme for the evolution of major LHCs and their relatives in the major photosynthetic eukaryotic groups. An oval indicates an endosymbiotic event. The primary (1°) symbol indicates that the endosymbiont was a cyanobacterium, while secondarily derived plastids (2°) originated from eukaryotic algae as indicated by the solid arrow. A yellow circle denotes a gain of a specific LHC or LHC relative. White rectangles indicate a gene-specific loss. Dotted lines indicate the presence of an alternate timing of an acquisition event. Dashed lines indicate alternative evolutionary pathways. OHP, One-helix protein; PBP, phycobilin protein.
PSII Antenna System
In considering the composition of an early PSII antenna system, CP29 is clearly an important component as orthologs to this LHC are present in all classes of green algae examined here, and are also present in the species with secondary plastids (Figs. 2 and 4). Although we did not find a Micromonas sp. CP29 homolog it has recently been identified in another member of the Mamiellales, O. tauri (Six et al., 2005), and is also present in M. viride, suggesting that it is likely present but that we simply did not identify it after examining 4,214 ESTs. Since the Mamiellales and M. viride are some of the earliest diverging green algal lineages (Lewis and McCourt, 2004), CP29 must have originated prior to their emergence. Of particular significance is the presence of CP29 in B. natans and E. gracilis, which lack specific orthologs to other algal LHC antenna proteins, with the possible exception of LI818 in B. natans. The other minor PSII antenna CP26 is present in all the green algae with primary plastids. Though we found no evidence for this complex in E. gracilis or B. natans, this could be due to the depth of library sequencing. Alternatively for E. gracilis, the deeply branching LHCII sequences could be CP26 like but just not resolved in this analysis. We did not detect any orthologs to CP24 (Lhcb6) in any of the green algae examined, nor was it detected in C. reinhardtii (Elrad and Grossman, 2004) or O. tauri (Six et al., 2005), suggesting this minor antenna is a recent invention restricted to land plants.
The minor antennae (CP29 and 26) are clearly ancestral and their presence in most Chl a/b-containing organisms suggests they have important roles in the structure and/or function of the light-harvesting antenna system. These minor antennae bind higher amounts of carotenoid than LHCII and act as the interface between LHCII trimers and PSII and it has also been proposed that they are the sites of nonphotochemical quenching (NPQ; Bassi et al., 1993; Dall'Osto et al., 2005). There is not a general agreement for this role, however, as CP29 and CP26 antisense Arabidopsis plants did not show the predicted changes in NPQ (Andersson et al., 2001). In E. gracilis at least, there is no evidence for NPQ as it lacks the xanthophyll cycle (Doege et al., 2000), despite having a clear CP29 homolog. Thus, in an evolutionary sense, CP29 (Lhcb4) presence is not correlated with a functional capability for NPQ.
An important role for CP29 in the early antenna system may have been energy redistribution between the reaction centers. In land plants and green algae, overexcitation of PSII induces LHCII phosphorylation and an association of the LHCII antenna with PSI in an attempt to balance electron flow; a process called state transitions (Haldrup et al., 2001). Recent biochemical characterization of PSI complexes locked in state 2 indicated that CP29 was among the PSII antennae that migrated to PSI (Takahashi et al., 2006). Modeling studies by Kargul et al. (2003) also proposed that CP29 was in close contact with the PsaH subunit of PSI and possibly coupled LHCII with PSI during state transitions. If CP29 has a significant role in this process then this suggests that state transitions likely developed before green algal diversification as a photoacclimation mechanism. It must be noted, however, that the presence of orthologous Lhcb4 (CP29) sequences does not necessarily indicate that the encoded protein retains the same role and functions as in C. reinhardtii and Arabidopsis and that this would have to be tested experimentally. The presence and absence of specific orthologs is also complicated by the apparent flexibility and potential redundant functionality. In Arabidopsis, knockdowns of the major trimeric LHCII resulted in CP26 forming trimers to compensate (Ruban et al., 2003), thus emphasizing such flexibility in antennae organization.
In this study, for all of the Chl a/b-containing organisms that possess LHCII homologs, there is evidence for the trimerization motif (WYGPDR; Hobe et al., 1995; Fig. 2), suggesting that trimerization of LHCII was an early adaptation for facilitating light harvesting. Trimers are more thermally and structurally stable than monomers, but the reason for trimerization is unknown (Wentworth et al., 2004). The prasinophyte-like antenna (Lhcp) appears to be an exception as it lacks a recognizable trimerization motif, suggesting differences in the organization of the PSII antennae. It would appear that the Mamiellales lost the typical LHCII-like antenna altogether in lieu of the Lhcp type since all other major green algae and green algal-derived plastids contain paralogs that cluster within the LHCII clade. The Lhcp type of antenna that binds Chl a, b, and a Chl c-like pigment may have become dominant in the process of spectral tuning to optimize light absorption. In the process, these proteins have changed considerably, masking any phylogenetic signal that would suggest from which paralog they arose. The presence of both LHCII and prasinophyte LHC types in M. viride suggests that the Lhcp type appeared early during the evolution of green algae, but was lost in both the UTC clade and the land plants.
