Abstract
Members of the epsin family of proteins (epsins) are characterized by the presence of an epsin N-terminal homology (ENTH) domain. Epsins have been implicated in various protein-trafficking pathways in animal and yeast (Saccharomyces cerevisiae) cells. Plant cells also contain multiple epsin-related proteins. In Arabidopsis (Arabidopsis thaliana), EPSIN1 is involved in vacuolar trafficking of soluble proteins. In this study, we investigated the role of Arabidopsis EpsinR2 in protein trafficking in plant cells. EpsinR2 contains a highly conserved ENTH domain but a fairly divergent C-terminal sequence. We found that the N-terminal ENTH domain specifically binds to phosphatidylinositol-3-P in vitro and has a critical role in the targeting of EpsinR2. Upon transient expression in protoplasts, hemagglutinin epitope-tagged EpsinR2 was translocated primarily to a novel cellular compartment, while a minor portion localized to the Golgi complex. Protein-binding experiments showed that EpsinR2 interacts with clathrin, AtVTI12, and the Arabidopsis homologs of adaptor protein-3 δ-adaptin and adaptor protein-2 α-adaptin. Localization experiments revealed that hemagglutinin epitope-tagged EpsinR2 colocalizes primarily with δ-adaptin and partially colocalizes with clathrin and AtVTI12. Based on these findings, we propose that EpsinR2 plays an important role in protein trafficking through interactions with δ-adaptin, AtVTI12, clathrin, and phosphatidylinositol-3-P.
Clathrin is a coat protein involved in several distinct intracellular transport steps, including the internalization of plasma membrane proteins by receptor-mediated endocytosis and the transport of lysosomal enzymes from the trans-Golgi network (TGN). In the process of generating a clathrin-coated vesicle (CCV), membrane proteins such as cargo receptors interact with a class of molecules called adaptors. The adaptors link the membrane proteins with clathrin molecules that form the coat of a lipid vesicle. Adaptors include a diverse group of proteins recognizing different classes of cargo receptor. The best characterized are a family of closely related proteins called the adaptor proteins (APs). To date, four different types of AP complexes have been identified, and each of them is thought to be responsible for the recruitment of clathrin to specific donor compartments (Kirchhausen, 1999; Robinson and Bonifacino, 2001).
In addition to AP complexes, alternate APs have recently been identified in a variety of eukaryotic cells. Among these is the epsin family of proteins. Epsin 1 was originally identified as a protein that plays a critical role in endocytosis (Chen et al., 1998). Subsequently, a large number of epsin-related proteins were identified in a variety of organisms (De Camilli et al., 2002; Wendland, 2002; Overstreet et al., 2003; Legendre-Guillemin et al., 2004; Song et al., 2006). A common feature of all epsin-related APs is the presence of a highly conserved epsin N-terminal homology (ENTH) domain. Epsin-related proteins can be divided into two functional categories: group one is involved in endocytosis from the plasma membrane and includes animal epsin1 and Lqf, and yeast (Saccharomyces cerevisiae) Ent1p and Ent2p (Chen et al., 1998; De Camilli et al., 2002; Wendland, 2002); group two is involved in lysosomal/vacuolar trafficking pathways from the TGN or endosomes and includes animal EpsinR/clint/enthoprotin, yeast Ent3p and Ent4p, and Arabidopsis (Arabidopsis thaliana) EPSIN1 (Kalthoff et al., 2002; Wasiak et al., 2002; Hirst et al., 2003; Chidambaram et al., 2004; Eugster et al., 2004; Saint-Pol et al., 2004; Song et al., 2006). Although the epsin proteins are divided into two functional groups, they may play essentially the same role in different pathways, that is, facilitation of CCV formation at donor membrane compartments.
Epsins facilitate formation of CCVs in two ways. First, they bind to donor lipid membranes through interactions with phospholipids, and the ENTH domain is subsequently inserted into the lipid bilayer. This introduces curvature into the membrane, which in turn facilitates the formation of the CCV (Legendre-Guillemin et al., 2004). This curvature-inducing activity may be common to all members of the epsin family. Second, epsins recruit clathrin, as well as other accessory proteins, to the donor membrane compartment through protein-protein interactions. Epsins have been shown to bind to a number of proteins, including clathrin, AP complexes, and vesicle-associated SNARE (Rosenthal et al., 1999; Drake et al., 2000; Chidambaram et al., 2004; Song et al., 2006). Interestingly, rat epsin1, and yeast Ent1p and Ent2p, which participate in endocytosis, have binding motifs for AP-2, the hetrotetrameric adaptor complex for endocytosis (Chen et al., 1998; Legendre-Guillemin et al., 2004). Animal EpsinR/enthoprotin/clint, yeast Ent3p, and Arabidopsis EPSIN1, which are involved in vacuolar/lysosomal trafficking, have binding motifs for AP-1, the hetrotetrameric adaptor for lysosomal/vacuolar trafficking (Duncan et al., 2003; Mills et al., 2003; Chidambaram et al., 2004; Song et al., 2006). Thus, it appears that the interaction of epsins with specific AP complexes may determine the specificity of their role in protein trafficking pathways. An additional element that may also regulate epsin function is the specificity of their interaction with phospholipids. Animal epsin1, which is involved in endocytosis, binds to phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2], whereas EpsinR, which localizes to the TGN, and yeast Ent3, which localizes to the multivesicular body (MVB), bind to phosphatidylinositol-4-P [PtdIns(4)P] and phosphatidylinositol-3,5-bisphosphate [PtdIns(3,5)P2], respectively (Itoh et al., 2001; Friant et al., 2003; Hirst et al., 2003). Epsin-related proteins are also recruited to donor membrane compartments via the interaction with cargo proteins, either directly or through a ubiquitin moiety. Thus, epsin family proteins facilitate assembly of CCVs at the plasma membrane, TGN, or MVB in a cargo-dependent manner (Friant et al., 2003).
Sequence analysis of the Arabidopsis genome revealed several epsin-related proteins, in addition to EPSIN1 (also termed EpsinR1 to reflect its functional similarity to animal EpsinR), which we recently determined was involved in trafficking of soluble protein cargo from the TGN to the central vacuole (Song et al., 2006). In this article, we investigated the biological role of EpsinR2. We demonstrated that EpsinR2 interacts with clathrin, Arabidopsis δ-adaptin, α-adaptin, AtVTI12, and phosphatidylinositol-3-P [PtdIns(3)P], and colocalizes with δ-adaptin and clathrin.
RESULTS
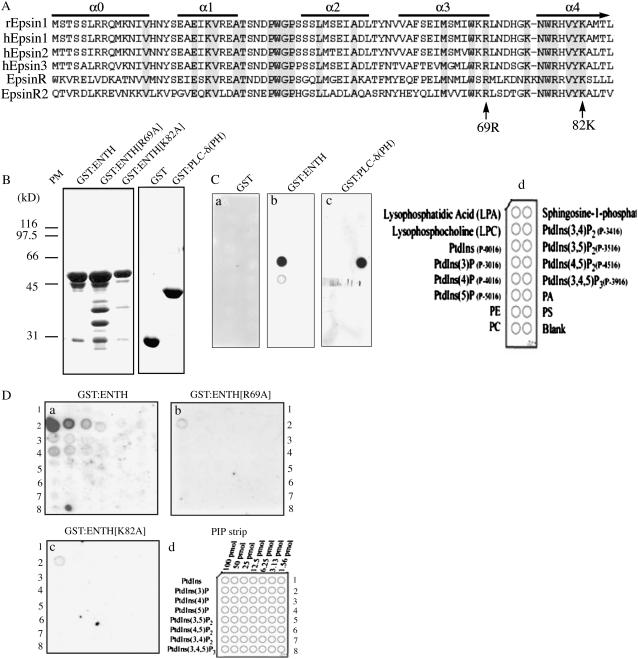
EpsinR2 Binds to PtdIns(3)P through Its N-Terminal ENTH Domain
The Arabidopsis genome encodes three closely related proteins that are highly homologous to rat epsin1 and EpsinR (Chen et al., 1998; Holstein and Oliviusson, 2005). Previously, we demonstrated that EPSIN1/EpsinR1 (EpsinR1) is involved in vacuolar trafficking of soluble proteins in Arabidopsis. In this study, we examined the function of the Arabidopsis EpsinR2 isoform. EpsinR2 displayed a high degree of amino acid sequence similarity to rat epsins and EpsinR, particularly in the highly conserved ENTH domain (Fig. 1A; Chen et al., 1998; Hirst et al., 2003; Legendre-Guillemin et al., 2004; Song et al., 2006).
Figure 1.
EpsinR2 binds to PtdIns(3)P through the N-terminal ENTH domain. A, Amino acid sequence alignment of various epsin family proteins, aligned according to their ENTH domains. α0 to α4 indicate α-helixes. B, Expression of the EpsinR2 ENTH domain. The indicated GST fusion proteins were expressed in E. coli and affinity purified to near homogeneity, then subjected to SDS-PAGE analysis. The gel was stained using Coomassie Blue. Note that expression of GST:ENTH[R69A] yielded minor degradation products, likely due to introduction of the mutation. C and D, Lipid-binding assay. The indicated purified GST fusion proteins were incubated with PIP lipid strips, followed by immunostaining with anti-GST antibody. Equal amounts (2 μg/mL) of protein were used for each assay.
