Abstract
The levels of endogenous brassinosteroids (BRs) and the expression of the biosynthesis/metabolism/perception genes involved have been investigated during the development and germination of pea (Pisum sativum) seeds. When seeds were rapidly growing, the level of biologically active BRs (brassinolide [BL] and castasterone [CS]) and the transcript levels of two BR C-6 oxidases (CYP85A1 and CYP85A6) reached a maximum, suggesting the significance of BL and CS in seed development. In the early stages of germination, CS, but not BL, appeared and its level increased in the growing tissues in which the transcript level of CYP85A1 and CYP85A6 was high, suggesting the significance of CS in seed germination and early seedling growth of pea. 6-Deoxocathasterone (6-deoxoCT) was the quantitatively major BR in mature seeds. At the early stage of germination, the level of 6-deoxoCT was specifically decreased, whereas the levels of downstream intermediates were increased. It seems that 6-deoxoCT is the major storage BR and is utilized during germination and early growth stages. The level of the mRNAs of BR biosynthesis and perception genes fluctuated during seed development. In mature seeds, most of mRNAs were present, but the level was generally lower compared with immature seeds. However, CYP90A9 mRNA rapidly increased during seed development and reached the maximum in mature seeds. The mRNAs stored in mature pea seeds seem to be utilized when seeds germinate. However, it was found that de novo transcription of mRNAs also starts as early as during seed imbibition.
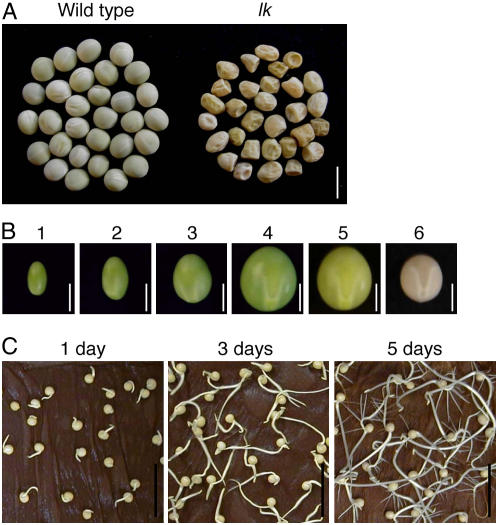
Generally, immature seeds of plants contain high levels of plant hormones, GAs, cytokinins (CKs), auxins, abscisic acid (ABA), and brassinosteroids (BRs). A number of plants contain both GAs and auxins at the highest levels during early to mid-embryo development, when CKs are decreasing rapidly and there is little or no ABA detectable (Rock and Quatrano, 1995). The increases in GAs and auxins in pea (Pisum sativum) and wheat (Triticum aestivum) are correlated with increases in pod and grain length. It thus seems that CKs have physiological roles in the early stage of embryo development, whereas GAs and auxins elicit their activity in the later stages. The level of ABA reaches the highest levels at the middle and late periods of seed development prior to returning to lower levels in the dry seed, resulting in acceleration of a buildup of nutrient reserves, an arrest of tissue growth, development of desiccation tolerance, and prevention of precocious germination (Rock and Quatrano, 1995). Choe et al. (2000) found that the dwarf5 (dwf5) mutant of Arabidopsis (Arabidopsis thaliana), which is BR deficient due to a defect in the sterol Δ7 reductase, produces aberrantly shaped seeds. The lk mutant of pea, which is severely BR deficient due to the impaired sterol 5α-reductase, also produces irregularly shaped seeds (Fig. 1; Nomura et al., 2004). A BR-deficient Vicia faba bean also produces small seeds (Fukuta et al., 2005). These facts indicate that sterols or BRs may be required for normal seed development.
Figure 1.
Pea seeds. A, Irregularly shaped seeds of the dwarf mutant lk of pea. The lk mutant is BR deficient due to a defect in a steroid 5α-reductase (see Fig. 2; Nomura et al., 2004). Bar = 1 cm. B, Plant materials for BR analyses during pea seed maturation. Seeds collected were grouped into six stages according to their size and appearance. Seed weights are shown in “Materials and Methods.” Bar = 5 mm. C, Plant materials for BR analyses during pea seed germination. Seeds were imbibed for 16 h and then incubated in the dark for 1, 3, or 5 d. Weights and lengths of seedlings are shown in “Materials and Methods.” Bar = 5 cm.
Germination of seeds is known to be retarded by ABA (Merlot and Giraudat, 1997) and, in lettuce (Lactuca sativa), accelerated by the de novo synthesis of GA triggered by red light irradiation (Toyomasu et al., 1998). The synthesis of α-amylase associated with seed germination is also accelerated by GAs (Richards et al., 2001). It has been known that germination of aged rice (Oryza sativa) seeds (Yamaguchi et al., 1987) as well as clover broomrape (Orobanche minor Sm.) seeds (Takeuchi et al., 1997) is stimulated by BR. Steber and McCourt (2001) showed that BR rescues the germination phenotype of severe GA biosynthetic mutants and of a GA-insensitive mutant by overcoming ABA-induced inhibition of germination. Furthermore, the germination rate of the dwf5 mutant is lower than that of the wild-type plants (Choe et al., 2000). These observations raise the possibility that BR is needed for normal germination.