PSI Antenna System
In Arabidopsis, there are six distinct LHCI genes (Lhca1–Lhca6), of which five (Lhca1–Lhca5) are expressed (Jansson, 1999; Ganeteg et al., 2004). There are four major protein types that are generally recognized in land plants (encoded by the genes Lhca1–Lhca4) that form heterodimers composed of the Lhca1/Lhca4- and Lhca2/Lhca3-encoded proteins (Croce et al., 2002) and are arranged in a half-moon-shaped belt bound to one side of PSI (Ben-Shem et al., 2003). LHCI dimerization is thought to aid in energy migration, pigment reorganization, and allowing for a more pronounced red absorption tail (Schmid et al., 1997; Ben-Shem et al., 2003). Of the five plant LHCI sequences, the Lhca3-encoded protein is the only clear homolog identified in all the primary plastid-containing green algae examined (Fig. 2; Table I). Phylogenetic analysis of C. reinhardtii (Tokutsu et al., 2004) and O. tauri (Six et al., 2005) Lhca3 also supported the presence of plant Lhca3 orthologs in green algae. In C. reinhardtii, N-terminal processing of the Lhca3-encoded proteins is involved with LHCI remodeling during iron deficiency and Lhca3 may act as a linker in the formation of a PSI-LHCI supercomplex (Naumann et al., 2005). Considering the retention of an Lhca3 ortholog in a diverse group of green algae and land plants, it is likely that these important regulatory and structural roles appeared early and have been maintained.
We identified two green algal-specific Lhca genes that are labeled Lhca2 (which is different from the plant Lhca2 gene) and Lhca9 (Fig. 2; Table I). This means there are three well-defined LHCI classes in green algae and it seems likely that the PSI belt of green algae consists of the plant homolog of Lhca3 and the green algal-specific proteins encoded by the Lhca2 and 9 genes. When an LHCI oligomeric complex was purified from a psaB deletion mutant of C. reinhardtii it was deficient in proteins encoded by the Lhca2, 3, and 9 genes (Takahashi et al., 2004). Because these only accumulate when the PSI core is present, they suggested that these subunits were in direct contact with PSI and may function to transfer excitation energy from the oligomeric LHCI antenna to PSI. Our phylogenetic analysis supports this proposal and indicates that further LHCI antenna diversification in other green algal lineages likely involved a modular addition to the core Lhca2, 3, and 9 subunits. In Arabidopsis, the absence of the green algal type of Lhca2 and 9 sequences indicates that they were replaced during plant evolution. We consistently resolve an Lhca1 group in the best tree using different methods that contain plant and green algal sequences, as in other studies (Tokutsu et al., 2004; Six et al., 2005), implying that Lhca1 may be the fourth LHCI making up the LHCI belt in green algae. Additionally, all Lhca1-encoded sequences, like all Lhca2/9-encoded sequences, have a conserved, shortened lumenal loop between their second and third membrane-spanning helices, and therefore have been proposed to have a unique structural/functional role in LHCI (Tokutsu et al., 2004). However, an Lhca1-specific group is not supported by resampling techniques and thus such a conclusion may not be warranted at this time.
While there are a total of five to six distinct LHCI proteins in O. tauri (Six et al., 2005), the LHCI antenna system in C. reinhardtii is composed of 11 proteins (Stauber et al., 2003; Elrad and Grossman, 2004) and has been estimated to be almost twice the size of the LHCI in land plants (Germano et al., 2002). This emphasizes the independent divergence of the LHCI antenna system in different algal groups. The changing complexity of LHCI is also evident in the detection of various taxon-specific LHCI proteins, including the Mamiellales-specific LHCI proteins.
We did not detect any plant or green algal LHCI homologs in the libraries from the Chl a/b-containing algae with secondary plastids E. gracilis and B. natans. The E. gracilis sequences presumed to be LHCI were clearly part of the Chl a/b-binding protein clade and always form a monophyletic group. In B. natans, the non-LHCII proteins were all excluded from the Chl a/b-binding clade. While the majority of these sequences were LI818 related, we also found a single LHC protein that was strongly related to cryptomonad and haptophyte Lhc sequences (Lhcz clade) and one weakly associated with the Lhcf genes (Lhcy1). The discovery of cryptomonad/haptophyte-like genes in a green lineage might suggest that in addition to LI818-like proteins, these homologs existed before the red and green algal lineages diverged. However, given the propensity of lateral gene transfer reported in chlorarachniophytes, such a conclusion would be premature (Archibald et al., 2003). Nevertheless, given the lack of LHCI orthologs, we would predict that the LI818 and cryptomonad/haptophyte-like sequences are functioning as the PSI antenna in B. natans, though this will have to be confirmed through biochemical analyses.
As E. gracilis and B. natans acquired their plastids secondarily from green algae, it is curious that LHCI homologs have not been detected. We cannot, however, rule out the possibility that such homologs were not detected due to the depth of EST sampling or the conditions under which the organisms were grown. With E. gracilis 25,595 ESTs were clustered, while the B. natans project was considerably smaller (3,462 ESTs). Nevertheless, we found considerable diversity in other organisms with similarly sized EST projects, indicating that if LHCI homologs are present, they are poorly expressed. Regardless, it is likely, especially with E. gracilis, that there were dramatic changes in the antennae system following secondary plastid acquisition. An explanation for these changes hinges on when and from which organisms E. gracilis and B. natans acquired their plastids, of which there is little information. Recent evidence from B. natans suggests that its plastid was acquired relatively late from within the UTC clade (Rogers et al., 2007), which would imply a loss and replacement of LHCI-like genes during the transfer of genetic information from the green algal endosymbiont to the host during plastid evolution. Something similar could have occurred during plastid acquisition in Euglena, but since these LHCI sequences group within the Chl a/b family (Fig. 1), then the acquisition of its plastid could have predated LHCI diversification in the early green algae, a hypothesis that would have to be tested by further sequence comparisons. Alternatively, there may have been rapid diversification of these sequences during plastid acquisition, thus masking their true relationships. In either of these scenarios, there appears to have been substantial reorganization of the PSI antenna, which may have been stimulated by a PSI trimeric to monomeric transition that occurred during some point in the evolution of the plastid (Ben-Shem et al., 2004).
