Abstract
Different classes of biotic (e.g. plant hormones) and abiotic (e.g. different wavelengths of light) signals act through specific signal transduction mechanisms to coordinate higher plant development. While a great deal of progress has been made, full signal transduction chains have not yet been described for most blue light- or abscisic acid-mediated events. Based on data derived from T-DNA insertion mutants and yeast (Saccharomyces cerevisiae) two-hybrid and coprecipitation assays, we report a signal transduction chain shared by blue light and abscisic acid leading to light-harvesting chlorophyll a/b-binding protein expression in etiolated Arabidopsis (Arabidopsis thaliana) seedlings. The chain consists of GCR1 (the sole Arabidopsis protein coding for a potential G-protein-coupled receptor), GPA1 (the sole Arabidopsis Gα-subunit), Pirin1 (PRN1; one of four members of an iron-containing subgroup of the cupin superfamily), and a nuclear factor Y heterotrimer comprised of A5, B9, and possibly C9. We also demonstrate that this mechanism is present in imbibed seeds wherein it affects germination rate.
Different classes of plant hormones and biologically active wavelengths of light act through specific signal transduction mechanisms to coordinate many, if not all, aspects of higher plant development (Borevitz et al., 2002). The paucity of signaling components and the abundance of transcription factors in the Arabidopsis (Arabidopsis thaliana) and other plant genomes is in contrast to that observed in most animal systems and suggests that signaling specificity may reside at the level of effectors, modifications of signaling components, and/or gene expression itself. Heterotrimeric G-protein-mediated cell signaling is one of the most highly conserved signaling mechanisms in eukaryotes (Hamm, 1998). Analysis of Arabidopsis seeds, seedlings, and plants mutant in the GPA1 gene, coding for the sole canonical Gα-subunit within the Arabidopsis genome, reveals that G proteins are likely to be involved in many processes critical for proper development, including germination and postgermination development, particularly root (Chen et al., 2006; Pandey et al., 2006), hypocotyl and leaf formation (Ullah et al., 2001), stomatal aperture (Wang et al., 2001), responses to abscisic acid (ABA; Colucci et al., 2002; Ullah et al., 2002; Lapik and Kaufman, 2003; Chen et al., 2006; Warpeha et al., 2006), sugar signaling (Chen et al., 2006; Huang et al., 2006), and blue light (BL) induction of Phe production (Warpeha et al., 2006).
In contrast to animal systems wherein there may be hundreds of genes coding for G-protein-coupled receptors and tens of genes coding for G-protein subunits, plant G-protein-coupled receptors and G-protein subunits are coded by only one or sometimes two genes (Ma et al., 1990; Weiss, et al., 1994; Josefsson and Rask, 1997; Marsh and Kaufman, 1999; Mason and Botella, 2000; Mason and Botella, 2001). It remains unclear how the sole Gα in Arabidopsis can account for all of the characterized responses for which it is presumed responsible. One answer may be divergence at the level of effectors and the ability of the one Gα to direct the activity of several different effector systems.
At least three classes of higher plant photoreceptors can perceive BL: phytochrome, phototropin, and cryptochrome (Briggs and Huala, 1999; Lin, 2000; Chen et al., 2004). There is a great deal of evidence for genetic, biochemical, and developmental interaction among the receptors or their respective signal transduction chains (Kaufman, 1993; Lin, 2000). There are also several BL-regulated phenomena that are not easily explained by the actions of these three classes of receptors (Briggs and Huala, 1999; Lin, 2000; Warpeha et al., 2006).
Irradiation of dark-grown seedlings with a single pulse of low-fluence BL equivalent in intensity to 1 s of full moonlight results in the induction of several nuclear-coded genes, including specific members of the light-harvesting chlorophyll a/b-binding protein (Lhcb) gene family (e.g. in Arabidopsis, the AtLhcb1*3 gene is activated, but not the AtLhcb1*1 and AtLhcb1*2 genes; Kaufman, 1993; Gao and Kaufman, 1994). This blue low-fluence (BLF) system exhibits reciprocity, has an immediate effect on transcription, and functions in the absence of protein synthesis (Marrs and Kaufman, 1991). Mutations in the known plant photoreceptors alone or in various combinations have no major effect on the photobiology of the BLF system (Gao and Kaufman, 1994; K.M. Folta, M.B. Anderson, and L.S. Kaufman, unpublished data), leaving the signaling mechanism an open question. In peas (Pisum sativum), irradiation with low-fluence BL is known to activate one or both of the two Gα-subunit proteins encoded within the pea genome (Warpeha et al., 1991; Marsh and Kaufman, 1999).
The DNA element responsible for the BLF system activation of the Lhcb transcription has been narrowed to a 10-bp sequence containing a perfect CCAAT box and ACT upstream context (Folta and Kaufman, 1999), strongly suggesting the involvement of a CCAAT box binding protein (nuclear factor Y [NF-Y]). The NF-Y proteins are heterotrimeric protein complexes consisting of NF-Y-A, -B, and -C subunits. For vertebrates, each subunit is generally coded by one gene. In contrast, the number of genes for each subunit varies across plants species (Yang et al., 2005). DNA and protein sequence analyses indicate that the Arabidopsis genome contains nine or 10 genes encoding each subunit (Gusmaroli et al., 2001, 2002).
ABA is known to affect many plant physiological and development processes including stomatal aperture and rate of germination. Mutations in either GCR1, the Arabidopsis putative G-protein-coupled receptor, and/or GPA1, the sole Arabidopsis Gα subunit, exhibit increased sensitivity to ABA (Lapik and Kaufman, 2003; Perfus-Barbeoch et al., 2004), which in some assays mutants have shown ABA sensitivity in specific cells and tissues, or even insensitivity (Wang et al., 2001; Mishra et al., 2006). Pandey et al. (2006) recently demonstrated that GCR1, GPA1, and AGB1, the sole Gβ-subunit in Arabidopsis, each negatively regulates ABA signaling in germination and young seedling development. In the case of germination, activation of the GPA1-mediated pathway interferes with the well documented ABA-mediated delay in seed germination (Lapik and Kaufman, 2003; Perfus-Barbeoch et al., 2004). How ABA regulates developing cells or is involved in cell signaling is under intense scrutiny, and recently published data has greatly improved understanding of some of the mechanisms involved (Weatherwax et al., 1996; Rohde et al., 2000; Gampala et al., 2002; Pandey and Assmann, 2004; Finkelstein et al., 2005; Chen et al., 2006; Mishra et al., 2006; Pandey et al., 2006; Warpeha et al., 2006; for review, see Finkelstein, 2006).
The Assmann lab has demonstrated a specific physical interaction between GCR1 and GPA1 (Pandey and Assmann, 2004). Insertion mutants in both genes suggest a role in ABA responses. These data were confirmed by further work using mutants of GCR1, GPA1, and AGB1, wherein each protein was demonstrated to be a component of ABA signaling, affecting germination and early development particularly in roots (Pandey et al., 2006).
Recently we have demonstrated that ABA can mimic BL effects in the etiolated cotyledon in the Phe production pathway (Warpeha et al., 2006). The signal transduction mechanism responsible is comprised of GCR1, GPA1, and prephenate dehydratase 1 (PD1; responsible for the conversion of prephenate to phenylpyruvate, the immediate precursor to Phe) where PD1 acts as the effector. Activated GPA1 was shown to both interact with and cause activation of PD1 protein, resulting in increased Phe synthesis and subsequently, several products derived from the phenylpropanoid pathway. How ABA or BL interacts with, activates, or excites the GCR1-GPA1 signaling systems in developing seeds or etiolated cotyledons is still unknown.