However, little knowledge is available that correlates endogenous BR levels with seed growth and maturation, as well as with seed germination. To get such information, we quantified endogenous BR levels in pea seeds and seedlings at the various growth and development stages. We also quantified the transcript levels of BR-related genes. To this end we have cloned various pea genes related to the biosynthesis and metabolism of BRs (see Fig. 2). Previously, some of those genes had been isolated as mutated genes in the lka, lkb, and lk mutants of pea by us. The LKB gene encodes sterol C-24 reductase (Nomura et al., 1997, 1999; Schultz et al., 2001) and the LK gene encodes sterol 5α-reductase (Nomura et al., 2004). The LKA gene encodes a BR receptor component that is the pea homolog of Arabidopsis BRI1 (Nomura et al., 2003). The DDWF1 gene of pea, cloned as a gene regulated by a dark-induced G protein, has been claimed to encode BR C-2 hydroxylase (Kang et al., 2001), but we could not justify this claim because its homolog is not present in the Arabidopsis genome. In this study we further cloned pea homologs of cytochrome P450 monooxygenases (P450s) that have already been characterized on BR biosynthesis in Arabidopsis, tomato (Solanum lycopersicum), and rice: CYP85A1 (DWARF)/C-6 oxidase (Bishop et al., 1999), CYP90A1 (CPD)/putative C-23 hydroxylase (Szekeres et al., 1996; Ohnishi et al., 2006), CYP90B1 (DWF4)/C-22 hydroxylase (Choe et al., 1998), CYP90D1/C-23 hydroxylase (Ohnishi et al., 2006), and CYP734A1 (BAS1)/C-26 hydroxylase (Neff et al., 1999). We discuss the roles of BRs in growth, maturation, and germination of pea seed on the basis of the level of endogenous BRs and of BR-related mRNAs.
Figure 2.
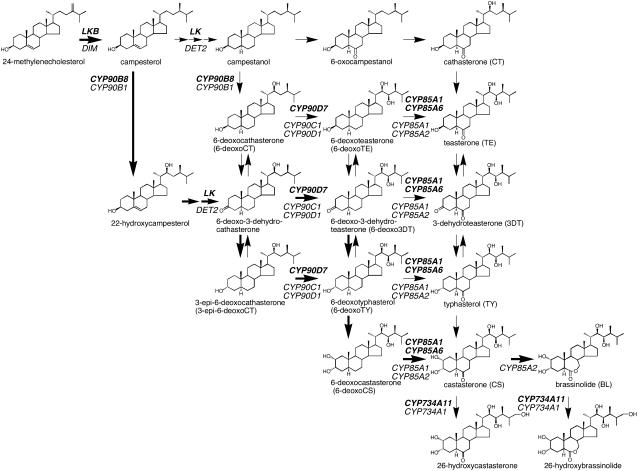
Proposed BR biosynthetic pathway in pea. The pathway was drawn based on Fujioka and Yokota (2003), Fujita et al. (2006), and Ohnishi et al. (2006). Possible major biosynthetic pathways are indicated by bold arrows. Possible BR biosynthetic genes of pea are indicated by bold letters. The corresponding Arabidopsis genes are indicated by normal italic letters.
RESULTS
Identification of Endogenous BRs and Their Quantitative Changes in Developing Pea Seeds
Immature to mature seeds of pea were harvested at six growth stages (Fig. 1). The levels of endogenous BRs were examined at five of these stages except stage 5, fully expanded “yellowed” seeds, because it has been difficult to harvest a large amount of seed materials exactly in stage 5. From these seeds, two biologically active BRs, castasterone (CS) and brassinolide (BL), were identified together with 6-deoxoBRs belonging to the late C-6 oxidation. 6-Oxo intermediate BRs belonging to the early C-6 oxidation pathway were not detected at any growth stage (see Fig. 2).
Quantitative changes of the endogenous BRs in these seeds are shown in Table I. The amounts of BL and CS increased toward stage 3 in parallel to the seed weight increase, but drastically decreased in the fully expanded green seeds (stage 4). Both BRs were not detectable in mature dried seeds (stage 6). The level of 6-deoxocastasterone (6-deoxoCS) rapidly increased and reached the highest level at stage 4, whereas it drastically decreased at stage 6. No such dramatic changes were observed for the levels of the upstream biosynthetic intermediate BRs. However, interestingly, high levels of 6-deoxoBRs were still contained in mature seeds. Among these BRs, 6-deoxocathasterone (6-deoxoCT) is the predominant BR, with the amount accounting for 80% of the total BR content.
Table I.
Endogenous levels of BRs during seed growth of pea
Cloning of BR Biosynthesis/Metabolism Genes from Pea Seed
Pea cytochrome P450 genes related to BR biosynthesis and metabolism were cloned by PCR-based approaches using oligonucleotide primers derived from the conserved nucleotide sequences in the expressed sequence tags of legumes Glycine max, Lotus japonicus, and Medicago truncatula, as well as in the known genes of Arabidopsis and tomato. The genes we cloned were CYP85A1 and CYP85A6 as DWARF homologs (Jager et al., 2007), CYP90A9 and CYP90A10 as CPD homologs, CYP90B8 as a DWF4 homolog, CYP90D7 as a CYP90D1 homolog, and CYP734A11 as a BAS1 homolog. Identity and/or similarity of these genes and encoding proteins in reference to Arabidopsis counterparts are shown in Table II.
Table II.