Evolution of PsbS
PsbS is an LHC-related protein that is predicted to have four MSRs (Kim et al., 1992) and is essential for efficient NPQ in land plants (Li et al., 2002). Though PsbS proteins possess some of the conserved amino acids implicated in Chl binding in the LHCs, the Chl-binding ability of PsbS is uncertain, though it can bind zeaxanthin in vitro (Aspinall-O'Dea et al., 2002; Dominici et al., 2002). Most PsbS proteins, including those of C. reinhardtii, S. oleracea, and A. acetabulum possess two conserved glutamic acid residues, E122 and E226 (data not shown), that become protonated with the light-dependent change in pH that is important in PsbS-mediated high-energy state quenching efficiency (Li et al., 2004). The O. tauri PsbS protein has the conserved E226, but the glutamic acid residue at 122 was conservatively substituted with an aspartic acid. In Arabidopsis, PsbS is essential for NPQ (Dominici et al., 2002; Li et al., 2004), though its role in this process is not fully understood. NPQ involves the quenching of Chl fluorescence through thermal dissipation (Elrad et al., 2002) and involves three components: PsbS-mediated high-energy state quenching through delivery and removal of active zeaxanthin (Horton and Ruban, 2005), photoinhibition, and state transitions (Holt et al., 2004). Identification of PsbS homologs in C. reinhardtii (Elrad and Grossman, 2004) and now A. acetabulum (Ulvophyceae) and O. tauri (Mamiellales) indicates that this protein evolved early and prior to green algal diversification. Its conspicuous absence from the diatom Thalassiosira pseudonana (Armbrust et al., 2004) and red alga Cyanidioschyzon merolae (Matsuzaki et al., 2004) genomes, suggests it is specific to the green lineages (Fig. 4). Though the presence of PsbS in green algae would suggest a similar dissipation strategy, the relative importance of this mechanism appears to be quite different in C. reinhardtii (Elrad and Grossman, 2004) and thus extrapolating function to other organisms has to be done cautiously. The absence of a PsbS homolog in other organisms in this study is likely due to the depth of sequencing the libraries and the expression level of the PsbS.
Tracing the Evolution of Antenna Complexity
Figure 4 summarizes the evolution of the LHC family in the context of diversity within the green line with a specific emphasis on antennae evolution in Arabidopsis, C. reinhardtii, and O. tauri. There are no LHC homologs yet identified in cyanobacteria, indicating that the LHCs are a eukaryotic invention. The glaucophytes, however, are the only eukaryotic group from which true LHC proteins have not yet been found, tentatively indicating that they appeared following the divergence of the glaucophytes from the green/red algal line. The glaucophytes do, however, possess a carotenoid-rich protein that shares epitopes with the LHCs but this may be structurally unique (Rissler and Durnford, 2005). Thus, the earliest we can date the appearance of the LHCs is prior to the separation of the red and green algae and these eukaryote-specific proteins likely arose from the high light inducible proteins that are present in cyanobacteria (Dolganov et al., 1995). Transfer of the high light inducible protein-encoding genes to the nucleus likely gave rise to the related one-helix proteins that were first identified in Arabidopsis (Andersson et al., 2003). A series of gene duplications and fusions could have occurred to give rise to the first classic LHC possessing three MSRs. Many of these LHC-like genes are induced in response to excess light exposure, thus making a connection between light stress and antenna evolution. Therefore, the dominant selective pressure for eventual LHC evolution was likely photoprotective and a fine tuning of excitation energy distribution between the photosystems that would allow greater plasticity in acclimating to environmental stress.
Unlike all other LHCs, LI818-like homologs are broadly distributed in algal groups with different pigmentation, spanning the classic red and green lines (Fig. 4), thus it is one of the earliest diverging LHCs that can be identified with the current evidence and its extant distribution indicates that it probably appeared prior to the red/green split (Richard et al., 2000). The separation of the red and green algae is characterized by retention of phycobilisomes in the red line and a loss in the green. The loss of phycobilisomes coincided with an extensive expansion of the LHC family, giving rise to a number of major antenna complexes, including the inner antennae, CP26 (Lhcb5) and CP29 (Lhcb4), and various LHCI genes (Lhca3, Lhca2, Lhca9, and possibly Lhca1). This occurred prior to green algal diversification, indicating that the antenna structure and complexity were well developed early in the evolution of green algae and land plants.
Following the emergence of different green algal groups, the peripheral LHCII and LHCI antennae system continued to change independently in different lineages, driven by gene duplications and divergence as these organisms adapted to varying niches in an attempt to optimize light harvesting and photoprotection. This is particularly evident with LHCII where the major antenna genes radiated independently in different groups. Evidence of the changing complexity of the antennae system is also present in LHCI where a number of family-specific LHCI genes were identified, indicative of a late emergence. In both cases, it seems that a core antennae were retained and further modifications built upon this existing structure.
The antenna structures of E. gracilis and B. natans are particularly interesting as the secondary origin of these plastids seemed to affect the evolution of the antennae systems, particularly for LHCI for which the available evidence is suggestive of a replacement of this antenna system. Though this must be a tentative conclusion until a genome project determines the exact complement of Lhc genes, it does appear as though the massive transfer of genes to the nucleus during plastid evolution was not without its share of gene losses and/or rapid divergence as the organism adapted to a photosynthetic lifestyle.