Our lab has previously demonstrated that Pirin1 (PRN1), an iron-containing member of the cupin superfamily, has a specific interaction with GPA1 and acts as the Gα effector in the signaling mechanism that inhibits the ABA-mediated delay in germination (Lapik and Kaufman, 2003). PRN was originally identified through its interactions with NF-Y, the heterotrimeric CCAAT box binding proteins (Wendler et al., 1997), suggesting a role for NF-Y in the GPA1-mediated interference in the ABA-induced delay of germination and a potential link between GPA1 and Lhcb expression.
The documented physical and genetic interactions between GCR1 and GPA1 (Pandey and Assmann, 2004; Pandey et al., 2006; Warpeha et al., 2006), between GPA1 and PRN1 (Lapik and Kaufman, 2003), and between human PRN and human NF-Y (Wendler et al., 1997; Dunwell et al., 2004; Pang et al., 2004), suggest that these four proteins could form a signal transduction mechanism: from potential receptor to potential gene-specific DNA-binding transcription factor. Because of the characterized role of the CCAAT box in the BLF system regulation of Lhcb expression and the known roles for GPA1 and PRN1 in effecting the ABA-mediated delay in seed germination (Lapik and Kaufman, 2003), it seemed reasonable to test Arabidopsis insertion mutants in GCR1, GPA1, PRN1, and individual NF-Y subunits for the BLF system regulation of Lhcb expression, the potential for ABA to regulate Lhcb expression in etiolated seedlings, and their respective effect on the ABA-mediated delay in seed germination.
The data presented herein indicate that the four proteins: GCR1 (a protein coding for a canonical G-protein-coupled receptor; Josefsson and Rask, 1997; Pandey and Assmann, 2004), GPA1 (the sole Arabidopsis Gα-subunit; Ma et al. 1990), PRN1 (a member of the cupin superfamily; Lapik and Kaufman, 2003), and NF-Y subunit B9 (also known as LEC1; Lee et al., 2003), subunit A5 (Gusmaroli et al., 2001, 2002), and possibly C9 (Gusmaroli et al., 2001, 2002) define a signal transduction chain leading to ABA- or BL-mediated regulation of nuclear gene transcription of Lhcb in etiolated Arabidopsis, and that this same signal transduction chain may also be responsible for attenuating the ABA-mediated delay in seed germination.
RESULTS
GCR1, GPA1, and PRN1 Are Required for BL Induction of Lhcb
We have previously demonstrated that PRN1 can act as an effector for GPA1 in a signaling mechanism that acts to inhibit the ABA-induced delay in seed germination (Lapik and Kaufman, 2003); the two proteins can interact and mutants in both share the same phenotype. Pandey and Assmann demonstrated that GPA1 can interact with GCR1 (Pandey and Assmann, 2004), and later confirmed that GCR1 and GPA1 were in a direct signaling mechanism affecting germination and early development (Pandey et al., 2006). We recently identified a positive signaling link between GCR1 and GPA1 regarding their shared role in BL- and ABA-induced activation of PD1 in the etiolated cotyledon, the immediate production of Phe, and subsequent production of products of the phenylpropanoid pathway (Warpeha et al., 2006).
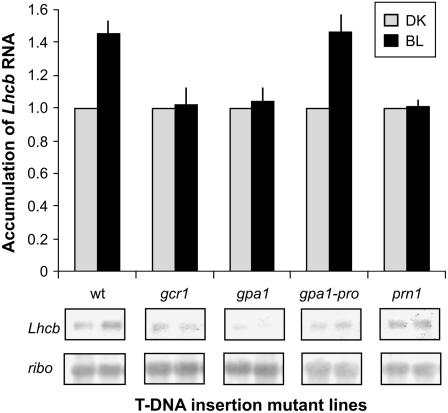
We sought to determine if GCR1, GPA1, and PRN1 have defined roles in the immature cotyledon, namely induction of Lhcb expression by the BLF system. Six-day-old etiolated wild type (Columbia [Col]) and multiple T-DNA insertion mutants for GCR1, GPA1, and PRN1 were irradiated with a single pulse of BL in the low-fluence range with a total fluence of 102 μmol m−2 or no light (DK). The resulting Lhcb RNA levels were determined by northern analysis (Lapik and Kaufman, 2003). The data presented in Figure 1 indicate that the single pulse of BL, unlike in wild-type seedlings, has no effect on Lhcb RNA levels in lines with T-DNA insertions in GCR1, GPA1, and PRN1. The loss of activity occurs regardless of ecotype (Col or Wassilewskija [Ws]; data not shown) and is replicated, for most genes, in at least two independent insertion lines for each gene. Insertions distal to the promoter region for GPA1 (approximately 600–680 bp upstream from the start of transcriptional initiation) do not result in the loss of BLF system activity. Induction of Lhcb expression was also monitored by quantitative PCR (qPCR) to confirm the fold level of induction by BL. The resulting range of BL induction compares well to that observed using northern analysis, as confirmed by Student's t test described in “Materials and Methods” (data not shown).
Figure 1.
Loss of BLF system-mediated Lhcb expression in T-DNA insertion lines for GCR1, GPA1, and PRN1. Six-day-old dark-grown seedlings from matched seed lots of wild-type Col Arabidopsis and T-DNA insertions in genes in GCR1, GPA1, and PRN1 were irradiated with a single pulse of low-fluence BL (102 μmol m−2) or DK, placed back in the dark for 2 h after which total RNA was extracted and used for northern-blot analysis (Lapik and Kaufman, 2003) for hybridization with labeled Lhcb. For each sample tested, an induction ratio (BL/DK) was defined where DK = 1.0. Each lane is standardized for loading by normalizing to rRNA levels (Warpeha and Kaufman, 1990) and is shown in the figure (ribo). Northern-blot analyses of mutants and wild type shown are representative. Induction ratios derive from at least three independent replicates, error shown is the se of the mean. Data representative of gcr1, gpa1, gpa1-pro (insertion upstream of Promoter), and prn1 mutants are shown in the figure.
Expression of NF-Y Subunits Is Limited in Etiolated Tissue
NF-Y is a heterotrimeric protein comprised of A, B, and C subunits. The Arabidopsis genome codes for 10, 10, and nine potential copies of the A, B, and C subunits, respectively, as determined by DNA and protein sequence analyses (Gusmaroli et al., 2001, 2002). The NF-Y family is poorly studied in higher plants, with most of the work focusing on specific members of the NF-Y-B subunit family, particularly NF-Y-B9 (LEC1; Lee et al., 2003) and the closely related NF-Y-B6 (LEC1 LIKE; Kwong et al., 2003) in Arabidopsis. There is some evidence for differential expression in plants of the NF-Y subunits (Gusmaroli et al., 2001, 2002).
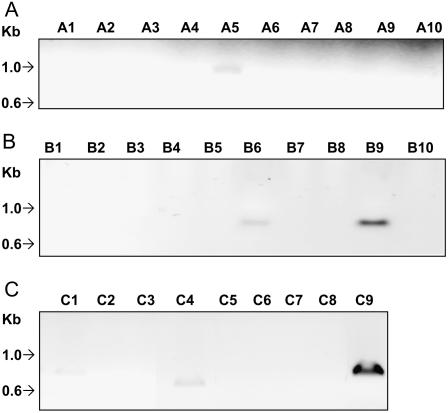
To better define the NF-Y-A, -B, and -C family members that have a potential for participation in the signaling mechanism of etiolated seedlings, we performed reverse transcription (RT)-PCR, using RNA from etiolated wild-type seedlings and primer pairs specific to each of the 29 NF-Y members. The results, shown in Figure 2, indicate that the NF-Y-A5, NF-Y-B6, NF-Y-B9, and NF-Y-C1, -C4, and -C9 subunits are expressed in the 6-d-old etiolated seedlings, suggesting that these NF-Y subunits have a potential role as signal carriers in the BLF system regulation of Lhcb transcription.