Comparison of BR-related genes/proteins between pea and Arabidopsis
| Pea | Arabidopsis | DNA
|
Protein
|
|
|---|---|---|---|---|
| Identity (%) | Identity (%) | Similarity (%) | ||
| CYP85A1 | CYP85A2 | 71 | 71 | 84 |
| CYP85A6 | CYP85A2 | 67 | 67 | 80 |
| (Pea CYP85A1 versus CYP85A6) | 80 | 77 | 86 | |
| CYP90A9 | CYP90A1 | 69 | 72 | 81 |
| CYP90A10 | CYP90A1 | 71 | 76 | 84 |
| (Pea CYP90A9 versus CYP90A10) | 77 | 77 | 85 | |
| CYP90B8 | CYP90B1 | 68 | 70 | 81 |
| CYP90D7 | CYP90D1 | 63 | 61 | 75 |
| CYP734A11 | CYP734A1 | 70 | 76 | 87 |
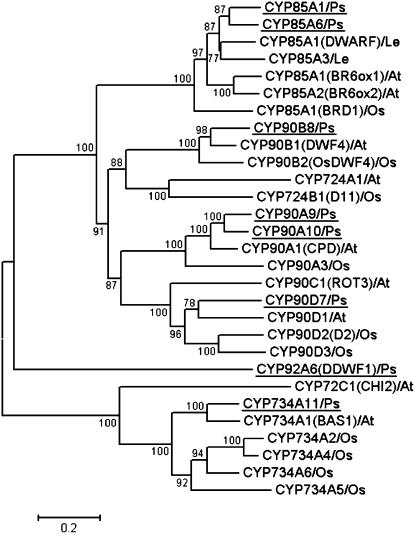
A phylogenetic tree of BR biosynthesis/metabolism-related P450s of pea, Arabidopsis, tomato, and rice was constructed using their amino acid sequences (Fig. 3). These analyses suggest that the pea P450 genes cloned will have the same functions as the indicated Arabidopsis, tomato, and rice genes. Recently, it was found that CYP85A1 of Arabidopsis and tomato catalyzes the conversion of 6-deoxoCS to CS, whereas Arabidopsis CYP85A2 and tomato CYP85A3 catalyze the conversion of 6-deoxoCS to BL (Kim et al., 2005; Nomura et al., 2005). However, we found that both pea CYP85A1 and CYP85A6 catalyze the conversion of 6-deoxoCS to CS but do not produce a significant amount of BL (Jager et al., 2007). It is interesting that pea has two CPD homologs, CYP90A9 and CYP90A10. It has been proposed that the Arabidopsis CYP90A1 gene encodes BR C-23 hydroxylase because its loss-of-function mutant cpd shows a severe dwarfism that is rescued by exogenous applications of BRs, including 23-hydroxylated precursors (Fig. 2; Szekeres et al., 1996). However, recently, Ohnishi et al. (2006) have suggested that CYP90A1 catalyzes a step different from C-23 hydroxylation but an unknown reaction in BR biosynthesis using in vitro enzyme assay. A putative 2-hydroxylase, DDWF1 (CYP92A6), did not fall into any phylogenetic clan of plant steroid hydroxylases, indicating that DDWF1 could be involved in a reaction other than BR biosynthesis.
Figure 3.
Phylogenetic relationships of BR-related P450s. The phylogenetic tree was constructed by the neighbor-joining method using the deduced full-length protein sequences of pea genes (Ps, underlined) and of BR-related P450 families in Arabidopsis (At), rice (Os), and tomato (Le). Gene name is indicated in parentheses. Bootstrap mode (1,000 replications) was used for estimating the confidence that can be assigned to particular nodes in the tree.
Fluctuation of Transcript Levels of BR Biosynthesis/Metabolism/Receptor Genes in Developing Pea Seeds
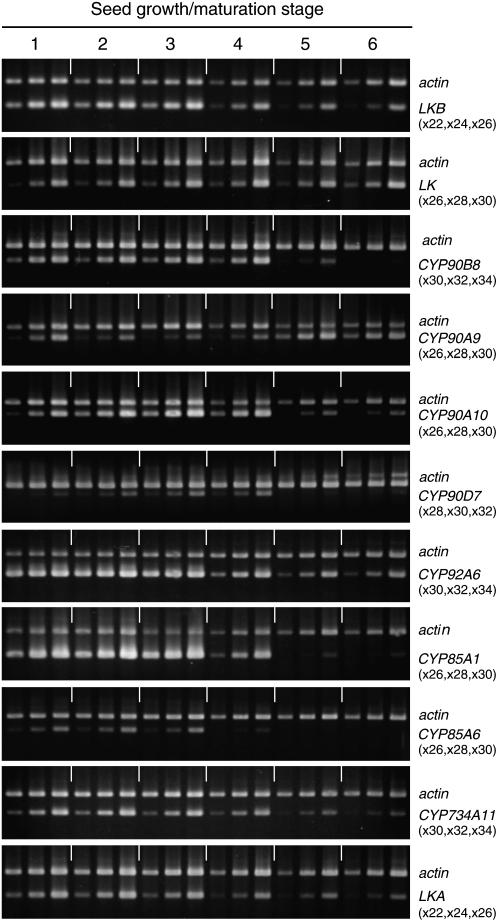
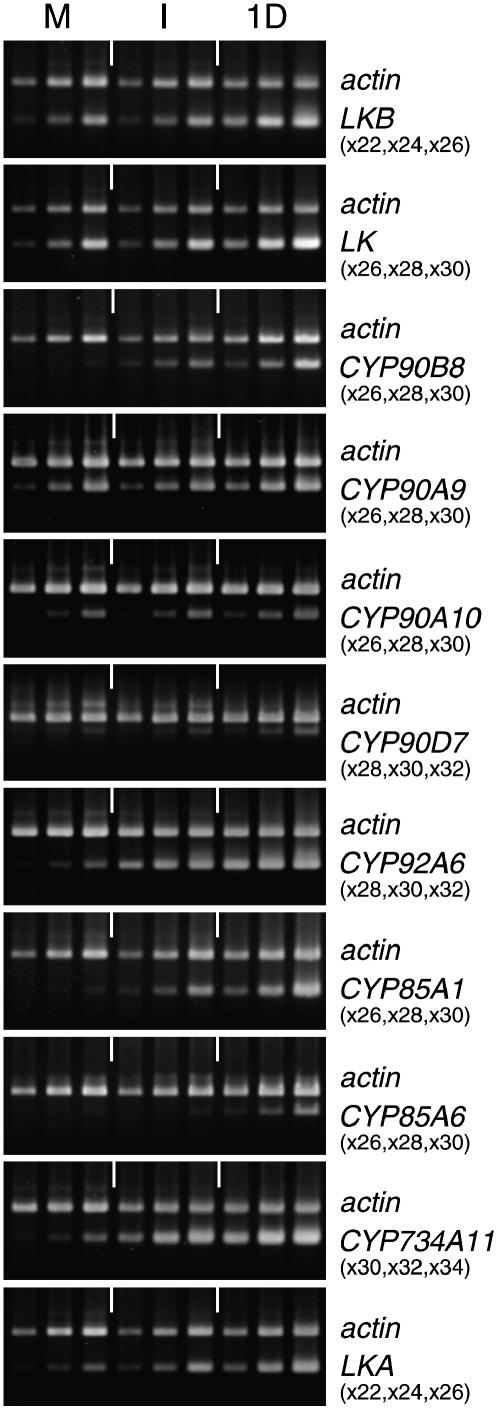
The transcript levels of the genes were examined at immature to mature stages 1 to 6 (Fig. 4; Supplemental Fig. S1). The transcripts of the CYP85A1 and CYP85A6 genes reached the highest level at stage 3, but then decreased in the fully expanded green seeds (stage 4) and vanished by stage 6. Interestingly, the CYP85A1 gene was more highly expressed than the CYP85A6 gene through all stages of growth (Fig. 4).
