Abstract
Phase II of germination represents a key developmental stage of plant growth during which imbibed seeds either enter stage III of germination, completing the germination process via radicle protrusion, or remain dormant. In this study, we analyzed the influence of the peroxisomal ATP-binding cassette transporter COMATOSE (CTS) on the postimbibition seed transcriptome of Arabidopsis (Arabidopsis thaliana) and also investigated interactions between gibberellin (GA) and CTS function. A novel method for analysis of transcriptome datasets allowed visualization of developmental signatures of seeds, showing that cts-1 retains the capacity to after ripen, indicating a germination block late in phase II. Expression of the key GA biosynthetic genes GA3ox1 and 2 was greatly reduced in cts seeds and genetic analysis suggested that CTS was epistatic to RGL2, a germination-repressing DELLA protein that is degraded by GA. Comparative analysis of seed transcriptome datasets indicated that specific cohorts of genes were influenced by GA and CTS. CTS function was required for expression of the flavonoid biosynthetic pathway. Confocal imaging demonstrated the exclusive accumulation of flavonoids in the epidermis of wild-type seeds. In contrast, flavonoids were absent from cts and kat2-1 mutant seeds, but accumulated following the application of sucrose, indicating an essential role for β-oxidation in inducing flavonoid biosynthetic genes. These results demonstrate that CTS functions very late in phase II of germination and that its function is required for the expression of specific gene sets related to an important biochemical pathway associated with seedling establishment and survival.
Germination is classically described by three phases of water uptake during which the seed imbibes (phase I) and reinitiates metabolic processes (phase II) prior to the emergence of the radicle through the endosperm and testa (Bewley and Black, 1994; Bewley, 1997; Muller et al., 2006). Subsequently, phase III, which is concurrent with radicle elongation, occurs before seedling establishment. Phase II represents a key development stage following which radicle elongation commences or a nongerminating dormant (D) state is maintained. Dry seeds require a period of after ripening for this capacity for dormancy to be lost, and both genetic and environmental cues are integrated by the seed to determine dormancy/germination status (Bewley and Black, 1994; Koornneef et al., 2002).
It is unclear precisely how developmental signaling results in decisions either to maintain dormancy or to initiate radicle protrusion, although the phytohormones abscisic acid and gibberellin (GA) have been shown to play opposing roles in repressing or increasing germination potential, respectively. Members of the DELLA protein domain family (in particular RGL2) specifically repress germination (Lee et al., 2002) and biochemical studies have shown that GA promotes germination by activating the degradation of DELLA proteins via the 26S proteasome pathway (Silverstone et al., 2001; Fu et al., 2004; Tyler et al., 2004). GA synthesis in imbibed seeds is activated by phytochrome and cold treatment, leading to the up-regulation of germination-associated genes (Yamaguchi et al., 1998; Yamauchi et al., 2004). Application of exogenous GA to the ga1-3 mutant that is disrupted in GA biosynthesis and cannot complete germination led to enhanced germination potential and up-regulation of more than 200 genes (Ogawa et al., 2003). Proteomic analysis of Arabidopsis (Arabidopsis thaliana) seeds during germination also shows an important role for GA in regulating specific germination-related protein components (Gallardo et al., 2002; Rajjou et al., 2004). Experiments at both transcriptome and proteome levels indicate that GA plays a relatively late role in processes associated with germination (Gallardo et al., 2002; Ogawa et al., 2003).
The Arabidopsis COMATOSE (CTS) gene was originally identified in a genetic screen for loci that promote germination (Russell et al., 2000). CTS is a single-copy gene encoding a full-length ATP-binding cassette transporter that is required for the import of several biologically important molecules into the peroxisome, including not only very-long-chain fatty acids associated with breakdown of seed-storage lipids, but also precursors of auxin and jasmonic acid biosynthesis (Zolman et al., 2001; Footitt et al., 2002; Hayashi et al., 2002; Theodoulou et al., 2005). Severe mutant alleles of CTS demonstrate an inability to complete germination, suggesting a role for CTS in the dormancy to radicle protrusion transition (Russell et al., 2000; Footitt et al., 2002), and protein profiles of cts seeds resembled those of the D wild type. Therefore, we proposed that cts seeds may remain in a forever-D state (Russell et al., 2000). In this study, we employed genome-profiling approaches to understand the influence of CTS on gene expression in the imbibed seed and to define the developmental status of cts mutants at the transcriptome level.
Transcriptome analysis has previously been used to identify not only the stored (S) mRNA population and the effect of imbibition, but also the changes in gene expression in relation to dormancy status (Nakabayashi et al., 2005; Cadman et al., 2006). The latter study identified core sets of genes whose expression is specifically associated with either after-ripened (AR) or D states in Arabidopsis, indicating that changes in abundance of specific cohorts of transcripts are dependent on the developmental status of the seed (Cadman et al., 2006).
Here, we show that CTS action is required late in phase II of germination, following after ripening. We conclude that CTS function is required for the expression of key biosynthetic genes and that CTS function is epistatic to RGL2. We also demonstrate that CTS expression is absolutely required for the up-regulation of the flavonoid biosynthetic pathway in the epidermis of the imbibed AR embryo.
RESULTS
Transcriptome Analysis of Wild-Type and cts-1 Seeds
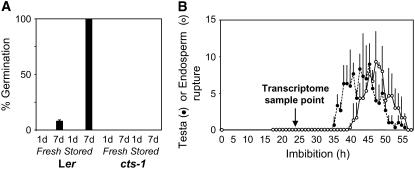
Transcriptome analysis was carried out using wild-type and cts-1 mutant seed samples to determine the influence of the mutation on seed transcript populations. The germination potential of Arabidopsis seeds consists of two time-dependent variables: dry after ripening and imbibition time. Whereas imbibed wild-type seeds will complete germination following a period of dry after ripening, imbibed, intact cts-1 seeds do not (Russell et al., 2000; Fig. 1A). We followed the frequency of testa and endosperm rupture in fully AR Landsberg erecta (Ler) wild-type seeds and showed that testa rupture occurs between 40 to 45 h and subsequent endosperm rupture (due to radicle elongation) occurs between 45 to 50 h (Fig. 1B). Transcriptomes of freshly harvested and S wild-type and mutant seeds were analyzed at 24-h imbibition, a time point well before testa and endosperm rupture, when all seeds were in phase II of germination (Fig. 1B). Freshly harvested wild-type samples are subsequently referred to as D and S as AR, whereas mutant samples are referred to as fresh (F) and S to avoid conferring any particular physiological status on the mutant seeds.
Figure 1.