MATERIALS AND METHODS
cDNA Libraries and Data Mining
Complementary DNA libraries for Bigelowiella natans (CCMP621), Euglena gracilis (strain Z), Micromonas sp. (CCMP490), and Mesostigma viride CCMP2046 (NIES 296) were commercially prepared from RNA provided from member labs of the protist EST project. The Acetabularia acetabulum cDNA library was provided by Dina Mandoli at the University of Washington. Bacterial plating, picking, DNA preparation, and sequencing were conducted at the Atlantic Genome Centre (Halifax, Nova Scotia) and the British Columbia Cancer Agency (Vancouver). Sequence traces were vector trimmed, clustered, and the processed sequence deposited into the Taxonomically Broad EST Database. Taxonomically Broad EST Database was mined for potential LHC-like genes using the in-house BLAST search program, Anabench BLAST (Badidi et al., 2003). Chlamydomonas reinhardtii LHCI, LHCII, CP29, and LI818 proteins, as well as the Chl-binding domain were used in tblastx searches for LHC-related sequences. A list of the LHC-related clusters found, the number of ESTs in each cluster, and suggested names are given in Table II. All clusters analyzed in this study can be accessed at http://tbestdb.bcm.umontreal.ca/searches/welcome.php. Individual ESTs have been deposited in dbEST (National Center for Biotechnology Information) and the annotated clusters have been deposited in GenBank (Table II).
Table II.
TBestDB cluster IDs; number of ESTs in each cluster; proposed gene and protein names for A. acetabulum, B. natans, E. gracilis, M. viride, and Micromonas sp. clusters; and GenBank (Third Party Annotation) accession numbers
Also included are newly described O. tauri LHC sequences. Asterisks indicate that the sequence is a polyprotein. Not all sequences present in table are included in all analyses.
| Species Name | Cluster ID | Number of ESTs | Proposed Gene Name | Proposed Protein Name | Accession Number |
|---|---|---|---|---|---|
| A. acetabulum | Aa0009 | 2 | Lhcbm13 | LHCII | BK005993 |
| Aa0027 | 4 | Lhcb4 | CP29 | BK005994 | |
| Aa0199 | 2 | Lhcbm1 | LHCII | BK005995 | |
| Aa0218 | 3 | Lhcbm9 | LHCII | BK005996 | |
| Aa0544 | 1 | Lhca9 | LHCI-9 | BK005997 | |
| Aa0891 | 1 | Lhca6 | LHCI-6 | BK005998 | |
| Aa0913 | 1 | Lhca7 | LHCI-7 | BK005999 | |
| Aa0942 | 1 | Lhcbm11 | LHCII | BK006000 | |
| Aa1077 | 2 | Lhca3 | LHCI-3 | BK006001 | |
| Aa1079 | 2 | Lhcbm12 | LHCII | BK006002 | |
| Aa1102 | 2 | Lhcbm8 | LHCII | BK006003 | |
| Aa1122 | 2 | Lhcbm4 | LHCII | BK006004 | |
| Aa1176 | 3 | Lhcbm10 | LHCII | BK006005 | |
| Aa1193 | 3 | Lhcbm3 | LHCII | BK006006 | |
| Aa1213 | 11 | Lhcbm6 | LHCII | BK006007 | |
| Aa1216 | 18 | Lhcbm5 | LHCII | BK006008 | |
| Aa2029 | 1 | Lhca2 | LHCI-2 | BK006009 | |
| Aa2098 | 2 | Lhcb5 | CP26 | BK006010 | |
| Aa2324 | 1 | Lhcbm2 | LHCII | BK006011 | |
| Aa2338 | 2 | Lhcbm7 | LHCII | BK006012 | |
| Aa0242 | 2 | PsbS1 | PSBS1 | BK006013 | |
| Aa0911 | 3 | PsbS2 | PSBS2 | BK006014 | |
| B. natans | Bn0444 | 4 | Lhcy1 | LHCY | BK005986 |
| Bn0464 | 4 | LI818;1 | LI818-1 | BK005987 | |
| Bn0525 | 8 | Lhcbm2 | LHCII | BK005988 | |
| Bn0551 | 26 | Lhcbm3 | LHCII | BK005989 | |
| Bn0553 | 27 | Lhcbm7 | LHCII | BK005990 | |
| Bn1177 | 1 | LI818;2.2 | LI818-2 | BK005991 | |
| Bn1678 | 1 | Lhcz1 | LHCZ | BK005992 | |
| E. gracilis | Eg0021 | 118 | Lhcb4 | CP29 | BK005977 |
| Eg0137* | 10 | Lhcbm1 | LHCII | BK005978 | |
| Eg1193* | 1 | Lhcbm3 | LHCII | BK005979 | |
| Eg1913* | 2 | Lhcbm4 | LHCII | BK005980 | |
| Eg2532 | 9 | Lhca3 | LHCI-6 | BK005981 | |
| Eg2554* | 12 | Lhcbm5 | LHCII | BK005982 | |
| Eg2577* | 14 | Lhcbm6 | LHCII | BK005983 | |
| Eg2590* | 27 | Lhca2 | LHCI-(1-4) | BK005984 | |
| Eg2596* | 34 | Lhca1 | LHCI-(1-4) | BK005985 | |
| M. viride | Mv0016 | 33 | Lhca3 | LHCI-3 | BK006015 |
| Mv0021 | 27 | Lhcp1.2 | LHCP | BK006016 | |
| Mv0039 | 14 | Lhca4 | LHCI-4 | BK006017 | |
| Mv0063 | 142 | Lhcbm1 | LHCII | BK006018 | |
| Mv0096 | 31 | Lhcb4 | CP29 | BK006019 | |
| Mv0099 | 17 | Lhca2 | LHCI-2 | BK006020 | |
| Mv0237 | 49 | Lhcbm4 | LHCII | BK006021 | |
| Mv0551 | 12 | Lhcbm2 | LHCII | BK006022 | |
| Mv0574 | 35 | Lhca5 | LHCI-4 | BK006023 | |
| Mv1486 | 13 | Lhca9 | LHCI-2 | BK006024 | |
| Mv1488 | 14 | Lhcb5 | CP26 | BK006025 | |
| Mv1492 | 25 | Lhca1 | LHCI-1 | BK006026 | |
| Mv1495 | 38 | Lhcp1.1 | LHCP | BK006027 | |
| Mv2259 | 14 | Lhcbm3 | LHCII | BK006028 | |
| Mv2412 | 2 | LI818;2 | LI818-2 | BK006029 | |
| Micromonas sp. | Ms0002 | 34 | Lhcp2.4 | LHCP | BK006030 |
| Ms0014 | 5 | Lhcp3 | LHCP | BK006031 | |
| Ms0015 | 33 | Lhcp2.5 | LHCP | BK006032 | |
| Ms0037 | 6 | Lhcb5 | CP26 | BK006033 | |
| Ms0056 | 19 | LI818;2 | LI818-2 | BK006034 | |
| Ms0066 | 49 | Lhcp2.3 | LHCP | BK006036 | |
| Ms0411 | 7 | Lhca3 | LHCI-3 | BK006035 | |
| Ms0669 | 3 | Lhca9 | LHCI-9 | BK006037 | |