Figure 4.
Transcript levels of BR-related genes during growth and maturation of pea seeds. The growth/maturation stages are explained in “Materials and Methods.” Ethidium bromide-stained agarose gels show the RT-PCR products from seeds. Primers for actin and the target gene were used in the same PCR. Sampling cycles of PCR are indicated in parentheses.
Intriguingly, the patterns of expression of the CYP90A9 and CYP90A10 transcripts were in striking contrast. The CYP90A10 transcript level increased toward stage 4, but thereafter sharply decreased. Its level in immature seed was low. However, the CYP90A9 transcript level was quite low until stage 4, but thereafter sharply increased and accumulated to a high level in mature, dry seed. These results suggest that the CYP90A9 and CYP90A10 genes function at quite different stages of growth.
The expression of the CYP90D7 gene, although not high at any stage of seed growth, was the highest at stages 3 and 4. It was not detectable at later stages (5 and 6).
The transcript levels of LKB, CYP90B8, CYP92A6, CYP734A11, and LKA were less markedly changed, but as a whole gradually decreased as seeds matured. However, the LK gene transcript gently increased and attained the highest level at stage 6. It is worth noting that mRNAs of LK, LKB, CYP90A9, CYP90A10, CYP92A6, CYP734A11, and LKA were still detectable even in mature, dry seeds of pea, but those of CYP90B8, CYP90D7, CYP85A1 and CYP85A6 were scarce or below the detection levels.
Quantitative Changes of Endogenous BRs in Germinating Pea Seeds
The levels of endogenous BRs fluctuated during seed germination as shown in Table III (Fig. 1). In the 16-h imbibed seed, the levels of endogenous BRs were comparable to those of mature seeds, and neither BL nor CS was detected. In the 1-d-old seedlings, CS appeared and its precursors, 6-deoxoCS, 6-deoxotyphasterol (6-deoxoTY) and 6-deoxo-3-dehydroteasterone (6-deoxo3DT), were markedly increased. In contrast, 6-deoxoCT was decreased to one-tenth.
Table III.
Endogenous levels of BRs during pea seed germination
| BRa | Seed Germination Stage
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Imbibed Seed | 1-d-Old Seedling | 3-d-Old Seedling
|
5-d-Old Seedling
|
|||||||
| Whole | Cotyledon | Shoot | Root | Whole | Cotyledon | Shoot | Root | |||
| ng kg−1 fresh weight | ||||||||||
| BL | ndb | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| CS | nd | 61 | 118 | 48 | 350 | 190 | 93 | 43 | 190 | 61 |
| 6-DeoxoCS | 73 | 600 | 1,507 | 1,030 | 1,830 | 3,410 | 890 | 830 | 910 | 1,040 |
| 6-DeoxoTY | 150 | 420 | 1,687 | 1,990 | 370 | 1,750 | 1,946 | 1,570 | 240 | 2,250 |
| 6-Deoxo3DT | 25 | 160 | 307 | 90 | 130 | 500 | 197 | 200 | 95 | 390 |
| 6-DeoxoTE | 81 | 94 | 158 | 310 | 210 | 330 | 113 | 100 | 85 | 210 |
| 6-DeoxoCT | 2,110 | 220 | 1,839 | 1,360 | 2,060 | 3,870 | 2,159 | 2,210 | 1,610 | 3,100 |
| Total | 2,439 | 1,555 | 5,616 | 4,828 | 4,950 | 10,050 | 5,398 | 4,953 | 3,130 | 7,051 |
Abbreviations are shown in Figure 2.
nd, Not detected.
The 3- and 5-d-old seedlings were dissected into cotyledons, shoots, and roots prior to extraction. CS, but not BL, was detected in these tissues. The level of 6-deoxoCT and downstream intermediates was high in these tissues. These findings indicate that de novo BR synthesis is already operating in cotyledons, shoots, and roots of the 3- and 5-d-old seedlings. Shoots were found to contain higher levels of CS than roots, in accord with the data reported for Arabidopsis, pea, and tomato (Bancos et al., 2002; Symons and Reid, 2004).
Fluctuations of Transcript Levels of BR Biosynthesis/Metabolism/Receptor Genes in Germinating Pea Seeds
The expression of CYP85A1, CYP90B8, CYP92A6, CYP734A11, and LKA was increased as early as during imbibition, whereas the expression of CYP90A9, CYP90A10, CYP85A6, and LKB was elevated in 1-d-old seedlings (Fig. 5; Supplemental Fig. S2). The increase observed in LK was less marked.
Figure 5.