The highly conserved ENTH domain binds to phospholipids (Legendre-Guillemin et al., 2004). In rats, plasma membrane-targeted epsin1 binds to PtdIns(4,5)P2, whereas EpsinR, which localizes to the TGN, binds to PtdIns(4)P (Itoh et al., 2001; Hirst et al., 2003). Yeast Ent3, which is targeted to MVBs, binds to PtdIns(3,5)P2 (Friant et al., 2003). Thus, organelle-specific targeting of epsins appears to correlate directly with their lipid-binding specificity. Conversely, the lipid-binding specificity may provide information about the role of epsin family members in protein-trafficking pathways. To determine the phospholipid-binding specificity of EpsinR2, we performed an in vitro lipid-binding assay (Stevenson et al., 1998; Kim et al., 2001b). Because the ENTH domain alone is sufficient for lipid binding, we expressed the N-terminal ENTH domain (encompassing amino acid residues 1–266) of EpsinR2 as a glutathione S-transferase (GST) fusion protein (GST:ENTH) in Escherichia coli. GST:ENTH was affinity purified from E. coli extracts (Fig. 1B) and incubated with nitrocellulose phosphatidyl inositol phosphate (PIP) strips that were spotted with different species of phospholipid. Proteins that bound to the PIP strip were detected using anti-GST antibody (Stevenson et al., 1998). EpsinR2 bound specifically to PtdIns(3)P (Fig. 1C, b). This was distinct from the ENTH domains of epsin1, EpsinR, and ENT3, which bind to PtdIns(4,5)P2, PtdIns(4)P, and PtdIns(3,5)P2, respectively (Itoh et al., 2001; Friant et al., 2003; Hirst et al., 2003). This result was also in contrast to the results of phospholipid-binding experiments with Arabidopsis EpsinR1, which did not bind to any phospholipids under these same conditions (data not shown; Song et al., 2006). The negative control, GST alone, did not bind to any lipids, and GST:phospholipase C-δ (pleckstrin homology), which was included as a positive control, displayed strong binding to PtdIns(4,5)P2, as previously observed (Fig. 1C, a and c; Garcia et al., 1995; Kim et al., 2001b). These results confirmed the specificity of lipid binding under these experimental conditions. To further characterize the interaction of EpsinR2 with phospholipids, we introduced two amino acid substitution mutations into the ENTH domain of EpsinR2: Arg (R)-69 to Ala (A; R69A) and Lys (K)-82 to Ala (K82A; Fig. 1A). According to the predicted structure of EpsinR2, R69 is located at the end of the fourth α-helix and is conserved in all ENTH domains. It is critical for binding of epsin1 to PtdIns(4,5)P2 (Ford et al., 2002). K82 is located in the middle of the fifth α-helix and is also conserved in all ENTH domains, although it is not directly involved in PtdIns(4,5)P2 binding. These mutants were expressed as GST fusion proteins in E. coli (GST:ENTH[R69A] and GST:ENTH[K82A]), affinity purified from bacterial cell extracts, and incubated with PIP strips. Bound proteins were detected using anti-GST antibody. The binding affinities of the substitution mutants to PtdIns(3)P were reduced to approximately one-tenth the level of binding of wild-type EpsinR2 (Fig. 1D, b and c), indicating that EpsinR2 binds specifically to PtdIns(3)P through its ENTH domain.
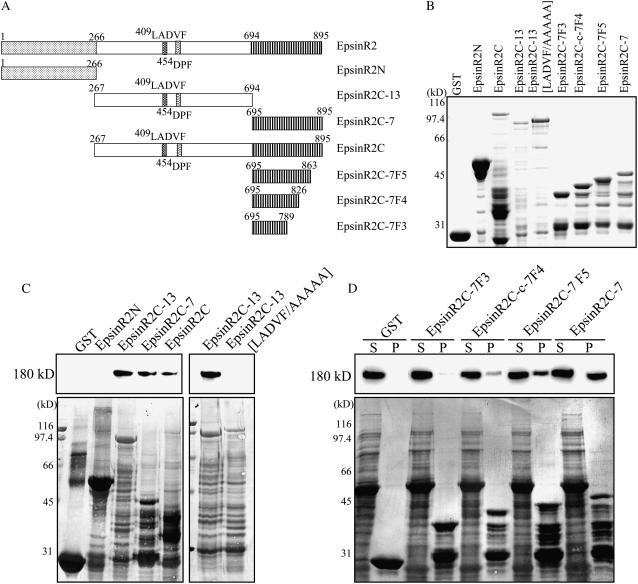
Clathrin Binds to Two Different Regions of EpsinR2
Epsins interact with a variety of proteins involved in vesicular trafficking, including clathrin (Chen et al., 1998; Rosenthal et al., 1999; Drake et al., 2000; Song et al., 2006). To examine whether EpsinR2 bound to clathrin, we performed a protein pull-down assay using recombinant EpsinR2. Full-length EpsinR2 was extremely sensitive to proteolytic degradation when expressed as a GST fusion protein in E. coli. It has been suggested that the C-terminal clathrin-binding region of epsin family proteins contains an unfolded region, which may be particularly sensitive to proteolytic degradation (Kalthoff et al., 2000). Therefore, we generated two fragments of EpsinR2 that we expressed as GST fusion proteins: the N-terminal ENTH domain (GST:EpsinR2N), and the remainder of the protein, comprising the middle and C-terminal domains (GST:EpsinR2C; Fig. 2A). GST:EpsinR2N and GST:EpsinR2C were affinity purified from E. coli extracts (Fig. 2B) and incubated with protein extracts from leaf tissues. GST:EpsinR2N- and GST:EpsinR2C-bound proteins were affinity purified using glutathione-agarose beads and analyzed by western blotting using anti-clathrin antibody (Song et al., 2006). A 180-kD protein was detected in GST:EpsinR2C protein complexes but not in GST:EpsinR2N or GST protein complexes (Fig. 2C), suggesting that EpsinR2 binds to clathrin through its C-terminal domain. To identify the clathrin-binding motif of EpsinR2, we divided EpsinR2C into two subdomains: EpsinR2C-13 and EpsinR2C-7 (Fig. 2A). These were expressed as GST fusion proteins in E. coli (GST:EpsinR2C-13 and GST:EpsinR2C-7) and used in the protein pull-down assay. We found that clathrin associated with both subdomains of EpsinR2C (Fig. 2C), indicating the presence of multiple clathrin-binding sites. Sequence analysis revealed the presence of a putative clathrin-binding motif in EpsinR2C-13, 409LADVF (Fig. 2A; Lafer, 2002). To confirm that the interaction of clathrin with EpsinR2C-13 was mediated by the 409LADVF motif, we generated a substitution mutant in which the amino acids of the LADVF motif were replaced with Ala (EpsinR2C-13[LADVF/AAAAA]; Fig. 2A). This mutant was expressed as a GST fusion protein (GST:EpsinR2C-13[LADVF/AAAAA]; Fig. 2B) in E. coli, and the purified protein was used in the protein pull-down assay. GST:EpsinR2C-13[LADVF/AAAAA] failed to associate with clathrin (Fig. 2C), indicating that the LADVF motif is responsible for binding. In contrast to EpsinR2C-13, EpsinR2C-7 did not appear to contain any known clathrin-binding motifs, suggesting the presence of a novel binding region. To define the clathrin-binding motif of EpsinR2C-7, we constructed a set of deletion mutants, gradually increasing the number of amino acids deleted from the C terminus of EpsinR2 (Fig. 2A). The deletion mutants were fused to GST, expressed in E. coli (Fig. 2B), and used in the protein pull-down assay. Interestingly, clathrin-binding affinity was proportional to the size of the C-terminal fragment (Fig. 2D), indicating that the Met-rich C terminus has multiple binding sites. Taken together, these results suggested that there is a C-terminal clathrin-binding region of EpsinR2 that is distinct from the LADVF motif located in the central region of the protein.
Figure 2.
EpsinR2 interacts with clathrin through two different binding regions. A, Schematic representation of the various EpsinR2 constructs used in this experiment. B, Expression of recombinant proteins. GST fusion proteins were affinity purified from E. coli extracts and subjected to SDS-PAGE analysis. The gel was stained with Coomassie Blue. Note that fusion proteins of the EpsinR2 C-terminal region were extremely sensitive to proteolytic degradation. C and D, Clathrin binding of EpsinR2. Purified GST alone (15 μg) or the indicated GST-fused EpsinR2 proteins (15 μg) were immobilized on glutathione-agarose beads and incubated with total protein extract (30 μg) from Arabidopsis leaf tissues in a 1-mL reaction volume. Protein complexes were collected by centrifugation and analyzed by western blotting using anti-clathrin antibody. P, Pellet; S, supernatant (one-tenth of the volume used in the pull-down experiment). Coomassie Blue-stained gels are shown in the bottom section.
