Abstract
A network of enzymatic and nonenzymatic antioxidants protects chloroplasts from photooxidative damage. With all enzymatic components being nuclear encoded, the control of the antioxidant capacity depends on chloroplast-to-nucleus redox signaling. Using an Arabidopsis (Arabidopsis thaliana) reporter gene line expressing luciferase under control of the redox-sensitive 2-cysteine peroxiredoxin A (2CPA) promoter, six mutants with low 2CPA promoter activity were isolated, of which five mutants show limitations in redox-box regulation of the 2CPA promoter. In addition to 2CPA, the transcript levels for other chloroplast antioxidant enzymes were decreased, although a higher oxidation status of the ascorbate pool, a higher reduction state of the plastoquinone pool, and an increased oxidation status of the 2-Cys peroxiredoxin pool demonstrated photooxidative stress conditions. Greening of the mutants, chloroplast ultrastructure, steady-state photosynthesis, and the responses to the stress hormone abscisic acid were wild type like. In the rosette state, the mutants were more sensitive to low CO2 and to hydrogen peroxide. Comparison of gene expression patterns and stress sensitivity characterizes the mutants as redox imbalanced in the regulation of nuclear-encoded chloroplast antioxidant enzymes and differentiates redox signaling cascades.
Photosynthesis provides a strong reducing power and a high risk for generation of reactive oxygen species (ROS) particularly under environmental constrains (Foyer et al., 1994). Accumulation of cytotoxic ROS is antagonized by superoxide dismutases, stromal- and thylakoid-bound ascorbate peroxidases (sAPxs and tAPxs), monodehydroascorbate and dehydroascorbate reductases (MDHARs and DHARs), glutathione reductases (GRs), peroxiredoxins (Prxs), and nonenzymatic reactions with the low Mr antioxidants ascorbate (Asc) and glutathione (GSH) (Baier and Dietz, 1999a; Asada, 2000; Dietz et al., 2006). Under unfavorable conditions, the biosynthesis and the activity of these antioxidants increase (Foyer et al., 1994; Karpinski et al., 1997; Kliebenstein et al., 1998; Baier et al., 2000; Rossel et al., 2002; Horling et al., 2003; Mittler et al., 2004) and stabilize the chloroplast redox poise (Foyer et al., 1994; Asada, 2000).
Sequence analysis of plant plastomes and genomes revealed that all genes for chloroplast antioxidant enzymes and all enzymes involved in the biosynthesis and the regeneration of low Mr antioxidants are nuclear encoded. Therefore, regulation of gene expression depends on chloroplast-to-nucleus signaling. Depending on the deviation from the cellular redox homeostasis different signaling pathways have been hypothesized (Baier and Dietz, 1999a, 2005; Pfannschmidt et al., 2001; Apel and Hirt, 2004; Foyer and Noctor, 2005). Class 1 signaling originates from specific redox pairs in the photosynthetic electron transport chain, such as reduced and oxidized plastoquinone. Class 2 signaling depends on the redox state of stromal redox pairs, such as the thioredoxin, GSH, and Asc pools, and class 3 signaling is triggered by ROS (Dietz, 2003; Pfannschmidt, 2003). Experimentally, nuclear expression of chloroplast proteins has been shown to tentatively correlate with redox shifts in the thioredoxin, NAD(P)/NAD(P) H, plastoquinone, GSH, and Asc pools (summarized in Baier and Dietz, 2005; Foyer and Noctor, 2005) on a high background level, which is coordinated with chloroplast development and light signals (Pena-Ahumada et al., 2006).
The Arabidopsis (Arabidopsis thaliana) gene for 2-Cys Prx-A (2CPA; Baier and Dietz, 1997) serves as a model in the analysis of chloroplast-to-nucleus redox signaling (Baier et al., 2004b). It encodes a chloroplast peroxidase (König et al., 2002), which protects the photosynthetic membrane against oxidative damage (Baier and Dietz, 1999b). The 314 bp promoter core coordinates 2CPA transcription with leaf development (Baier et al., 2004b). Upstream of the core promoter, a 216 bp cis-element provides responsiveness to redox signals (Baier et al., 2004b) and regulates 2CPA expression depending on the redox state of the electron acceptors of PSI (Baier et al., 2004b). Putative signals are the redox states of ferredoxin (Fd), stromal thioredoxins, NAD(P) H, and the Asc and the GSH systems (Baier and Dietz, 2005), which are class 2 redox signals according to the classifications by Pfannschmidt (2003) and Dietz (2003).
The induction of genes encoding antioxidant enzymes upon oxidative stress conditions was observed in various studies with transgenic plants and mutants (Baier et al., 2000; Mittler et al., 2004; Vandenabeele et al., 2004). In signal transduction, chloroplast redox signals interplay with the antioxidant activity of the regulated genes. For example, antisense suppression of 2CPA expression specifically increases expression of sAPx, tAPx, and MDHAR, which counteract the loss of 2-Cys Prx function (Baier et al., 2000). However, tight linkage between enzymatic and nonenzymatic reactions (Foyer et al., 1994; Fridlyand and Scheibe, 1999) limits experimental signal differentiation. Thus, the first models on chloroplast-to-nucleus signal transduction were based on selective experimental data that need further substantiation by genetic means (Pfannschmidt, 2003; Baier and Dietz, 2005; Foyer and Noctor, 2005; Baier et al., 2006).
To address the redox signaling pathway(s) involved in the expressional control of the chloroplast antioxidant system, a mutant screen was performed in Arabidopsis and mutants were isolated based on decreased transcriptional activity of 2CPA. Low transcript levels of other genes with compensatory function indicate expressional coregulation. Here, five mutants are presented in which redox regulation of 2CPA through the 216 bp redox active promoter domain is affected, and one with decreased activation of the core promoter. It is shown that the mutational defects impact on the regulation of various genes for chloroplast antioxidant enzymes and cause higher sensitivity of the mutants to hydrogen peroxide (H2O2) and low CO2.
RESULTS
Isolation of Mutants Affected in 2CPA Gene Expression
To isolate mutants affected in transcriptional regulation of a chloroplast antioxidant enzyme, a mutant screen was performed in transgenic Arabidopsis expressing luciferase under control of the 2CPA promoter (Baier et al., 2004b). Previous characterization of the reporter gene line (Baier et al., 2004b) demonstrated that transgene regulation copies 2CPA transcript abundance regulation. For mutagenesis, the line T19-2 was selected, which carries a single insertion of the reporter gene on the bottom arm of chromosome III close to bacterial artificial chromosome MQC12.
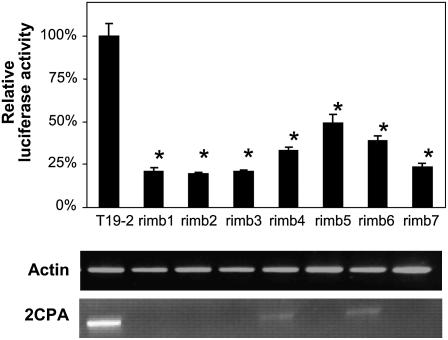
Seeds of the homozygous reporter gene line T19-2 were treated for 5 h with 50 mm ethyl methanesulfonate to introduce random point mutations. Twenty-thousand seedlings of the M2 progeny were grown aseptically in constant light. At 10 d, the seedlings were individually screened for decreased in vivo luciferase activity. At this stage of seedling development, in wild-type plants 2CPA promoter activity is strong in primary leaves and hardly detectable in cotyledons (Baier et al., 2004b). Plants with less than two-thirds of the reporter gene activity of the parental line were selected for further analysis (Fig. 1). To exclude artifacts due to lower luminescent areas or unspecific defects in chloroplast development, only plants were selected that were indistinguishable from the parental line at screening age in respect of size and color. The inheritance of the low reporter gene activity was tested in the progenies under the same conditions. In seven lines the decrease in luciferase expression was confirmed (Fig. 1).
Figure 1.
Gene expression in 10-d-old rimb mutants. The relative luciferase activity of 2CPA∷Luc and the transcript abundance of the endogenous 2CPA gene in 10-d-old seedlings expressing luciferase under control of the 2CPA promoter. The luciferase activity was determined in vivo 5 min after spraying the seedlings with luciferin (n = 28–45; mean ± se). Significant differences to T19-2 (Student's t test; P < 0.001) are indicated by asterisks. For RT-PCR analysis the cDNA samples were standardized on actin-2 transcript amount.
Scoring the luciferase activity in the F2 progeny of the backcrosses to the parental line T19-2 in 10-d-old seedlings demonstrated segregation of single recessive Mendelian loci (Supplemental Fig. S1). Strongest penetrance of the phenotype was observed in rimb1, rimb6, and rimb7 (Supplemental Fig. S1).
Because redox imbalanced (rimb) 2 and rimb3 were originally isolated from the same seed pool, they were tested for genetic independence by mapping the mutations to chromosome arms. One-thousand seedlings of the F2 population of the mutants with Arabidopsis var. Landsberg erecta (Ler) wild type were screened and rescreened for low luciferase activity and genotyped with simple sequence length polymorphic markers designed against sequence length polymorphisms between the Arabidopsis ecotypes Columbia-0 and Ler. Mapping of rimb3 to the middle section of the bottom arm of chromosome IV and rimb2 to the top of the top arm of chromosome III demonstrated genetic independence.
Trans-Activity of the Mutations
To test whether the mutations act in trans, reverse transcription (RT)-PCR analysis of 2CPA transcript amount was performed with cDNA samples standardized on actin-2 transcript amount (Fig. 1). Like the reporter gene activity, the transcript levels of the endogenous 2CPA gene were decreased in 10-d-old seedlings grown under screening conditions. Due to different locations of the reporter and the 2CPA gene on chromosome III and lack of transcript sequence similarity, it can be concluded that the mutations act on the regulatory elements of the 2CPA promoter, which the reporter gene and the endogenous gene have in common.