| Ms0696 | 4 | Lhca2 | LHCI-2 | BK006038 | |
| Ms0726 | 5 | Lhca4 | LHCI-4 | BK006039 | |
| Ms0739 | 11 | LI818;1 | LI818-1 | BK006040 | |
| Ms0741 | 13 | Lhcp2.2 | LHCP | BK006041 | |
| Ms0743 | 8 | Lhcp1 | LHCP | BK006042 | |
| Ms1341 | 1 | Lhca1 | LHCI-1 | BK006043 | |
| Ms2046 | 53 | Lhcp2.1 | LHCP | BK006044 | |
| O. tauri | Ot03g04770 | – | Lhca2 | LHCI-2 | – |
| Ot06g04910 | – | PsbS | PSBS | – |
The amino acid sequences inferred from EST clusters were aligned using version 1.83 of the Clustal W multiple sequence alignment program (Thompson et al., 1994) and manually adjusted using the BioEdit Sequence Alignment Editor (Hall, 1999). Additional LHC sequence data were added to augment the analyses, including data from C. reinhardtii (a chlorophyte), Arabidopsis (Arabidopsis thaliana; an embryophyte), Physcomitrella patens (a bryophyte), Tetraselmis sp. (a trebouxiophyte), and Ostreococcus tauri (a prasinophyte of the order Mamiellales). Additional red algal and the chromalveolate sequences were added to supplement the analysis. Truncated sequences were removed from the alignments, which are available upon request. The characters used in the subsequent analyses were located within and immediately flanking each of the three MSRs of the LHCs, thus excluding the ambiguously aligned regions, as previously described (Durnford et al., 1999). Where possible, the included characters started and ended with a conserved amino acid that usually delineated a structural feature as defined by the LHCII crystal structure. As the LHCs are integral membrane proteins, the dataset is dominated by amino acids that exist within the membrane. This may have a tendency to skew the evolutionary model, as hydrophobic amino acids would have a biased selection. This in combination with their relatively small size prevents effective resolution of the deeper relationships between major LHC classes, though this approach is valuable for identifying orthologs and distinct classes of proteins. For E. gracilis LHCs that were part of polyproteins, we manually trimmed individual proteins at the decapeptide linkers and included them in our analyses. In A. acetabulum, the conventional termination codons TAA and TAG code for the amino acid glutamine (Schneider et al., 1989) and the protein sequences were edited to account for these differences.
The nomenclature used in this manuscript was based on the guidelines established for land plants and green algae by Jansson et al. 1992 and 1999 and based on the accepted nomenclature of the LHC genes used for Arabidopsis (Jansson, 1999), C. reinhardtii (Elrad and Grossman, 2004), and O. tauri (Six et al., 2005). The conventions observed in previous studies dictate that the photosystem-specific antennae are differentiated with either an “a” for a PSI-associated light-harvesting protein (i.e. Lhca) or a “b” for a gene encoding a PSII-associated LHC (i.e. Lhcb). Different members of each family are differentiated with a numeral after the Lhca/b (i.e. Lhca1). For the LHCII sequences, we followed the nomenclature of Elrad and Grossman (2004) and distinguished the main antennae genes of PSII with Lhcbm plus a number identifying the specific sequence. Nearly identical isoforms, when present, were distinguished with a period and another numeral (i.e. Lhca1.1). A nomenclature of E. gracilis Lhc genes was a challenge as the genes are very large and encode polyproteins with multiple LHC types that are posttranslationally cleaved at conserved decapeptide linkers to yield the individual proteins (Houlné and Schantz, 1988). Any nomenclature has to refer to the specific gene and the order in the polyprotein; therefore, a single gene encoding a LHCI polyprotein with five individual protein units would be written as Lhca1;1 to Lhca1;5.
Phylogenetic and Hydrophobicity Analyses
Phylogenetic analyses used included Neighbor-Joining Distance (NJD), Bayesian algorithms, and maximum likelihood. The ProtTest program (Abascal et al., 2005) was used to select the best-fit model of amino acid substitution for each of the analyses for the different alignments based upon the Akaike Information Criterion framework (Akaike, 1974). The PROTDIST application of the Phylogenetic Inference Package (PHYLIP) version 3.65 (Felsenstein, 1989) was used to create distance matrices with the Jones, Taylor, and Thornton model for amino acid substitution and the coefficient of variation of substitution rates among positions. NJD trees were generated using the NEIGHBOR program in PHYLIP and consensus trees were generated with CONSENSE. Branch stability and the internal consistency of the data sets were assessed by bootstrap analysis using the SEQBOOT program and 1,000 replicates. Bootstrap values that support a node on the resulting tree in greater than 50% of the pseudosamples are shown.