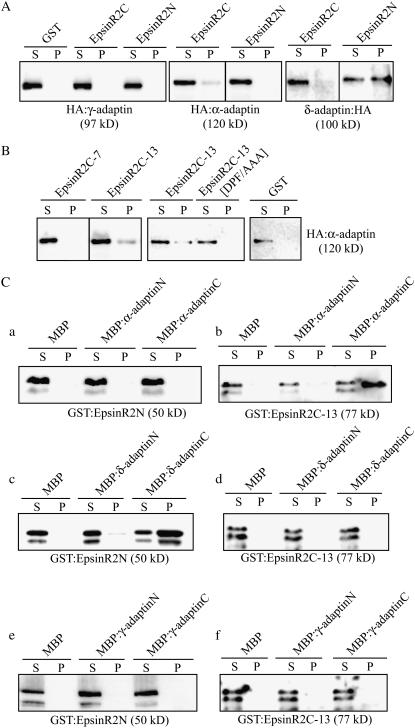
EpsinR2 Binds to α-Adaptin and δ-Adaptin
Epsin family proteins bind to AP complexes (Legendre-Guillemin et al., 2004). Epsin1 has multiple α-adaptin-binding motifs (Chen, et al., 1998), whereas EpsinR has multiple γ-adaptin-binding motifs (Mills et al., 2003). In plant cells, EpsinR1 binds preferentially to the Arabidopsis homolog of γ-adaptin and has weak binding affinity for the α-adaptin homolog (Song et al., 2006). To determine whether EpsinR2 has a role in intracellular trafficking, we examined EpsinR2 binding to AP complexes. In plant cells, the presence and biological significance of AP complexes remain to be clarified. Therefore, we first isolated several cDNAs encoding the Arabidopsis proteins that displayed the highest amino acid sequence homology to animal γ-adaptin, α-adaptin, and δ-adaptin of the AP-1, AP-2, and AP-3 complexes, respectively (Sanderfoot and Raikhel, 2003). We designated these Arabidopsis proteins γ-adaptin, α-adaptin, and δ-adaptin. We generated C- or N-terminal hemagglutinin (HA) fusion proteins of the three Arabidopsis adaptins, and they were transiently expressed in protoplasts. Extracts of transformed protoplasts were then incubated with GST:EpsinR2N or GST:EpsinR2C. Protein complexes were precipitated with glutathione agarose beads and analyzed by western blotting using anti-HA antibody. As seen in Figure 3A, EpsinR2 bound to both α-adaptin and δ-adaptin, but did not bind to γ-adaptin (Fig. 3A). We also observed that different domains of EpsinR2 mediated its interaction with the two adaptins: the C-terminal region bound to α-adaptin and the N-terminal region bound to δ-adaptin. To further define the binding site for α-adaptin, we examined the binding properties of GST:EpsinR2C-13 and GST:EpsinR2C-7 in transformed protoplast extracts using the protein pull-down assay. As seen in Figure 3B, EpsinR2C-13 bound to α-adaptin. Sequence analysis revealed that EpsinR2C-13 contains a DPF motif, which is the motif in animal epsin 1 that mediates binding to α-adaptin (Brett et al., 2002). To determine whether this motif was responsible for binding to α-adaptin, we generated an EpsinR2C-13 mutant in which the amino acids of the DPF motif were replaced with Ala (EpsinR2[DPF/AAA]). EpsinR2C[DPF/AAA] failed to bind to α-adaptin (Fig. 3B), indicating that the C-terminal DPF motif of EpsinR2 is responsible for binding to α-adaptin. These results suggested that EpsinR2 interacts with α-adaptin and with relatively higher affinity to δ-adaptin, via distinct binding motifs, and does not associate with γ-adaptin.
Figure 3.
EpsinR2 binds to α-adaptin and, with higher affinity, to δ-adaptin. A and B, Binding of EpsinR2 to δ-adaptin and α-adaptin but not to γ-adaptin. Protoplasts were transformed with the indicated constructs, and total protein extract (30 μg) was used in a protein pull-down assay with the indicated GST fusion proteins (15 μg) or GST alone (15 μg). Protein complexes were precipitated with glutathione agarose beads and analyzed by western blotting using the anti-HA antibody. S, Supernatant (10% of total); P, pellet. C, Reciprocal protein pull-down experiments. The indicated MBP fusion proteins or MBP alone (10 μg) was incubated with GST:EpsinR2N (2 μg) or GST:EpsinR2C (2 μg) in a 1-mL reaction volume. Proteins were precipitated with amylose resin and analyzed by western blotting using an anti-GST antibody. S, Supernatant (20% of total); P, pellet.
To confirm the interaction of EpsinR2 with α-adaptin and δ-adaptin and define the binding domains of α-adaptin and δ-adaptin that mediate the interaction, reciprocal protein pull-down experiments were performed. Both α-adaptin and δ-adaptin were divided into two regions, a C-terminal ear domain (α-adaptinC and δ-adaptinC) and an N-terminal trunk domain (α-adaptinN and δ-adaptinN), and expressed as maltose-binding protein (MBP) fusion proteins in E. coli. Purified α-adaptin or δ-adaptin fusion proteins were then incubated with GST:EpsinR2N or GST:EpsinR2C-13. MBP alone was also included as a negative control. Protein complexes were precipitated with amylose resin and analyzed by western blotting using anti-GST antibody. GST:EpsinR2C-13 associated specifically with MBP:α-adaptinC (Fig. 3C, b), confirming that the interaction is mediated by the EpsinR2C-13 region and the C-terminal ear domain of α-adaptin. In the case of δ-adaptin, GST:EpsinR2N was found in association with MBP:δ-adaptinC, but not MBP:δ-adaptinN (Fig. 3C, c), indicating that the C-terminal region of δ-adaptin binds to the N-terminal region of EpsinR2. These results demonstrated that EpsinR2 binds to the C-terminal ear domains of α-adaptin and δ-adaptin. We also performed reciprocal protein pull-down experiments with γ-adaptin. The N- and C-terminal regions of γ-adaptin were expressed as MBP fusion proteins and purified from E. coli. MBP:γ-adaptinN and MBP:γ-adaptinC were then incubated with GST:EpsinR2N or GST:EpsinR2C-13. Neither domain of γ-adaptin bound to GST:EpsinR2N or GST:EpsinR2C-13, confirming that there is no association between EpsinR2 and γ-adaptin.
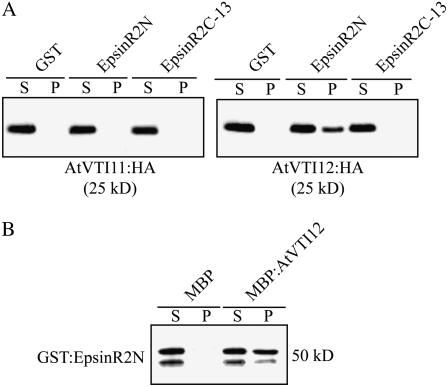
EpsinR2 Binds to AtVTI12
EpsinR and Ent3p bind to the SNARE proteins Vti1b and Vti1p, respectively (Chidambaram et al., 2004; Hirst et al., 2004). In Arabidopsis, EpsinR1 interacts specifically with AtVTI11 (Song et al., 2006). We were interested in whether EpsinR2 bound to AtVTI11 and AtVTI12, which localize primarily to the TGN, with a minor portion localized at the prevacuolar compartment (PVC; Zheng et al., 1999; Bassham et al., 2000; Surpin et al., 2003). We generated C-terminal HA fusion proteins of AtVTI11 and AtVTI12 (Song et al., 2006) and transiently expressed them in protoplasts. Protein extracts obtained from transformed protoplasts were incubated with purified GST:EpsinR2N or GST:EpsinR2C-13, and protein complexes were precipitated with glutathione agarose beads and then analyzed by western blotting using anti-HA antibody. AtVTI12, but not AtVTI11, coprecipitated with GST:EpsinR2N (Fig. 4A), indicating that, while AtVTI11 and AtVTI12 are highly homologous, EpsinR2 preferentially binds to AtVTI12 through its ENTH domain.
Figure 4.
EpsinR2 binds to AtVTI12. A, Specific binding of EpsinR2 to AtVTI12. Total protein extracts (30 μg) were prepared from protoplasts transformed with HA:AtVTI11 or HA:AtVTI12 and used in pull-down assays with the indicated GST fusion proteins (15 μg). Protein complexes were precipitated as described for Figure 3 and analyzed by western blotting using an anti-HA antibody. S, Supernatant (10% of total); P, pellet. B, Reciprocal protein pull-down assay. GST:EpsinR2N (2 μg) was incubated with 10 μg of purified MBP:AtVTI12 or MBP alone. Proteins were precipitated with amylose resin and analyzed by western blotting using an anti-GST antibody. S, Supernatant (20% of total); P, pellet.
To confirm these results, we performed a reciprocal protein pull-down assay. We expressed an MBP fusion protein of AtVTI12 in E. coli, and purified MBP:AtVTI12, or MBP alone, was incubated with GST:EpsinR2N. Protein complexes were precipitated with amylose resin and analyzed by western blotting using anti-GST antibody. As seen in Figure 4B, GST:AtVTI12 coprecipitated with MBP:AtVTI12, but not MBP alone, confirming that AtVTI12 interacts directly with the N-terminal ENTH domain of EpsinR2.