Germination characteristics of wild-type and cts-1 mutant seeds. A, Germination potential of freshly harvested or S (AR) wild-type and cts-1 mutant seeds used for microarray analysis. Germination was assessed at 1 d (24 h) and 7 d on WA media. Values shown are averages (+sd) of triplicate experiments using seeds derived from different plants. B, Frequency of testa (black circles, dotted line) and endosperm (white circles, solid line) rupture of AR Ler wild-type seeds. Values shown are average of triplicate experiments using seeds derived from different plants. Error bars indicate sd.
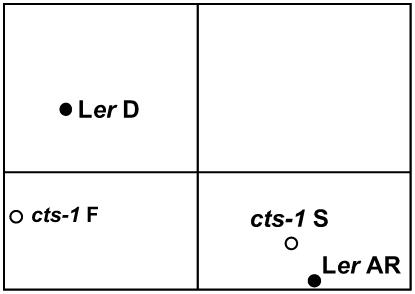
Total RNA was extracted from three biological replicates and transcriptome profiles were obtained using the Affymetrix ATH1 24K Genechip array (derived gene sets are presented in Supplemental Table S1). Total numbers of differentially expressed genes are shown in Table I. As expected, large numbers of genes were differentially expressed in D wild-type seeds in comparison to AR seeds. Differential gene expression was also observed for transcriptomes of cts-1 F compared to S seeds. A smaller number of genes were also differentially expressed between Ler AR and cts-1 S seeds. This latter comparison included CTS as the second most differentially regulated transcript (28-fold more highly expressed in wild type than cts-1; Supplemental Table S1). Principal component analysis (PCA) was used to analyze differences between the Ler and mutant seed transcriptome datasets (Fig. 2). This analysis showed clear differences between the Ler D and AR and cts-1 S and F states, which were well separated in the first dimension (2,377 genes accounting for 66% of the variation), and identified similarities between the cts-1 S and Ler AR and the cts-1 F and Ler D states. This suggests that at the transcriptome level, cts-1 S seeds closely resemble those of Ler AR.
Table I.
Numbers of genes differentially regulated between RNA populations derived from imbibed wild-type and cts-1 seeds
Gene lists are presented in Supplemental Table S1.
| Comparison | Comparison Up-Regulated Gene Seta | No. Differentially Up-Regulated Genesa |
|---|---|---|
| Ler D versus AR | D up | 1,595 |
| AR up | 1,945 | |
| cts-1 F versus cts-1 S | cts-1 F up | 1,111 |
| cts-1 S up | 1,465 | |
| Ler AR versus cts-1 S | cts-1 S up | 561 |
| Ler AR up | 465 |
Filter on fold change >2.
Figure 2.
PCA applied to seed transcriptome datasets. Black circles indicate Ler wild-type D or AR seeds; white circles cts-1 mutant F or S seeds.
Using Transcriptome Signatures to Define Developmental States
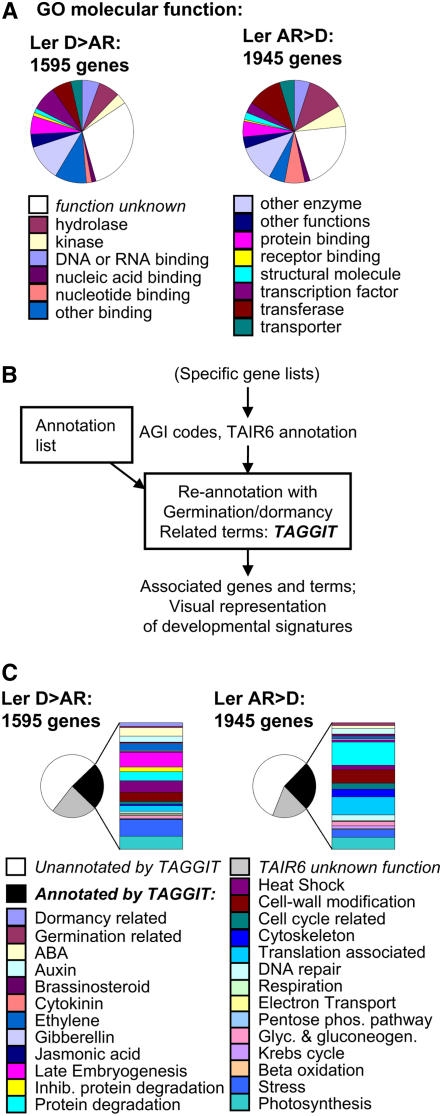
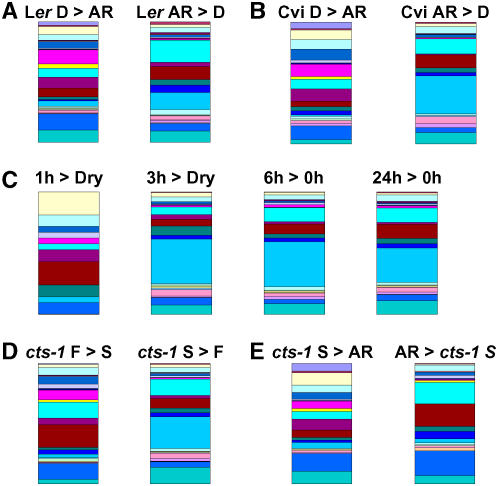
Previous transcriptome studies have employed the Gene Ontology (GO) annotations of molecular function in primary analyses of gene datasets. Analysis of gene lists using this approach provided information for between 70% and 80% of genes (Fig. 3A). However, we found that the ontological classification supplied by GO terms failed to provide much useful biological information in relation to known processes involved in dormancy and germination that could be used to understand the developmental status of the seed (Fig. 3A). Therefore, an approach was adopted whereby lists of differentially expressed genes obtained from transcriptome experiments were analyzed by annotating genes in relation to previously reported functions associated with embryo maturation, dormancy, or germination. A workflow was developed (TAGGIT; Fig. 3B) for reannotation of gene lists using more than 1,000 specific Arabidopsis Genome Initiative (AGI) codes and The Arabidopsis Information Resource (TAIR) 6 annotations derived from a review of the germination literature (Supplemental Table S2; Supplemental Dataset S1). It is important to note that this analysis is intended to provide information about the relative representation of genes with confirmed functions associated with dormancy and germination and thus indicate the developmental status of the transcriptome. For all analyses, this annotation provided information for between one-fourth and one-half of TAIR6-annotated, differentially expressed genes (Fig. 3C). Signatures from Ler D or AR samples were easily distinguishable (Figs. 3C and 4A). As expected, AR-specific transcriptomes are dominated by genes encoding proteins associated with processes including cell wall modification, translation, cytoskeleton function, and mobilization of storage lipids. The D-specific transcriptome includes transcripts encoding proteins associated with embryo maturation/storage, heat shock, dehydrins, and protease inhibitors. Publicly available Affymetrix array datasets from experiments carried out by other researchers were also used to validate this approach. The recently published transcriptome analysis of different dormancy states in the Arabidopsis accession Cape Verde Islands (Cvi; Cadman et al., 2006) was analyzed using the TAGGIT workflow (Fig. 4B, Supplemental Table S4). Resulting developmental signatures of both Cvi D- and AR-related transcriptomes showed similar profiles and representations of functional groups as those produced from transcriptome datasets of Ler. To provide a temporal analysis of changes in developmental signatures associated within early stages of germination, a previously reported germination time-course transcriptome dataset available at Nottingham Arabidopsis Stock Centre (NASC) arrays was reanalyzed (Nakabayashi et al., 2005; Fig. 4C; Supplemental Table S4). These analyses show that, in comparison with dry seeds, an easily identifiable germination transcriptome signature is evident 3 h after the onset of imbibition. This profile does not change significantly over the subsequent 24 h of imbibition. Comparison of signatures obtained from transcripts differentially regulated in imbibed cts-1 S seeds compared with cts-1 F seeds showed that these gene sets contained transcripts encoding genes of very different functions (Fig. 4D; Supplemental Table S1). Comparison with Ler shows that the signature associated with the transcriptome up-regulated in cts-1 S seeds (compared to cts-1 F seeds) closely resembled the Ler AR seed signature and, for cts-1 F seeds (compared to cts-1 S seeds), resembled the Ler D seed signature. Analysis of genes differentially expressed between the Ler AR and cts-1 S transcriptomes (Fig. 4E) showed that imbibed cts-1 S seeds contain some transcripts associated with dormancy.