Figure 2.
Differential expression of NF-Y subunits in Arabidopsis. Dark-grown wild-type Col seedlings were grown for 6 d, after which total RNA was extracted. This RNA was used for first-strand cDNA synthesis and subsequent RT-PCR with primer pairs specific to each of the NF-Y-A, -B, and -C subunits. A, NF-Y-A subunit expression in dark-grown seedlings. Lanes are numbered for each NF-Y subunit (i.e. A1 = NF-Y-A1). B, NF-Y-B subunit expression in dark-grown seedlings. As with A, lane number corresponds to subunit number (i.e. B1 = NF-Y-B1). C, NF-Y-C subunit expression in dark-grown seedlings. As with A, lane number corresponds to subunit number (i.e. C1 = NF-Y-C1).
NF-Y-B9 (LEC1) and NF-Y-A5 Are Required for BLF System Regulation of Lhcb Expression
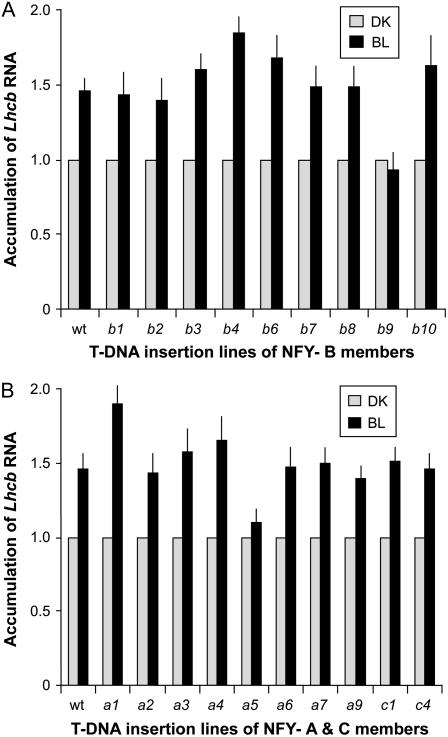
T-DNA insertion mutants are available in most members of the NF-Y-A family, the NF-Y-B family, and only a few members of the NF-Y-C family, including C1 and C4. There are no publicly available insertion mutants for the coding region of the C9 gene. To identify NF-Y members with a defined role in the BLF system-mediated Lhcb expression, we tested all available T-DNA insertion mutants of NF-Y-A, NF-Y-B, and NF-Y-C. The data presented in Figure 3 indicate that insertions in only one A family member, NF-Y-A5, and only one B family member, B9, exhibit a loss of BLF system-mediated Lhcb expression. Both NF-Y-A5 and NF-Y-B9 were identified by RT-PCR as being expressed in the etiolated tissue. T-DNA insertions in all other NF-Y-B members, including NF-Y-B6, which is closely related to the NF-Y-B9 protein, do not result in the loss of the BLF system-mediated Lhcb expression. Similarly, insertions in all other NF-Y-A members and NF-Y-C1 and C4 do not exhibit the loss of BLF system-mediated Lhcb expression.
Figure 3.
Northern analysis indicates the loss of BLF system-mediated Lhcb expression in T-DNA insertion lines for NF-Y-B9 and NF-Y-A5 subunits. A, NF-Y-B family. Seedlings were grown, treated, and accumulation of Lhcb RNA analyzed as described in Figure 1. Mutants available for the members of the NF-Y-B family were tested: nf-y-b1, nf-y-b2, nf-y-b3, nf-y-b4, nf-y-b6 (LEC1 LIKE), nf-y-b7, nf-y-b8, and nf-y-b9 (LEC1). See Supplemental Table S1 for the seed lines used. B, NF-Y-A and -C family. Seedlings were grown, treated, and analyzed as described in section A of this figure for available members of the NF-Y-A and -C families: nf-y-a1, nf-y-a2, nf-y-a3, nf-y-a4, nf-y-a5, nf-y-a6, nf-y-a7, nf-y-a9, nf-y-c1, and nf-y-c4. See Supplemental Table S1 for the seed lines used.
ABA Can Elicit Lhcb Expression in the Cotyledons of Etiolated Seedlings and Does So through a GPA1-Mediated Pathway
Based on the data presented in Figures 1 to 3, the BL signal transduction mechanism is minimally comprised of the GCR1, GPA1, PRN1, NF-Y-A5, NF-Y-B9, and potentially NF-Y-C9. We know from earlier published work that ABA, like BL, can elicit the GCR1-GPA1 effector signaling pathways in etiolated cotyledons, as was demonstrated for the PD1 signaling pathway (Warpeha et al., 2006). Thus, it was of interest to determine if ABA can activate Lhcb expression via the GCR1-GPA1-PRN1-NFY signaling chain in etiolated cotyledons. If so, it would establish that both ABA and low-fluence BL can use the exact same signaling mechanism, and, at the same time, provide evidence of ABA as a signal for a simple positive transduction mechanism.
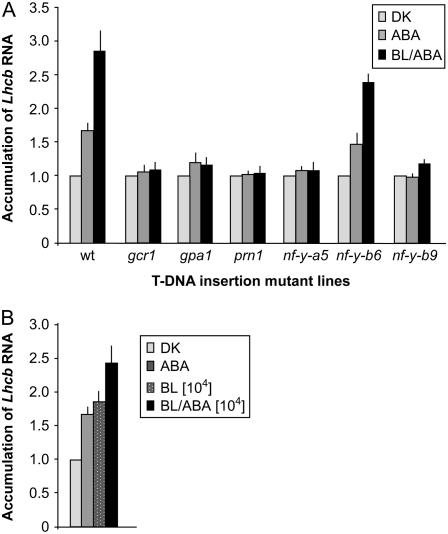
Lhcb RNA levels were measured in 6-d-old etiolated wild-type seedlings that had been treated with 800 nm ABA dissolved in ethanol (approximately 70% saturation for the germination response; Lapik and Kaufman, 2003) or with ethanol alone (Fig. 4). The ABA treatment in wild-type seedlings, just like low-fluence BL, induces Lhcb expression. We also tested the ability of ABA to induce Lhcb in T-DNA insertion mutants of the components of the BLF pathway: gcr1, gpa1, prn1, nf-y-b9, and nf-y-a5. ABA fails to induce Lhcb RNA accumulation in the mutant seedlings, indicating that ABA and low-fluence BL depend upon the same GPA1-based signaling pathway.
Figure 4.
ABA can utilize the same signaling pathway as BL to elicit Lhcb expression in etiolated seedlings. A, Response to 102 μmol m−2 BL and 800 nm ABA. Six-day-old dark-grown seedlings from matched seed lots of wild-type Col Arabidopsis and T-DNA insertions in GCR1 (SALK_027808), GPA1 (SALK_066823), PRN1 (SALK_006939), NF-Y-B9 (LEC1; SALK_000450), NF-Y-A5 (SALK_006559), and NF-Y-B6 (LEC1 LIKE; SALK_118236) were sprayed with ABA (800 nm) in ethanol, or ethanol alone (DK), or irradiated with a single pulse of low-fluence BL (102 μmol m−2) immediately followed by spraying with 800 nm ABA. Treated seedlings were placed back in the dark for 2 h after which total RNA was extracted and used for northern-blot analysis as described in Figure 1. Data shown represent induction levels (ABA/DK or BL-ABA/DK). Data derive from at least three independent replicates. Bars represent the se of the mean. B, Response to 104 μmol m−2 BL and 800 nm ABA. Seedlings were grown and treated the same as described for Figure 4A, except that a near saturating dose of BL (104 μmol m−2) was used in the treatment. RNA extraction and analysis was also the same as for samples in Figure 4A.