Transcript levels of BR-related genes during seed germination of pea. The growth/maturation stages are explained in “Materials and Methods.” Ethidium bromide-stained agarose gels show the RT-PCR products from mature dried (M), 16-h imbibed (I), and 1-d-old germinating seeds (1D).
Shoots, roots, and cotyledons of 3-d-old seedlings were also examined. It should be noted that expressions of CYP85A1 and CYP85A6 were high in shoots and roots, but scarce in cotyledons (Fig. 6). Expressions of the other genes were rather evenly distributed in shoots, roots, and cotyledons.
Figure 6.
Transcript levels of BR-related genes in 3-d-old pea seedlings. Ethidium bromide-stained agarose gels show the RT-PCR products from cotyledons (C), shoots (S), and roots (R) of 3-d-old seedlings.
DISCUSSION
The Late C-6 Oxidation Pathway Is Predominant In Pea as Well as in a Majority of Plants
Biosynthesis of CS from campestanol has been known to occur through the early C-6 oxidation pathway via cathasterone or the late C-6 oxidation pathway via 6-deoxoCT, 6-deoxoteasterone (6-deoxoTE), 6-deoxo3DT, 6-deoxoTY, and 6-deoxoCS (Fig. 2; Fujioka and Yokota, 2003). In addition, the early C-22 oxidation pathway from campesterol to 6-deoxoCT via 22-hydroxycampesterol and 6-deoxo-3-dehydroCT has been demonstrated by Fujioka et al. (2002) and by Fujita et al. (2006; Fig. 2). Quite recently, novel shortcut routes were discovered by Ohnishi et al. (2006), where 6-deoxo-3-dehydroCT and its reduction product, 3-epi-6-deoxoCT, are 23-hydroxylated by CYP90C1 and CYP90D1 to give 6-deoxo3DT and 6-deoxoTY, respectively (Fig. 2). The available evidence indicated that the campestanol-independent pathway from campesterol to CS involving the shortcut routes is the main biosynthesis stream in Arabidopsis. This study as well as earlier works (Bancos et al., 2002) showed that endogenous BRs in pea tissues are comparable to those in Arabidopsis and tomato. Thus, the campestanol-independent pathway is also predominant in pea and probably in a majority of higher plants (Fig. 2).
However, we earlier identified BRs belonging to the early C-6 oxidation pathway from pea tissues, although at very low levels, i.e. typhasterol (TY) from immature pea seeds (Yokota et al., 1996) and teasterone (TE), 3-dehydroteasterone (3DT), and TY from pea shoots (Nomura et al., 1999). The conversion of 6-deoxoTY to TY, 6-deoxo3DT to 3DT, and 6-deoxoTE to TE has been demonstrated to be catalyzed by Arabidopsis and tomato CYP85A1 (Shimada et al., 2001). Thus, it seems that TY, 3DT, and TE earlier identified from pea tissues are synthesized from those 6-deoxoBRs by C-6 oxidases such as CYP85A1 and/or CYP85A6 (Fig. 2). Both TE and 3DT are also likely to be derived from TY by a reverse reaction (Suzuki et al., 1994; Stündl and Schneider, 2001).
Growth and Development of Pea Seed May Be Controlled by CS and BL
The C-6 oxidation is a rate-limiting step in the production of bioactive BRs because 6-deoxoCS, a direct precursor of CS, commonly accumulates at relatively high levels in several plant species (Nomura et al., 2001). It has been shown that the formation of CS from 6-deoxoCS is catalyzed by CYP85A1 in tomato (Bishop et al., 1999), Arabidopsis (Shimada et al., 2001), and rice (Hong et al., 2002; Mori et al., 2002). Recently, we have found that CYP85A3 (tomato) and CYP85A2 (Arabidopsis) function as multifunctional enzymes that catalyze consecutive oxidation reactions at the C-6 of 6-deoxoCS to produce BL via CS (Kim et al., 2005; Nomura et al., 2005). We found that pea CYP85A1 and CYP85A6 functioned principally as BR C-6 oxidases that convert 6-deoxoCS to CS when expressed in yeast (Jager et al., 2007). No full BR C-6/Baeyer-Villiger oxidase that converts 6-deoxoCS to BL via CS has yet been identified from pea. The transcript levels of CYP85A1 and CYP85A6 correlate well with the endogenous levels of BL and CS during seed growth. The amounts of BL and CS reached a maximum at stage 3 when seeds were rapidly growing, but were drastically reduced when the seeds were fully expanded. In contrast, 6-deoxoCS built up in fully expanded seeds (stage 4) when the conversion of 6-deoxoCS to CS by CYP85A1 and CYP85A6 was decreased. These findings are consistent with BL and CS being required for growth and development of seeds.
The bas1-D mutant of Arabidopsis, in which CYP734A1 (previously named CYP72B1) encoded by the BAS1 gene is amplified, was reported to deactivate BL and CS by C-26 hydroxylation (Neff et al., 1999; Turk et al., 2003). However, in the bas1-D mutant, the levels of 6-deoxoCS, CS, and BL are much lower compared with wild-type plants, suggesting that the BAS1 protein can also degrade 6-deoxoCS. Inpea seeds, the CYP734A11 transcript is highly expressed in immature seeds, suggesting that it functions to control BR levels throughout seed growth.