Figure 3.
Analysis of transcriptome profiles using the GO molecular function or TAGGIT approaches. A, Ontological classification of genes in the Ler D > Ler AR or Ler AR > Ler D sets using GO molecular function. The proportional representation of the total gene set is shown in each case; genes of unknown function using this methodology are colored white. B, Flow diagram illustrating the workflow used to reannotate gene lists using the TAGGIT macro. C, Classification of genes in the Ler D > Ler AR or Ler AR > Ler D sets using the TAGGIT macro. The proportional representation of the total gene set is shown in each case, TAIR6 annotated genes of no known function are shown in gray, TAGGIT unannotated genes in white, and TAGGIT annotated genes in black, expanded by the side to show proportional representation of individual classes. Datasets derived from Supplemental Table S1. In each case, > indicates the up-regulated gene set in the comparison.
Figure 4.
Comparisons of developmental signatures of differentially expressed transcriptomes. A, Comparison of genes differentially expressed in Ler AR and D 24-h imbibed seed samples. B, Comparison of genes differentially expressed in accession Cvi AR and D samples (Cadman et al., 2006). Dataset obtained from NASCarrays. C, Comparison of genes differentially expressed from seeds imbibed for 0 to 24 h (Nakabayashi et al. 2005). According to the design of the reported experiments, early time points were compared to unimbibed (dry) seeds, later time points to 0-h imbibed seeds. D, Comparison of genes differentially expressed in cts-1 S and F 24-h imbibed seed samples. E, Comparison of genes differentially expressed in cts-1 S and Ler AR 24-h imbibed seed samples. Each graph shows gene-set name and the proportional representation of the genes identified using the TAGGIT workflow (see Table I for total numbers of differentially expressed genes; Fig. 3 for key to gene groups). Percentage of genes identified using the TAGGIT workflow in each comparison are given in corresponding Supplemental figures. In each case, > indicates the up-regulated gene set in the comparison.
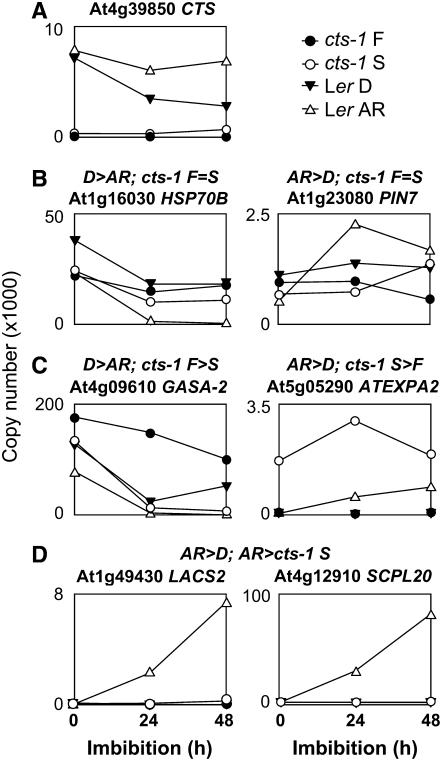
Quantitative reverse transcription (RT)-PCR was used to verify classes of expression patterns observed in transcriptome analyses and to provide a time-course analysis of gene expression in cts-1 and Ler seeds of both D/F and AR/S developmental states over 48-h imbibition (Fig. 5). The CTS gene has previously been shown to be under translational regulation (Footitt et al., 2002). Consistent with this observation, CTS transcripts were present in both D and AR Ler seeds (Fig. 5A). At least five patterns of expression were observed and representative genes showing these patterns in the original transcriptome datasets were analyzed. HSP70B and PIN7 represent genes showing opposite expression patterns in wild-type seeds, but that were expressed at similar levels in cts-1 F and S seeds at 24 h (Fig. 5B). In contrast, the genes GASA-2 and ATEXPA2 were regulated in a similar way in both wild-type and mutant seeds (Fig. 5C). Finally, many genes were highly up-regulated in wild-type AR seeds in comparison to cts-1 S (examples shown are LACS2 and SCPL20; Fig. 5D).
Figure 5.
Gene expression profiles during 48 h following the initiation of imbibition for Ler and cts-1 seeds. Quantitative RT-PCR analysis was carried out using RNA obtained from Ler D and AR and cts-1 S and F seeds imbibed on WA over 48 h. AGI codes and names of encoded proteins are indicated for each transcript. In each case, the expression characteristics of genes in transcriptome datasets at 24-h imbibition are given as titles above graphs. A, Expression of CTS. B, Transcripts showing no change in expression in cts-1 S or F seeds. C, Transcripts differentially regulated in cts-1 S or F seeds. D, Transcripts showing highly induced expression in Ler AR seeds only.