Photosynthetic Electron Transport in the rimb Mutants
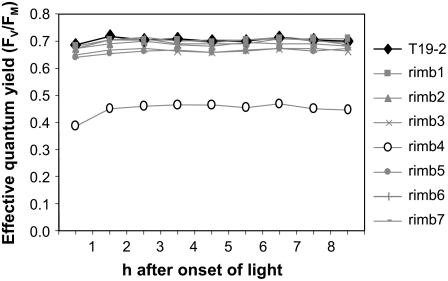
Regulation of 2CPA transcription correlates with the photosynthetic electron pressure at the acceptor site of PSI (Baier et al., 2004b). To analyze whether the mutants are affected in photosynthetic electron transport, steady-state chlorophyll-a fluorescence patterns were determined. The effective quantum yield (ΦPSII = FV′/FM′) reflects the efficiency by which PSII transforms excitation energy into chemical energy (Schreiber and Bilger, 1993). In 3-week-old rimb mutants, ΦPSII was determined throughout the day with single saturating light flashes (every 60 min, beginning 30 min after onset of light). All mutants, except the highly chlorotic rimb4, showed a wild-type-like ΦPSII around 0.7 (Fig. 2). In rimb4, it was reduced to 0.387 to 0.466 (Fig. 2), demonstrating a severe mutational impact on photosynthetic performance. As photosynthetic electron transport controls 2CPA promoter activity (Baier et al., 2004b), rimb4 was excluded from the further analysis.
Figure 2.
Photosynthetic performance of the rimb mutants. Effective quantum yield of PSII (FV′/FM′) in 3-week-old rimb1 to rimb7 and T19-2 plants grown on soil between 0.5 and 8.5 h after onset of light (n = 9).
Target Element in the 2CPA Promoter
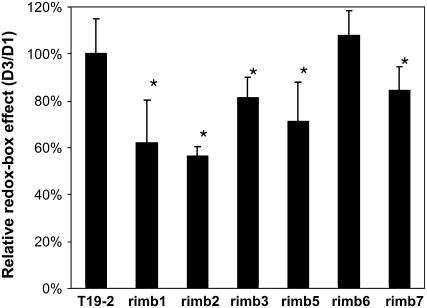
In the 2CPA promoter, regulation of the 314 bp minimal promoter coordinates the expression intensity with chloroplast development, while redox regulation takes place on the 216 bp redox box located upstream of the minimal promoter (Baier et al., 2004b). To distinguish whether the mutants are affected in regulation of the minimal promoter or in regulation of the redox box, mesophyll protoplasts isolated from 3-week-old mutants and the parental line were transfected with equal amounts of β-glucuronidase (GUS)-reporter gene plasmids encoding glucuronidase under control of either the 314 bp minimal promoter (D1) or the 530 bp promoter fragment (D3), which includes the 216 bp redox box (Baier et al., 2004b). The GUS activity was quantified after 6 h incubation at a light intensity of 50 μmol quanta m−2 s−1 (Fig. 3). For comparison, the protoplast preparation and DNA quality were standardized by setting the ratio of D3 and D1-GUS expression in T19-2 protoplasts to 100%. In rimb1, rimb2, rimb3, rimb5, and rimb7 less activation of the longer promoter fragment than the 314 bp promoter of the redox box resulted in lower D3/D1 ratios. In rimb6 the D3/D1 ratio was similar to T19-2, demonstrating that the mutational defects do not specifically affect regulation of the redox box (Fig. 3).
Figure 3.
Relative activation of the redox box. The relative GUS-reporter gene activity controlled by the 530 bp 2CPA promoter fragment containing the redox-sensitive promoter domain (D3) was quantified in protoplasts isolated from the parental line T19-2 and the mutants and standardized on the GUS-reporter gene activity measured under control of the 314 bp minimal promoter (D1) relative to the expression intensity in T19-2 (mean ± sd). Significant changes (Student's t test; P < 0.05) are indicated by an asterisk.
Abscisic Acid Responsiveness of the Mutants
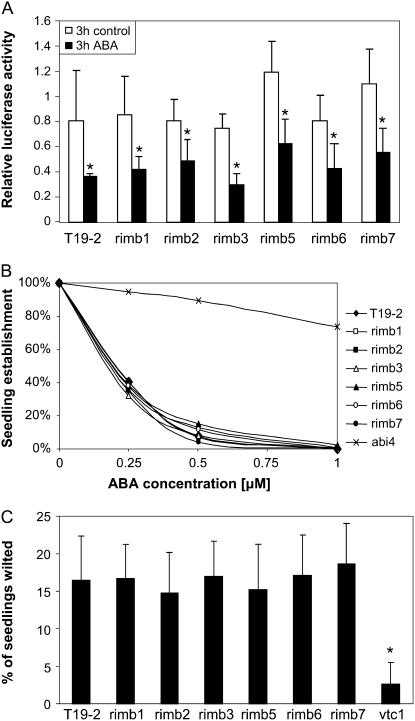
In wild-type plants, the 216 bp redox-sensitive promoter fragment also mediates abscisic acid (ABA) responsiveness (Baier et al., 2004b). To test whether low 2CPA expression resulted from increased ABA responsiveness, 9-d-old seedlings grown in 50 μmol quanta m−2 s−1 were screened for luciferase activity 3 h after application of 40 μm ABA to 100 μL medium. Like in T19-2, the luciferase activity was decreased in all mutants by approximately 60% (Fig. 4). In addition, no differences between T19-2 and the mutants were observed in physiological tests for ABA responsiveness, such as germination on 0 to 1 μm ABA (Koornneef et al., 1984) and wilting (Finkelstein, 1994), with six independent seed batches (Fig. 4).
Figure 4.
Screening of the rimb mutants for increased ABA sensitivity. A, In vivo luciferase activity after 4 h illumination at 50 μmol quanta m−2 s−1 in presence and absence of ABA. After 10 d growth on 100 μL solid Murashige and Skoog medium supplemented with 0.5% (w/v) Suc in microtiterplates, 20 μL of 40 μm ABA were applied to half of the seedlings 1 h after onset of light (mean ± sd; n = 36). Significant changes (Student's t test; P < 0.05) are indicated by asterisks. B, Sensitivity of seed germination to ABA. Root emergence was quantified after 7 d on media containing 0 to 1 μm ABA, which inhibits seed germination (six independent seed batches with at least 150 seeds). C, Percentage of seedlings with wilted cotyledons 4 h after opening the petri dishes at a relative humidity of 50%. The plant material was adapted to high humidity by growth on sterile media in water saturated petri dishes (n = 370–488 cultivated in eight batches; mean ± sd).
Light Regulation of 2CPA Promoter Activity
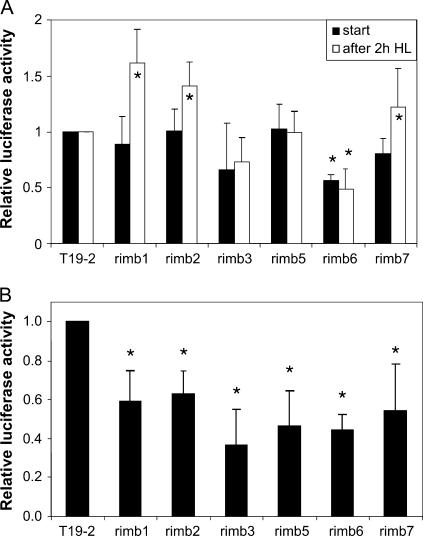
Under low light conditions of 50 μmol quanta m−2 s−1 1 h after onset of illumination, the reporter gene activity was not significantly different from that in the parental line T19-2 (Fig. 5A). Two hour illumination with 400 μmol quanta m−2 s−1 increased the relative reporter gene activity in rimb1, rimb2, and rimb7, demonstrating light intensity-dependent induction of gene expression (Fig. 5A).
Figure 5.
Light regulation of 2CPA promoter activity. A, Relative luciferase activity in the rimb mutants after 2 h illumination of 10-d-old seedlings adapted to 50 μmol quanta m−2 s−1 with 400 μmol quanta m−2 s−1 standardized on the luciferase activity observed in the parental line T19-2 (white bars) compared to the relative luciferase activity prior to the increased light treatment (black bars; mean ± sd). Significant changes (Student's t test; P < 0.05) are indicated by an asterisk. B, Relative luciferase activity in the rimb mutants after 5 h illumination of 10-d-old seedlings adapted to 100 μmol quanta m−2 s−1 with 400 μmol quanta m−2 s−1 standardized on the luciferase activity observed in the parental line T19-2 (mean ± sd). Significant changes (Student's t test; P < 0.05) are indicated by asterisks.
In seedlings adapted to 100 μmol quanta m−2 s−1, which is in our growth chambers a light intensity yielding optimal biomass production, 5 h illumination with 400 μmol quanta m−2 s−1 decreased the reporter gene activity stronger in all mutants than in T19-2, indicating stronger responses to photosynthetic signals (Fig. 5B).
Content and Redox State of Low Molecular Weight Antioxidants
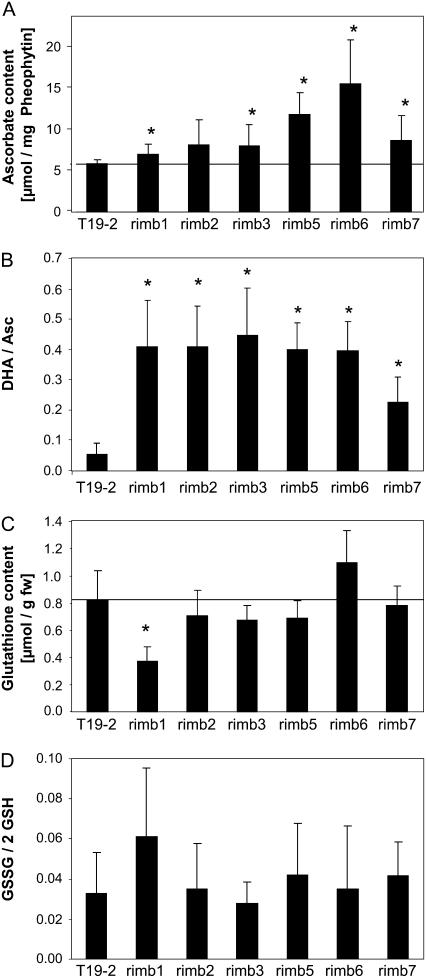
The redox state and the pool size of low Mr antioxidants and cellular thiols are indicators for the cellular redox state (Foyer et al., 1994). In all mutants with low 2CPA promoter activity the Asc pool was more oxidized than in the parental line (Fig. 6B). In rimb1, rimb3, rimb5, rimb6, and rimb7, the Asc pool size was increased (Fig. 6A). The GSH pool size was decreased in rimb1 (Fig. 6C), while in rimb2, rimb3, and rimb5 trends toward slightly less GSH were observed. However, in all mutants except rimb1, the GSH pool was kept reduced, demonstrating that the shift in the cellular redox poise was almost specific to Asc, while the redox state of the major cellular thiol pool was kept widely balanced.