The PHYML program (Guindon and Gascuel, 2003; Guindon et al., 2005) was used for the maximum-likelihood analyses (http://atgc.lirmm.fr/phyml/) utilizing the Whelan and Goldman amino acid substitution matrix, with γ correction (four categories) and accounting for the number of invariant sites. Due to the large number of sequences included in the analyses and to ensure a reasonable computation time, only 100 pseudoreplicates were used.
MrBayes v3.1.2 (Huelsenbeck and Ronquist, 2001) was used for the Bayesian analyses (http://cbsu.tc.cornell.edu). Blosum62 with four γ distribution categories and incorporating the number of invariant sites was used as the substitution matrix. The number of generations performed varied slightly with the analysis and was 4.41 × 106 for the global LHC superfamily analysis (Fig. 1), 5.07 × 106 for the Chl a/b superfamily analysis (Fig. 2), and 5.00 × 106 for the PsbS analysis (Fig. 3). For each, we used a sampling frequency of 100 with a 25% burnin value and the consensus type was allcompat. All phylogenetic trees are displayed using the TREEVIEW program (Page, 1996). LI818 sequences were used as an outgroup in all analyses since it is the only LHC-like sequence present in most of the major groups of photosynthetic eukaryotes.
Hydrophobicity plots were performed on the ExPaSy server (www.expasy.ch) using ProtScale (Gasteiger et al., 2005) with the Kyte and Doolittle amino acid scale and a window size of 19. Transit peptides were predicted with ChloroP (Emanuelsson et al., 1999).
Sequence data from this article can be found in the Third Party Annotation Section of the DDBJ/EMBL/GenBank databases under the accession numbers TPA: BK005977 to BK006046.
This work was supported by grants from Genome Canada, Genome Atlantic, and Genome British Columbia as part of the Protist EST Program and supported through the Natural Science and Engineering Research Council of Canada.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Dion G. Durnford (durnford@unb.ca).
Open Access articles can be viewed online without a subscription.
References
- Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21 2104–2105 [DOI] [PubMed] [Google Scholar]
- Akaike H (1974) New look at statistical-model identification. IEEE Transactions on Automatic Control 19 716–723 [Google Scholar]
- Andersson J, Walters RG, Horton P, Jansson S (2001) Antisense inhibition of the photosynthetic antenna proteins CP29 and CP26: implications for the mechanism of protective energy dissipation. Plant Cell 13 1193–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson U, Heddad M, Adamska I (2003) Light stress-induced one-helix protein of the chlorophyll a/b-binding family associated with photosystem I. Plant Physiol 132 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM, Rogers MB, Toop M, Ishida K, Keeling PJ (2003) Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc Natl Acad Sci USA 100 7678–7683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, et al (2004) The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306 79–86 [DOI] [PubMed] [Google Scholar]
- Aspinall-O'Dea M, Wentworth M, Pascal A, Robert B, Ruban A, Horton P (2002) In vitro reconstitution of the activated zeaxanthin state associated with energy dissipation in plants. Proc Natl Acad Sci USA 99 16331–16335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badidi E, De Sousa C, Lang BF, Burger G (2003) AnaBench: a web/CORBA-based workbench for biomolecular sequence analysis. BMC Bioinformatics 4 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi R, Pineau B, Dainese P, Marquardt J (1993) Carotenoid-binding proteins of photosystem II. Eur J Biochem 212 297–303 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Frolow F, Nelson N (2003) Crystal structure of plant photosystem I. Nature 426 630–635 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Frolow F, Nelson N (2004) Light-harvesting features revealed by the structure of plant photosystem I. Photosynth Res 81 239–250 [DOI] [PubMed] [Google Scholar]
- Croce R, Morosinotto T, Castelletti S, Breton J, Bassi R (2002) The Lhca antenna complexes of higher plants photosystem I. Biochim Biophys Acta 1556 29–40 [DOI] [PubMed] [Google Scholar]
- Dall'Osto L, Caffarri S, Bassi R (2005) A mechanism of nonphotochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. Plant Cell 17 1217–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane JA, Fraunholz M, Su V, Maier U-G, Martin W, Durnford DG, McFadden GI (2000) Evidence for nucleomorph to host nucleus gene transfer: light-harvesting complex proteins from cryptomonads and chlorarachniophytes. Protist 151 239–252 [DOI] [PubMed] [Google Scholar]
- Doege M, Ohmann E, Tschiersch H (2000) Chlorophyll fluorescence quenching in the alga Euglena gracilis. Photosynth Res 63 159–170 [DOI] [PubMed] [Google Scholar]
- Dolganov NA, Bhaya D, Grossman AR (1995) Cyanobacterial protein with similarity to the chlorophyll a/b binding proteins of higher plants: evolution and regulation. Proc Natl Acad Sci USA 92 636–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici P, Caffarri S, Armenante F, Ceoldo S, Crimi M, Bassi R (2002) Biochemical properties of the PsbS subunit of photosystem II either purified from chloroplast or recombinant. J Biol Chem 277 22750–22758 [DOI] [PubMed] [Google Scholar]
- Durnford DG, Deane JA, Tan S, McFadden GI, Gantt E, Green BR (1999) A phylogenetic assessment of the eukaryotic light-harvesting antenna proteins, with implications for plastid evolution. J Mol Evol 48 59–68 [DOI] [PubMed] [Google Scholar]