Analysis of the Relationship between CTS Function and GA during Germination
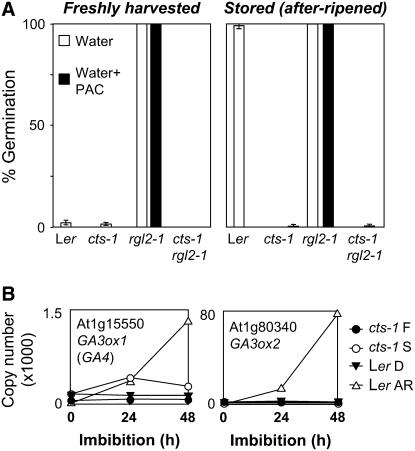
Previously published transcriptome and physiological analyses suggested that GA regulates gene expression late in phase II of germination (Ogawa et al., 2003). To investigate the relationship between CTS and GA, we used a mutant in the GA signal transduction pathway (rgl2-1) that lacks a DELLA domain protein known to be a major repressor of GA function during germination (Lee et al., 2002). We analyzed the influence of cts-1 on the germination potential of rgl2-1 mutant seeds using plants homozygous for both mutations. We analyzed germination using both freshly harvested and AR seed populations to define the germination potential of single- and double-mutant seeds (Fig. 6A). Only freshly harvested rgl2-1 seeds showed a high germination potential. Following after ripening, Ler seeds could complete germination on media without paclobutrazol (PAC; an inhibitor of GA biosynthesis), as did rgl2-1 seeds. Both F and AR rgl2-1 seeds were resistant to PAC, whereas AR Ler seeds were not. Double-mutant rgl2-1 cts-1 seeds showed very low germination potential on all media tested, even following storage that allowed complete after ripening of Ler seeds (Fig. 6A). This suggests that CTS is epistatic to RGL2. Expression of the GA biosynthesis genes AtGA3ox1 and 2 has been shown to be activated during germination (Yamaguchi et al., 1998; Yamauchi et al., 2004). In agreement with this result, quantitative RT-PCR analysis showed that the expression of both genes was increased in Ler AR in comparison with D seeds and this occurs maximally at 48 h in association with testa and endosperm rupture (Figs. 1B and 6B). Analysis of cts-1 showed that expression of GA3ox1 initially mirrored that of wild-type AR seeds, but subsequently declined, and expression of GA3ox2 was very low and did not increase during imbibition.
Figure 6.
Germination potential of cts-1 rgl2-1 seeds and expression of GA biosynthesis genes in wild-type and cts-1 seeds. A, Germination potential after 7 d is shown for freshly harvested and S/AR seed lots assayed on WA media either with or without exogenous PAC (10 μm). Data represent mean and sd. B, Quantitative RT-PCR analysis of genes encoding AtGA3ox1 and 2 in Ler and cts-1 seeds. RNA was obtained from Ler D and AR and cts-1 S and F seeds imbibed on WA over 48 h. AGI numbers and function of encoded proteins are indicated for each transcript.
Using Comparative Transcriptomics to Define Subsets of Transcripts Requiring CTS and GA for Expression
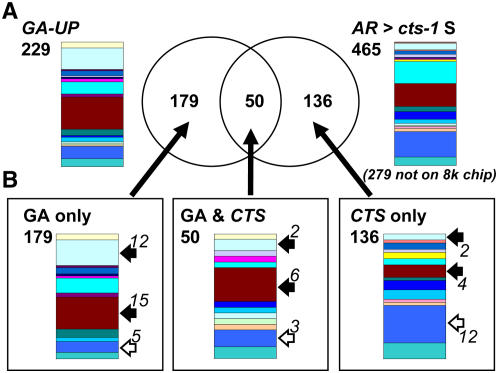
Previously, Ogawa et al. (2003) used a small 8,200 gene array to define more than 200 genes up-regulated by GA4 within 12 h of application. Analysis of these genes using the TAGGIT workflow demonstrated that this gene set had a developmental signature that was similar to that of the sets Ler AR > cts-1 S, Ler AR > Ler D, and cts-1 S > cts-1 F (Figs. 4 and 7A; Supplemental Table S3).
Figure 7.
Comparison of transcriptomes up-regulated by GA4 treatment of ga1-3 seeds and up-regulated in Ler in comparison to cts-1. A, Venn diagram resulting from comparison of GA4 up-regulated gene set (229 genes) with the Ler AR > cts-1 S gene set (465 genes, 279 not on the Affymetrix 8K Genechip). Transcriptome dataset for GA4 treatment of ga1-3 seeds (GA up-regulated) was obtained from Ogawa et al. (2003). Differential gene expression is represented by Venn diagrams. Original developmental signatures and total numbers of genes from these gene sets are shown next to the Venn diagram. Numbers of genes represented in both gene sets are shown within the intersections of the two sets. B, Representations of the developmental signatures of genes from the different Venn diagram sets. Specific functional groups are highlighted with white (increased representation in the CTS set) or black (increased representation in the GA set) arrows. Numbers next to arrows indicate absolute numbers of genes in each functional category of each set. Percentage of genes identified using the TAGGIT workflow in each comparison are given in Supplemental figures. Colors correspond to functional categories shown in Figure 3.