EpsinR2 Localizes Primarily to a Novel Cellular Compartment, and a Minor Portion Localizes to the Golgi Complex
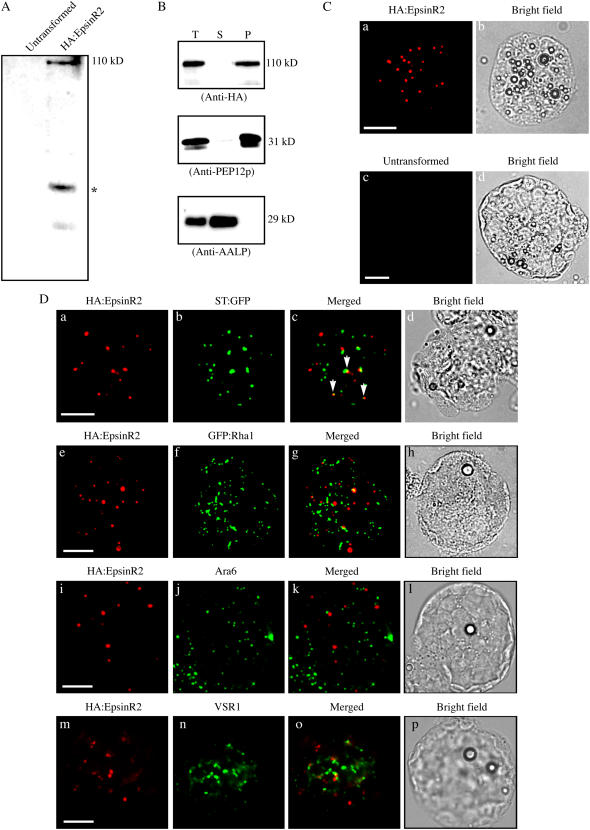
We next analyzed the localization of EpsinR2 to determine whether it plays a role in intracellular trafficking. We generated an expression construct encoding an HA fusion protein of EpsinR2 under the cauliflower mosaic virus 35S promoter (HA:EpsinR2), which was introduced into protoplasts obtained from Arabidopsis leaf tissues (Jin et al., 2001). When transformed protoplast extracts were examined by western-blot analysis using anti-HA antibody, we observed a 110-kD molecular mass protein in transformed protoplasts that was absent in untransformed protoplasts (Fig. 5A). This protein, representing HA:EpsinR2, was larger than the expected molecular mass of HA:EpsinR2 (95 kD), consistent with previous results showing that other epsin homologs migrate slower than expected during SDS-PAGE (Chen et al., 1998; Song et al., 2006). Initially, we examined the subcellular distribution of HA:EpsinR2 using fractionation by ultracentrifugation. Protein extracts obtained from transformed protoplasts were separated into soluble and membrane fractions, then analyzed by western blotting using anti-HA antibody. The majority of HA:EpsinR2 was detected in the pelleted (insoluble) fraction, and a minor portion was present in the soluble fraction (Fig. 5B), suggesting that EpsinR2 is membrane associated. Arabidopsis aleurain-like protein (AALP) and AtPEP12p were also examined as controls for fractionation, using anti-AALP and anti-PEP12p antibodies, respectively (Sohn et al., 2003). AALP is a soluble vacuolar protein, and AtPEP12p is a t-SNARE that localizes to the PVC (da Silva Conceição et al., 1997; Ahmed et al., 2000). As expected, AALP and AtPEP12p were detected in the soluble and membrane fractions, respectively (Fig. 5B). These results indicated that EpsinR2 behaves similarly to EpsinR1 in Arabidopsis and to other epsin-related proteins in animal cells (Legendre-Guillemin et al., 2004; Song et al., 2006).
Figure 5.
EpsinR2 primarily localizes to a novel compartment, and a minor proportion localizes to the Golgi complex. A, Expression of HA:EpsinR2 in protoplasts. Protoplasts from leaf tissues were transformed with HA:EpsinR2, and protein extracts were analyzed by western blotting using anti-HA antibody. Protein extracts of untransformed protoplasts were included as a negative control. *, Proteolytic degradation product of HA:EpsinR2. B, Subcellular distribution of HA:EpsinR2. Protoplasts were transformed with HA:EpsinR2, and protein extracts were separated into soluble and membrane fractions by ultracentrifugation. Fractions were analyzed by western blotting using anti-HA, anti-AALP, and anti-PEP12p antibodies. S, Supernatant; P, pellet fraction containing membrane proteins. C, Immunostaining of HA:EpsinR2. Protoplasts transformed with HA:EpsinR2 were immunostained with anti-HA antibody, followed by detection using trimethylrhodamine isothiocyanate-labeled anti-rat IgG secondary antibody. Untransformed protoplasts were included as a negative control. Scale bar = 20 μm. D, Partial colocalization of HA:EpsinR2 and ST:GFP. Protoplasts were transformed with the indicated constructs. HA:EpsinR2, Ara6, and VSR were detected by immunohistochemistry using anti-HA, anti-Ara6, and anti-VSR antibodies, respectively. GFP was observed directly. Arrows indicate colocalization of HA:EpsinR2 and ST:GFP. Scale bar = 20 μm.
We then performed immunocytochemistry on Arabidopsis protoplasts using anti-HA antibody. HA:EpsinR2 displayed a punctate staining pattern in a majority (over 95%) of transformed protoplasts (Fig. 5C, a), but not in untransformed cells (Fig. 5C, c). Based on sequence analysis, EpsinR2 is more closely related to EpsinR than Epsin1 (Legendre-Guillemin et al., 2004). In animal cells, EpsinR localizes to the Golgi complex and endosomes (Hirst et al., 2004). In addition, Arabidopsis EpsinR1, a close homolog of EpsinR2 in plant cells, also localizes primarily to the Golgi complex, with a minor portion localizing to the PVC (Song et al., 2006). Thus, we examined whether EpsinR2 localized to the Golgi complex. Protoplasts were cotransformed with HA:EpsinR2 and a construct encoding a chimeric protein consisting of rat sialyltransferase (ST) and green fluorescent protein (GFP; ST:GFP), which localizes to the Golgi complex in plant cells (Boevink et al., 1998; Jin et al., 2001; Lee et al., 2002). HA:EpsinR2 was detected using an anti-HA antibody, and ST:GFP was observed directly using green fluorescence. Both HA:EpsinR2 and ST:GFP were present in punctate staining patterns (Fig. 5D), but only a small proportion (10%–20%) of total HA:EpsinR2-positive staining appeared to overlap that of ST:GFP (Fig. 5D, a–c). This pattern of expression was distinct from that of EpsinR1 in Arabidopsis (Song et al., 2006).
We then examined whether EpsinR2 localized to endosomes. Two different types of endosome exist in Arabidopsis cells: Ara6- and Ara7-positive endosomes (Ueda et al., 2001). As a marker protein for Ara7-positive endosomes, we used Rha1, a close homolog of Ara7. Rha1 was previously shown to localize to the PVC and colocalize with Ara7 in leaf protoplasts (Sohn et al., 2003; Lee et al., 2004). To determine if EpsinR2 localized to Ara7-positive endosomes, we generated an expression construct encoding a fusion protein of the small GTP-binding protein Rha1 and GFP (GFP:Rha1). Protoplasts were cotransformed with HA:EpsinR2 and GFP:Rha1, then examined by immunohistochemistry using anti-HA antibody. Both EpsinR2 and GFP:Rha1 displayed nonoverlapping, punctate staining patterns (Fig. 5D, e–g), indicating that HA:EpsinR2 does not localize to a Rha1-positive cellular compartment, the PVC. Next, we examined the localization of EpsinR2 and Ara6. Protoplasts transformed with HA:EpsinR2 were coimmunostained with anti-HA and anti-Ara6 antibodies. Both proteins displayed nonoverlapping punctate staining patterns (Fig. 5D, i–k), indicating that HA:EpsinR2 does not localize to Ara6-positive endosomes. Next, we examined colocalization of EpsinR2 and A. thaliana vacuolar sorting receptor (AtVSR). Previously, AtVSR localized to the PVC with a minor portion to the TGN (Ahmed et al., 2000; Tse et al., 2004). Protoplasts transformed with HA:EpsinR2 were immunostained with anti-VSR antibody. Both proteins displayed nonoverlapping punctate staining patterns (Fig. 5D, m–o), consistent with the data showing that EpsinR2 did not colocalize with GFP:Rha1. These results strongly suggested that HA:EpsinR2 localizes to a novel cellular compartment.
EpsinR2 Colocalizes with Clathrin, AtVTI12, and δ-Adaptin, with Varying Degrees of Overlap
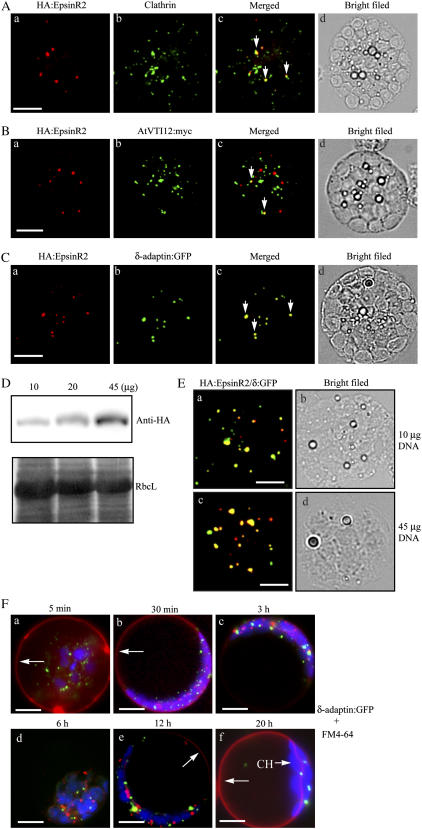
The observed interaction between EpsinR2 and clathrin (see Fig. 2) led us to investigate whether the two proteins also colocalized in cells. Protoplasts were transformed with HA:EpsinR2, and coimmunostained with anti-HA and anti-clathrin antibodies. Both clathrin and HA:EpsinR2 displayed punctate staining patterns; clathrin, in particular, localized to numerous punctate loci. The majority (95%) of HA:EpsinR2-positive staining overlapped with approximately 20% of the clathrin-positive loci (Fig. 6A), consistent with earlier results showing that EpsinR2 interacted with clathrin. The partial colocalization of clathrin and EpsinR2 is consistent with clathrin's involvement in multiple steps of protein trafficking in plant cells (Holstein, 2002; Song et al., 2006).