6-DeoxoCT May Be a Major Storage BR Utilized during Germination
In mature seeds, the biologically active BRs CS and BL were not detected. However, their biologically inactive precursors were stored in mature seeds. Among them, 6-deoxoCT was predominant in mature seeds. This profile of endogenous BRs in mature seeds was not changed after 2 years of storage at 4°C (data not shown), indicating that these precursors may work as storage forms for years. The endogenous BR levels were not affected by imbibition (Table III), but de novo synthesis of CS and its precursors was clearly observed in 1-d-old seedlings. In contrast, only 6-deoxoCT was drastically reduced in 1-d-old seedlings, indicating that CS and its precursors were synthesized at the expense of 6-deoxoCT stored in seeds. This suggests that 6-deoxoCT is used as a storage BR and that de novo synthesized 6-deoxoCT is not fully supplied at this stage. The level of 6-deoxoCT was largely restored in shoots, roots, and seeds of 3- and 5-d-old seedlings, indicating that de novo synthesis of 6-deoxoCT and upstream intermediates starts within 3 d of the start of germination.
Transcripts of Biosynthesis, Metabolism, and Receptor Genes of BRs Are Stored in Pea Seeds
Transcripts of biosynthesis genes LKB, LK, CYP90A9, and CYP90A10, as well as of the receptor gene LKA, were detected in mature seeds, suggesting that these mRNAs may be promptly utilized to generate and perceive BRs as soon as seeds germinate. Interestingly, the level of CYP90A9 mRNA was increased during maturation and reached the highest level in mature seeds (Fig. 4; Supplemental Fig. S1). Furthermore, high levels of LKB and LK were maintained in mature seeds. In accord with this, the LK gene (as well as its Arabidopsis homolog DET2) is known as a steady-state gene whose transcription is little affected by growth stage (Li et al., 1996; Nomura et al., 2004). The high mRNA levels of LKB, LK, and CYP90A9 suggest that these genes play especially important roles in generating BRs in the early stages of germination.
Germination of Pea Seeds and Early Seedling Growth May Be Controlled by the Synthesis of CS
In imbibed pea seeds, no change was observed in the profile of endogenous BR levels as compared with that of mature seed. However, the mRNAs of CYP90B8, CYP85A1, and CYP734A11 had already started to increase. In 1-d-old seedlings all mRNAs examined were largely increased (Fig. 5; Supplemental Fig. S2) and de novo synthesis of CS was observed (Table III). As seen in 3- and 5-d-old seedlings, cotyledons, shoots, and roots were all found to synthesize CS. In 3-d-old seedlings, transcription of CYP85A1 and CYP85A6 was observed in the shoot and root, but was very scarce in the cotyledons (Fig. 6), reflecting the higher level of CS in shoots and roots than in cotyledons. BL was not detected in any tissues of the dark-grown seedlings in this study, suggesting that only CS is required for early growth of seedlings. However, significant amounts of BL, in addition to CS, have been detected in shoots of light-grown young seedlings (Symons et al., 2002) and of natural light-grown older plants (36 d old and 6 months old) of pea (Nomura et al., 1997; Supplemental Table S1), suggesting that both BL and CS are required for light growth and development of pea. If there is no other member of the CYP85A family in pea, it is possible that CYP85A1 and/or CYP85A6 may produce BL in different cellular environment in plants from in yeast cells (Jager et al., 2007).
In tomato, only CS is required for shoot growth, whereas both CS and BL are responsible for the development of fruits (Nomura et al., 2005). Recently, Kim et al. (2005) and Nomura et al. (2005) pointed out that both CS and BL are required for the full growth of Arabidopsis seedlings. These findings, together with this work, suggest that the roles of CS and BL are varied spatially and temporally depending on species.
MATERIALS AND METHODS
Plant Materials
We used the pure wild-type cultivar of garden pea (Pisum sativum), Torsdag. Immature seeds were harvested from plants grown in a field under natural conditions in May to June, 2001. Seeds collected were grouped according to their size and appearance (Fig. 1; Table I). The number and total weight (g) of harvested seeds as well as averaged seed weight (g) were as follows (in parentheses): stage 1 (335, 9.7, 0.029), stage 2 (148, 16.3, 0.111), stage 3 (175, 33.0, 0.189), stage 4 (330, 158.2, 0.480; fully expanded, green), stage 5 (10, 4.88, 0.488; fully expanded, yellow), and stage 6 (200, 56.2, 0.281; mature, dry).
As for germinating seedlings, 200 mature pea seeds were imbibed in running tap water for 16 h, then incubated in wet paper towels at 25°C in the dark for 1, 3, or 5 d (Fig. 1). The imbibed seeds weighed 409 mg, whereas 1-d-old seedlings weighed 508 mg. Three-day-old seedlings were dissected into cotyledons (459 mg), shoots (110 mg, 13–26 mm), and roots (96 mg, 20– 70 mm). Five-day-old seedlings were also dissected into cotyledons (467 mg), shoots (290 mg, 80–135 mm), and roots (144 mg, 80–125 mm).
Quantification of Endogenous BRs
The methanol extract of the harvested materials was spiked with 2H6-labeled BRs (Nomura et al., 1999) as internal standards before reduction to an aqueous residue. The aqueous residue was partitioned between ethyl acetate and 0.5 m K2HPO4 buffer. The ethyl acetate phase was evaporated to dryness and partitioned between hexane and 80% methanol. The 80% methanol phase was evaporated to dryness and the residual solid was purified on a column of charcoal (chromatography grade; Wako Pure Chemicals), which was eluted with methanol:water (6:4 and 8:2, v/v), methanol, methanol:chloroform (9:1, 7:3, 5:5, 3:7, and 1:9, v/v), and chloroform. To monitor the biological activity of BRs, sample aliquots were assayed by the rice (Oryza sativa) lamina inclination test (Yokota et al., 1996). The methanol:chloroform (9:1–3:7) fractions were chromatographed on a column of Sephadex LH-20 (bed volume, 500 mL; Pharmacia) using methanol:chloroform (4:1, v/v) as a mobile phase. Successive 10-mL fractions were collected. Fractions 37 to 41 were combined, dissolved in methanol, and passed through columns of diethylaminopropyl silica (Bondesil; Varian) and octadecylsilica (ODS-SS-1020-T; Senshu Science). Reversed-phase HPLC was carried out on a Senshu Pak ODS-3251-D column (250 × 8 mm i.d.; Senshu Science) eluted with the following acetonitrile-water gradient at a flow rate of 2.5 mL min−1: 0 to 20 min, 45% acetonitrile; 20 to 40 min, 45% to 100% acetonitrile; and 40 to 60 min, 100% acetonitrile. Fractions were collected every min. The column oven temperature was maintained at 40°C. BR fractions were collected based on bioassay and the retention times of authentic BRs. The quantification of endogenous BRs was performed by GC-selected ion monitoring according to Nomura et al. (2001).