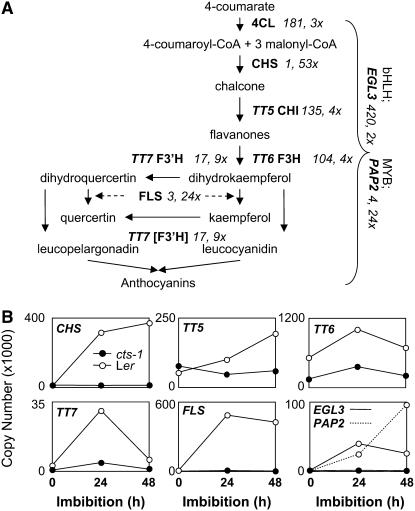
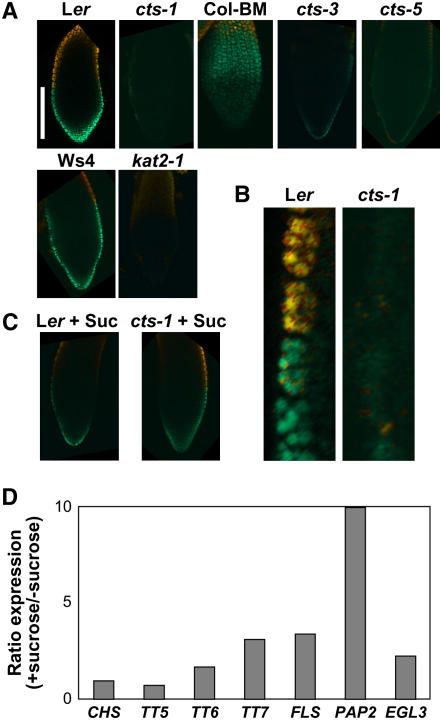
To understand the relationship between GA and CTS regulation of the imbibed seed transcriptome, we compared the Ogawa GA up-regulated genes with the Ler AR > cts-1 S seed gene set (Fig. 7). A total of 465 genes are up-regulated in the Ler AR compared to cts-1 S seeds and, of these, 186 were represented by probes on the 8K Affymetrix chip used by Ogawa et al. (2003). Comparison of these gene sets revealed cohorts of commonly and independently expressed sets of genes (Fig. 7B; Supplemental Table S3). Quantitative RT-PCR analysis demonstrated that genes requiring both GA and CTS for expression showed strong germination-related gene expression (e.g. LACS2; Fig. 5D) and were not expressed either in Ler D seeds or either cts-1 F or S seeds. The 179 genes of the GA-only class represent transcripts that require GA for expression but do not require CTS (e.g. ATEXPA2; Fig. 5C), and the remaining 136 genes within the CTS-only class represent those requiring only CTS for expression. Different protein functional groups were overrepresented in each class; for example, more genes encoding cell wall modification enzymes or functions associated with auxin biology were present in the GA-only class (Fig. 7B), whereas more genes associated with stress were present in the CTS-only class. Further analysis revealed that this gene set contained genes associated with flavonoid biosynthesis and transcription factors associated with anthocyanin gene regulation. Analysis of the original Ler AR > cts-1 S seed gene set revealed that these genes represent some of the most highly differentially regulated genes (Fig. 8A; Supplemental Table S1). For example, the CHS gene was the most differentially regulated transcript in the Ler AR > cts-1 S seed gene set. Quantitative RT-PCR analysis showed that these genes were all strongly induced during phase II germination between 24 to 48 h, but not in S cts-1 seeds (Fig. 8B). The accumulation of flavonoids preceding germination was analyzed using diphenyl boric acid (DPBA) fluorescence for cts-1, cts-3, cts-5, kat2-1, and respective wild-type seeds (Fig. 9; Peer et al., 2001). Both cts-3 and cts-5 represent strong nongerminating missense alleles (H. Schmuths, F. Theodoulou, A. Baker, and M. Holdsworth, unpublished data). The kat2-1 mutation results in a lack of peroxisomal thiolase activity (Germain et al., 2001), preventing peroxisomal β-oxidation and germination (Pinfield-Wells et al., 2005; Footitt et al., 2006). Previous analysis has shown that different flavonoid-DPBA conjugates exhibit unique fluorescence colors (Peer et al., 2001); we have improved this technique using confocal laser-scanning microscopy followed by metaanalysis to examine accumulation and distribution of flavonols. Flavonoid accumulation appeared to be restricted to the epidermis of the radicle and cotyledons of imbibed wild-type embryos. Both quercetin and kaempferol were observed in the epidermis of the radicle of all three wild-type accessions at 24-h imbibition (Ler, Wassilewskija [Ws]-4, and Columbia [Col]-BM; Fig. 9A), with some variation in distribution patterns between accessions. For example, in Ler, kaempferol accumulation was observed throughout the epidermis, with quercetin fluorescence observed in a discrete boundary present approximately halfway up from the radicle tip (Fig. 8B). In the mutants analyzed, either no or very faint fluorescence corresponding to quercetin and kaempferol was observed. Addition of Suc to the media allowed accumulation of flavonoids in the cts-1 mutant by 24 h and led to up-regulation of several flavonoid biosynthetic genes (Fig. 9, C and D).
Figure 8.
Expression of genes of the flavonoid biosynthesis pathway in wild-type and cts-1 seeds. A, Schematic representation of flavonoid biosynthesis (redrawn from Peer et al., 2001). Enzymes are shown with associated position in the Ler AR > cts-1 S seed gene set and fold differential up-regulation in AR seeds (see Supplemental Table S1). 4CL, 4-Coumarate-CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone-3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; FLS, flavonol synthase; TT, transparent testa; PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2; EGL3, ENHANCER OF GLABRA 3. B, Quantitative RT-PCR analysis of gene expression in wild-type Ler and cts-1 S seeds imbibed on WA.
Figure 9.
Visualization of flavonoid accumulation in 24-h imbibed radicles of mutant and wild-type seeds. Orange fluorescence corresponds to quercetin-DPBA, green fluorescence to kaempferol-DPBA, and yellow fluorescence to colocalization of flavonoids. Respective wild-type seedlings are shown next to mutants (Ler, cts-1; Col-BM, cts-3, 5; Ws4, kat2-1). A, Bar represents 100 μm. B, Closeup of epidermis of Ler and cts-1. C, Visualization of flavonoids in Ler and cts-1 seed radicles incubated in the presence of 0.5% Suc. D, Quantitative RT-PCR analysis of flavonoid-associated gene expression in cts-1 seeds imbibed 24 h on WA in the presence of Suc (germination after 7 d: +Suc 11%, −Suc 0%).
DISCUSSION
This article reports the characterization of gene expression associated with phase II of the germination pathway in Arabidopsis prior to radicle emergence and investigates the effect of the peroxisomal ATP-binding cassette transporter CTS on the imbibed seed transcriptome. Other transcriptome comparisons have recently been carried out in relation to seed maturation, dry seeds, GA-related control of postimbibition gene expression, and dormancy status (Ruuska et al., 2002; Ogawa et al., 2003; Yamauchi et al., 2004; Nakabayashi et al., 2005; Cadman et al., 2006). Our analysis focused on seeds with defined temporal characteristics, both for storage time (and therefore after-ripening status), and imbibition time (and therefore germination phase) to understand the nature of the developmental defect of the cts-1 mutation.