Figure 6.
HA:EpsinR2 colocalizes with δ-adaptin:GFP, clathrin, and AtVTI12:myc. A, Partial colocalization of EpsinR2 and clathrin. Protoplasts were transformed with HA:EpsinR2, and the localization of clathrin and HA:EpsinR2 was examined by immunohistochemistry using anti-clathrin and anti-HA antibodies. Arrows indicate colocalization of HA:EpsinR2 and clathrin. Scale bar = 20 μm. B, Partial colocalization of EpsinR2 and AtVTI12. Protoplasts were transformed with HA:EpsinR2 and AtVTI12:myc and examined by immunohistochemistry using anti-HA and anti-myc antibodies. Arrows indicate colocalization of HA:EpsinR2 and AtVTI12:myc. Scale bar = 20 μm. C, Colocalization of EpsinR2 and δ-adaptin:GFP. Protoplasts were transformed with HA:EpsinR2 and δ-adaptin:GFP and examined by immunohistochemistry using anti-HA antibody. GFP signals were observed directly. Arrows indicate colocalization of HA:EpsinR2 and δ-adaptin:GFP. Scale bar = 20 μm. D and E, Localization of EpsinR2 is unaffected by its expression level. Increasing amounts of HA:EpsinR2 expression plasmid were introduced into protoplasts, together with 10 μg of δ-adapatin:GFP, and the expression level and localization of these two proteins were examined by western-blot analysis and immunohistochemistry using anti-HA antibody. RbcL, Large subunit of the Rubisco complex stained with Coomassie Blue as a loading control; δ:GFP, δ-adaptin:GFP. Scale bar = 20 μm. F, The δ-adaptin:GFP-positive cellular compartment is not stained with FM4-64. Protoplasts transformed with δ-adaptin:GFP were incubated with 30 μm FM4-64 dye 24 h after transformation. Images represent majority (50%–60%) of protoplasts at the indicated time points after staining with FM4-64. Arrows in a and b, FM4-64 at the plasma membrane; arrows in e and f, FM4-64 at the tonoplast of the central vacuole. CH, Chloroplast; red, FM4-64; green, δ-adaptin:GFP; blue, chloroplast. Scale bar = 20 μm.
We next examined the localization of EpsinR2 relative to AtVTI12, which localizes to the Golgi complex (Bassham et al., 2000; Surpin et al., 2003). We generated an expression construct encoding a myc epitope-tagged form of AtVT112 (AtVTI12:myc), and protoplasts were cotransformed with HA:EpsinR2 and AtVTI12:myc. When protoplasts were coimmunostained with anti-HA and anti-myc antibodies, AtVTI12:myc displayed a punctate staining pattern, and approximately 10% to 20% of AtVTI12:myc-positive loci colocalized with a portion (10%–20%) of ectopic HA:EpsinR2 (Fig. 6B), consistent with earlier results showing an interaction between these two proteins.
To examine the localization of EpsinR2 and δ-adaptin, we generated an expression construct encoding a GFP fusion protein of δ-adaptin (δ-adaptin:GFP) and cotransformed protoplasts with δ-adaptin:GFP and HA:EpsinR2. Cells were then immunostained with anti-HA antibody, and δ-adaptin:GFP was observed directly. Both proteins displayed closely overlapping punctate staining patterns (Fig. 6C). The degree of overlap was over 80%, indicating that there was a strong interaction in vivo between HA:EpsinR2 and δ-adaptin. To examine whether the level of expression of HA:EpsinR2 affected its localization, δ-adaptin:GFP and varying amounts of HA:EpsinR2 were introduced into protoplasts, and the localization of HA:EpsinR2 was examined. As expected, protein levels of HA:EpsinR2 increased with increasing amounts of HA:EpsinR2 DNA (Fig. 6D). HA:EpsinR2 colocalized with δ-adaptin:GFP in cells that were transformed with either 10 or 45 μg of HA:EpsinR2 (Fig. 6E), indicating that up to a 5-fold increase in the level of expression of HA:EpsinR2 did not affect its localization.
To investigate whether EpsinR2 localized to endosomes involved in endocytosis, we subjected cells to analysis using FM4-64. Because FM4-64 is imaged in live cells, we also examined the localization of δ-adaptin:GFP, as it colocalized closely with HA:EpsinR2 (Fig. 6C). FM4-64 did not stain δ-adaptin:GFP-positive cellular compartments during internalization from the plasma membrane to the central vacuole (Fig. 6F), demonstrating that EpsinR2 is not targeted to endosomes involved in endocytosis. This was consistent with earlier results showing that HA:EpsinR2 does not colocalize with Ara6 and Rha1. These results provided additional evidence that EpsinR2 predominantly localizes to a novel cellular compartment.
Binding of the ENTH Domain to PtdIns(3)P Is Critical for the Localization of EpsinR2
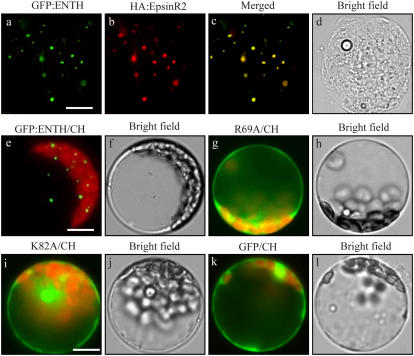
To investigate the targeting mechanism of EpsinR2 to the novel cellular compartment we observed in the previous experiments, we examined the role of lipid binding to EpsinR2. We generated expression constructs encoding GFP fusion proteins of the wild-type EpsinR2 ENTH domain (GFP:ENTH) and the two ENTH domain Ala substitution mutants (GFP:ENTH[R69A] and GFP:ENTH[K82A]) and examined the localization of these proteins in protoplasts. Protoplasts were transformed with GFP:ENTH and full-length HA:EpsinR2, and then cells were subjected to immunohistochemistry using anti-HA antibody. GFP was observed directly. GFP:ENTH and HA:EpsinR2 displayed closely overlapping, punctate staining patterns (Fig. 7, a–c), indicating that the N-terminal ENTH domain was sufficient for targeting of EpsinR2. To confirm this, the localization of GFP:ENTH[R69A] and GFP:ENTH[K82A] was examined. Both mutants displayed a diffuse pattern that was similar to GFP alone (Fig. 7, g–l). These results strongly suggested that the ENTH domain, and most likely PtdIns(3)P binding to the ENTH domain, is critical for localization of EpsinR2 to a novel compartment. We termed this novel compartment “δ compartment.”
Figure 7.
An intact ENTH domain is critical for proper targeting of EpsinR2. Protoplasts were transformed with the indicated constructs and their localization was examined directly using GFP signals or by immunohistochemistry using anti-HA antibody. CH, Chloroplast. Scale bar = 20 μm.
DISCUSSION
Binding Partners of EpsinR2
The primary structure and biochemical properties of Arabidopsis EpsinR2 clearly indicate that it belongs to the epsin family of proteins (Chen et al., 1998; Holstein and Oliviusson, 2005). In this study, we demonstrated that, similar to other epsins, EpsinR2 binds to phospholipids through its ENTH domain. However, the lipid-binding specificity of EpsinR2 was different from other epsins. Epsin1, Ent3p, and EpsinR bind to PtdIns(4,5)P2, PtdIns(3,5)P2, and PtdIns(4)P, respectively (Itoh et al., 2001; Friant et al., 2003; Hirst et al., 2003). In contrast, we showed that EpsinR2 binds specifically to PtdIns(3)P (Fig. 1C). Thus, EpsinR2 may be recruited to a specific compartment rich in PtdIns(3)P in plant cells, as suggested previously for other epsin-related proteins (Itoh et al., 2001; Friant et al., 2003; Hirst et al., 2003). Similar to its role in animal and yeast cells (Wurmser and Emr, 1998; Gillooly et al., 2000; Simonsen et al., 2001), PtdIns(3)P plays a critical role in vacuolar trafficking in plant cells (Kim et al., 2001a). Both the PtdIns(3)P-specific binding domain of early endosome antigen 1 and wortmannin, an inhibitor of PtdIns 3-kinase, strongly suppress vacuolar trafficking at the Golgi complex in BY-2 and Arabidopsis leaf cells (Matsuoka et al.,1995; Kim et al., 2001a), and wortmannin treatment affects MBV formation in plant cells (Tse et al., 2004).
We demonstrated that EpsinR2 interacts with several proteins, including clathrin. Analogous to other epsin family proteins, we showed that EpsinR2 has two clathrin-binding regions (Fig. 2, C and D): an LADVF motif, located in the middle region of the molecule, which conforms to the canonical clathrin-binding motif (Lafer, 2002); and a C-terminal region that does not contain a defined clathrin-binding motif, similar to EpsinR1 (Song et al., 2006). The finding that clathrin-binding affinity was proportional to the length of the C-terminal region strongly suggests the presence of multiple binding motifs in this portion of the protein, similar to AP180 in animal cells (Fig. 2; Morgan et al., 2000). AP180 contains multiple DLL motifs in its C-terminal region, and clathrin assembly by AP180 is proportional to the number of DLL motifs. The C-terminal region of EpsinR2 contains multiple MGM motifs, but it is not clear whether the MGM motif binds clathrin. One possibility is that both the LADVF and the C-terminal binding region of EpsinR2 are involved in recruiting clathrin during the formation of CCVs at the TGN, similar to other epsin proteins (Ford et al., 2002; Mills et al., 2003). However, we cannot rule out the possibility that the two clathrin-binding regions of EpsinR2 have different functional roles (Ford et al., 2002; Mills et al., 2003; Chen and De Camilli, 2005).