If low-fluence BL and ABA utilize the same signal transduction chain, then the combination of a near-saturating BL treatment (104 μmol m−2; Anderson et al., 1999) and a near-saturating ABA treatment (800 nm; Lapik and Kaufman, 2003) would not be expected to exceed saturation, where the two responses would not be completely additive. The data shown in Figure 4B show the results of treatment with BL and/or ABA. The near-saturating doses of BL and ABA are not additive for Lhcb accumulation. The difference between the individual BL treatment response plus the ABA treatment response (additive) is significantly different from the combined treatments single response, according to the Student's t test (95% confidence level, P = 0.012). These data indicate that BL and ABA share all or part of a signal transduction chain where each BL and ABA are capable of activating expression. The data are consistent with a shared signal transduction chain of GCR1-GPA1-PRN1-NF-Y.
GCR1, GPA1, PRN1, NF-Y-A5, NF-Y-B6, NF-Y-B9, and NF-Y-C4 Are Required for the Signaling Mechanism That Acts to Inhibit the ABA-Mediated Delay in Seed Germination
The data presented in Figures 1 to 4 establish that ABA can act as a signal for and that PRN1 can act as a signal carrier in the BL-mediated GCR1, GPA1 signal transduction pathway leading to the expression of Lhcb in etiolated seedlings. We have previously demonstrated that PRN1 acts as the effector for GPA1 in the signaling mechanism that inhibits the ABA-mediated delay in seed germination (Lapik and Kaufman, 2003). It appears that PRN1 participates in different signaling mechanisms in different tissues and there is redundancy among signaling components in the germinating seed and young seedling. We tested the effect of ABA on germination rate in the same T-DNA insertion mutants as those described in the experiments for BL regulation of Lhcb expression in etiolated cotyledons.
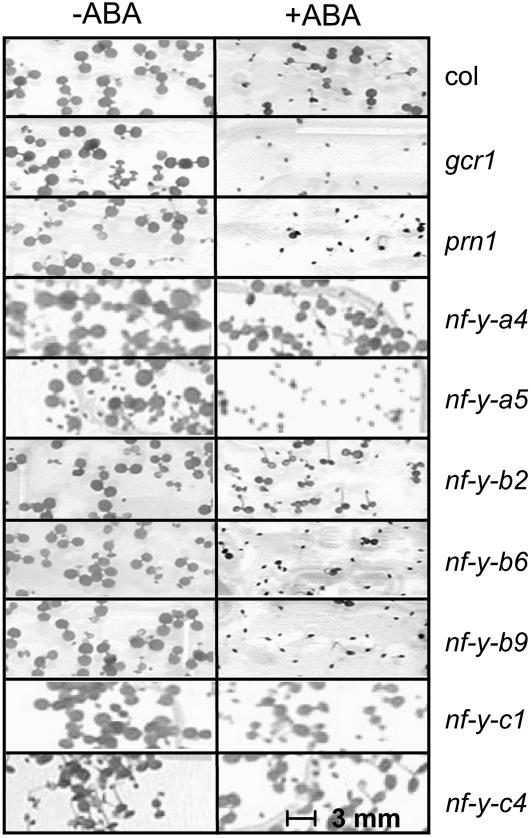
The results shown in Figure 5 indicate that T-DNA insertion mutants of GCR1, NF-Y-A5, NF-Y-B9, and the closely related NF-Y-B6 show a marked increase in the delay in germination in response to ABA (i.e. appear to be hypersensitive to ABA). Insertions in other NF-Y-A or NF-Y-B family members, like NF-Y-A4 or NF-Y-B2 (both shown as examples), appear similar to wild type and therefore are not likely to have a role in this response. We also tested the insertion mutants in NF-Y-C1 and NF-Y-C4 since these components are expressed in etiolated seedlings (Fig. 2). The nf-y-c4 mutant seed demonstrates a measurable increase in germination delay when compared to wild-type levels of germination, suggesting a role for NF-Y-C4 in the germination process. Graphical representation of the data can be viewed in Supplemental Figure S1.
Figure 5.
Seed germination in T-DNA insertion mutants of GCR1, PRN1, NF-Y- B9 (LEC1), NF-Y-B6 (LEC1 LIKE), and NF-Y-C4 are hypersensitive to ABA. Matched seed lots of wild-type Col and T-DNA insertions in genes: GCR1 (SALK_027808), PRN1 (SALK_006939), NF-Y-B9 (LEC1; SALK_000450), NF-Y-B6 (LEC1 LIKE; SALK_118236), NF-Y-B2 (SALK_025666), NF-Y-A5 (SALK_006559), NF-Y-A4 (SALK_003337), NF-Y-C1 (SALK_086334), and NF-Y-C4 (SALK_032163) were planted on media supplemented with (+) 600 nm ABA or without (−) ABA, stratified, then placed at 22°C in continuous white light as described (Lapik and Kaufman, 2003). Germinating seeds were counted every day after planting as described (Lapik and Kaufman, 2003). Representative photographs of germination and early seedling development taken 7 d after stratification are shown.
Both NF-Y-B9 and NF-Y-B6 Can Physically Interact with PRN1
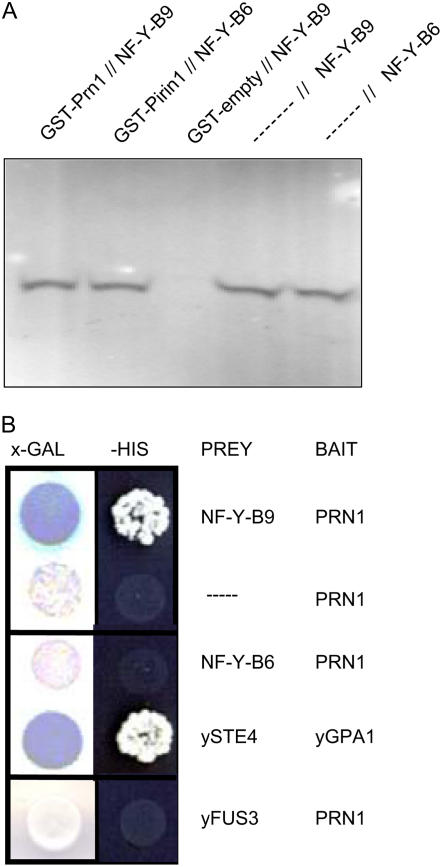
The data shown in Figures 1 to 4 establish roles for GCR1, GPA1, PRN1, NF-Y-A5, NF-Y-B9, and potentially NF-Y-C9 in the BLF system activation of Lhcb transcription and, in Figure 5, roles for GCR1, GPA1, PRN1, NF-Y-A5, NF-Y-B6, NF-Y-B9, and NF-Y-C4 in the signaling mechanism responsible for inhibiting the ABA-mediated delay in seed germination. GCR1 and GPA1 (Pandey and Assmann, 2004) and GPA1 and PRN1 (Lapik and Kaufman, 2003) are known to have specific interactions. Mammalian PRN is known to directly interact with mammalian NF-Y for transcription activation (Romier et al., 2003). Yeast (Saccharomyces cerevisiae) two-hybrid and in vitro coprecipitation assays were used to determine if specific interactions could occur between PRN1 and NF-Y-B9 or between PRN1 and NF-Y-B6. NF-Y-B9 is active in both Lhcb and germination responses; NF-Y-B6, although a very similar protein to NF-Y-B9, is only active in the germination response. The results of the in vitro coprecipitation experiments (Fig. 6) indicate that PRN1 has the potential for a specific interaction with both NF-Y-B9 and NF-Y-B6.
Figure 6.