Previously reported transcriptome studies of dormancy and germination used conventional tags for functional analysis of genes (Nakabayashi et al., 2005; Cadman et al., 2006; MIPS or KEGG annotation according to the GO consortium classification of genes). We found this form of analysis unsatisfactory because these classification systems do not reflect specific processes previously shown to be important for seed biology. Therefore, we developed a workflow based on the Excel macro TAGGIT that reannotated gene lists produced from transcriptome comparisons according to user-defined search terms (including AGI codes and the TAIR6 annotation of the Arabidopsis genome [http://www.arabidopsis.org]). This analysis produces a visual description of the representation of specific groups of genes in lists and can be used to deduce developmental status. As the functions of more genes are elucidated, this methodology can be expanded to accommodate relevant novel information, thereby increasing the proportion of genes that can be annotated. Using this system, wild-type AR- and D-specific transcriptomes can be easily discriminated (Fig. 4, A and B). In addition, we could use this system with previously published datasets to generate useful new information (e.g. that a germination transcriptome profile becomes evident within the first 3 h, information not readily apparent by conventional transcriptome analysis; Fig. 4C). In combination with PCA of transcriptome datasets, the TAGGIT workflow showed that cts-1 seeds do after ripen at the transcriptome level, as suggested by previous physiological analyses (Footitt et al., 2006). Therefore, CTS does not influence after ripening, but is involved in germination-specific processes following after ripening. The TAGGIT workflow was also used to show that there were no differences in the representation of transcripts encoding genes for cell expansion (Dolan and Davies, 2004) between cts-1 S and Ler AR seed transcriptomes (data not shown), demonstrating that cts-1 S seeds are blocked after the accumulation of gene sets associated with postgerminative processes. Analysis of developmental signatures also showed that cts-1 S seeds contain a small number of transcripts associated with the D state (Fig. 4E), indicating that some aspects of the D transcriptome are retained in the mutant seeds. These transcripts may require the action of biochemical pathways associated with CTS function for removal.
Quantitative RT-PCR analysis revealed at least five different gene expression patterns and validated the observed gene chip hybridization results (Fig. 5). In agreement with other germination transcriptome studies (Ogawa et al., 2003; Toorop et al., 2005; Cadman et al., 2006), we observed both D- and AR-specific expression patterns in wild-type seeds, most notably the up-regulation of genes associated with translation, cytoskeleton function, cell wall modification, and auxin transport in AR seeds (Fig. 5; Supplemental Table S1). Our analysis of the D transcriptome corresponds to the PD24h (primary dormancy 24 h imbibed) state investigated by Cadman et al. (2006), who identified in the accession Cvi a core set of 442 genes specific to five D seed states. Of the genes shown to be more highly expressed in D than AR Ler seeds in this study, 275 were shared with this 442 gene set, and of the 779 shown by Cadman et al. (2006) to be AR-specific in Cvi, 358 were shared in common with the Ler AR-specific transcripts reported here. In both cases, and for Col-0 at 24-h imbibition (Nakabayashi et al., 2005), developmental signatures were highly similar, indicating that transcriptome analyses were comparable across different accessions.
The relationship between GA promotion of germination and CTS function was analyzed to determine the interaction between these two activators of germination. It has recently been suggested that GA acts late in phase II (Gallardo et al., 2002; Ogawa et al., 2003; Yamauchi et al., 2004). Previously, we showed that cts-1 mutant seeds were insensitive to the germination enhancement of applied GA, indicating that CTS function may be required later than GA action in phase II (Russell et al., 2000). To understand the genetic relationships between CTS and GA signaling, we analyzed the germination potential of cts-1 rgl2-1 double-mutant seeds. The cts-1 rgl2-1 double mutant demonstrated a nongerminating phenotype even after dry seed storage (Fig. 6). Freshly harvested rgl2-1 seeds were shown to be non-D, suggesting that RGL2 is involved in the activation of dormancy-associated processes, in addition to repression of germination, as previously shown (Lee et al., 2002). One function of CTS may therefore be required after GA signal transduction in phase II. However, quantitative RT-PCR analysis showed that the genes encoding GA3ox1/2 were not up-regulated in cts-1 S seeds as they were in Ler AR seeds (Fig. 6B). The AtGA3ox1 transcript is known to be regulated by negative feedback following GA treatment of ga1-3 seeds, whereas AtGA3ox2 transcripts appear to show altered timing of expression (Yamaguchi et al., 1998). Our results demonstrated that, in wild-type seeds, both genes showed maximal activation at 48-h imbibition, at around the time of testa and endosperm rupture. The lack of transcript accumulation in cts-1 suggests that CTS is required for the expression of these genes and suggests positive feedback of CTS function.
A comparative approach using transcriptomics datasets was taken to define the relationship between CTS and GA-regulated gene expression during germination (Fig. 7). The developmental signature of the GA up-regulated gene set presented in Ogawa et al., (2003) resembled that of Ler AR seeds, demonstrating that GA treatment induces a germination-like gene expression pattern (Fig. 7A). The developmental signature of the cts-1 S > cts-1 F seed gene set also resembled that of Ler AR seeds. This indicates that CTS functions very late in phase II, after the germination transcriptome is activated, and within a similar developmental window to that of GA. It was noted by Ogawa et al. (2003) that temporal differences in the expression of GA4 up-regulated genes may be related to the involvement of other factors. CTS may represent one of these factors for the gene set common to both. Transcripts that require both CTS and GA for expression were present in all but one of the temporal classes defined by Ogawa et al. (2003; data not shown). This may indicate a close association between GA and CTS action or that the application of GA4 to ga1-3 seeds used by Ogawa et al. (2003) represents an experimental system in which imbibed seeds are already competent to respond to CTS action via after ripening. All functional categories associated with germination were represented in these three gene sets, indicating that both GA and CTS influence the expression of genes representing the same functional categories. Several categories had higher representation in specific groups. In particular, genes encoding enzymes and transcriptional regulators of the flavonoid pathway were exclusively represented in the CTS-only set (Fig. 8). Whereas Ler AR embryos accumulated flavonoids in the epidermal layer of the radicle, no accumulation was observed in several cts mutant alleles, indicating that peroxisomal β-oxidation may be required for flavonoid accumulation before germination. In agreement with this, reduced accumulation was also observed in kat2-1 mutant seeds. Previous work showed that the flavonoid pathway is coordinately regulated following Arabidopsis radicle protrusion in response to light (Kubasek et al., 1992). Growth of Arabidopsis seedlings on sugar induces the anthocyanin biosynthetic pathway and Suc appears to be the most effective inducer of this pathway (Teng et al., 2005; Solfanelli et al., 2006). The observation that peroxisomal β-oxidation may be required for induction of flavonoid biosynthesis in the absence of external sugar suggests that breakdown of stored lipids may provide the source of sugar for the induction of the enzymes of this pathway and for the CoA substrate used for flavonoid biosynthesis. It is known that at least one gene associated with β-oxidation (an acyl-CoA oxidase) is induced by UV light in parsley (Petroselinum crispum), which was presumed to aid the production of precursors for secondary metabolites (Logemann et al., 2000). We previously showed that endogenous Suc levels increase prior to completion of germination in wild-type Ler and Ws-2 seeds (Footitt et al., 2002, 2006), but not in severe cts mutants or other mutants associated with lipid breakdown. It may therefore be that Suc, produced from stored lipid, induces the flavonoid biosynthetic genes specifically in the endosperm. Imbibition of cts-1 in the presence of Suc led to the up-regulation of early flavonoid biosynthetic genes and flavonol accumulation (Fig. 9). We observed increased accumulation of RNAs for the basic helix-loop-helix (bHLH) and MYB transcription factors EGL3 and PAP2 in Ler AR imbibed seeds in comparison to cts-1 S seeds (Fig. 8). Previous studies have revealed an important role for the PAP1 MYB transcription factor in seedling anthocyanin production and that PAP1 is Suc induced following germination (Teng et al., 2005). Both PAP1 and PAP2 are up-regulated in the suc2 Suc transporter mutant, which accumulates high levels of Suc (Lloyd and Zakhleniuk, 2004). Our transcriptome datasets taken at an earlier, pregermination time point did not reveal differential PAP1 expression between Ler AR and cts-1 S, although PAP2 was the fourth most differentially regulated gene (24-fold differentially expressed in the wild type; Supplemental Table S1). This apparent discrepancy in inducibility may be due to different experimental conditions, which included exogenous addition of sugars to the growth medium at high levels and analysis at different developmental stages. It has been postulated that a MYB-basic helix-loop-helix-WDR (MBW) transcription factor complex is required for activation of flavonoid promoters (Lepiniec et al., 2006); therefore, up-regulation of EGL3 and PAP2 would provide two members of this complex. Although we did not detect up-regulation of the WDR protein TTG1, which has been associated with the MBW complex (Baudry et al., 2004), we did observe up-regulation of the WDR-containing gene At3g27640.