AtVTI12, a vesicle-associated SNARE that localizes to the TGN in plant cells, also bound to EpsinR2 (Fig. 4; Bassham et al., 2000; Surpin et al., 2003). Surpin et al. (2003) showed that atvti12 mutants display an accelerated senescence phenotype on nutrient-poor medium. However, the role of AtVTI12 in protein trafficking remains to be elucidated. Arabidopsis EpsinR1 and animal EpsinR bind to AtVTI11 and vtib, respectively (Chidambaram et al., 2004; Hirst et al., 2004; Song et al., 2006). AtVI11 and AtVTI12 are 60% identical at the amino acid sequence level and have partially redundant functions (Surpin et al., 2003). The specificity of binding of EpsinR1 and EpsinR2 to AtVTI11 and AtVTI12, respectively, implies that there are functional differences between the two isoforms. They may, however, partially complement each other under specific conditions. Consistent with this theory is that the zig mutant, which carries a mutation in AtVTI11, displays a defect in the gravitropic response (Kato et al., 2002).
Finally, we demonstrated that EpsinR2 binds to AP complexes. Interestingly, EpsinR2 displayed strong binding affinity to δ-adaptin, the Arabidopsis homolog of δ-adaptin (Fig. 3). This is different from EpsinR1, which displays strong binding affinity to γ-adaptin, the Arabidopsis homolog of γ-adaptin. Previously, we demonstrated that EpsinR1 is involved in trafficking of soluble proteins to the central vacuole (Song et al., 2006). In animal cells, EpsinR, which also binds strongly to γ-adaptin of AP-1, is involved in trafficking to the lysosome (Hirst et al., 2003). The interaction between δ-adaptin and EpsinR2 is strongly suggestive of a role for EpsinR2 in AP-3-mediated trafficking pathways. However, the biological role of AP-3 remains to be established in plant cells. In yeast and animal cells, membrane proteins are transported by AP-3 to the vacuole and lysosome, respectively (Cowles et al., 1997; Hirst and Robinson, 1998; Honing et al., 1998; Bonifacino and Traub, 2003). Both EpsinR1 and EpsinR2 contain a DPF motif, which mediates binding to Arabidopsis α-adaptin in vitro. However, the functional importance of the interaction between α-adaptin and epsins remains the subject of future studies.
Localization of EpsinR2
Previous studies have shown that different epsins localize to different cellular compartments, including the plasma membrane, Golgi complex, and endosomes (Itoh et al., 2001; Hirst et al., 2003; Song et al., 2006). Ectopically expressed HA-EpsinR2 in protoplasts was transported primarily to a novel compartment to which δ-adaptin:GFP also localized (Fig. 6B). We named this compartment the δ compartment. The localization of EpsinR2 to the δ compartment was strongly dependent on an intact ENTH domain. This result suggested that the δ compartment is rich in PtdIns(3)P. Previous studies have shown that PtdIns(3)P plays a critical role in protein trafficking from endosomes in both animal and plant cells (Wurmser and Emr, 1998; Gillooly et al., 2000; Kim et al., 2001a; Simonsen et al., 2001). In animal cells, AP-3 is located in endosomes (Peden et al., 2004). However, other than δ-adaptin, EpsinR2 did not colocalize to the δ compartment with any of the organellar proteins we examined. Furthermore, the δ compartment was not labeled with FM4-64 (Fig. 6D), indicating that it is not involved in endocytosis. In addition to the δ compartment, a minor proportion of EpsinR2 localized to the Golgi complex, and possibly the TGN, based on its partial colocalization with the Golgi marker ST:GFP and the TGN marker AtVTI12 (Fig. 5C; Kim et al., 2001a). The localization pattern of EpsinR2 was distinct from that of EpsinR1 in Arabidopsis, which localizes primarily to the Golgi complex, with a minor proportion localizing to the PVC (Song et al., 2006). Thus, the two epsin homologs in Arabidopsis localize to two different compartments, suggesting that they participate in different pathways. The localization of EpsinR2 was analogous to that of AtVSR1, which is primarily located in the PVC, although it binds to cargo proteins at the TGN (Ahmed et al., 2000; Tse et al., 2004). By analogy to AtVSR1, the presence of a minor portion of EpsinR2 in the Golgi complex supports the theory that EpsinR2 cycles between the Golgi complex and a novel cellular compartment in the vacuolar trafficking of cargo proteins.
MATERIALS AND METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was cultured on B5 plates in a growth chamber for 1 to 2 weeks at 20°C under a 16-h-light/8-h-dark cycle. Leaf tissue was harvested from plants and used immediately for protoplast isolation.
Construction of Plasmids
An EpsinR2 cDNA was amplified from an Arabidopsis cDNA library using the specific primers 5′-GAATTCATGAAGAAAGTCTTCGGACAAACTGTTAGA-3′ and 5′-CTCGAGTTACCGGTATCCACCACCATAGGATTGTTG-3′. To generate GST:ENTH, the EpsinR2 cDNA was digested with EcoRI and XhoI, and the resultant 780-bp fragment was ligated into the corresponding restriction sites of pGEX5X-1. The ENTH domain Ala substitution mutants ENTH[R69A] and ENTH[K82A] were generated using two sequential PCR reactions. Initially, 5′ and 3′ fragments were amplified by PCR, and the resulting fragments were then combined into a single fragment by a second round of PCR. The following primers were used: 5′-GTCATATGGAAAGCGCTTAGCGACACC-3′ and 5′-ATTATCATCAGCTCGAGCTTAACTGCCCCG-3′ for the 5′ fragment of ENTH[R69A]; 5′-GGTGTCGCTAAGCGCTTTCCATATGAC-3′and 5′-GAATTCATGAAGAAAGTCTTCGGACAAACTGTTAGA-3′ for the 3′ fragment of ENTH[R69A]; 5′-GAATTCATGAAGAAAGTCTTCGGACAAACTGTTAGA-3′ and 5′-ATTATCATCAGCTCGAGCTTAACTGCCCCG-3′ for the second amplification of ENTH[R69A]; 5′-CGGCATGTCTATGCGGCTTTGACAGTT-3′ and 5′-ATTATCATCAGCTCGAGCTTAACTGCCCCG-3 for the 5′ fragment of ENTH[K82A]; and 5′-AACTGTCAAAGCCGCATAGACATGCCG-3′ and 5′-GAATTCATGAAGAAAGTCTTCGGACAAACTGTTAGA-3 for the 3′ fragment of ENTH[K82A]. The 5′ and 3′ fragments of ENTH[K82A] were ligated as described for ENTH[R69A]. ENTH[R69A] and ENTH[K82A] were fused to GST similarly to the wild-type ENTH domain. To generate GST:EpsinR2C, the EpsinR2 cDNA was digested with XhoI, and the resultant 1,900-bp fragment was ligated into pGEX5X-2. To generate GST:EpsinR2C-13, the EpsinR2 cDNA was digested with XhoI and BglII, and the resulting fragment was ligated into pGEX5X-2. For GST:EpsinR2C-7, the EpsinR2C-7 fragment was prepared by PCR using the primers 5′-AATTCAGATCTCACACCTTTAACAGGA-3′ and 5′-CTCGAGTTACCGGTATCCACCACCATA-3′. The resulting fragment was ligated into pGEX5X-2. GST:EpsinR2C-7F3, GST:EpsinR2C-7F4, and GST:EpsinR2C-7F5 were generated by PCR amplification using the following primers: forward primers 5′-CTCGAGTTAAGGTGGAGGTCTCAGAGCAGTTGC-3′ for 7F3, 5′-CTCGAGCTATCCGGCCCCCATACCCATACCCAT-3′ for 7F4, and 5′-CTCGAGTTATGGGTTTTGTGGTTGCATCGGATA-3′for 7F5. The primer 5′-AATTCAGATCTCACACCTTTAACAGGA-3′ was used as a reverse primer for all three constructs. The C-terminal fragments were then ligated into pGEX5X-2. To generate GST:EpsinR2C-13[LADVF/AAAAA], EpsinR2C-13[LADVF/AAAAA] was prepared by two sequential PCR reactions using the following oligonucleotide primers: T3 primer and 5′-GGTTCCGCTGCAGCCGCAGCTTCATCA-3′ for the 5′ fragment, and 5′-ATTATCATCAGCTCGAGCTTAACTGCCCCG-3′ and 5′-CCAAGGCGACGTCGGCGTCGAAGTAGT-3′ for the 3′ fragment. The two PCR fragments were combined by a second round of PCR using T3 primer and 5′-CCAAGGCGACGTCGGCGTCGAAGTAGT-3′, then the resulting fragment was fused to GST. GST:EpsinR2C-13[DPF/AAA] was generated similarly to GST:EpsinR2C-13[LADVF/AAAAA] using the T3 primer and 5′-AGGAGAGTCACCAGCTGCCGCATCAAATGACTG-3′ for the 5′ fragment; 5′-ATTATCATCAGCTCGAGCTTAACTGCCCG-3′ and 5′-CAGTCATTTGATGCCGCAGCTGGTGACTCTCCT-3′ for the 3′ fragment; and T3 primer and 5′-CCAAGGCGACGTCGGCGTCGAAGTAGT-3′ for the second round of PCR.