Cloning of BR Biosynthesis Genes from Pea
Primer sequences used in this study are shown in Supplemental Table S2. First primers were designed based on highly conserved nucleotide sequences between legume expressed sequence tags and BR biosynthesis genes of Arabidopsis (Arabidopsis thaliana) and tomato (Solanum lycopersicum). PCR amplification was carried out with the Expand High Fidelity PCR system (Roche). Templates were from single-strand cDNA libraries that were made from immature seeds or 7-d-old shoots of pea. The resulting products were sequenced using Long-Read Tower sequencer (Amersham Biosciences). Based on those sequences, gene-specific primers were designed to amplify the 5′ and 3′ ends of each gene. Primers listed in Supplemental Table S2 were used sequentially for 5′- and 3′-RACE reactions according to the 5′/3′ RACE kit (Roche). The resulting products were sequenced and these fragments were assembled to construct an open reading frame. Primers were designed to amplify the full-length cDNA and the full-length clones were sequenced to check the PCR errors. Sequence analysis was performed using MacVector software (Oxford Molecular). The phylogenetic tree was constructed by neighbor-joining with p-distance and bootstrap replication (1,000 replications) using MEGA Version 3.1 software (http://www.megasoftware.net/). Amino acid sequences of the P450 members in Arabidopsis, tomato, and rice were obtained from the Cytochrome P450 home page (http://drnelson.utmem.edu/CytochromeP450.html).
Determination of Gene Transcript Levels
Primer sequences used in this study are shown in Supplemental Table S3. The transcript levels of genes were analyzed by semiquantitative reverse transcription (RT)-PCR. Total RNA was extracted from pools of 10 to 20 immature seeds harvested from five to 10 pods and from shoots, roots, and cotyledons excised from pools of 10 to 40 individuals of 3- and 5-d-old seedlings using RNeasy Plant Mini kit with RNase-Free DNase set (Qiagen). Single-strand cDNA was synthesized using 2.5 μg of total RNA, SuperScript II reverse transcriptase (Invitrogen), and oligo(dT) primer according to the manufacturer's instructions. PCR amplification was performed using 500 nm gene-specific primers and 50 to 100 nm actin primers (as a control) with the same concentration of template cDNAs in the same tube. The product was sequenced to confirm the identical band. RT-PCR products obtained by three different amplification cycles were run on an agarose gel. Gels stained with ethidium bromide were digitized and analyzed using luminescent image analyzer GENEGENIUS (Syngene).
Sequence data from this article have been deposited with the DDBJ/EMBL/GenGank data libraries under the following accession numbers: AB218761 (CYP90A9), AB218762 (CYP90A10), AB218763 (CYP90B8), AB277551 (CYP90D7), and AB218764 (CYP734A11).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The intensity of each band in Figure 4.
Supplemental Figure S2. The intensity of each band in Figure 5.
Supplemental Table S1. Endogenous BR levels in pea tissues.
Supplemental Table S2. Primer sequences used for cloning.
Supplemental Table S3. Primer sequences used for RT-PCR.
Supplementary Material
Acknowledgments
We thank Kyomi Shibata for technical assistance. We also thank Prof. James B. Ried (University of Tasmania, Australia) for originally providing pea seeds and critical reading of the manuscript, and Dr. David Nelson (University of Tennessee) for the P450 designation.
This work was supported by a grant-in-aid for scientific research from the Japan Society for the Promotion of Science (grant no. 1146007 to T.Y.) and by a Research Fellowship from the Japan Society for the Promotion of Science for Young Scientists (to T.N.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Takao Yokota (yokota@nasu.bio.teikyo-u.ac.jp).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bancos S, Nomura T, Sato T, Molnar G, Bishop GJ, Koncz C, Yokota T, Nagy F, Szekeres M (2002) Regulation of transcript levels of the Arabidopsis cytochrome p450 genes involved in brassinosteroid biosynthesis. Plant Physiol 130 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Nomura T, Yokota T, Harrison K, Noguchi T, Fujioka S, Takatsuto S, Jones JDG, Kamiya Y (1999) The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc Natl Acad Sci USA 96 1761–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA (1998) The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Tanaka A, Noguchi T, Fujioka S, Takatsuto S, Ross AS, Tax FE, Yoshida S, Feldmann KA (2000) Lesions in the sterol Δ7 reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J 21 431–443 [DOI] [PubMed] [Google Scholar]
- Fujioka S, Takatsuto S, Yoshida S (2002) An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol 130 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54 137–164 [DOI] [PubMed] [Google Scholar]
- Fujita S, Ohnishi T, Watanabe B, Yokota T, Takatsuto S, Fujioka S, Yoshida S, Sakata K, Mizutani M (2006) Arabidopsis CYP90B1 catalyses the early C-22 hydroxylation of C27, C28 and C29 sterols. Plant J 45 765–774 [DOI] [PubMed] [Google Scholar]
- Fukuta N, Fukuzono K, Kawaide H, Abe H, Nakayama M (2005) Physical restriction of pods causes seed size reduction of a brassinosteroid-deficient faba bean (Vicia faba). Ann Bot (Lond) 97 65–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y, et al (2002) Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J 32 495–508 [DOI] [PubMed] [Google Scholar]