We showed that CTS was required for flavonoid accumulation prior to completion of germination. It has been suggested that flavonoid accumulation at the seedling developmental stage provides several important functions, including protection from reactive oxygen species, UV irradiation, pathogens, and modulation of auxin transport. It is possible that flavonols produced at this time point modulate auxin transport (Murphy et al., 2000; Brown et al., 2001) because several key auxin transporters and MDR/PGP proteins are highly up-regulated before germination (Fig. 5; Ogawa, et al. 2003), and flavonoid accumulation in the epidermis of the radicle and cotyledons is consistent with epidermal localization of up-regulated auxin transporters. However, our previous analysis showed that AR ttg-1 cts-1 and tt4-1 cts-1 double mutants supplied with Suc complete germination (Russell et al., 2000) and indicate that flavonoids are not required for completion of germination (Debeaujon et al., 2000). Because one role of flavonoids is antioxidant activity, perhaps their primary role during germination may be to protect against membrane lipid peroxidation due to β-oxidation of seed storage lipids in concert with the described role for vitamin E (Sattler et al., 2004).
In conclusion, we have shown that CTS influences the germination transcriptome at a late stage of phase II prior to testa and endosperm rupture. Comparative transcriptome analysis showed evidence of interactions between CTS and GA in the expression of subsets of the germination transcriptome and that CTS is required for epidermal accumulation of flavonoids and genes regulating flavonoid biosynthesis prior to germination.
MATERIALS AND METHODS
Plant Growth, Seed Storage, and Germination Assays
All plants were grown in controlled-environment rooms (Footitt et al., 2006). All plants used in single experiments were grown at the same time and germination assays and statistical treatment carried out as previously described (Footitt et al., 2006). Freshly harvested D seeds were obtained from yellow siliques. To obtain AR or S material, the seeds were stored in the dark in a controlled-environment incubator at 24°C for the required time period. All seed material was tested for germination potential on water-agarose (WA) media or other media as indicated in the figure legends. For determination of germination potential, seeds on media plates were incubated at 22°C under continuous light for 7 d. The percentage of seeds that had completed germination was recorded using radicle protrusion through the endosperm as the criterion. PAC (10 μm) was added to plant growth media where indicated. For analysis of flavonoids, mutant dry cts-1 seeds were briefly vortexed with glass beads prior to plating, which promotes the completion of germination and permeability of seeds to Suc, which normally does not influence cts-1 germination (Footitt et al., 2006).
RNA Extraction, Affymetrix Microarray Analysis, and Quantitative RT-PCR Analysis
RNA Extraction and Hybridization
For transcriptome analysis, total RNA was extracted from seed material collected after 24-h imbibition on WA using the borate isolation method (modification of protocol from the Cotton Genome Center, University of California, Davis [http://cottongenomeceter.ucdavis.edu/protocols/RNA]). Seed lots were taken from three independent plants constituting three biological replicates. In all cases, germination potential of a companion seed sample was analyzed for 7 d to determine the final germination potential of the sample used for transcriptome analysis. cRNA labeling and hybridizations were performed by NASC Microarray facilities (University of Nottingham). Three biological repetitions were carried out (i.e. each seed lot used was obtained from different plants grown under the same controlled conditions) for each genotype (original expression data for all hybridizations are available at NASC through the NASCarrays service [http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl]). Gene expression levels were measured by detection calls and signal intensities using Microarray Suite 5.0 software (Affymetrix). Further analysis was performed with GeneSpring 7.2 software (Agilent Technologies). Genes showing 2-fold differential expression between samples, with a P value < 0.05 in one-way ANOVA (parametric test without the assumption of equal variances) were used for further analyses. PCA was performed with GeneSpring 7.2 software using mean centering of all genes. RNA for quantitative RT-PCR analysis was isolated as described above. Real-time quantitative PCR of derived cDNA was carried out using the Universal ProbeLibrary system (Roche Diagnostics) in triplicate. Primers and probes for each gene assay were designed using the Universal ProbeLibrary Assay Design Center (https://www.roche-applied-science.com/sis/rtpcr/upl/adc.jsp; Roche Applied Science). A normalization factor was obtained from the geometric mean of three control reference genes chosen from analysis of the microarray dataset (At3g58530, At4g28990, and At5g39710) and was used to obtain relative expression levels (Vandesompele et al., 2002).
TAGGIT Work Flow Analysis
Developmental signatures were obtained using the Microsoft Excel TAGGIT macro to reannotate gene lists (see Supplemental Fig. S1 for workflow; Supplemental Dataset S1 for TAGGIT macro; Supplemental Table S2 for description of gene and search string tag list). Other datasets analyzed here, derived from Cadman et al. (2006) and Nakabayashi et al. (2005), were obtained either from NASCarrays or Dr. Eiji Nambara (RIKEN) and analyzed as described above.