PRN1 interacts with both NF-Y-B9 and NF-Y-B6. A, In vitro protein association assays. Full-length PRN1–GST fusions and radiolabeled translated NF-Y-B9 or NF-Y-B6 were coincubated in varying combinations. The PRN1-GST fusion, along with bound proteins were washed and resolved on 4% to 20% gradient SDS-PAGE gels as described (Lapik and Kaufman, 2003). Results were visualized using a Phosphor-Imager (Lapik and Kaufman, 2003). Lane 1: Full-length PRN1 interaction with full-length NF-Y-B9. Lane 2: Full-length PRN1 interaction with full-length NF-Y-B6. Lane 3: Negative control GST empty plasmid expressed protein does not interact with full-length NF-Y-B9 (LEC1). Lane 4: Full-length NF-Y-B9 alone (approximately 24 kD) as a molecular mass marker. Lane 5: Full-length NF-Y-B6 alone (approximately 24 kD) as a molecular mass marker. B, Yeast two-hybrid experiments. The potential for interaction between PRN1 and NF-Y-B9 or the closely related NF-Y-B6 was tested in a yeast two-hybrid assay (Lapik and Kaufman, 2003). The indicated combinations of the bait (pGBT9) and prey (pGAD424) constructs were transformed into the yeast reporter strain AH109. Empty vector (pGAD424) and the yeast mating response protein, yFUS3, were used as negative bait controls as shown. The interaction between the yeast Gα (yGPA1) and Gβ (ySTE4) was used as a positive interaction control. Initial transformants were plated on 9-cm sd plates (Trp−, Leu−, Lys−, His−) to select for His prototrophy. Positive transformants were also streaked on X-α-Gal-containing selective media where LacZ reporter gene activity was monitored visually. Blue color indicates positive interaction.
The yeast two-hybrid data, confirm by a second, independent assay, the potential for a positive physical interaction between PRN1 and NF-Y-B9 (Fig. 6). The yeast cells expressing NF-Y-B6 undergo two to three cell divisions but fail to form viable colonies. Thus, we are unable to confirm that the PRN1-NF-Y-B6 interaction is either specific or strong in the yeast two-hybrid assay.
DISCUSSION
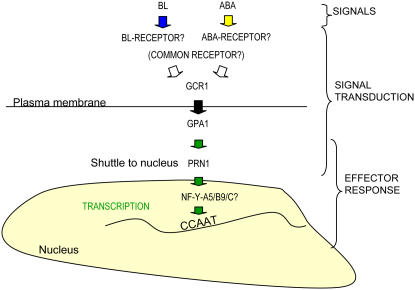
The data presented herein demonstrate: (1) both BL and ABA can induce Lhcb expression in etiolated Arabidopsis seedlings; (2) the physical interaction of PRN1 and NF-Y-B9; (3) the expression of NF-Y-A5, NF-Y-B9, and NF-Y-C9 in etiolated tissue; and (4) the shared loss of BLF system induction of Lhcb in T-DNA insertion mutant lines of GCR1, GPA1, PRN1, NF-Y-A5, and NF-Y-B9. Considered with previously published data that the CCAAT box as the BLF system DNA regulatory element in the Lhcb gene (Folta and Kaufman, 1999), along with the capability of GCR1-GPA1 interaction (Pandey and Assmann, 2004; Pandey et al., 2006), allows us to define a potentially complete signal transduction mechanism for the BLF system-mediated gene expression of Lhcb: GCR1, GPA1, PRN1, NF-Y, and the CCAAT box of Lhcb. A proposed model is shown in Figure 7.
Figure 7.
A proposed model for the GCR1, GPA1, PRN1, NF-Y signal transduction mechanism responsible for BL-induced accumulation of Lhcb RNA. The figure depicts the potential interface between the BL- or ABA-activated GCR1, GPA1, PRN1, NF-Y signal transduction chain and the 5′ untranslated region of Lhcb. The PRN1 protein is depicted as a shuttle protein which upon being activated by BL-excited GPA1, would be capable of entering the nucleus to interact with NF-Y-B9 and NF-Y-A5 and a particular NF-Y-C subunit to facilitate transcription.
We also report that the same signal transduction mechanism, with the potential involvement of the additional or alternative NF-Y components NF-Y-B6 and NF-Y-C4, is involved in inhibiting the ABA-mediated delay in seed germination.
The possibility of abiotic (BL) and biotic (ABA) signals stimulating the same pathway is not a new concept (Brocard-Gifford et al., 2003; Fan et al., 2004; Zhang et al., 2004; Warpeha et al., 2006) and BL and ABA cross talk is known to exist (Weatherwax et al., 1996; Yadav et al., 2005; Warpeha et al., 2006). We recently demonstrated that GCR1 and GPA1, both critical to the BL- and ABA-mediated Lhcb expression in etiolated seedlings, are also critical to the mechanism through which BL and ABA act to increase PD1 activity, Phe synthesis, and phenylpropanoid pathway activity in etiolated seedlings (Warpeha et al., 2006).
Single genes code for the Gα-subunit (GPA1) and the putative G-protein-coupled receptor in Arabidopsis (GCR1), so it is not surprising that identical signal transduction mechanisms are used by different signals to control either similar or different responses in same or different tissues, or in tissues of different developmental states under specific conditions. Our finding that the same signal transduction mechanism would be used by two different signals such as BL and ABA, to achieve the same response in the same tissue is less intuitive and requires further exploration. Given the large number of systems and phenomena thought to rely on G-protein-mediated signaling it is likely that the sole Arabidopsis Gα-subunit GPA1 interacts with many effector proteins (Assmann, 2002; Koornneef et al., 2002; Jones and Assmann, 2004; Perfus-Barbeoch et al., 2004). However, to date, only a few partners have been identified, including PRN1 (Lapik and Kaufman, 2003), Phospholipase C (Apone et al., 2003), Phospholipase Eα1 (Zhao and Wang, 2004), PD1 (Warpeha et al., 2006), and THF1 (Huang et al., 2006).
It is unclear how BL and ABA stimulate the GCR1, GPA1-based pathways in the cotyledons of etiolated seedlings. Given the very different nature of the two signals and therefore the chemical/structural nature of the receptors that would detect the two signals, it would be reasonable to assume the existence of two separate receptors. However, because ABA is derived from an asymmetric cleavage of the BL-absorbing carotenoids (i.e. zeaxanthin, antheraxanthin, violaxanthin) and because of the structural relationship of the heterocyclic head group on ABA and the carotenoid from which it derives, it is not unreasonable to speculate that a single receptor could bind, depending on the specific situation, either ABA or a BL-absorbing carotenoid. BL-absorbing carotenoids have been implicated as receptors in other phenomena in plants (Quinones and Zeiger, 1994; Quinones et al., 1998). The excitatory molecule responsible for the signaling mechanism that acts to interfere with the ABA-mediated delay in seed germination is not certain.
GCR1 belongs to the secretin (so-called 7-TM2) class of G-protein-coupled receptors. The secretin class tends to bind ligands on the extracellular surface in part through an N-terminal extracellular extension. There has been some similarity shown to the GPCR transmembrane structure in Dictyostelium and Drosophila (Chen et al., 2004; Pandey and Assmann, 2004). The lack of structure and the presence of several sites for potential secondary modification and the extracellular nature of the ligand-binding region, suggests that GCR1 could have many potential ligands in plants. While GCR1 has the characteristics of a classic heterotrimeric G protein, including sequence similarity with predicted seven transmembrane spanning domains (Josefsson and Rask, 1997), direct interaction with Gα (Pandey and Assmann, 2004; Pandey et al., 2006) and localization to the plasma membrane fraction (Pandey and Assmann, 2004), and, while the phenotypes generated by the overexpression of GCR1 are consistent with its being a receptor (Colucci et al., 2002), there are no direct data proving that GCR1 functions as a G-protein-coupled receptor.
