Abstract
Alternative oxidase (AOX) is encoded in small multigene families in plants. Functional analysis of the Arabidopsis (Arabidopsis thaliana) alternative oxidase 1c (AtAOX1c) promoter, an AOX gene not induced by oxidative stress, indicated that regulation of expression was complex, with the upstream promoter region containing positive and negative response regions. Comparison to the promoter region of soybean (Glycine max) alternative oxidase 2b (GmAOX2b), another AOX gene not induced by oxidative stress, revealed that they contained seven sequence elements in common. All elements were active in the promoter region of AtAOX1c in suspension cells and in leaf tissue from Columbia and mutant plants, where a mitochondrial protein import receptor was inactivated. Analysis of coexpressed and putatively coregulated genes, the latter defined as containing five or more sequence elements functional in AtAOX1c, indicated that AtAOX1c was coregulated with components involved with cell division and growth. Consistent with this analysis, we demonstrated that site II elements, previously shown to regulate the proliferating cell nuclear antigen, are present in the upstream promoter region of AtAOX1c and were strong negative regulators of AtAOX1c expression. It was demonstrated that NDB4, a gene encoding an external NAD(P)H dehydrogenase, displayed strong coexpression with AtAOX1c. Overall, these results indicate that AtAOX1c is regulated by growth and developmental signals.
Alternative oxidase (AOX) is a cyanide-insensitive terminal oxidase present on the inner mitochondrial membrane that oxidizes ubiquinone to reduce oxygen to water (Umbach et al., 2006). It is intensively studied in plants due to its ability to dissociate electron transport from ATP synthesis (Finnegan et al., 2004). In higher plants, it is encoded by a small gene family, with five genes in Arabidopsis (Arabidopsis thaliana; Saisho et al., 1997; Thirkettle-Watts et al., 2003), four in sugarcane (Saccharum officinarum), and tomato (Lycopersicon esculentum; Holtzapffel et al., 2003; Navet et al., 2003; Borecky et al., 2006), and three in maize (Zea mays), rice (Oryza sativa), and soybean (Glycine max; Whelan et al., 1996; Ito et al., 1997; Karpova et al., 2002). Genes encoding AOX can be divided into two groups, AOX1 and AOX2, with AOX1 genes showing higher protein sequence similarity between species than AOX2 genes within a species (Considine et al., 2002). An examination of the promoters combined with gene expression analysis of all soybean and Arabidopsis AOX genes indicated that the expression pattern observed between species was not conserved with gene orthology (Thirkettle-Watts et al., 2003). For example, in soybean, AOX2a and AOX2b (GmAOX2a and GmAOX2b) are the predominantly expressed genes in a variety of organs at different growth stages, whereas, in Arabidopsis, AOX1a and AOX1c (AtAOX1a and AtAOX1c) display the highest expression levels (Thirkettle-Watts et al., 2003). Furthermore, even when closely related species are compared, such as soybean and Vigna, there appears to be differences in how the single AOX1 gene in these species is regulated (Costa et al., 2007).
Although AOX is widely expressed in plants, the majority of studies have focused on the stress-inducible nature of AOX transcript abundance. This is due, in part, to the fact that it is highly responsive to a variety of treatments that induce oxidative stress (Finnegan et al., 2004; Clifton et al., 2006; Rhoads et al., 2006). Induction of AOX by various treatments has become a model for the study of retrograde regulation from the mitochondrion to the nucleus in plants. Overall, it can be concluded from numerous studies in various plants that it is often only a single AOX gene that is induced under stress (Rhoads and Vanlerberghe, 2004; Rhoads et al., 2006). Utilizing the stress-inducible nature of AOX, the promoter region of AtAOX1a has been dissected to define stress-responsive regions (Dojcinovic et al., 2005). Using a variety of different treatments and inhibitors to protein kinases or phosphatases, induction of AOX1 in soybean and tobacco (Nicotiana tabacum) has been shown to occur via more than one signal transduction pathway (Vanlerberghe and McIntosh, 1996, 1997; Djajanegara et al., 2002). Generally, it has been concluded that AOX is responsive to oxidative stress and plays a central role in preventing the accumulation of reactive oxygen species (ROS; Finnegan et al., 1997; Vanlerberghe and McIntosh, 1997; Rhoads et al., 2006). However, analysis of AOX gene expression in a variety of plants indicates that it is also expressed in a wide variety of organs and growth stages, suggesting roles not associated with oxidative stress (Vanlerberghe and McIntosh, 1997; Thirkettle-Watts et al., 2003; Finnegan et al., 2004; Clifton et al., 2006).