Confocal Laser-Scanning Microscopy and Flavonol Localization
Embryos were excised from seeds imbibed 24 h on WA plates or seeds were nicked and imbibed on WA + 0.5% Suc plates and then stained with 5 mm DPBA (Sigma). Samples were analyzed with an upright Zeiss LSM 510 metaconfocal laser-scanning microscope, Plan-Neofluar 25×/0.8 1 mm corr differential interference contrast objective, using a 405-nm diode laser with 405/488/543-nm dichroic and 410- to 752-nm band pass in λ mode. Detector gain was set to avoid saturated pixels in images and detector offset was set to avoid pixels of zero value in unstained tissue. Data were deconvoluted using fluorescence spectra from genuine flavonoid standards. Background fluorescence was subtracted based on fluorescence spectrum of a DPBA-stained tt4 mutant, which accumulates no flavonoids. Data were acquired in 12-bit depth. Images were processed with LSM 2.8 or 3.2 (Zeiss) and Photoshop 5.0 (Adobe Systems) software.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Dataset S1. TAGGIT Microsoft Excel macro.
Supplemental Table S1. Detailed descriptions of differentially up-regulated genes in comparisons used in this article.
Supplemental Table S2. Representative references associated with gene developmental stage classifications, AGI code, and search strings used in conjunction with the TAGGIT macro and TAIR6 annotations of genes.
Supplemental Table S3. Lists of genes related to comparison of Ler AR > cts-1 S seed gene set with the Ogawa et al. (2003) GA-induced gene set.
Supplemental Table S4. Lists of genes related to comparisons of Cvi AR and D seed samples (Cadman et al., 2006) and Col-0 germination time course (Nakabayashi et al., 2005).
Supplementary Material
Acknowledgments
We are grateful to Dr. Jinrong Peng (Proteos, Singapore) for the kind gift of rgl2-1 mutant seeds. We thank Dr. Eiji Nambara (Plant Science Centre, RIKEN) for sharing transcriptome datasets with us, Dr. Cliff Bray (University of Manchester) for help with identification of DNA repair-associated genes, Dan Zadik (University of Nottingham) and Dr. Nick James (NASC, University of Nottingham) for help with analysis of transcriptomics datasets, and Simon Holdsworth (Andover, UK) for the development of several Microsoft Excel macros used for transcriptome analysis. We also thank Dr. Alison Baker (University of Leeds) for many helpful discussions.
This work was supported by the Exploiting Genomics Initiative, Biotechnology and Biological Research Council (BBSRC; grant to E.C.), by a Lawes Trust Ph.D. studentship (to T.H.), and by the University of Nottingham (grant to A.M. and H.S.). Rothamsted Research receives grant-aided support from the BBSRC (UK). W.P. acknowledges a National Science Foundation grant to Angus S. Murphy and an Underwood/BBSRC grant.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Michael J. Holdsworth (michael.holdsworth@nottingham.ac.uk).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39 366–380 [DOI] [PubMed] [Google Scholar]
- Bewley JD (1997) Seed germination and dormancy. Plant Cell 9 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Black M (1994) Seeds; Physiology of Development and Germination. Plenum Press, New York
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol 126 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE (2006) Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J 46 805–822 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Davies J (2004) Cell expansion in roots. Curr Opin Plant Biol 7 33–39 [DOI] [PubMed] [Google Scholar]
- Footitt S, Marquez J, Schmuths H, Baker A, Theodoulou FL, Holdsworth M (2006) Analysis of the role of COMATOSE and peroxisomal beta-oxidation in the determination of germination potential in Arabidopsis. J Exp Bot 57 2805–2814 [DOI] [PubMed] [Google Scholar]
- Footitt S, Slocombe SP, Larner V, Kurup S, Wu YS, Larson T, Graham I, Baker A, Holdsworth M (2002) Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J 21 2912–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Richards DE, Fleck B, Xie DX, Burton N, Harberd NP (2004) The Arabidopsis mutant sleepy1(gar2-1) protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2002) Proteomics of Arabidopsis seed germination: a comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol 129 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain V, Rylott EL, Larson TR, Sherson SM, Bechtold N, Carde JP, Bryce JH, Graham IA, Smith SM (2001) Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid beta-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J 28 1–12 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Nito K, Takei-Hoshi R, Yagi M, Kondo M, Suenaga A, Yamaya T, Nishimura M (2002) Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid beta-oxidation. Plant Cell Physiol 43 1–11 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5 33–36 [DOI] [PubMed] [Google Scholar]
- Kubasek WL, Shirley BW, McKillop A, Goodman HM, Briggs W, Ausubel FM (1992) Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell 4 1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Cheng H, King KE, Wang WF, He YW, Hussain A, Lo J, Harberd NP, Peng JR (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57 405–430 [DOI] [PubMed] [Google Scholar]
- Lloyd JC, Zakhleniuk OV (2004) Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J Exp Bot 55 1221–1230 [DOI] [PubMed] [Google Scholar]
- Logemann E, Tavernaro A, Schulz WG, Somssich IE, Hahlbrock K (2000) UV light selectively coinduces supply pathways from primary metabolism and flavonoid secondary product formation in parsley. Proc Natl Acad Sci USA 97 1903–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller K, Tintelnot S, Leubner-Metzger G (2006) Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol 47 864–877 [DOI] [PubMed] [Google Scholar]
- Murphy A, Peer WA, Taiz L (2000) Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 211 315–324 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41 697–709 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwalhara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS (2001) Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol 126 536–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinfield-Wells H, Rylott EL, Gilday AD, Graham S, Job K, Larson T, Graham I (2005) Sucrose rescues seedling establishment but not germination of Arabidopsis mutants disrupted in peroxisomal fatty acid catabolism. Plant J 43 861–872 [DOI] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D (2004) The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell L, Larner V, Kurup S, Bougourd S, Holdsworth MJ (2000) The Arabidopsis COMATOSE locus regulates germination potential. Development 127 3759–3767 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D (2004) Vitamin E is essential for seed longevity, and for preventing lipid peroxidation during germination. Plant Cell 16 1419–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13 1555–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoulou FL, Job K, Slocombe SP, Footitt S, Holdsworth M, Baker A, Larson TR, Graham IA (2005) Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants: implications for transport of jasmonate precursors into peroxisomes. Plant Physiol 137 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toorop PE, Barroco RM, Engler G, Groot SPC, Hilhorst HWM (2005) Differentially expressed genes associated with dormancy or germination of Arabidopsis thaliana seeds. Planta 221 637–647 [DOI] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu JH, Dill A, Alonso JM, Ecker JR, Sun TP (2004) DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3 RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RGS, Kamiya Y, Sun TP (1998) Phytochrome regulation and differential expression of gibberellin 3 beta-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S (2004) Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B (2001) The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol 127 1266–1278 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.