Pandey and Assmann (2004) and Pandey et al. (2006) have utilized T-DNA mutants for G-protein pathway components to study guard cell function and germination and postgermination development. They demonstrated that GCR1 and GPA1 seem to have some opposing activities with regard to stomatal opening (Pandey and Assmann, 2004), but act in concert in ABA signaling during germination and development (Pandey et al., 2006). Our data both for Lhcb expression in the etiolated cotyledons and for the ABA effect on seed germination indicate a standard or an in common type of receptor for both the BL and ABA G-protein relationship in that the phenotypes of the gcr1 and the gpa1 mutants are the same, suggesting that GCR1 and GPA1 act in concert.
Very little is known about PRN proteins in plants (for review, see Dunwell et al., 2004). PRN, initially identified as an NF-Y interacting protein in animals, was subsequently identified as also capable of binding to the ankyrin domain of Bcl-3, a nuclear-localized member of the IκB protooncogene family (Dechend et al., 1999). IκB normally functions to sequester dimerized members of the NF-κB/REL family of transcription factors in the cytoplasm (thereby preventing their action as transcription factors) and release of the various NF-κB/REL factors occurs in response to specific signaling mechanisms including a variety of G-protein-mediated pathways (Gao et al., 2004). The GCR1, GPA1, PRN1, NF-Y-A5, B9, and C(?) signal transduction mechanism requires a means to traffic its signal into the nucleus. PRN1 is an excellent candidate for that role.
In animals, there is evidence that during cell and tissue differentiation there can be large changes and differences in expression of individual subunits of NF-Y (Gurtner et al., 2003). Little data exist on the specific roles for NF-Y in plants. There are data to indicate differential expression, splicing, and tissue distribution for NF-Y-A, -B, and -C subunits (as determined by sequence information), suggesting NF-Y may play diverse roles in Arabidopsis (Edwards et al., 1998; Gusmaroli et al., 2001, 2002). NF-Y-A subunits show much more sequence conservation than that of the B and C subunits, which show some evidence of asymmetric evolution in plants (Yang et al., 2005). Recently, in sunflower (Helianthus annuus; Fambrini et al., 2006) and flax (Linum usitatissimum; Gutierrez et al., 2006), NF-Y factors have been expressed and shown to be involved in embryogenesis and young cotyledons. In tomato (Lycopersicon esculentum), nuclear regulators of gene expression are able to bind to an NF-Y-C factor that may act as a recruitment factor effecting gene expression (Ben-Naim et al., 2006).
The RT-PCR data presented herein suggest that only the NF-Y-A5, NF-Y-B9, NF-Y-C1, NF-Y-C4, and NF-Y-C9 subunits are expressed in 6-d-old etiolated seedlings.
NF-Y-B9, identified herein as a member of the BLF system and the signaling mechanism inhibiting the ABA-mediated delay in seed germination, was originally described as a desiccation embryonic lethal which, if helped past the desiccation issue, could produce a leafy cotyledon phenotype (Lee et al., 2003). NF-Y-B9, more commonly known as LEC1, is critical in the developing seed and LEC1 regulates many aspects of late embryogenesis (West et al., 1994; Kwong et al., 2003; Lee et al., 2003).
The closely related member NF-Y-B6 (LEC1 LIKE) was identified by its ability to partially restore wild-type features to plants mutated in the coding region of NF-Y-B9 (Kwong et al., 2003). Our finding that NF-Y-B6 has a role in the signaling mechanism responsible for interfering with the ABA-mediated delay in seed germination, but does not seem to participate in the BLF system induction of Lhcb expression in the etiolated cotyledons is consistent with the previous observations regarding the relationship between NF-Y-B9 and NF-Y-B6 (Kwong et al., 2003). Other embryonic tissue-active proteins involved with LEC1 and LEC1 LIKE can be found in tissues other than seed, like FUS3 (Reidt et al., 2001), which may be involved in vegetative processes. ABI3 also is reported to be active in nonseed tissues including the shoot apex (Rohde et al., 1999; Kurup et al., 2000), as is ABI5 and ABF3 in seeds and seedling stress responses (Finkelstein et al., 2005). Brocard-Gifford et al. (2003) report many loci involved with ABA responses, like LEC1 and the abscisic insensitive genes, which also appear to mediate responses to light in postgermination development.
The variety of tissues and developmental stages where GCR1 and GPA1 interaction is important to physiology (guard cells, etiolated cotyledons, roots, and embryonic tissue in the seed) may be reflective of the paucity of signal transduction genes in higher plants and demonstrate that early signaling mechanisms can have shared components. Arabidopsis may redeploy this signaling mechanism, albeit with different effectors and modifiers at many stages of plant development, or in many tissues of the plant to accommodate the fact there is only one G-protein-coupled receptor and one Gα-subunit in the genome. In plants in general, these data may serve to underscore the importance of understanding the network of possibilities in G-protein signaling in plant development.
MATERIALS AND METHODS
Plant Material
Matched seed lots of wild-type Col Arabidopsis (Arabidopsis thaliana) and siblings carrying T-DNA insertions within coding regions of these genes are listed in Supplemental Table S1. The accessions were obtained from the Arabidopsis Biological Resource Center (Alonso et al., 2003) or from the Arabidopsis Knockout Facility, University of Wisconsin, Madison. All lines used herein are in the Col background except for pir1-1 (Lapik and Kaufman, 2003) and CS6539 that are in the Ws background. Seeds stocks were obtained from plants grown in Scott Metromix 200 in continuous white light. All seeds are homozygous for the reported insertion except for accessions of NF-Y-B9 (all accessions; Kwong et al., 2003; Lee et al., 2003) and the pir1-1 (in the Ws background only; data not shown; Lapik and Kaufman, 2003), as they are embryonic lethals. Where available, T-DNA seed lines with insertions located in the promoter (approximately 600–680 bp upstream from the coding region; GPA1: SALK_057120, SALK_59035) were also tested. All homozygous insertion seeds appear to represent null mutants based on RT-PCR data.
BL and ABA Treatment/Northern Analysis
Six-day-old dark-grown Arabidopsis seedlings Col wild type or insertion mutants were grown on 0.8% agarose plates containing only 0.5× Murashige and Skoog media as described in Lapik and Kaufman (2003). The growth media contains no additional sugar, hormones, vitamins, or other nutrients. Seedlings were irradiated with a single pulse (10 s) of low-fluence BL (total fluence of 102 μmol m−2), or a single pulse (100 s) of saturation fluence BL (total fluence of 104 μmol m−2), or DK, and/or treated with 800 nm ABA dissolved in ethanol, or ethanol alone, placed back in the dark for 2 h after which total RNA was extracted and used for northern-blot analysis (Lapik and Kaufman, 2003). Lhcb and rDNA probes used, quantitation methods, and means of normalization to rRNA levels are described elsewhere (Marrs and Kaufman, 1991). Induction ratios (BL/DK or ABA/ETOH or BL + ABA/ETOH) derive from at least three independent replicates. All induction data is correct for loading through the quantitation of the rRNA as described (Warpeha and Kaufman, 1990). Error bars represent se of the mean. Real-time qPCR was used to measure the level of Lhcb RNA induction in response to a single pulse of low-fluence BL as a confirmation of the northern-blot analysis methods. This PCR was performed using iCycler protocols and the SYBR kit from Invitrogen (first-strand synthesis from RNA and real-time PCR) from wild-type untreated (dark or DK) and BL-treated seedlings. The qPCR-derived information (four different RNA sets of dark and BL treated) data was analyzed using (ΔΔCoT). The resulting range of BL induction is 1.54 to 2.19 (data not shown). This compares well to the induction level range in replicates we observe using northern analysis (1.40–2.18). The Student's t test indicates the P value for the BL induction levels by comparing the two methods (northern versus PCR) is 0.526; similarly the P value for the DK controls is 0.765, so the range/means of the two methods are not different (95% confidence level). Thus, the northern-blot method of analysis is a good indicator of induced gene expression.