Furthermore, it is usually only a single member of the AOX gene family that is induced by oxidative stress (Vanlerberghe and McIntosh, 1997; Considine et al., 2002; Finnegan et al., 2004; Clifton et al., 2005, 2006; Rhoads et al., 2006). In soybean, GmAOX1 is induced by oxidative stress, whereas GmAOX2a and GmAOX2b display organ and developmental regulation (Finnegan et al., 1997; McCabe et al., 1998; Djajanegara et al., 2002). In Arabidopsis, AtAOX1a is the predominantly expressed AOX and it is also the most highly induced in terms of magnitude of response and number of treatments that result in its induction (Clifton et al., 2005; Elhafez et al., 2006). In contrast, whereas AtAOX1c is also widely expressed in Arabidopsis (Thirkettle-Watts et al., 2003; Clifton et al., 2006; Elhafez et al., 2006), it is largely unchanged by a variety of treatments that induce oxidative stress (Thirkettle-Watts et al., 2003; Clifton et al., 2005). Thus, analysis of the regulation of AtAOX1c may provide insight into the regulation of AOX expression under normal conditions and also a general model for nuclear-encoded mitochondrial genes.
RESULTS
The AtAOX1c Promoter Contains Positive and Negative Response Regions and Elements
To characterize the promoter region of AtAOX1c, we tested for functional regions in the promoter using two approaches. The first was to carry out a deletion analysis of the promoter as used to define functional regions in other AOX promoters (Thirkettle-Watts et al., 2003; Dojcinovic et al., 2005) and for nuclear genes encoding other mitochondrial components (Zabaleta et al., 1998; Welchen et al., 2004; Ribeiro et al., 2005; Welchen and Gonzalez, 2005). The second approach was to predict elements and test their function by deletion. However, several hundred elements can be predicted in a 1-kb promoter sequence using a variety of commonly used algorithms (Tompa et al., 2005). To overcome this problem, we compared the AtAOX1c promoter to the promoter region of GmAOX2b. This promoter was chosen because GmAOX2b is expressed in soybean in a variety of tissues and is not induced by oxidative stress (Thirkettle-Watts et al., 2003). A comparison between the promoter regions of AtAOX1c and GmAOX2b revealed seven sequence elements in common, all within 500 bp of the transcriptional start site of AtAOX1c (Fig. 1; Supplemental Fig. S1). To test the predicted elements, we utilized Arabidopsis suspension cell culture and leaf tissue because previous expression analysis indicated that AtAOX1c was expressed in both of these tissues (Thirkettle-Watts et al., 2003).
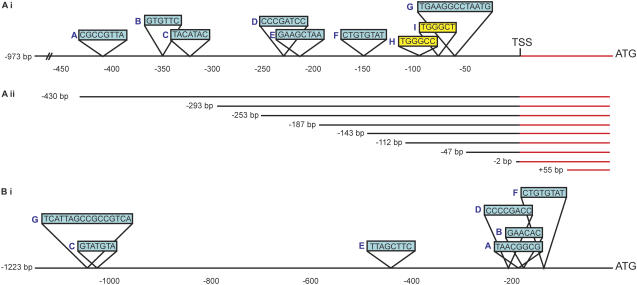
Figure 1.
Schematic representation of the AtAOX1c and GmAOX2b promoters. A (i), Region of the AtAOX1c promoter examined 973 bp upstream of the transcriptional start site (TSS), with the position of predicted sequence elements shown. Sequence elements in common with the GmAOX2b promoter regions are highlighted in blue and designated A to G. Site II elements are highlighted in yellow and designated H and I. The 5′-untranslated region (UTR) is highlighted in red. A (ii), Diagrammatic representation of the deletion constructs of the AtAOX1c promoter. The 5′-UTR is highlighted in red. All numbering is with respect to the TSS. B, Schematic representation of the putative GmAOX2b promoter region. The 1,223 bp upstream of the TSS of GmAOX2b showing common sequence elements with AtAOX1c are shown highlighted in blue. Letter designations (A–G) follow Arabidopsis counterparts in A. Diagrammatic representations are not to scale. [See online article for color version of this figure.]