Figure 6.
Content and redox states of Asc and GSH in 3-week-old rimb mutants grown on soil under long-day illumination (14 h light/10 h dark; n = 6–10; mean ± sd). Significant differences to T19-2 (Student's t test; P < 0.05) are indicated by asterisks.
Content and Redox State of 2-Cys Prx
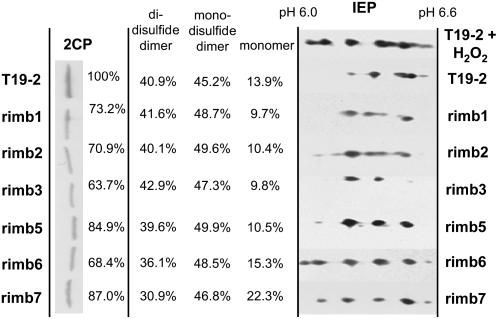
In all rimb mutants, consistent with decreased transcript abundance, the 2-Cys Prx protein level was decreased. As a marker for oxidative stress inside chloroplasts, the redox state of 2-Cys Prx was determined by one-dimensional (1-D) and two-dimensional (2-D) gel electrophoresis. Depending on the redox state and the regeneration potential by thiol-disulfide isomerases such as thioredoxins, 2-Cys Prx dimers can be covalently linked by none, one, or two disulfide bonds (König et al., 2002). For analysis of intermolecular disulfide bonds, proteins were immediately extracted in SDS-containing buffer and analyzed on 20% polyacrylamide gels. Compared to the parental line T19-2, in the rimb mutants increased relative amounts of dimers were detected, which are linked by only one disulfide bond (monodisulfide dimer; Fig. 7). In rimb1, rimb2, rimb3, and rimb5, the amount of noncovalently linked 2-Cys Prx (dissociated into monomers) was decreased, demonstrating a higher propensity to form intermolecular disulfide bonds, such as by insufficient reductive regeneration of the active site (König et al., 2002, 2003). In contrast, in rimb6 and rimb7 the amount of the noncovalently linked 2-Cys Prx form was increased, indicating either higher 2-Cys Prx regeneration or accumulation of sulfinic acid derivates of 2-Cys Prx protein.
Figure 7.
Content and redox states of 2-Cys Prx in 3-week-old rimb mutants. Left: The total 2-Cys Prx protein amount was determined in DTT-treated plant samples by western-blot analysis with 2-Cys Prx antibody and quantification of the band intensities. Middle: Intermolecular disulfide bond formation was analyzed in protein samples extracted immediately in SDS-containing buffer by 20% SDS-PAGE, western-blot analysis with 2-Cys Prx-specific antibodies, and quantification of band intensities. Right: The pI of 2-Cys Prx was determined by 2-D PAGE with immobilized pH gradients (pH 4–7) and western-blot analysis with 2-Cys Prx-specific antibodies.
For differentiation between reduced and oxidized 2-Cys Prx, the proteins were separated electrophoretically according to their pI after reductive cleavage of the disulfide bonds with dithiothreitol (DTT) and freezing of the thiol groups by iodoacetamide (Sheehan, 2006). For reduced 2-Cys Prx an pI (IEP) of 6.54 was determined. In the parental line, 45% of the protein had an IEP of 6.32 and a small portion of 2-Cys Prx one of about 6.21. Oxidation of fresh protein extracts with H2O2 partially oxidized the protein, resulting in IEPs of 6.21, 6.12, and 6.05.
In all rimb mutants the amount of the oxidized 2-Cys Prx form with an IEP of 6.21 was increased and that of the reduced form immobilizing at pH 6.54 was decreased. In addition, in rimb2, rimb5, rimb6, and rimb7 oxidized 2-Cys Prx forms accumulated, which were only detected in H2O2-treated protein samples of the parental line T19-2, but not in untreated T19-2 samples.
Growth Habit of the Mutants
Although indistinguishable at screening age, from 2 weeks onwards the mutants grew slower than the parental line under long days (14 h light). In plants older than 3 to 4 weeks, under long-day illumination, early leaf senescence (rimb1) and chlorosis (rimb3 and rimb5) developed randomly and starch (especially in rimb5 and rimb6) and anthocyanins (especially in rimb6) accumulated (Table I). The strongest phenotype was observed in rimb4, which turned highly chlorotic soon after expansion of the primary leaves. Indicating severe pleiotropic defects, including decreased photosynthetic activity (Fig. 2), rimb4 was excluded from further analysis. By contrast, when grown under short-day illumination (10 h light), only subtle phenotypic differences were observed in the other rimb mutants (data not shown).
Table I.
Protein, chlorophyll, anthocynanin and starch contents of 3-week-old rimb mutants grown under long-day illumination (14 h light/10 h dark; harvest: 3 h after onset of light)
Significant differences to T19-2 (Student's t test; P < 0.05) are indicated by an asterisk.
| Protein | Chlorophyll | Anthocyanin E530 corr | Starch | |
|---|---|---|---|---|
| mg g−1 fresh weight | mg g−1 fresh weight | g−1 fresh weight | μmol Glc g−1 fresh weight | |
| T19-2 | 12.58 ± 0.62 | 0.769 ± 0.084 | 19.40 ± 1.36 | 5.21 ± 1.39 |
| rimb1 | 12.52 ± 0.38 | 0.761 ± 0.046 | 21.36 ± 0.95* | 6.60 ± 1.53* |
| rimb2 | 12.85 ± 0.19 | 0.722 ± 0.063 | 21.01 ± 1.43* | 7.08 ± 1.50* |
| rimb3 | 12.29 ± 0.57 | 0.641 ± 0.057* | 21.07 ± 2.57 | 12.54 ± 3.15* |
| rimb5 | 12.28 ± 1.02 | 0.604 ± 0.101* | 23.87 ± 2.46* | 23.31 ± 3.37* |
| rimb6 | 12.82 ± 1.27 | 0.727 ± 0.055 | 32.44 ± 4.27* | 29.16 ± 4.92* |
| rimb7 | 13.02 ± 0.91 | 0.766 ± 0.099 | 20.00 ± 1.29 | 8.68 ± 2.61* |
| n | 7 | 5 | 8 | 10 |
Chloroplast Ultrastructure
Redox imbalances can affect chloroplast development, which can influence gene expression of chloroplast proteins (Rizhsky et al., 2003). Therefore, the chloroplast ultrastructure was analyzed in the rimb mutants. In young mutant leaves, which show the most pronounced differences in 2CPA transcript amount compared to the parental line, the chloroplast ultrastructure was wild type like. Small starch granules were observed in chloroplasts of rimb1 and rimb5 (data not shown). At the mature stage, transitory starch accumulated in all rimb mutants. Most starch was observed in chloroplasts of rimb5, in which one big starch granule with low electron density was formed, and in rimb6, in which several granules with high electron density were observed. In parallel to starch accumulation, thylakoid stacking was decreased by one to three layers in rimb5 and rimb6, while rimb2 and rimb3 showed slightly stronger (2–3 layers) grana stacking than T19-2 (data not shown).
Performance of the Mutants in Low CO2
To test the performance of the mutants under stress conditions, 4-week-old plants were exposed to 50 ppm CO2 for 8 and 16 h, respectively. After the treatment, the plants were transferred to standard growth conditions (350 ppm CO2). Reduced CO2 availability restricts ribulose-1,5-bisphosphate carboxylation and favors ribulose-1,5-bisphosphate oxygenation. Here, the low CO2 treatment was started with the onset of light to avoid carbohydrate biosynthesis prior to activation of ribulose-1,5-bisphosphate oxygenation.
Low CO2 supports H2O2 and superoxide production by photorespiration (Ogren, 1984) and photoreduction of O2 (Fridlyand and Scheibe, 1999). Increased ROS generation demands for extra antioxidant protection. Consistently, the expression of many antioxidant enzymes, including 2CPA, is induced in wild-type plants and protects the leaves against photooxidative damage (Baier et al., 2004b; Wormuth et al., 2006).
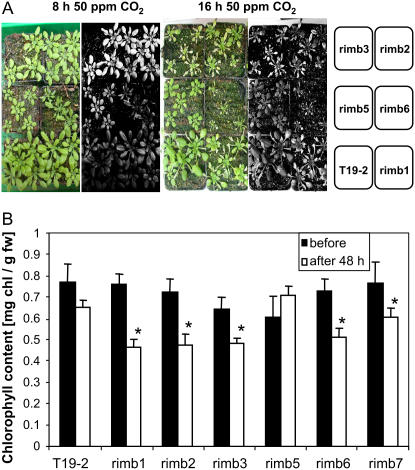
Two days after the 8 h CO2 deprivation treatment, strong chlorosis was observed in rimb2 and rimb3 (Fig. 8A, left). To a lesser extent the chlorophyll content decreased all over the leaf blades in rimb1, rimb5, and rimb6. Under the same conditions, it only decreased in the leaf tips in T19-2. Forty-eight hours after the 16 h treatment with 50 ppm CO2 (Fig. 8A, right) in rimb1, rimb2, rimb3, and rimb6, all leaves, which were exposed to low CO2 in the premature state, were highly chlorotic. Ninety-six hours after the treatment (data not shown) the leaves were completely bleached, while less leaves were affected in the parental line. Leaves formed during the postfumigation period and the leaves of rimb5, which had highest starch contents (Table I), developed no visible damage.
Figure 8.