RT-PCR Experiments
RNA of dark-grown or light-grown 6-d-old Arabidopsis was extracted as described in Warpeha and Kaufman (1990). First-strand and PCR synthesis was achieved by using SuperScriptTM First-Strand Synthesis system for RT-PCR (Invitrogen). PCR results were confirmed with Invitrogen High Fidelity PCR kit. Gene-specific primers were obtained by comparing sequence via BLAST and selecting 18- to 23-mers that were specific to each NF-Y subunit (IDT). Results were visualized by ethidium bromide staining.
ABA Hypersensitivity Germination Experiments
Matched seed lots were planted as described for northern-blot assays, except that the media was supplemented with ABA (Sigma) ranging from 0 nm up to 1,000 nm (Lapik and Kaufman, 2003). Seedlings were grown at 22°C in continuous white light. Germination was measured as described previously (Lapik and Kaufman, 2003), assessed visually on a daily basis, and photographed 7 d poststratification.
In Vitro Protein Association Assays
Full-length PRN1 was subcloned into the glutathione S-transferase (GST)-fusion expression vector pGEX-4T-1 (Amersham Pharmacia Biotech), prepared, expressed, and purified as described (Lapik and Kaufman, 2003). Radiolabeled NF-Y-B9 and NF-Y-B6 protein were produced by coupled in vitro transcription/translation using TNT T7 Coupled Wheat Germ Extract system (Promega) as directed and as modified (Lapik and Kaufman, 2003). In vitro association assays were conducted as described previously (Lapik and Kaufman, 2003). Samples were resolved on 4% to 20% gradient SDS-PAGE gels. Results were visualized using a Phosphor-Imager (Molecular Dynamics; Lapik and Kaufman, 2003).
Yeast Two-Hybrid Experiments
The potential for interaction between PRN1 and either NF-Y-B9 or the closely related protein NF-Y-B6 was tested in a yeast (Saccharomyces cerevisiae) two-hybrid assay as described (Lapik and Kaufman, 2003). Bait and prey vectors, pGBT9 and pGAD424, respectively, were obtained from CLONTECH (Palo Alto) and used as directed. The indicated combinations of the bait (pGBT9) and prey (pGAD424) constructs were transformed into the yeast reporter strain AH109. The same inserts were also cloned in the opposite vectors (bait and prey switch) with similar results (data not shown). Empty vector (pGAD424) and the yeast mating response protein yFUS3 were used as negative bait controls. Other negative controls included NF-Y-B9 and NF-Y-B6 with empty prey vector (data not shown). The interaction between the yeast Gα (yGPA1) and Gβ (ySTE4) was used as a positive interaction control. Initial transformants were plated on 9-cm sd plates (Trp−, Leu−, Lys−, His−) to select for His prototrophy. Positive transformants were also streaked on X-α-Gal-containing selective media where LacZ reporter gene activity was monitored visually; blue color indicating a positive interaction.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Graphical representation of the ABA hypersensitivity germination experiments is shown in this figure.
Supplemental Table S1. Accessions of null T-DNA insertion lines for potential BL-induced Lhcb signal transduction pathway components were tested for a variety of genes as described in “Materials and Methods” under “Plant Material.”
Supplementary Material
Acknowledgments
We thank M. Metodiev for assistance with the yeast two-hybrid assays, Oliver Appelbe for assistance with PCR, Alan M. Jones for supplying GPA1 insertions lines in Ws, and M. Metodiev and K. Folta for careful reading of this manuscript.
This work was supported by the U.S. Department of Agriculture (grant no. CREES 2005–02389 to L.S.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Lon S. Kaufman (lkaufman@uic.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Huaming Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson MB, Folta KM, Warpeha KM, Gibbons J, Gao J, Kaufman LS (1999) Blue light-directed destabilization of the pea Lhcb1*4 transcript depends upon sequences within the 5′-UTR. Plant Cell 11 1579–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apone F, Alyeshmerni N, Wiens K, Chalmers D, Chrispeels MJ, Colucci G (2003) The G-protein-coupled receptor GCR1 regulates DNA synthesis through activation of phosphatidylinositol-specific phospholipase C. Plant Physiol 133 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann SM (2002) Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell (suppl) 14 S355–S373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, Samach A, Lifschitz E (2006) The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J 46 462–476 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Maloof JN, Lutes J, Dabi T, Redfern JL, Trainer GT, Werner JD, Asami T, Berry CC, Weigel D, et al (2002) Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana. Genetics 160 683–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Huala E (1999) Blue light photoreceptors in higher plants. Annu Rev Cell Dev Biol 15 33–62 [DOI] [PubMed] [Google Scholar]
- Brocard-Gifford IM, Lynch TJ, Finkelstein RR (2003) Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol 131 78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-G, Pandey S, Huang J, Alonso JM, Ecker JR, Assmann SM, Jones AM (2006) GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol 135 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38 87–117 [DOI] [PubMed] [Google Scholar]
- Colucci G, Apone F, Alyeshmerni N, Chalmers D, Chrispeels MJ (2002) GCR1, the putative Arabidopsis G protein-coupled receptor gene is cell cycle-regulated, and its overexpression abolishes seed dormancy and shortens time to flowering. Proc Natl Acad Sci USA 99 4736–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechend R, Hirano F, Lehmann K, Heissmeyer V, Ansieau S, Wulczyn FG, Scheidereit C, Leutz A (1999) The Bcl-3 oncoprotein acts as a bridging factor between NF-kappa B/Rel and nuclear co-regulators. Oncogene 18 3316–3323 [DOI] [PubMed] [Google Scholar]
- Dunwell JM, Purvis A, Khuri S (2004) Cupins: the most functionally diverse protein superfamily? Phytochemistry 65 7–17 [DOI] [PubMed] [Google Scholar]
- Edwards D, Murray JAH, Smith AG (1998) Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol 117 1015–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fambrini M, Durante C, Cionini G, Geri C, Giorgetti L, Michelotti V, Salvini M, Pugliesi C (2006) Characterization of LEAFY COTYLEDON1-LIKE gene in Helianthus Annuus and its relationship with zygotic and somatic embryogenesis. Dev Genes Evol 216 253–264 [DOI] [PubMed] [Google Scholar]
- Fan LM, Zhao ZX, Assmann SM (2004) Guard cells: a dynamic signaling model. Curr Opin Plant Biol 7 537–546 [DOI] [PubMed] [Google Scholar]
- Finkelstein R (2006) Studies of abscisic acid perception finally flower. Plant Cell 18 786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Gampala SSL, Lynch TJ, Thomas TL, Rock CD (2005) Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol Biol 59 253–267 [DOI] [PubMed] [Google Scholar]
- Folta KM, Kaufman LS (1999) Regions of the pea Lhcb1*4 promoter necessary for blue-light regulation in transgenic Arabidopsis. Plant Physiol 120 747–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala SSL, Finkelstein RR, Sun SSM, Rock CD (2002) ABI5 interacts with abscisic acid signaling effectors in rice protoplasts. J Biol Chem 277 1689–1694 [DOI] [PubMed] [Google Scholar]
- Gao H, Sun Y, Wu Y, Luan B, Wang Y, Qu B, Pei G (2004) Identification of beta-arrestin2 as a G protein-coupled receptor-stimulated regulator of NF-kappaB pathways. Mol Cell 14 303–317 [DOI] [PubMed] [Google Scholar]
- Gao J, Kaufman LS (1994) Blue-light regulation of the Arabidopsis thaliana Cab1 gene. Plant Physiol 104 1251–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner A, Manni I, Fuschi P, Mantovani R, Guadagni F, Sacchi A, Piaggio G (2003) Requirement for down-regualtion of the CCAAT-binding activity of the NF-Y transcription factor during skeletal muscle differentiation. Mol Biol Cell 14 2706–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusmaroli G, Tonelli C, Mantovani R (2001) Regulation of the CCAAT-binding NF-Y subunits in Arabidopsis thaliana. Gene 264 173–185 [DOI] [PubMed] [Google Scholar]