- Elrad D, Grossman AR (2004) A genome's-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii. Curr Genet 45 61–75 [DOI] [PubMed] [Google Scholar]
- Elrad D, Niyogi KK, Grossman AR (2002) A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14 1801–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppard M, Krumbein WE, von Haeseler A, Rhiel E (2000) Characterization of fcp4 and fcp12, two additional genes encoding light harvesting proteins of Cyclotella cryptica (Bacillariophyceae) and phylogenetic analysis of this complex gene family. Plant Biol 2 283–289 [Google Scholar]
- Felsenstein J (1989) PHYLIP—Phylogeny inference package (version 3.2). Cladistics 5 164–166 [Google Scholar]
- Ganeteg U, Klimmek F, Jansson S (2004) Lhca5—an LHC-type protein associated with photosystem I. Plant Mol Biol 54 641–651 [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins M, Appel R, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In JM Walker, ed, The Proteomics Protocols Handbook, Ed 1, Vol 1. Humana Press Inc., Totowa, NJ, pp 571–607
- Germano M, Yakushevska AE, Keegstra W, van Gorkom HJ, Dekker JP, Boekema EJ (2002) Supramolecular organization of photosystem I and light-harvesting complex I in Chlamydomonas reinhardtii. FEBS Lett 525 121–125 [DOI] [PubMed] [Google Scholar]
- Green BR (2003) The evolution of light-harvesting antennas. In BR Green, WW Parson, eds, Advances in Photosynthesis and Respiration: Light-Harvesting Antennas In Photosynthesis, Ed 1, Vol 13. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 129–168
- Green BR, Durnford DG (1996) The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 47 685–714 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52 696–704 [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O (2005) PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33 W557–W559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldrup A, Jensen PE, Lunde C, Scheller HV (2001) Balance of power: a view of the mechanism of photosynthetic state transitions. Trends Plant Sci 6 301–305 [DOI] [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41 95–98 [Google Scholar]
- Hobe S, Förster R, Klingler J, Paulsen H (1995) N-proximal sequence motif in light-harvesting chlorophyll a/b-binding protein is essential for the trimerization of light-harvesting chlorophyll a/b complex. Biochemistry 34 10224–10228 [DOI] [PubMed] [Google Scholar]
- Holt NE, Fleming GR, Niyogi KK (2004) Toward an understanding of the mechanism of nonphotochemical quenching in green plants. Biochemistry 43 8281–8289 [DOI] [PubMed] [Google Scholar]
- Horton P, Ruban A (2005) Molecular design of the photosystem II light-harvesting antenna: photosynthesis and photoprotection. J Exp Bot 56 365–373 [DOI] [PubMed] [Google Scholar]
- Houlné G, Schantz R (1988) Characterization of cDNA sequences for LHCI apoproteins in Euglena gracilis—the messenger-RNA encodes a large precursor containing several consecutive divergent polypeptides. Mol Gen Genet 213 479–486 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17 754–755 [DOI] [PubMed] [Google Scholar]
- Jansson S (1999) A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci 4 236–240 [DOI] [PubMed] [Google Scholar]
- Jansson S, Green B, Grossman AR, Hiller R (1999) A proposal for extending the nomenclature of light-harvesting proteins of the three transmembrane helix type. Plant Mol Biol Rep 17 221–224 [Google Scholar]
- Jansson S, Pichersky E, Bassi R, Green BR, Ikeuchi M, Melis A, Simpson DJ, Spangfort M, Staehelin LA, Thornber JP (1992) A nomenclature for the genes encoding the chlorophyll a/b-binding proteins of higher plants. Plant Mol Biol Rep 10 242–253 [Google Scholar]
- Kargul J, Nield J, Barber J (2003) Three-dimensional reconstruction of a light-harvesting complex I-photosystem I (LHCI-PSI) supercomplex from the green alga Chlamydomonas reinhardtii: insights into light harvesting for PSI. J Biol Chem 278 16135–16141 [DOI] [PubMed] [Google Scholar]
- Karol KG, McCourt RM, Cimino MT, Delwiche CF (2001) The closest living relatives of land plants. Science 294 2351–2353 [DOI] [PubMed] [Google Scholar]
- Keeling PJ (2004) Diversity and evolutionary history of plastids and their hosts. Am J Bot 91 1481–1493 [DOI] [PubMed] [Google Scholar]
- Kim S, Sandusky P, Bowlby NR, Aebersold R, Green BR, Vlahakis S, Yocum CF, Pichersky E (1992) Characterization of a spinach psbS cDNA encoding the 22 kDa protein of photosystem II. FEBS Lett 314 67–71 [DOI] [PubMed] [Google Scholar]
- Kouril R, Zygadlo A, Arteni AA, de Wit CD, Dekker JP, Jensen PE, Scheller HV, Boekema EJ (2005) Structural characterization of a complex of photosystem I and light-harvesting complex II of Arabidopsis thaliana. Biochemistry 44 10935–10940 [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W, Wang DN, Fujiyoshi Y (1994) Atomic model of plant light-harvesting complex by electron crystallography. Nature 367 614–621 [DOI] [PubMed] [Google Scholar]
- Lemieux C, Otis C, Turmel M (2007) A clade uniting the green algae Mesostigma viride and Chlorokybus atmophyticus represents the deepest branch of the Streptophyta in chloroplast genome-based phylogenies. BMC Biol 5 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LA, McCourt RM (2004) Green algae and the origin of land plants. Am J Bot 91 1535–1556 [DOI] [PubMed] [Google Scholar]
- Li XP, Gilmore AM, Caffarri S, Bassi R, Golan T, Kramer D, Niyogi KK (2004) Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. J Biol Chem 279 22866–22874 [DOI] [PubMed] [Google Scholar]