Performance of the mutants under photorespiratory conditions. A, Habitus of five rimb mutants and T19-2 48 h after 8 or 16 h fumigation with 50 ppm CO2. The digital color images were taken 48 h after the end of the low CO2 treatment. The signal intensity of the green channel was squared using Photoshop CS and visualized in gray scale images. Chlorophyll contents (B) prior to 16 h fumigation (black bars) with 50 ppm CO2 and 48 h after the treatment (white bars; n = 6; mean ± sd). Significant changes (Student's t test; P < 0.05) are indicated by asterisks.
Effect of H2O2 Application on the Mutants
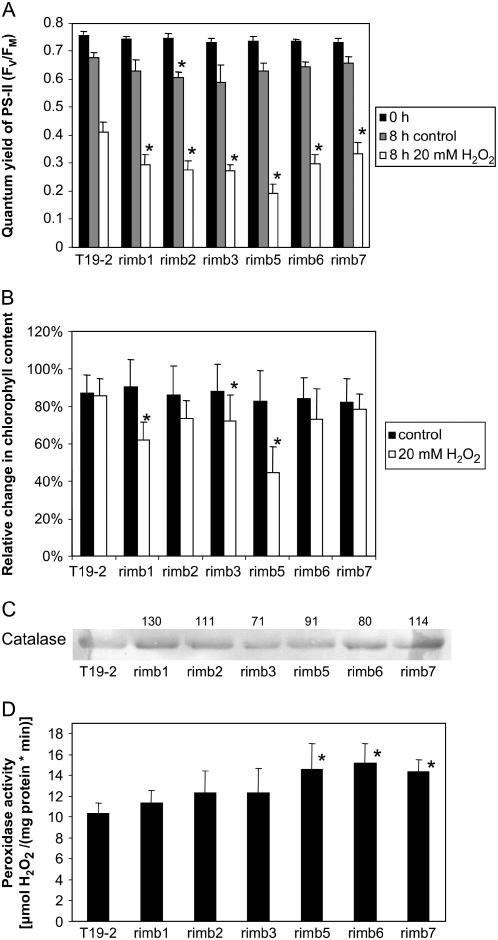
Because low CO2 availability can increase H2O2 production (Ogren, 1984), the mutants were tested for their sensitivity to exogenously supplied H2O2. Leaf discs of 5-week-old plants were floated for 8 h on 20 mm H2O2. ΦPSII was analyzed as an indicator for chloroplast damage. In the mutants, H2O2 application decreased ΦPSII stronger than in the parental line (Fig. 9A). In addition, chlorophyll degradation was enhanced (except in rimb7; Fig. 9B), demonstrating that the mutants are more sensitive to H2O2. Rimb5, which barely responded to low CO2 (Fig. 7), showed highest sensitivity to H2O2.
Figure 9.
Effect of H2O2 on the quantum yield of PSII (A) and the chlorophyll content (B). Leaf discs of 9 mm diameter were floated for 8 h on 20 mm H2O2 under constant illumination with 100 μmol quanta m−2 s−1. The catalase protein levels were analyzed immunochemically by western-blot analysis of 10 μg protein extracted from mature leaves of 3-week-old plants (C) and the protein amount was calculated relative to T19-2. D, Guaiacol peroxidase activity as indicator for extracellular and cytosolic peroxidase activity in 3-week-old rimb mutants harvested 1 h after onset of light (n = 6). Significant differences to T19-2 (Student's t test; P < 0.05; mean ± sd) are indicated by asterisks.
To analyze whether the higher sensitivity to H2O2 might be caused by decreased catalase levels, catalase protein amounts were detected immunochemically in samples standardized on the same protein amount. In the rimb mutants the catalase levels were wild type like or slightly increased (Fig. 9C). In addition, guaiacol peroxidase activity was increased in the mutants, demonstrating higher extracellular and cytosolic peroxidase activity (Fig. 9D), while 2-Cys Prx protein levels were decreased (Fig. 7).
Gene Expression in Mature Leaves
Transcript Amount Regulation Patterns in Wild-Type Arabidopsis
To identify candidate genes for transcript analysis in the mutants, transcript amount data for 223 putatively redox regulated genes were selected from 249 publicly available Affimetrix Chip experiments with Arabidopsis premature and mature leaves. Pearson correlation coefficients were calculated for all combinations of selected genes, which cover all genes for Halliwell-Asada-Cycle enzymes (chloroplast superoxide dismutases, Asc peroxidases, GR, and MDHAR and DHAR), Prxs, the small electron carrier proteins thioredoxins, glutaredoxins, and cyclophilins and nuclear-encoded components of the photosynthetic electron transport chain. In addition, a set of genes for cytosolic enzymes with expressional response to the carbohydrate availability (e.g. triose phosphate isomerase and Glc-6-P dehydrogenase) and extraplastidic antioxidant enzymes (e.g. cytosolic GR and the three catalases) were included as references.
Closest coregulation with 2CPA (At3g11630) was observed for chloroplast cyclophilin AtCyp20-3 (At3g62030; Kcorr 2CPA = 96%; Supplemental Fig. S2), which supports the peroxidase activity of 2-Cys Prx (Laxa et al., 2007), followed by two cytochrome b6f subunits (PetC [At4g03280] and PetM [At2g26500]), plastocyanin PetE2 (At1g20340), two cab transcripts (Lhca2 [At3g61470] and Lhcb4.1 [At5g01530]), various thioredoxins (At1g43560, At4g03520, At1g50320, At3g15360, At3g02730, At3g02730, and At1g69880), two chloroplast glutaredoxins (Grx-CxxS14 [At3g54900] and Grx-CxxS12 [At2g20270]), Fd-thioredoxin reductase (At5g08410), and NADP-malate dehydrogenase (At5g58330). The only other antioxidant enzyme with a Kcorr 2CPA higher than 60% was PrxIIE (At3g52960; Kcorr 2CPA = 69.6%), which encodes, like 2CPA, a chloroplast Prx.
The transcript abundances of components of the Halliwell-Asada-Cycle, e.g. CuZn-SOD Csd2 (At2g28190) and tAPx (At1g77490) and sAPx (At4g08390), were negatively correlated with 2CPA transcript levels. This indicates either independent regulation or strong compensatory responses similar to gene expression regulation observed in transgenic and mutant plants with decreased activity of one specific antioxidant enzyme (Baier et al., 2000; Mittler et al., 2004; Vandenabeele et al., 2004). A distance tree drawn by equivalently weighting the correlation coefficients for all pairs of genes clustered the transcript abundance regulation of these genes into several distantly related groups (Supplemental Fig. S2). Bioinformatic comparison of promoter sequences by sequence similarity analysis by Cis-Regulatory Element Detection Online (http://mips.gsf.de/proj/regulomips) and comparison of putative transcription factor binding sites after MatInspector analysis (Cartharius et al., 2005) supported the hypothesis that each gene is regulated by an individual pattern of signals and, thus, is suited for mutant-based signal transduction analysis.
Regulation of Redox-Regulated Genes in the rimb Mutants
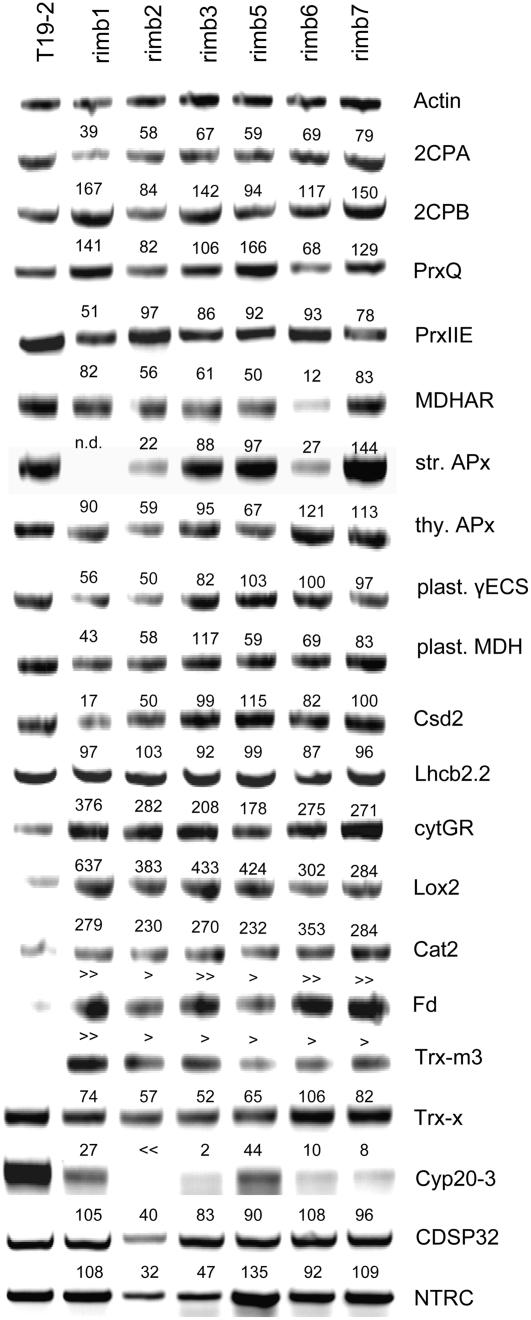
To study the impact of the mutations on gene expression regulation, RT-PCR was performed 1 h after onset of light with cDNA of 3-week-old plants adapted to 100 μmol quanta m−2 s−1. Samples from three independently grown batches of plant material were standardized on actin-2 transcript amount (At5g09810). The specificity of the primers (Table II) and the amplification reaction was confirmed by sequence analysis of the amplified products. The transcript levels of the four chloroplast Prxs 2CPA (At3g11630), 2CPB (At5g06290), PrxQ (At3g26060), PrxIIE (At3g52960), and that of sAPx (At4g08390) and tAPx (At1g77490), MDHAR (At1g63940), chloroplast γECS (At4g23100), chloroplast NADP-malate dehydrogenase (NADP-MDH; At5g58330), chloroplast CuZn-superoxide dismutase (Csd2; At2g28190), cytosolic GR (cytGR; At3g54660), lipoxygenase-2 (Lox2; At3g45140), Fd (PetF1; At1g60950), Lhcb2.2 (At2g05070), and catalase 2 (Cat2; At4g35090), and the genes for several proteins supporting 2-Cys Prx activity (Dietz et al., 2006), namely chloroplast thioredoxin-m3 (Trx-m3; A2g15570), thioredoxin-x (At1g50320; Collin et al., 2003), CDSP32 (At1g76080; Broin et al., 2002), and the NADPH-thioredoxin reductase C (NTRC; At2g41680; Moon et al., 2006), and the cyclophilin Cyp20-3 (At3g62030; Laxa et al., 2007) were tested for their transcript abundance in 3-week-old rimb mutants (Fig. 10).