- Gusmaroli G, Tonelli C, Mantovani R (2002) Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene 283 41–48 [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Conejero G, Castelain M, Guenin S, Verdeil J-L, Thomasset B, Van Wuytswinkel O (2006) Identification of new gene expression regulators specifically expression during seed maturation. J Exp Bot 57 1919–1932 [DOI] [PubMed] [Google Scholar]
- Hamm HE (1998) The many faces of G protein signaling. J Biol Chem 273 669–672 [DOI] [PubMed] [Google Scholar]
- Huang J, Taylor JP, Chen J-G, Uhrig JF, Schnell DJ, Nakagawa T, Korth KL, Jones AM (2006) The plastid protein THYLAKOID FORMATION 1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell 18 1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Assmann SM (2004) Plants: the latest model system for G-protein research. EMBO Rep 5 572–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson LG, Rask L (1997) Cloning of a putative G-protein-coupled receptor from Arabidopsis thaliana. Eur J Biochem 249 415–420 [DOI] [PubMed] [Google Scholar]
- Kaufman LS (1993) Transduction of blue-light signals. Plant Physiol 102 333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5 33–36 [DOI] [PubMed] [Google Scholar]
- Kurup S, Jones HD, Holdsworth MJ (2000) Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J 21 143–156 [DOI] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapik Y, Kaufman LS (2003) The Arabidopsis cupin domain protein AtPirin1 and AtGPA1, the Arabidopsis Gα subunit interact with each other and regulate seed germination and early seedling development. Plant Cell 15 1578–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Fischer RL, Goldberg RB, Harada JJ (2003) Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc Natl Acad Sci USA 100 2152–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C (2000) Plant blue-light receptors. Trends Plant Sci 5 337–341 [DOI] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM (1990) Molecular cloning and characterization of GPA1, a G protein alpha subunit gene from Arabidopsis thaliana. Proc Natl Acad Sci USA 87 3821–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs KA, Kaufman LS (1991) Rapid transcriptional regulation of the Cab and pEA207 gene families by blue light in the absence of cytoplasmic protein synthesis. Planta 183 327–333 [DOI] [PubMed] [Google Scholar]
- Marsh JF, Kaufman LS (1999) Cloning and characterization of PGA1 and PGA2, two G protein alpha-subunits from pea that promote growth in the yeast Saccharomyces cerevisiae. Plant J 19 237–247 [DOI] [PubMed] [Google Scholar]
- Mason MG, Botella JR (2000) Completing the heterotrimer: isolation and characterization of an Arabidopsis thaliana G protein gamma-subunit cDNA. Proc Natl Acad Sci USA 97 14784–14788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Botella JR (2001) Isolation of a novel G-protein gamma-subunit from Arabidopsis thaliana and its interaction with G beta. Biochim Biophys Acta 1520 147–153 [DOI] [PubMed] [Google Scholar]
- Mishra G, Zhang W, Deng F, Zhao J, Wang X (2006) A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312 264–266 [DOI] [PubMed] [Google Scholar]
- Pandey S, Assmann SM (2004) The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein α subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16 1616–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Chen J-G, Jones AM, Assmann SM (2006) G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol 141 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pang H, Bartlam M, Zeng QH, Miyatake H, Hisano T, Miki K, Wong LL, Gao GF, Rao ZH (2004) Crystal structure of human pirin—an iron-binding nuclear protein and transcription cofactor. J Biol Chem 279 1491–1498 [DOI] [PubMed] [Google Scholar]
- Perfus-Barbeoch L, Jones AM, Assmann SM (2004) Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Curr Opin Plant Biol 7 719–731 [DOI] [PubMed] [Google Scholar]
- Quinones MA, Lu ZM, Zeiger E (1998) Genetic variation of stomatal conductance, blue light sensitivity and zeaxanthin content in guard cells of Pima cotton (Gossypium barbadense). Physiol Plant 103 560–566 [Google Scholar]
- Quinones MA, Zeiger E (1994) A putative role of the xanthophyll, zeaxanthin, in blue light photoreception of corn coleoptiles. Science 264 558–561 [DOI] [PubMed] [Google Scholar]
- Reidt W, Ellerstrom M, Kolle K, Tewes A, Tiedemann J, Altschmied L, Baumlein H (2001) FUS-3 dependent gene regulation during late embryogenesis. J Plant Physiol 158 411–418 [Google Scholar]
- Rohde A, De Rycke R, Beeckman T, Engler G, Van Montagu M, Boerjan W (2000) ABI3 affects plastid differentiation in dark-grown Arabidopsis seedlings. Plant Cell 12 35–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Van Montagu M, Boerjan W (1999) The ABSCISIC ACID-INSENSITIVE 3 (ABI3) gene is expressed during vegetative quiescence processes in Arabidopsis. Plant Cell Environ 22 261–270 [Google Scholar]
- Romier C, Cocchiarella Mantovani R, Moras D (2003) The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J Biol Chem 278 1336–1345 [DOI] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Wang SC, Jones AM (2002) Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol 129 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Young JC, Im KH, Sussman MR, Jones AM (2001) Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292 2066–2069 [DOI] [PubMed] [Google Scholar]
- Wang XQ, Ullah H, Jones AM, Assmann SM (2001) G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292 2070–2072 [DOI] [PubMed] [Google Scholar]
- Warpeha KM, Lateef SS, Lapik Y, Anderson MB, Lee B-S, Kaufman LS (2006) G-protein-coupled receptor 1, g-protein Gα-subunit 1, and prephenate dehydratase 1 are required for blue light-induced production of phenylalanine in etiolated Arabidopsis1. Plant Physiol 140 844–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warpeha KMF, Hamm HE, Rasenick MM, Kaufman LS (1991) A blue-light activated GTP binding protein in the plasma membrane of etiolated pea. Proc Natl Acad Sci USA 88 8925–8929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warpeha KMF, Kaufman LS (1990) 2 distinct blue-light responses regulate the levels of transcripts of specific nuclear-coded genes in pea. Planta 182 553–558 [DOI] [PubMed] [Google Scholar]
- Weatherwax SC, Ong MS, Degenhardt J, Bray EA, Tobin EM (1996) The interaction of light and abscisic acid in the regulation of plant gene expression. Plant Physiol 111 363–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss CA, Garnaat CW, Mukaik K, Hu Y, Ma H (1994) Isolation of cDNAs encoding guanine-nucleotide-binding protein beta-subunit homologs from maize (ZGB1) and Arabidopsis (AGB1). Proc Natl Acad Sci USA 91 9554–9558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler WMF, Kremmer E, Forster R, Winnacker EL (1997) Identification of Pirin, a novel highly conserved nuclear protein. J Biol Chem 272 8482–8489 [DOI] [PubMed] [Google Scholar]
- West MAL, Yee KM, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ (1994) LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6 1731–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S (2005) A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zengyan X, Glover BJ (2005) Asymmetric evolution of duplicate genes encoding the CCAAT-binding factor NF-Y in plant genomes. New Phytol 165 623–632 [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang HB, Takemiya A, Song CP, Kinoshita T, Shimazaki KI (2004) Inhibition of blue light-dependent H+ pumping by abscisic acid through hydrogen peroxide-induced dephosphorylation of the plasma membrane H+-ATPase in guard cell protoplasts. Plant Physiol 136 4150–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Wang XM (2004) Arabidopsis phospholipase D alpha 1 interacts with the heterotrimeric G-protein alpha-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J Biol Chem 279 1794–1800 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.