To fuse EpsinR2, EpsinR2N, EpsinR2C, EpsinR2C-13, and EpsinR2C-13[LADVF/AAAAA] with HA, fragments were isolated from the respective GST fusion constructs by digestion with BamHI and XhoI, then inserted into the appropriate vector to generate an N-terminal HA epitope tag.
Isolation of Arabidopsis γ-adaptin (At1g23900) was described previously (Song et al., 2006). The Arabidopsis homologs of α-adaptin and δ-adaptin were isolated by PCR using the following primers: 5′-CGGGGTACCATGTCGTCGTCTTCCACTTCTATAATG-3′ and 5′-CGGGGTACCTCCCAAGAGAAAATCTGGAATTATAAC-3′ for δ-adaptin (At1g48760); 5′-CGGGGTACCATGTCGTCGTCTTCCACTTCTATA-3′ and 5′-AGCGTAATCTGGAACATCGTATGGGTACAAGAGAAAATCTGGAATTATACCTTG-3′ for δ-adaptin:HA; and 5′-CGGGGTACCATGACCGGAATGAGAGGTCTCTCCGTA-3′ and 5′-CGGGGTACCTTTAAGTAAGCCAGCGAGCATAGCTCC-3′ for α-adaptin (At5g22770). Amplified products were fused to the N or C terminus of GFP or ligated into the appropriate HA-tagging vector. To generate MBP:δ-adaptinN and MBP:δ-adaptinC, the C- and N-terminal regions of δ-adaptin were generated by PCR using the following primers: 5′-GAATTCATGTCGTCGTCTTCCACTTCTATA-3′ and 5′-GAATTCTCACTCAACGTATTCACCTGAAAC-3′ for δ-adaptinN, and 5′-GGATCCATGTTCTCTAAGAATCCGTATGAA-3′ and 5′-GGATCCTCACAAGAGAAAATCTGGAATTAT-3′ for δ-adaptinC. Amplified products were ligated into pMAL-C2X.
To generate MBP:AtVTI12, the AtVTI12 cDNA was amplified using the primers 5′-GGATCCTTAATGAGAAAGCTTGTATGAGATGATCAA-3′ and 5′-GGATCCATGAGCGACGTATTTGAAGGGTACGAGCGT-3′, and the resultant fragment was ligated into pMAL-C2X. The myc epitope was fused to the C terminus of AtVTI12 using PCR and the primers 5′-ATGAGCGACGTATTTGAAGGGTACGAGCGT-3′ and 5′-TTACAGATCCTCTTCTGAGATGAGTTTTTGTTCATGAGAAAGCTTGTATGAGATGACAA-3. All PCR products were confirmed by sequencing.
To generate MBP:α-adaptinN, MBP:α-adaptinC, MBP:γ-adaptinN, and MBP:γ-adaptinC, the N- and C-terminal regions of α-adaptin and γ-adaptin were PCR amplified using following primers: 5′-GGATCCATGACCGGAATGAGAGGTCTCTCCGTA-3′ and 5′-GGATCCTTAAGTCTCATGTATTGCGATTTTGTCCAA-3′ for α-adaptinN (1–484 amino acids); 5′-GAATTCATGGTGATCTTACATGAGAAGCTTCCT-3′ and 5′-GAATTCTTAAAGTAAGCCAGCGAGCATAGCTCCTGG-3′ for α-adaptinC (485–1,012 amino acids); 5′-GAATTCATGAATCCCTTTTCTTCTGGTACT-3′ and 5′-GAATTCTCACTGATCAATGTACCATAGTTTTTC-3′ for γ-adaptinN (1–425 amino acids); and 5′-GAATTCATGCTCAAGGTTCTGTGTGAGGCT-3′ and 5′-GAATTCTCACAACCCGCGAGGGAAGTTGCT-3′ for γ-adaptinC (426–876 amino acids).
Transient Expression and in Vivo Targeting of Reporter Cargo Proteins
Plasmids were introduced by polyethylene glycol-mediated transformation (Jin et al., 2001; Lee et al., 2002) into Arabidopsis protoplasts prepared from leaf tissues. Expression of fusion proteins was monitored at the indicated time points after transformation, and images were captured with a cooled CCD camera and a Zeiss Axioplan fluorescence microscope (Jin et al., 2001).
Protein Preparation and Gel-Blot Analysis
To prepare cell extracts, transformed protoplasts were lysed by repeated freeze-thaw cycles and centrifuged at 7,000g for 5 min at 4°C in a microfuge to remove cell debris (Jin et al., 2001). Proteins present in the medium (protoplast culture supernatants) were prepared by TCA precipitation. Western-blot analysis was performed as described previously (Jin et al., 2001). Immunoblots were detected with LAS3000 (FUJIFILM).
Immunohistochemistry
Transformed protoplasts were placed onto poly-l-Lys-coated glass slides, then incubated in 3% paraformaldehyde in fixing buffer (10 mm HEPES, pH 7.2, 154 mm NaCl, 125 mm CaCl2, 2.5 mm maltose, 5 mm KCl) for 1 h at room temperature (Frigerio et al., 1998; Park et al., 2004). Fixed cells were incubated with anti-HA, anti-myc, anti-Ara6, or anti-clathrin antibodies at 4°C overnight and washed with buffer (10 mm Tris-HCl, pH 7.2, 154 mm NaCl, 0.25% gelatin, 0.1% Triton X-100, and 0.01% SDS) three times. Subsequently, cells were incubated with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (Sigma) or trimethylrhodamine isothiocyanate-conjugated anti-rat IgG (Zymed) secondary antibodies. Images were captured as described above.
Expression of Recombinant Proteins in Escherichia coli
Expression constructs were introduced into the E. coli strain BL21(DE3), and expression of recombinant proteins was induced with 1 mm isopropyl-d-thiogalactopyranoside for 4 h at 28°C or for 1 h at 37°C. Cells were harvested by centrifugation at 5,000g for 5 min at 4°C and resuspended in ice-cold buffer (20 mm Tris-HCl, pH 7.4, 200 mm NaCl, 1 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride) containing protease inhibitors (1 μg/mL aprotinin, 1 μg/mL antipain). Protoplasts were lysed by sonication at 4°C. Cell debris was removed by centrifugation at 18,000g for 15 min at 4°C. For purification of recombinant proteins, the cleared supernatant was incubated with 1/100th volume of preequilibrated amylose resin (New England Biolabs) or glutathione agarose 4B (Peptron) on an orbital shaker for 2 h at 4°C. Beads were collected by centrifugation at 1,000g for 1 min and washed three times with ice-cold suspension buffer. Fusion proteins were eluted by adding 10 mm maltose, 50 mm Tris-HCl, pH 8.0, or 5 mm reduced glutathione, and 50 mm Tris-HCl, pH 8.0.
In Vitro Lipid-Binding Assay
PIP arrays were obtained from Echelon. Lipid binding was performed according to the manufacturer's protocol, using 2 μg/mL purified protein in 100 mm Tris-HCl, pH 7.6, 0.1% (v/v) Tween 20, and 154 mm NaCl at 4°C overnight. Proteins bound to lipids were detected using the anti-GST antibody diluted 1:1,000 dilution in Tris-buffered saline plus Tween 20.
In Vitro Protein Pull-Down Assay
Total protein extracts obtained from protoplasts were incubated with MBP fusion proteins or MBP alone, and GST fusion proteins or GST alone, immobilized on amylose-Sepharose beads or glutathione agarose beads, respectively, in binding buffer (150 mm NaCl, 10 mm Tris-HCl, pH 7.4, 1 mm EDTA, 1 mm EGTA, 0.5% to 0.1% Triton X-100, protease inhibitor cocktail [Roche Diagnotics]) on ice for 2 h. Sepharose or agarose beads and the associated proteins were washed with ice-cold binding buffer three times. Proteins in the precipitates were analyzed by western blotting using anti-GST antibody (Calbiochem).
Acknowledgments
The authors thank Dr. Akihiko Nakano (RIKEN, Japan) for the anti-Ara6 antibody.
This work was supported by the Ministry of Science and Technology (Korea) National Creative Research Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Inhwan Hwang (ihhwang@postech.ac.kr).
Open Access articles can be viewed online without a subscription.