Table II.
List of primers
| Gene | Gene Code | Sense Primer | Antisense Primer |
|---|---|---|---|
| Actin | At5g09810 | GAGAAGATGACTCAGATC | ATCCTTCCTGATATCGAC |
| Luciferase | ATTGACAAGGATGGATGGCTAC | AGGGAGACCGGCCTCGAGATCG | |
| 2CPA | At3g11630 | CTCTCCATCTGTTTCTTT | GTACCTTTTTCGTATCAT |
| 2CPB | At5g06290 | ATAGCTTCTTCTTCTTCC | CATGTGTTCAATCTTAGC |
| PrxQ | At3g26060 | TGGCTCCACACTCACTCA | TCAGGCTGGAACTGGTTG |
| PrxIIE | At3g52960 | ATGGCGACTTCTCTCTCC | AGTCCCACAGGCTTATCC |
| sAPx | At4g08390 | TGTTCCAGTTAGCTAGTG | GGTTGAGTAAATTAGGTGC |
| tAPx | At1g77490 | AATAGTTGCCTTGTCTGG | GGAATATATGATCACCACG |
| Cat2 | At4g35090 | AGCAACTTGCTTTCTGTC | TTAGATGCTTGGTCTCAC |
| Csd2 | At2g28190 | CTCCGTTCCTCTTTCAGC | GCGTCAAGCCAATCACAC |
| γ-ECS | At4g23100 | CCTATGTACTTTGCCTAC | CCTTATTCCGGAGACTCG |
| Fd | At1g60950 | CCACAAGCCATAATCCTC | AAAGCTGGTGAGTAGGTG |
| Cyt. GR | At3g54660 | GGATTTGTGTGAGTCTGC | TAATAGCCAGCTCATGTC |
| Lox2 | At3g45140 | CCTGATGAAGAACTGATC | AAGAGACAGAGATACAG |
| MDH | At5g58330 | ATGAAGAGATCCAAGAGC | CTACTTCAATGCCATGTTC |
| MDHAR | At1g63940 | TTGGAATTGGAGCTAAGC | TTCTTCGACTGAAGATGC |
| CDSP32 | At1g76080 | CCGGACCAGTTAATGGGGAG | AAGTGACACGAACGCCTGAG |
| Trx-m3 | At2g15579 | TTCTGCTCTTCAAGAACG | AGGCAACAACAGTAGTAAC |
| Trx-x | At1g50320 | CTAGTTCGGTGATAAGATGC | ATTGAGTTCAAGAGACCATC |
| Cyp20-3 | At3g62030 | TGACCGATGCATTGTCTTTG | AAGCTCGAGCCAATTGTTCA |
| NTRC | At2g41680 | ATCGTCAGTTATTGGAAGGC | CTCTTTCTTCATCTTCACAC |
Figure 10.
Transcript analysis in the rimb mutants by gene-specific RT-PCR. The transcript abundances of selected genes were analyzed in the rimb mutants and the parental line T19-2 by RT-PCR in cDNA samples standardized on actin-2 transcript amount. The numbers give the means of the percentage of signal intensity relative to the T19-2 in three independent experiments. For Fd and Trx-m3 very strong induction was detected, tentatively quantified and labeled with ≫ and >, respectively.
The 2CPA transcript amount was decreased in all rimb mutants demonstrating persistence of the mutational defects in the rosette stage. However, consistent with a low expression intensity of 2CPA in older tissues (Baier et al., 2000), the relative differences between T19-2 and the mutants were less than at screening age (Fig. 1). In parallel to low 2CPA expression, the transcript levels of Cyp20-3, Trx-x, PrxIIE, chloroplast MDHAR, and NADPH-MDH were decreased in all rimb mutants (highest Spearman correlation). The mRNA levels of Lhcb2.2 were unchanged and that of Fd (At1g60950), Trx-m3 (At2g15579), cytGR (At3g54660), Lox2 (At3g45140), and peroxisomal Cat2 (At4g35090) were increased. The other genes showed mutant-specific expression regulation (Fig. 10). The strongest relative decrease in transcript abundance in all rimb mutants was observed for Cyp20-3. The Pearson correlation coefficient, which quantitatively weights the similarity in transcript abundance, was highest for γECS (Kcorr 2CPA = 0.841).
Comparing the mutants for transcript abundance regulation, Pearson correlation coefficients higher than 0.97 were observed between rimb1, rimb2, and rimb3, between rimb3 and rimb7, and between rimb6 and rimb7. Considering only the genes for chloroplast antioxidant enzymes the transcript abundance regulation of rimb1 and rimb2 (Kcorr = 0.627) separated from that of rimb3, rimb5, and rimb7 (Kcorr = 0.713–0.742), while rimb6 showed the most specific gene expression pattern.
DISCUSSION
To address pathways involved in the expressional regulation of chloroplast antioxidant enzymes, a mutant screen was performed in Arabidopsis, expressing luciferase under control of the 2CPA promoter. Six mutants were isolated with trans-active mutational defects in activation of 2CPA transcription (Fig. 1). All mutations were recessive (Supplemental Fig. S1), showed gradual differences in penetrance (Supplemental Fig. S1), and resulted in individually different phenotypes (Table I) and expression patterns (Fig. 10).
The rimb Mutants Represent a Novel Type of Mutant
Screening for decreased expression of a chloroplast antioxidant enzyme distinguishes the rimb mutants from mutants like rax1 (Ball et al., 2004) and axl8 (Rossel et al., 2006), which express cytosolic Asc peroxidase APx2 stronger than wild type. Rax1 carries a mutation in γ-glutamyl-cysteine synthase (Ball et al., 2004), limiting the first step of GSH biosynthesis (Mullineaux and Rausch, 2005). Thus, APx2 expression is induced by the decreased availability of the low Mr antioxidant GSH (Ball et al., 2004). In contrast, in the rimb mutants 2CPA promoter activity was decreased despite a higher oxidation state of the Asc pool (Fig. 6) and of oxidation of 2-Cys Prx protein (Fig. 7), which is a chloroplast-specific marker for oxidative stress (Baier and Dietz, 1997; König et al., 2003), demonstrating that gene expression regulation is uncoupled from redox sensing.
In knockdown lines of chloroplast CuZn superoxide dismutase (SOD-kd; Rizhsky et al., 2003) the expression of various nuclear-encoded chloroplast proteins involved in photosynthesis was decreased in response to oxidative chloroplast damage. In contrast, in the rimb mutants expression of cab genes (e.g. Lhcb2.2; Fig. 10) and chloroplast ultrastructure (data not shown) were unaffected. The quantum yields of PSII (ΦPSII = FV/FM) were wild type like, indicating high photosynthetic electron transport efficiency and a high oxidation state of intrinsic components of the photosynthetic electron transport chain (Schreiber and Bilger, 1993; Fig. 2). Consistent with not or much less inhibited photosynthesis in the rimb mutants, unlike the SOD-kd, only a subset of nuclear-encoded chloroplast proteins was affected in gene expression regulation (Fig. 10). Five mutants, namely rimb1, rimb2, rimb3, rimb5, and rimb7, were specifically limited in the activation of the recently identified 216 bp redox-sensitive promoter domain (Baier et al., 2004b; Fig. 3). The low activation of the redox box in five rimb mutants and the limitation in regulation of the minimal promoter in rimb6 resulted in decreased transcript abundance of several, but not all tested nuclear-encoded chloroplast enzymes (Fig. 10), suggesting that both types of mutants represent novel classes of Arabidopsis mutants suited for studying chloroplast-to-nucleus signaling.
rimb6
Transcript abundance regulation of rimb6 was most distinct from that of the other rimb mutants (Fig. 10). Since the activity of the 530 bp promoter element (D3) was decreased to the same extend as the 314 bp distal promoter element (D1; Fig. 3), rimb6 is not primarily affected in redox-box regulation. Comparison with transgene regulation of the parental line (Figs. 1 and 3) suggests regulatory defects in activation of the 314 bp promoter core, which contains R, G, and I boxes known for coordination of nuclear gene expression with chloroplast development upon greening and in dependence of light availability (Donald and Cashmore, 1990).
Chloroplast-nucleus coordination in greening Arabidopsis seedlings has been intensively studied in recent years using mutant approaches. However, unlike, for example, the genomes uncoupled mutants (Mochizuki et al., 2001), cab-underexpressed1 (Li et al., 1995), and immutans (Wu et al., 1999), rimb6 was not affected in greening (Fig. 9B) and chloroplast development (data not shown), and, unlike the fluorescence mutant (Meskauskiene et al., 2001), rimb6 did not show defects in chlorophyll biosynthesis upon greening of etiolated seedlings (data not shown). rimb6, like the other rimb mutants, is also distinct from Arabidopsis mutants with altered regulation of nuclear expression of chloroplast proteins, such as the high chlorophyll fluorescence mutants, which were screened for high chlorophyll fluorescence (Meurer et al., 1996), because photosynthetic electron transport (Fig. 2) was wild type like. It is therefore concluded that rimb6 represents a novel mutant type with disturbed coordination of nuclear expression of specific chloroplast proteins, such as MDHAR, Cyp20-3, and sAPx (Fig. 10).
Five of the rimb Mutants Are Affected in Redox Regulation of Gene Expression
The five other rimb mutants were limited in activation of the promoter elements that provide responsiveness to redox signals correlating with the electron pressure at the acceptor site of PSI (Baier et al., 2004b; Fig. 3). Consistent with this conclusion, a 5-fold increase of light on top of limited light availability (Fig. 5A) activated 2CPA promoter activity by stimulating the promoter core, while increased light availability on top of the regular growth light intensity (Fig. 5B) resulted in a lower level of relative reporter gene activation in rimb1, rimb2, rimb3, rimb5, and rimb7 than in the parental line.