- Li XP, Muller-Moule P, Gilmore AM, Niyogi KK (2002) PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc Natl Acad Sci USA 99 15222–15227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W (2004) Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428 287–292 [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Nishida K, et al (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428 653–657 [DOI] [PubMed] [Google Scholar]
- Muchhal US, Schwartzbach SD (1992) Characterization of a Euglena gene encoding a polyprotein precursor to the light-harvesting chlorophyll a/b-binding protein of photosystem II. Plant Mol Biol 18 287–299 [DOI] [PubMed] [Google Scholar]
- Naumann B, Stauber EJ, Busch A, Sommer F, Hippler M (2005) N-terminal processing of Lhca3 is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J Biol Chem 280 20431–20441 [DOI] [PubMed] [Google Scholar]
- Nield J, Kruse O, Ruprecht J, da Fonseca P, Büchel C, Barber J (2000) Three-dimensional structure of Chlamydomonas reinhardtii and Synechococcus elongatus photosystem II complexes allows for comparison of their oxygen-evolving complex organization. J Biol Chem 275 27940–27946 [DOI] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12 357–358 [DOI] [PubMed] [Google Scholar]
- Rhiel E, Mörschel E (1993) The atypical chlorophyll a/b/c light-harvesting complex of Mantoniella squamata: molecular cloning and sequence analysis. Mol Gen Genet 240 403–413 [DOI] [PubMed] [Google Scholar]
- Richard C, Ouellet H, Guertin M (2000) Characterization of the LI818 polypeptide from the green unicellular alga Chlamydomonas reinhardtii. Plant Mol Biol 42 303–316 [DOI] [PubMed] [Google Scholar]
- Rissler HM, Durnford DG (2005) Isolation of a novel carotenoid-rich protein in Cyanophora paradoxa that is immunologically related to the light-harvesting complexes of photosynthetic eukaryotes. Plant Cell Physiol 46 416–424 [DOI] [PubMed] [Google Scholar]
- Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ (2007) The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol Biol Evol 24 54–62 [DOI] [PubMed] [Google Scholar]
- Ruban AV, Wentworth M, Yakushevska AE, Andersson J, Lee PJ, Keegstra W, Dekker JP, Boekema EJ, Jansson S, Horton P (2003) Plants lacking the main light-harvesting complex retain Photosystem II macro-organization. Nature 421 648–652 [DOI] [PubMed] [Google Scholar]
- Savard F, Richard C, Guertin M (1996) The Chlamydomonas reinhardtii LI818 gene represents a distant relative of the cabI/II genes that is regulated during the cell cycle and in response to illumination. Plant Mol Biol 32 461–473 [DOI] [PubMed] [Google Scholar]
- Schmid VH, Cammarata KV, Bruns BU, Schmidt GW (1997) In vitro reconstitution of the photosystem I light-harvesting complex LHCI-730: heterodimerization is required for antenna pigment organization. Proc Natl Acad Sci USA 94 7667–7672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider SU, Leible MB, Yang XP (1989) Strong homology between the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase of two species of Acetabularia and the occurrence of unusual codon usage. Mol Gen Genet 218 445–452 [DOI] [PubMed] [Google Scholar]
- Six C, Worden AZ, Rodriguez F, Moreau H, Partensky F (2005) New insights into the nature and phylogeny of prasinophyte antenna proteins: Ostreococcus tauri, a case study. Mol Biol Evol 22 2217–2230 [DOI] [PubMed] [Google Scholar]
- Stauber EJ, Fink A, Markert C, Kruse O, Johanningmeier U, Hippler M (2003) Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryot Cell 2 978–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulli C, Schwartzbach SD (1996) A soluble protein is imported into Euglena chloroplasts as a membrane-bound precursor. Plant Cell 8 43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Iwai M, Takahashi Y, Minagawa J (2006) Identification of the mobile light-harvesting complex II polypeptides for state transitions in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 103 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Yasui TA, Stauber EJ, Hippler M (2004) Comparison of the subunit compositions of the PSI-LHCI supercomplex and the LHCI in the green alga Chlamydomonas reinhardtii. Biochemistry 43 7816–7823 [DOI] [PubMed] [Google Scholar]
- Teramoto H, Nakamori A, Minagawa J, Ono TA (2002) Light-intensity-dependent expression of Lhc gene family encoding light-harvesting chlorophyll-a/b proteins of photosystem II in Chlamydomonas reinhardtii. Plant Physiol 130 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutsu R, Teramoto H, Takahashi Y, Ono TA, Minagawa J (2004) The light-harvesting complex of photosystem I in Chlamydomonas reinhardtii: protein composition, gene structures and phylogenic implications. Plant Cell Physiol 45 138–145 [DOI] [PubMed] [Google Scholar]
- Wentworth M, Ruban AV, Horton P (2004) The functional significance of the monomeric and trimeric states of the photosystem II light harvesting complexes. Biochemistry 43 501–509 [DOI] [PubMed] [Google Scholar]
- Wolfe GR, Cunningham FX, Durnford DG, Green BR, Gantt E (1994) Evidence for a common origin of chloroplasts with light-harvesting complexes of different pigmentation. Nature 367 566–568 [Google Scholar]
- Yakushevska AE, Keegstra W, Boekema EJ, Dekker JP, Andersson J, Jansson S, Ruban AV, Horton P (2003) The structure of photosystem II in Arabidopsis: localization of the CP26 and CP29 antenna complexes. Biochemistry 42 608–613 [DOI] [PubMed] [Google Scholar]