References
- Ahmed SU, Rojo E, Kovaleva V, Venkataraman S, Dombrowski JE, Matsuoka K, Raikhel NV (2000) The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J Cell Biol 149 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, Sanderfoot AA, Kovaleva V, Zheng H, Raikhel NV (2000) AtVPS45 complex formation at the trans-Golgi network. Mol Biol Cell 11 2251–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C (1998) Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J 15 441–447 [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72 395–447 [DOI] [PubMed] [Google Scholar]
- Brett TJ, Traub LM, Fremont DH (2002) Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure 10 797–809 [DOI] [PubMed] [Google Scholar]
- Chen H, De Camilli P (2005) The association of epsin with ubiquitinated cargo along the endocytic pathway is negatively regulated by its interaction with clathrin. Proc Natl Acad Sci USA 102 2766–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Fre S, Slepnev VI, Capua MR, Takei K, Butler MH, Di Fiore PP, De Camilli P (1998) Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature 394 793–797 [DOI] [PubMed] [Google Scholar]
- Chidambaram S, Mullers N, Wiederhold K, Haucke V, von Mollard GF (2004) Specific interaction between SNAREs and epsin N-terminal homology (ENTH) domains of epsin-related proteins in trans-Golgi network to endosome transport. J Biol Chem 279 4175–4179 [DOI] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD (1997) The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell 91 109–118 [DOI] [PubMed] [Google Scholar]
- da Silva Conceição A, Marty-Mazars D, Bassham DC, Sanderfoot AA, Marty F, Raikhel NV (1997) The syntaxin homolog AtPEP12p resides on a late post-Golgi compartment in plants. Plant Cell 9 571–582 [PMC free article] [PubMed] [Google Scholar]
- De Camilli P, Chen H, Hyman J, Panepucci E, Bateman A, Brunger AT (2002) The ENTH domain. FEBS Lett 513 11–18 [DOI] [PubMed] [Google Scholar]
- Drake MT, Downs MA, Traub LM (2000) Epsin binds to clathrin by associating directly with the clathrin-terminal domain. Evidence for cooperative binding through two discrete sites. J Biol Chem 275 6479–6489 [DOI] [PubMed] [Google Scholar]
- Duncan MC, Costaguta G, Payne GS (2003) Yeast epsin-related proteins required for Golgi-endosome traffic define a gamma-adaptin ear-binding motif. Nat Cell Biol 5 77–81 [DOI] [PubMed] [Google Scholar]
- Eugster A, Pecheur EI, Michel F, Winsor B, Letourneur F, Friant S (2004) Ent5p is required with Ent3p and Vps27p for ubiquitin-dependent protein sorting into the multivesicular body. Mol Biol Cell 15 3031–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT (2002) Curvature of clathrin-coated pits driven by epsin. Nature 419 361–366 [DOI] [PubMed] [Google Scholar]
- Friant S, Pecheur EI, Eugster A, Michel F, Lefkir Y, Nourrisson D, Letourneur F (2003) Ent3p is a PtdIns(3,5)P2 effector required for protein sorting to the multivesicular body. Dev Cell 5 499–511 [DOI] [PubMed] [Google Scholar]
- Frigerio L, de Virgilio M, Prada A, Faoro F, Vitale A (1998) Sorting of phaseolin to the vacuole is saturable and requires a short C-terminal peptide. Plant Cell 10 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P, Gupta R, Shah S, Morris AJ, Rudge SA, Scarlata S, Petrova V, McLaughlin S, Rebecchi MJ (1995) The pleckstrin homology domain of phospholipase C-delta 1 binds with high affinity to phosphatidylinositol 4,5-bisphosphate in bilayer membranes. Biochemistry 34 16228–16234 [DOI] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H (2000) Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J 19 4577–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Miller SE, Taylor MJ, von Mollard GF, Robinson MS (2004) EpsinR is an adaptor for the SNARE protein Vti1b. Mol Biol Cell 15 5593–5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Motley A, Harasaki K, Peak Chew SY, Robinson MS (2003) EpsinR: an ENTH domain-containing protein that interacts with AP-1. Mol Biol Cell 14 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Robinson MS (1998) Clathrin and adaptors. Biochim Biophys Acta 1404 173–193 [DOI] [PubMed] [Google Scholar]
- Holstein SE (2002) Clathrin and plant endocytosis. Traffic 3 614–620 [DOI] [PubMed] [Google Scholar]
- Holstein SE, Oliviusson P (2005) Sequence analysis of Arabidopsis thaliana E/ANTH-domain-containing proteins: membrane tethers of the clathrin-dependent vesicle budding machinery. Protoplasma 226 13–21 [DOI] [PubMed] [Google Scholar]
- Honing S, Sandoval IV, von Figura K (1998) A di-leucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. EMBO J 17 1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Koshiba S, Kigawa T, Kikuchi A, Yokoyama S, Takenawa T (2001) Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science 291 1047–1051 [DOI] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong G-W, Hwang I (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalthoff C, Alves J, Urbanke C, Knorr R, Ungewickell EJ (2000) Unusual structural organization of the endocytic proteins AP180 and epsin 1. J Biol Chem 277 8209–8216 [DOI] [PubMed] [Google Scholar]
- Kalthoff C, Groos S, Kohl R, Mahrhold S, Ungewickell EJ (2002) Clint: a novel clathrin-binding ENTH-domain protein at the Golgi. Mol Biol Cell 13 4060–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Morita MI, Fukaki H, Yoshiro Y, Uehara M, Nihama M, Tasaka M (2002) SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell 14 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Eu YJ, Yoo CM, Kim YW, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I (2001. a) Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YW, Park DS, Park SC, Kim SH, Cheong GW, Hwang I (2001. b) Arabidopsis dynamin-like 2 that binds specifically to phosphatidylinositol 4-phosphate assembles into a high-molecular weight complex in vivo and in vitro. Plant Physiol 127 1243–1255 [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T (1999) Adaptors for clathrin-mediated traffic. Annu Rev Cell Dev Biol 15 705–732 [DOI] [PubMed] [Google Scholar]
- Lafer EM (2002) Clathrin-protein interactions. Traffic 3 513–520 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Sohn EJ, Lee MH, Hwang I (2004) The Arabidopsis rab5 homologs rha1 and ara7 localize to the prevacuolar compartment. Plant Cell Physiol 45 1211–1220 [DOI] [PubMed] [Google Scholar]
- Lee MH, Min MK, Lee YJ, Jin JB, Shin DH, Kim DH, Lee KH, Hwang I (2002) ADP-ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and maintenance of endoplasmic reticulum morphology in Arabidopsis. Plant Physiol 129 1507–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS (2004) ENTH/ANTH proteins and clathrin-mediated membrane budding. J Cell Sci 117 9–18 [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Bassham DC, Raikhel NV, Nakamura K (1995) Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J Cell Biol 130 1307–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills IG, Praefcke GJ, Vallis Y, Peter BJ, Olesen LE, Gallop JL, Butler PJ, Evans PR, McMahon HT (2003) EpsinR: an AP-1/clathrin interacting protein involved in vesicle trafficking. J Cell Biol 160 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JR, Prasad K, Hao W, Augustine GJ, Lafer EM (2000) A conserved clathrin assembly motif essential for synaptic vesicle endocytosis. J Neurosci 20 8667–8676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overstreet E, Chen X, Wendland B, Fischer JA (2003) Either part of a Drosophila epsin protein, divided after the ENTH domain, functions in endocytosis of delta in the developing eye. Curr Biol 13 854–860 [DOI] [PubMed] [Google Scholar]
- Park M, Kim SJ, Vitale A, Hwang I (2004) Identification of the protein storage vacuole and protein targeting to the vacuole in leaf cells of three plant species. Plant Physiol 134 625–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden AA, Oorschot V, Hesser BA, Austin CD, Scheller RH, Klumperman J (2004) Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J Cell Biol 164 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS, Bonifacino JS (2001) Adaptor-related proteins. Curr Opin Cell Biol 13 444–453 [DOI] [PubMed] [Google Scholar]
- Rosenthal JA, Chen H, Slepnev VI, Pellegrini L, Salcini AE, Di Fiore PP, De Camilli P (1999) The epsins define a family of proteins that interact with components of the clathrin coat and contain a new protein module. J Biol Chem 274 33959–33965 [DOI] [PubMed] [Google Scholar]
- Saint-Pol A, Yelamos B, Amessou M, Mills IG, Dugast M, Tenza D, Schu P, Antony C, McMahon HT, Lamaze C, et al (2004) Clathrin adaptor epsinR is required for retrograde sorting on early endosomal membranes. Dev Cell 6 525–538 [DOI] [PubMed] [Google Scholar]
- Sanderfoot A, Raikhel N (2003) The secretory system of Arabidopsis. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0098, http://www.aspb.org/publications/arabidopsis/
- Simonsen A, Wurmser AE, Emr SD, Stenmark H (2001) The role of phosphoinositides in membrane transport. Curr Opin Cell Biol 13 485–492 [DOI] [PubMed] [Google Scholar]
- Sohn EJ, Kim ES, Zhao M, Kim SJ, Kim H, Kim Y-W, Lee YJ, Hillmer S, Sohn U, Jiang L, et al (2003) Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Lee MH, Lee G-J, Yoo CM, Hwang I (2006) Epsin1 plays an important role in the vacuolar trafficking of soluble cargo proteins in plant cells via its interactions with clathrin, AP1, AtVTI11, and AtVSR1. Plant Cell 18 2258–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JM, Perera IY, Boss WF (1998) A phosphatidylinositol 4-kinase pleckstrin homology domain that binds phosphatidylinositol 4-monophosphate. J Biol Chem 273 22761–22767 [DOI] [PubMed] [Google Scholar]
- Surpin M, Zheng H, Morita MT, Saito C, Avila E, Blakeslee JJ, Bandyopadhyay A, Kovaleva V, Carter D, Murphy A, et al (2003) The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15 2885–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse YC, Mo B, Hillmer S, Zhao M, Lo SW, Robinson DG, Jiang L (2004) Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16 672–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Yamaguchi M, Uchimiya H, Nakano A (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J 20 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasiak S, Legendre-Guillemin V, Puertollano R, Blondeau F, Girard M, de Heuvel E, Boismenu D, Bell AW, Bonifacino JS, McPherson PS (2002) Enthoprotin: a novel clathrin-associated protein identified through subcellular proteomics. J Cell Biol 158 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B (2002) Epsins: adaptors in endocytosis? Nat Rev Mol Cell Biol 3 971–977 [DOI] [PubMed] [Google Scholar]
- Wurmser AE, Emr SD (1998) Phosphoinositide signaling and turnover: PtdIns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J 17 4930–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, von Mollard GF, Kovaleva V, Stevens TH, Raikhel NV (1999) The plant vesicle-associated SNARE AtVTI1a likely mediates vesicle transport from the trans-Golgi network to the prevacuolar compartment. Mol Biol Cell 10 2251–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]