Previously it was shown that redox regulation of the 2CPA redox box depends on ABA (Baier et al., 2004b). ABA-dependent suppression of 2CPA promoter activity (T19-2 in Fig. 4A) suggested that in the rimb mutants low 2CPA expression may result from increased responsiveness to ABA. However, no difference in ABA responsiveness of 2CPA promoter activity (Fig. 4A) and ABA-regulated phenotypes such as wilting (Fig. 4C) and germination (Fig. 4B) were observed in any rimb mutant.
The rimb mutants showed higher sensitivity to an increase in the light intensity (Fig. 5B), to low CO2 (Fig. 8, A and B), and to H2O2 (Fig. 9A) and developed stress phenotypes with leaf age (Table I). Consistently, increased oxidation of the Asc pool (Fig. 6B) and accumulation of oxidized 2-Cys Prx (Fig. 7) mirrored redox imbalances. In rimb1 and rimb3 the portion of noncovalently linked 2-Cys Prx was decreased, demonstrating that the redox imbalances mainly impacted on reductive regeneration of the thiol residues by thiol-disulfide isomerase, such as thioredoxin (König et al., 2002). In contrast, in rimb6 and rimb7, determination of the dimerization status and the pIs indicated increased amounts of monomers, in which the cysteinyl residues are hyperoxidized and cannot form disulfide bonds (König et al., 2003).
In rimb5, rimb6, and rimb7 increased amounts of 2-Cys Prx forms with IEPs between 6.05 and 6.22 are consistent with accumulation of Asc, which is an alternative marker for oxidative stress (Foyer et al., 1994). Stronger oxidation of the Asc pool and increased oxidation of 2-Cys Prx, but decreased transcript levels of various chloroplast antioxidant enzymes in rimb1, rimb2, rimb3, rimb5, and rimb7 (Fig. 10), illustrates defects in redox signaling.
Pearson correlation analysis of the transcript levels of nuclear-encoded chloroplast antioxidant enzymes grouped rimb1 with rimb2 and rimb3 with rimb5 and rimb7, indicating two subtypes of mutants. Stronger decreased transcript levels of 2CPA, sAPx, tAPx, γECS, and Csd2 suggest that the penetrance of the mutational defects on transcript abundance regulation is stronger in rimb1 and rimb2. It is concluded that rimb1 and rimb2 are mutated in more central elements of the signal transduction pathway or affected in stronger regulators than rimb3, rimb5, and rimb7.
Defects in Redox-Box Regulation Do Not Affect Typical ROS Pathways
Depending on the extent of deviation from cellular homeostasis, various kinds of signaling cascades have been postulated to mediate chloroplast-to-nucleus redox signaling. These range from sensing of the redox state of the plastoquinone pool to responsiveness to ROS signals (summarized in Pfannschmidt et al., 2001; Baier and Dietz, 2005; Foyer and Noctor, 2005). To date, most attention has been given to ROS-induced signaling cascades, which are involved in responses to pathogens and photooxidative stress (Mahalingam et al., 2003; Mateo et al., 2004; Suzuki and Mittler, 2005). Results from genome array hybridizations allowed selection of marker genes for specific types of ROS signals (op den Camp et al., 2003; Suzuki and Mittler, 2005; Gadjev et al., 2006). The increased oxidation states of the Asc pool (Fig. 6) and of 2-Cys Prx protein (Fig. 7) display deviation from the cellular redox homeostasis (Fig. 6). Up-regulation of transcript levels of Cat2 and Lox2 (Fig. 10; Desikan et al., 2001; Kiddle et al., 2003), of the singlet oxygen marker BAP1 (At3g61190) and of the superoxide/H2O2 marker gene Fer1 (At5g01600; op den Camp et al., 2003; data not shown) demonstrated activation of stress-induced genes in all rimb mutants. In addition, the transcript levels of Fd (At1g60950) and thioredoxin m3 (At2g15570) were increased, which encode chloroplast redox proteins involved in distribution of electrons from the photosynthetic electron transport chain, while 2CPA, PrxIIE, Cyp20-3, MDHAR, and Trx-x transcript levels were decreased (Fig. 10).
Lox2 expression is induced, for example, by insufficient antioxidant protection by Asc (Kiddle et al., 2003) and in light regimes promoting PSI activity (Fey et al., 2005), stimulating superoxide formation in the Mehler reaction (Mehler, 1951), and resulting in oxidation of the GSH pool (Fey et al., 2005). In signal transduction, jasmonate transmits the redox signals and activates Lox2 via the NPR1-dependent signaling pathway (Spoel et al., 2003). Consistent with the conclusion on independent redox signal transduction mechanisms drawn from the transcript abundance pattern depicted in Figure 10, 2CPA transcription is insensitive to jasmonate (and salicylate) (Baier et al., 2004b).
Compared to ROS signaling in pathogenesis and under excess photosynthetic excitation energy (summarized in Mullineaux et al., 2006; Torres et al., 2006), little is known about the mechanisms controlling nuclear expression of chloroplast antioxidant enzymes upon less severe redox imbalances (Baier and Dietz, 2005). Recently, Davletova et al. (2005) reported that in response to insufficient cytosolic H2O2 detoxification, the chloroplastic H2O2 scavenging system collapses by oxidative inhibition of the enzyme function. However, the steady-state level of APx activity was elevated during early light stress (Davletova et al., 2005). Since the activity of sAPx and tAPx is highly sensitive to H2O2 (Asada, 2000; Miyake et al., 2006), it implicates expressional induction of chloroplast APx. In the rimb mutants, the redox shift in the Asc pool (Fig. 6) and increased oxidation of 2-Cys Prx protein (Fig. 7) demonstrated intracellular redox imbalances. Like in 2-Cys Prx antisense lines, which resemble the rimb mutants in respect to (1) low 2CPA expression (Figs. 1, 8C, and 10), (2) the shift in the redox state of the Asc pool (Fig. 6), (3) an unchanged redox state of the GSH pool (Fig. 6), and (4) wild-type-like chlorophyll and protein levels in young tissues (Table I), an induction of sAPx, MDHAR, and/or tAPx transcript levels can be expected (Baier et al., 2000). However, only in rimb7 higher transcript levels were observed for the two APx genes (Fig. 10). In parallel, this mutant showed highest 2CPA transcript levels and strongest 2-Cys Prx reduction in the rosette state (Figs. 7 and 10), demonstrating that in rimb7 the (developmental) penetrance of the mutational defects is weak.
In the other rimb mutants induction of sAPx, tAPx, and MDHAR expression was either missing or only the transcript level of one of the genes was increased (Fig. 10). As shown in the Asc biosynthetic mutant vtc1 (Kiddle et al., 2003), tissue levels of Asc correlate with sAPx transcript levels and Asc decreases tAPx transcript abundance. Comparing vtc1 with wild-type plants, and adg1 and pgr1 mutants, Wormuth et al. (2006) described recently an 86% correlation of 2CPA transcript abundance with the oxidation state of the Asc pool. The Asc level was at least slightly increased in the rimb mutants and the Asc pool more oxidized (Fig. 6). Consequently, in the rimb mutants the correlation between Asc availability and the expression of sAPx, tAPx, and 2CPA (Kiddle et al., 2003; Wormuth et al., 2006) was broken. In contrast, the correlation between the Asc pool size and redox state was maintained for Lox2 expression (Kiddle et al., 2003). It is concluded that a specific signaling pathway is affected in the rimb mutants, which is distinct from induction of Lox2, Cat2, Fd, and Trx-m3.
Regulation of Several Chloroplast Antioxidant Enzymes Is Coupled
Even stronger than the 2CPA transcript levels, the relative transcript levels of Cyp20-3 were decreased, which encodes a cyclophilin promoting 2-Cys Prx activity (Laxa et al., 2007). The relative changes in transcript abundance were subjected to Pearson correlation analysis. Only a correlation coefficient of +0.491 was observed between 2CPA and Cyp20-3 in 3-week-old rimb mutants (Fig. 10), while it was 0.960 in wild-type plants (Supplemental Fig. S2). The difference suggests that the two genes are transcriptionally coregulated, but independently posttranscriptionally regulated.
In addition to Cyp20-3, the transcript levels of thioredoxin-x, MDHAR, PrxIIE, and tAPx were decreased in the five mutants that are affected in redox-box regulation (Figs. 3 and 10). Lack of sequence similarities excludes posttranscriptional regulation, such as control by microRNAs or transcript stabilizing proteins (Lee et al., 2006; Sunkar et al., 2006) and, therefore, demonstrates transcriptional coregulation.
The transcript levels of most other tested nuclear-encoded chloroplast antioxidant enzymes (2CPB, PrxQ, sAPx, and Csd2) and redox-active proteins (MDH, CDSP32, and NTRC) were differentially regulated in the various mutants. For 2CPB, PrxQ, sAPx, Csd2, MDH, CDSP32, and NTRC increased transcript levels in some mutants showed that the transcript levels are not exclusively dependent on the mutated signaling pathway, consistent with hypothesis on regulatory networks by transcript correlation analysis in wild-type Arabidopsis (Supplemental Figs. S2). For example, the 2CPB promoter contains strong ethylene response elements (data not shown), by which gene expression can secondarily and independently be induced (Fig. 10). Thus, comparative transcriptome analysis of the rimb mutants provides insight into the cross talk between parallel induced, compensatory signaling cascades in addition to demonstrating common links between the expressional regulation of nuclear encoded chloroplast proteins.