- Jager CE, Symons GM, Nomura T, Yamada Y, Smith JJ, Yamaguchi S, Kamiya Y, Weller JL, Yokota T, Reid JB (2007) Characterization of two brassinosteroid C-6 oxidase genes in pea. Plant Physiol 143 1894–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JG, Yun J, Kim DH, Chung KS, Fujioka S, Kim JI, Dae HW, Yoshida S, Takatsuto S, Song PS, et al (2001) Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell 105 625–636 [DOI] [PubMed] [Google Scholar]
- Kim TW, Hwang JY, Kim YS, Joo SH, Chang SC, Lee JS, Takatsuto S, Kim SK (2005) Arabidopsis CYP85A2, a cytochrome P450b, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 17 2397–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272 398–401 [DOI] [PubMed] [Google Scholar]
- Merlot S, Giraudat J (1997) Genetic analysis of abscisic acid signal transduction. Plant Physiol 114 751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Nomura T, Ooka H, Ishizaka M, Yokota T, Sugimoto K, Okabe K, Kajiwara H, Satoh K, Yamamoto K, et al (2002) Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol 130 1152–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S, et al (1999) BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA 96 15316–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Bishop GJ, Kaneta T, Reid JB, Chory J, Yokota T (2003) The LKA gene is a Brassinosteroid Insensitive 1 homolog of pea. Plant J 36 291–300 [DOI] [PubMed] [Google Scholar]
- Nomura T, Jager CE, Kitasaka Y, Takeuchi K, Fukami M, Yoneyama K, Matsushita Y, Nyunoya H, Takatsuto S, Fujioka S, et al (2004) Brassinosteroid deficiency due to truncated steroid 5α-reductase causes dwarfism in the lk mutant. Plant Physiol 135 2220–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Kitasaka Y, Takatsuto S, Reid JB, Fukami M, Yokota T (1999) Brassinosteroid/sterol synthesis and plant growth as affected by lka and lkb mutations of pea. Plant Physiol 119 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Kushiro T, Yokota T, Kamiya Y, Bishop GJ, Yamaguchi S (2005) The last reaction producing brassinolide is catalyzed by cytochrome P-450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. J Biol Chem 280 17873–17879 [DOI] [PubMed] [Google Scholar]
- Nomura T, Nakayama M, Reid JB, Takeuchi Y, Yokota T (1997) Blockage of brassinosteroid biosynthesis and sensitivity causes dwarfism in garden pea. Plant Physiol 113 31–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Sato T, Bishop GJ, Kamiya Y, Takatsuto S, Yokota T (2001) Accumulation of 6-deoxocathasterone and 6-deoxocastasterone in Arabidopsis, pea and tomato is suggestive of common rate-limiting steps in brassinosteroid biosynthesis. Phytochemistry 57 171–178 [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Szatmari A-M, Watanabe B, Fujita S, Bancos S, Koncz C, Lafos M, Shibata K, Yokota T, Sakata K, et al (2006) C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18 3275–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards DE, King KE, Ait-ali T, Harberd N (2001) How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signalling. Annu Rev Plant Physiol Plant Mol Biol 52 67–88 [DOI] [PubMed] [Google Scholar]
- Rock CD, Quatrano RS (1995) The role of hormones during seed developments. In PJ Davies, ed, Plant Hormones—Physiology, Biochemistry and Molecular Biology, Ed 2. Kluwer, Utrecht, The Netherlands, pp 671–697
- Schultz L, Kerckhoffs LH, Klahre U, Yokota T, Reid JB (2001) Molecular characterization of the brassinosteroid-deficient lkb mutant in pea. Plant Mol Biol 47 491–498 [DOI] [PubMed] [Google Scholar]
- Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, Yokota T, Kamiya Y, Bishop GJ, Yoshida S (2001) Bassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol 126 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber CM, McCourt P (2001) A role for brassinosteroids in germination in Arabidopsis. Plant Physiol 125 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stündl U, Schneider B (2001) 3-β-Brassinosteroid dehydrogenase activity in Arabidopsis and tomato. Phytochemistry 58 989–994 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Inoue T, Fujioka S, Takatsuto S, Yanagisawa T, Yokota T, Murofushi N, Sakurai A (1994) Possible involvement of 3-dehydroteasterone in the conversion of teasterone to typhasterol in cultured cells of Catharanthus roseus. Biosci Biotechnol Biochem 58 1186–1188 [Google Scholar]
- Symons GM, Reid JB (2004) Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol 135 2196–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons GM, Schultz L, Kerckhoffs LHJ, Davies NW, Gregory D, Reid JB (2002) Uncoupling brassinosteroid levels and de-etiolation in pea. Physiol Plant 115 311–319 [DOI] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85 171–182 [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Omigawa Y, Ogasawara M, Yoneyama K, Konnai M, Worsham AD (1997) Effects of brassinosteroids on conditioning and germination of clover broomrape seeds. Plant Growth Regul 16 153–160 [Google Scholar]
- Toyomasu T, Kawaide H, Mitsuhashi W, Inoue Y, Kamiya Y (1998) Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol 118 1517–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Denzel MA, Torres QI, Neff MM (2003) CYP72B1 inactivates brassinosteroid hormones: an intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol 133 1643–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Wakizuka T, Hirai K, Fujii S, Fujita A (1987) Stimulation of germination in aged rice seed by pre-treatment with brassinolide. Proceedings of the 14th Annual Plant Growth Regulation Society of America Meeting. Plant Growth Regulation Society of America, Honolulu, pp 26–27
- Yokota T, Matsuoka T, Koarai T, Nakayama M (1996) 2-Deoxybrassinolide, a brassinosteroid from Pisum sativum seed. Phytochemistry 42 509–511 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.