CONCLUSION
This report describes the isolation and biochemical characterization of novel mutants with decreased expression of 2CPA and other genes for chloroplast antioxidant enzymes. Low expression of various genes with compensatory regulation in wild-type plants (Figs. 1 and 10; Supplemental Fig. S2) demonstrates coregulation by common signaling pathways. For rimb1, rimb2, rimb3, rimb5, and rimb7 the mutational defects limited redox activation of the 2CPA promoter. Comparison of transcript abundance regulation in rimb1 and rimb2 with rimb3, rimb5, and rimb7 showed tight coregulation of MDHAR, γECS, Cyp20-3, Trx-x, and tAPx with 2CPA than sAPx and Csd2. The novel type of mutant presented here gives genetic evidence for the independence of lately defined redox signaling pathways (e.g. Pfannschmidt et al., 2001; Baier and Dietz, 2005; Foyer and Noctor, 2005; Gadjev et al., 2006) and differentiate signaling cascades. The cosuppression of genes for various chloroplast antioxidant enzymes under oxidizing conditions and the separation of redox-box regulation from induction of marker genes for ROS signaling, such as Cat2, Lox2, BAP1, and Fer1 (Kiddle et al., 2003; op den Camp et al., 2003), represent two steps toward understanding intercompartment redox signaling.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) wild-type plants and mutants were grown on soil under controlled conditions (long-day illumination: 14 h light [70–100 μmol quanta m−2 s−1] and 23°C/10 h dark and 18°C; 45% to 55% relative humidity) unless otherwise specified. Leaf discs were cut with an 8 mm cork screw from the leaf blades of mature leaves of 5-week-old plants and incubated upside up on the indicated medium in the light (100 μmol quanta m−2 s−1). For the CO2 fumigation experiment, the plants were transferred to a 28 L perspex chamber and fumigated with synthetic air controlled by a Millipore Tylan RO 7030 system. For in vitro growth, seeds were surface sterilized with 80% ethanol followed by 20% (v/v) household bleach, washed at least five times with sterile water, stratified at 4°C for 2 d in the dark, and germinated on Murashige and Skoog medium (Duchefa Biochemie BV) containing 0.5% or 1% (w/v) Suc, respectively. Plants were grown in constant light at 20°C and 10 to 400 μmol quanta m−2 s−1 or in cycling light as described for soil-grown plants. Effectors were applied to the medium.
Isolation of the rimb Mutants and Genetic Analysis
Approximately 200 mg seeds of a homozygous 2CPA:Luc reporter gene line (Baier et al., 2004b) were treated with a 50 mm ethyl methanesulfonate (Sigma) for 5 h. The mutagenized M1 seeds were sown in 100 pools on soil. The harvested M2 seeds were surface sterilized and grown in microtiter plates on Murashige and Skoog medium containing 1% Suc in continuous light. After 10 d growth a total of 20,000 m2 seedlings, representing 2,000 m1 plants were screened using a Victor Luminometer (Wallac, Perkin-Elmer). Five minutes after spraying with a 1 mm luciferin (Duchefa), 0.01% (v/v) Triton X-100 solution, luciferase activity was measured for 1 s. Seedlings showing lower luminescence than the parental line were selected for further analysis. The progenies were rescreened under the same conditions for confirmation of the inheritance of the low luciferase phenotype. To show the independence of the mutants, the mutations were mapped to chromosome arms from the F2 population of crosses to Arabidopsis Ler according to Jander et al. (2002). Selection and confirmation of low luciferase lines was performed as described for isolation from the mutagenized seedling population.
Gene Expression Analysis
RNA isolation and RT-PCR was performed as described in Baier et al. (2000). The cDNA samples were standardized on actin-2 transcript amount. The transcripts of the genes of interest were amplified using gene-specific primers. The primer sequences and Arabidopsis Genome Initiative codes are given in Table I. The signal intensities were quantified from the fluorogrammes of ethidium bromide stained DNA separated on agarose gels using the GELSCAN software package (BIOSCITECH) and normalized to the transcript abundance observed in the parental line. Representative examples from three independent experiments are shown.
Protein Extraction, 2-D and 1-D Gel Electrophoresis, and Western-Blot Analysis
For 1-D western-blot analysis mature leaves were extracted in 1.44% (w/v) glycine, 0.303% (w/v) Tris, 2% (w/v) SDS either in the presence or absence of 2% (v/v) 2-mercaptoethanol, separated in 20% resolving gels after addition of 1 volume 2× loading buffer and analyzed as described in Baier and Dietz (1997). Band intensities were quantified using the GELSCAN software package (BIOSCITECH).
For 2-D gel electrophoresis the plant material was extracted at the ratio of 1:10 with 50 mm Tris-HCl pH 8.0, 1 mm phenylmethylsulfonyl fluoride supplemented with 20 mm DTT or 20 mm H2O2, respectively. Following precipitation with trichloroacetic acid (Amme et al., 2006) the proteins were dissolved in sample buffer (8 m urea; 4% [w/v] CHAPS [3-{(3-cholamidopropyl) dimethylamonio}-1-propanesulfonate]; 60 mm DTT; 2% [v/v] Pharmalyte 4 to 7 [GE-Healthcare/Amersham]; and 0.002% [w/v] bromphenol blue). For each genotype, 350 μg protein were separated on 18-cm strips with immobilized pH gradient of 4 to 7 (GE Healthcare/Amersham) in an IPGphor unit (GE Healthcare/Amersham) using the following settings: 12 h 30 V, 2 h 60 V, 1 h 500 V, 1 h 1,000 V and 8,000 V for a total of about 60 kVh. After two 15 min equilibration steps in 50 mm Tris-HCl (pH 8.8), 6 m urea, 30% (v/v) glycerol, 2% (w/v) SDS, 0.01% (w/v) bromphenol blue, first supplemented with 1% (w/v) DTT and then with 2.5% (w/v) iodoacetamide, the stripes were mounted on 12% SDS-polyacrylamide gels (Baier and Dietz, 1997) and separated for 45 min at 50 mA. 2-Cys Prx protein was detected after western blotting (Baier and Dietz, 1997) with anti-2-Cys Prx antibody and SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Analysis of Expressional Regulation Patterns
For analysis of coregulation of 223 putatively redox-regulated genes, from the signal intensities published in 249 publicly available Affimetrix cDNA Chip experiments with premature and mature Arabidopsis leaf tissues (http://affymetrix.arabidopsis.info/AffyWatch.html), the Pearson correlation coefficients were calculated for all possible pairs of genes. For the distance plot (Supplemental Fig. S1) the data ranging from −1 to +1 were transformed by f(x) = (1 + x)/2 and analyzed with PAUP matrix using PHYLIP online (http://evolution.genetics.washington.edu/phylip/phylipweb.html) and plotted into a distance tree.
Metabolite Analysis
Asc, GSH, chlorophyll, pheophytin, and protein contents were determined as described in Baier et al. (2000) and guaiacol peroxidase activity as described in Baier and Dietz (1999b). Starch levels were measured as Glc after digestion with α-amylase and amylo-glucosidase as described in Baier et al. (2004a), with the exception that the starch content was standardized on fresh weight. H2O2 amounts were quantified enzymatically at 436 nm in a reaction mixture containing 100 mm K2HPO4/KH2PO4 pH 6.5 and 0.05% (v/v) guaiacol (Sigma) with 4 units horseradish peroxidase (Sigma).
Protoplast Isolation and Transfection
Protoplasts were isolated from 3-week-old T19-2 and the mutants as described in Seidel et al. (2004) and transfected in parallel with the reporter gene constructs D1 and D3 (Baier et al., 2004b) according to Seidel et al. (2005). After 6 h incubation at 20 μmol quanta m−2 s−1, the glucuronidase activity was quantified according to Abel and Theologis (1998). To normalize protoplast and plasmid quality D3-driven glucuronidase activity was standardized on D1-driven activity and on the activity observed in the parental line T19-2.
Hormone Sensitivity Tests
For physiological ABA sensitivity tests, germination of 2- to 8-week-old seeds from six independent harvests was analyzed on Murashige and Skoog plates containing 1% (w/v) Suc and 0.4% (w/v) phytagel (Sigma) and supplemented with 0 to 1 μm ABA (Sigma). After 7 d the number of seeds with emerged roots was counted. Wilting was quantified by monitoring cotyledon wrinkling of seedlings grown in high humidity after exposure to 25% to 30% relative humidity.
Electron microscopy of Arabidopsis chloroplasts was performed with leaves of 5-week-old plants grown under long-day illumination as described by König et al. (2002) using the contrasting method B by Engels et al. (1997).
Chlorophyll-a Fluorescence Measurements
The quantum yield of PSII (ΦPSII = FV/FM) was measured every hour beginning 30 min after onset of light under growth conditions using the Mini-PAM Fluorometer (Walz). Light flashes of 2,000 μmol quanta m−2 s−1 were given to analyze the maximum fluorescence. Calculation of photosynthetic parameters was performed as described by Schreiber and Bilger (1993).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Segregation analysis of the low luciferase phenotype in the F2 generation of the backcross of the rimb mutants to the parental line T19-2.
Supplemental Figure S2. Transcript amount correlation analysis.
Supplementary Material
Acknowledgments
We thank Daniela Ledebrink, Lea Johanning, and Rachel Holman for technical assistance, Prof. Jürgen Feierabend for the catalase antibody, and Thorsten Seidel and Fred Rook for technical advice.
This work was supported by the Deutsch Forschungsgemeinschaft (grant nos. FOR 387 and Ba2011/2) and Bielefeld University.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Margarete Baier (margarete.baier@uni-bielefeld.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abel S, Theologis A (1998) Transient gene expression in protoplasts of Arabidopsis thaliana. Methods Mol Biol 82 209–217 [DOI] [PubMed] [Google Scholar]
- Amme S, Matros A, Schlesier B, Mock HP (2006) Proteome analysis of cold stress response in Arabidopsis thaliana using DIGE-technology. J Exp Bot 57 1537–1546 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55 373–399 [DOI] [PubMed] [Google Scholar]
- Asada K (2000) The water-water cycle as alternative photon and electron sink. Philos Trans R Soc Lond B Biol Sci 355 1419–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M, Dietz K-J (1997) The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J 12 179–190 [DOI] [PubMed] [Google Scholar]
- Baier M, Dietz K-J (1999. a) The costs and benefits of oxygen for photosynthesizing cells. Prog Bot 60 282–314 [Google Scholar]
- Baier M, Dietz K-J (1999. b) Prospective function of chloroplast 2-cysteine peroxiredoxin in photosynthesis: evidence from transgenic Arabidopsis. Plant Physiol 119 1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M, Dietz K-J (2005) Chloroplasts as source and target of cellular redox regulation: a discussion on chloroplast redox signals in the context of plant physiology. J Exp Bot 56 1449–1462 [DOI] [PubMed] [Google Scholar]
- Baier M, Hemmann G, Holman R, Corke F, Card R, Smith C, Rook F, Bevan MW (2004. a) Characterization of mutants in Arabidopsis showing increased sugar-specific gene expression, growth and developmental responses. Plant Physiol 134 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M, Kandlbinder A, Dietz K-J, Golldack D (2006) New insights into abiotic stress signalling in plants. Prog Bot 67 248–274 [Google Scholar]
- Baier M, Noctor G, Foyer CH, Dietz K-J (2000) Antisense suppression of 2-cysteine peroxiredoxin in Arabidopsis specifically enhances the activities and expression of enzymes associated with ascorbate metabolism but not glutathione metabolism. Plant Physiol 120 823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M, Ströher E, Dietz K-J (2004. b) The acceptor availability at photosystem I and ABA control nuclear expression of 2-Cys peroxiredoxin-A in Arabidopsis thaliana. Plant Cell Physiol 45 997–1006 [DOI] [PubMed] [Google Scholar]
- Ball L, Accotto G-P, Bechtold U, Creissen G, Funck D, Jimenez A, Kular B, Leyland N, Mejia-Carranza J, Reynolds H, et al (2004) Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 16 2448–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broin M, Cuine S, Eymery F, Rey P (2002) The plastidic 2-cysteine peroxiredoxin is a target for a thioredoxin involved in the protection of the photosynthetic apparatus against oxidative stress. Plant Cell 14 1417–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21 2933–2942 [DOI] [PubMed] [Google Scholar]
- Collin V, Issakidis-Bourguet E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, Miginiac-Maslow M (2003) The Arabidopsis plastidial thioredoxins—new functions and new insights into specificity. J Biol Chem 278 23747–23752 [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen network of Arabidopsis. Plant Cell 17 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Mackerness S, Hancock JT, Neill SJ (2001) Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol 127 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ (2003) Redox control, redox signaling, and redox homeostasis in plant cells. Int Rev Cytol 228 141–193 [DOI] [PubMed] [Google Scholar]
- Dietz K-J, Jacob S, Oelze M-L, Laxa M, Tognetti V, de Miranda SMN, Baier M, Finkemeier I (2006) The function of peroxiredoxins in plant organelle redox metabolism. J Exp Bot 57 1697–1709 [DOI] [PubMed] [Google Scholar]
- Donald RGK, Cashmore AR (1990) Mutation of either G-box or I-box sequences profoundly affect expression from the Arabidopsis rbcS-1A promoter. EMBO J 9 1717–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels A, Kahmann U, Ruppel HG, Pistorius EK (1997) Isolation, partial characterization and localization of a dihydrolipoamide dehydrogenase from the cyanobacterium Synechocystis PCC 6803. Biochim Biophys Acta 1340 33–44 [DOI] [PubMed] [Google Scholar]
- Fey V, Wagner R, Bräutigam K, Wirtz M, Hell R, Dietzmann A, Leister D, Oelmüller R, Pfannschmidt T (2005) Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J Biol Chem 280 5318–5328 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR (1994) Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J 5 765–771 [Google Scholar]
- Foyer CH, Lelandais M, Kunert KJ (1994) Photooxidative stress in plants. Physiol Plant 92 696–717 [Google Scholar]
- Foyer CH, Noctor G (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridlyand LE, Scheibe R (1999) Controlled distribution of electrons between acceptors in chloroplasts: a theoretical consideration. Biochim Biophys Acta 1413 31–42 [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horling F, Lamkemeyer P, König J, Finkemeier I, Kandlbinder A, Baier M, Dietz K-J (2003) Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis. Plant Physiol 131 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM (1997) Photosynthetic electron transport regulates expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9 627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiddle G, Pastori GM, Bernard S, Pignocchi C, Antoniw J, Verrier PJ, Foyer CH (2003) Effects of leaf ascorbate content on defense and photosynthesis gene expression in Arabidopsis thaliana. Antioxid Redox Signal 5 23–32 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Monde R-A, Last RL (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Baier M, Horling F, Kahmann U, Harris G, Schürmann P, Dietz K-J (2002) The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of PET. Proc Natl Acad Sci USA 99 5738–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Lotte K, Plessow R, Brockhinke A, Baier M, Dietz K-J (2003) Reaction mechanism of plant 2-Cys peroxiredoxin: role of the C-terminus and the quaternary structure. J Biol Chem 278 24409–24420 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid-insensitive mutants in Arabidopsis thaliana. Physiol Plant 61 377–383 [Google Scholar]
- Laxa M, König J, Dietz K-J, Kandlbinder A (2007) Role of the cysteinyl residues in Arabidopsis thaliana cyclophilin CYP20-3 in peptidyl-prolyl cis-trans isomerase and redox-related function. Biochem J 401 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B-h, Kapoor A, Zhu J, Zhu J-K (2006) STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell 18 1736–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H-m, Culligan K, Dixon RA, Chory J (1995) CUE1: a mesophyll cell-specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell 7 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam R, Gomez-Buitrago AM, Eckardt N, Shah N, Guevara-Garcia A, Day P, Raina R, Fedoroff NV (2003) Characterizing the stress/defense transcriptome of Arabidopsis. Genome Biol 4 R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo A, Muhlenbrock P, Rusterucci C, Chang CCC, Miszalski Z, Karpinska B, Parker JE, Mullineaux PM, Karpinski S (2004) Lesion simulating disease 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol 136 2818–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler A (1951) Studies on the reaction of illuminated chloroplasts. I. Mechanisms of the reduction of oxygen and other Hill reagents. Arch Biochem Biophys 33 65–77 [DOI] [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J, Meierhoff K, Westhoff P (1996) Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and Northern hybridization. Planta 198 385–396 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network in plants. Trends Plant Sci 9 490–498 [DOI] [PubMed] [Google Scholar]
- Miyake C, Shinzaki Y, Nishioka M, Horiguchi S, Tomizawa KI (2006) Photoinactivation of ascorbate peroxidase in isolated tobacco chloroplasts: Galdieria partia APX maintains the electron flux through the water-water-cycle in transplastomic tobacco plants. Plant Cell Physiol 47 200–210 [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (gun5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JC, Jang HH, Chae HB, Lee JR, Lee SY, Jung YJ, Shin MR, Lim HS, Chung WS, Yun DJ, et al (2006) The C-type Arabidopsis thioredoxin reductase ANRT-C acts as an electron donor to 2-Cys peroxiredoxins in chloroplasts. Biochem Biophys Res Commun 348 478–484 [DOI] [PubMed] [Google Scholar]
- Mullineaux PM, Karpinski S, Baker NR (2006) Spatial dependence for hydrogen peroxide-directed signaling in light-stressed plants. Plant Physiol 141 346–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux PM, Rausch T (2005) Glutathione, photosynthesis and the redox regulation of stress-responsive gene expression. Photosynth Res 86 459–474 [DOI] [PubMed] [Google Scholar]
- Ogren WL (1984) Photorespiration: pathways, regulation, and modification. Annu Rev Plant Physiol 35 415–442 [Google Scholar]
- op den Camp RGL, Przybla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hindeg E, Göbel C, Feussner I, et al (2003) Rapid induction of distinct stress responses after release of singlet oxygen. Plant Cell 15 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Ahumada A, Kahmann U, Dietz K-J, Baier M (2006) Regulation of peroxiredoxin expression versus expression of Halliwell-Asada-Cycle enzymes during early seedling development of Arabidopsis thaliana. Photosynth Res 89 99–112 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T (2003) Chloroplast redox signals: how photosynthesis controls its own genes. Trends Plant Sci 8 33–41 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Allen JE, Oelmüller R (2001) Principles of redox control in photosynthesis gene expression. Physiol Plant 112 1–9 [Google Scholar]
- Rizhsky L, Liang H, Mittler L (2003) The water-water-cycle is essential for chloroplast protection in the absence of stress. J Biol Chem 278 38921–38925 [DOI] [PubMed] [Google Scholar]
- Rossel JB, Walter PB, Hendrickson L, Chow WS, Poole A, Mullineaux PM, Pogson BJ (2006) A mutation affecting ascorbate peroxidase 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ 29 269–281 [DOI] [PubMed] [Google Scholar]
- Rossel JB, Wilson IW, Pogson BJ (2002) Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol 130 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber U, Bilger W (1993) Progress in chlorophyll fluorescence research: major developments during the past years in retrospect. Prog Bot 54 151–173 [Google Scholar]
- Seidel T, Golldack D, Dietz K-J (2005) Mapping of C-termini of V-ATPase subunits by in vivo-FRET measurements. FEBS Lett 579 4374–4382 [DOI] [PubMed] [Google Scholar]
- Seidel T, Kluge C, Hanitzsch M, Ross J, Sauer M, Dietz K-J, Golldack D (2004) Colocalization and FRET-analysis of subunits c and a of the vacuolar H+-ATPase in living plant cells. J Biotechnol 112 165–175 [DOI] [PubMed] [Google Scholar]
- Sheehan D (2006) Detection of redox-based modification in two-dimensional electrophoresis proteomic separation. Biochem Biophys Res Commun 349 455–462 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Koorneef A, Classens SMC, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Metraux JP, Brown R, Kazan K, et al (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu J-K (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18 2051–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Mittler R (2005) Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiol Plant 126 45–51 [Google Scholar]
- Torres MA, Jones JDG, Dangl JL (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S, Vanderauwera S, Vuylsteke M, Rombauts S, Langebartels C, Seidlitz HK, Zabeau M, Van Montagu M, Inzé D, Van Breusegem F (2004) Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J 39 45–58 [DOI] [PubMed] [Google Scholar]
- Wormuth D, Baier M, Kandlbinder A, Scheibe R, Hartung W, Dietz K-J (2006) Regulation of gene expression by photosynthetic signals triggered through modified CO2 availability. BMC Plant Biol 6 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Wright DA, Wetzel C, Voytas DF, Rodermel S (1999) The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homologue that functions during early chloroplast biogenesis. Plant Cell 11 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.