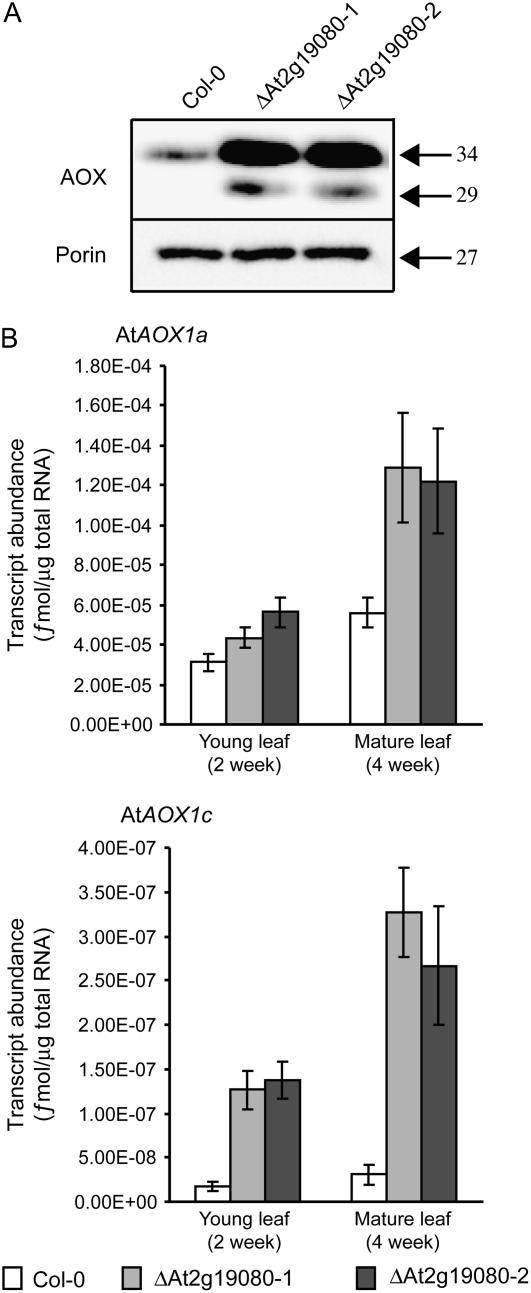
Additionally, we had previously identified a T-DNA insertional knockout of At2g19080 (ΔAt2g19080), which encodes a mitochondrial protein that plays a role in the import of proteins into mitochondria (R. Lister and J. Whelan, unpublished data). In these plants, transcript and protein levels of AtAOX1c are increased in abundance. Western-blot analysis of isolated mitochondria probed with antibodies to AOX from Arabidopsis Columbia-0 (Col-0) plants result in a single protein with an apparent molecular mass of 34 kD (Fig. 2A). Because this is in agreement with the apparent molecular mass previously reported for AtAOX1a (Fiorani et al., 2005; Umbach et al., 2005) and given that AtAOX1a transcript levels are at least 10-fold higher than AtAOX1c (Fig. 2B), the protein detected is proposed to be AtAOX1a. However, in ΔAt2g19080 lines, an additional cross-reacting protein was present with an apparent molecular mass of 29 kD (Fig. 2A). This is likely to represent the product of AtAOX1c because quantitative reverse transcription (QRT)-PCR analysis of ΔAt2g19080 lines indicates that transcript abundance of AtAOX1c was increased 5- to 10-fold compared to levels in Col-0 plants (Fig. 2B). However, it cannot be ruled out that the 29-kD protein detected may be a breakdown product of the 34-kD protein. Transcript abundance for the other three AOX genes in Arabidopsis was unchanged and expressed at low levels, as previously documented (Thirkettle-Watts et al., 2003; Clifton et al., 2006). Because ΔAt2g19080 plants showed enhanced expression of AtAOX1c, we also tested the ability of the AtAOX1c promoter to drive β-glucuronidase (GUS) activity in leaves from these insertional knockout lines.
Figure 2.
Expression of AtAOX1a and AtAOX1c in Arabidopsis leaf tissue. A, Western-blot analysis of isolated mitochondria from 4-week-old leaf tissue from Col-0 plants and plants with an insertional knockout of At2g19080 (ΔAt2g19080). Mitochondrial proteins were probed with monoclonal antibodies raised against AOX and porin. For AOX, an additional cross-reacting protein in mitochondrial samples from ΔAt2g19080 is present compared to wild type. Apparent molecular mass is indicated in kilodaltons. B, QRT-PCR analysis of transcript abundance for AtAOX1a and AtAOX1c in Col-0 and ΔAt2g19080 plants in young leaf (2 week) and mature leaf (4 week) tissue.
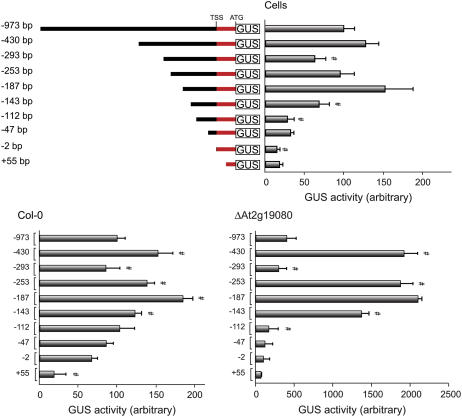
The ability of the −973-bp promoter and various deletions to drive GUS activity was tested in suspension cells and leaf tissue from Col-0 and ΔAt2g19080 4-week-old plants (Fig. 3). Overall, the analysis revealed several active regions that differed in the magnitude of their response between the three systems, and, notably, the activity of the regions was greatly enhanced in ΔAt2g19080. For wild-type cells and Col-0 leaf tissue, GUS activity driven by the −973-bp promoter was set to 100% and GUS activity driven by the other promoter regions was expressed relative to this. GUS values were similar between cells and leaves from Col-0 plants in agreement with a previous report that AtAOX1c is expressed at relatively similar levels in both (Thirkettle-Watts et al., 2003; Clifton et al., 2005). Because ΔAt2g19080 displayed large increases in GUS activity, these values were expressed relative to the activity of the −973-bp promoter from Col-0 plants. This allowed the differences in magnitude of GUS activity as well as fold changes with different constructs to be assessed.
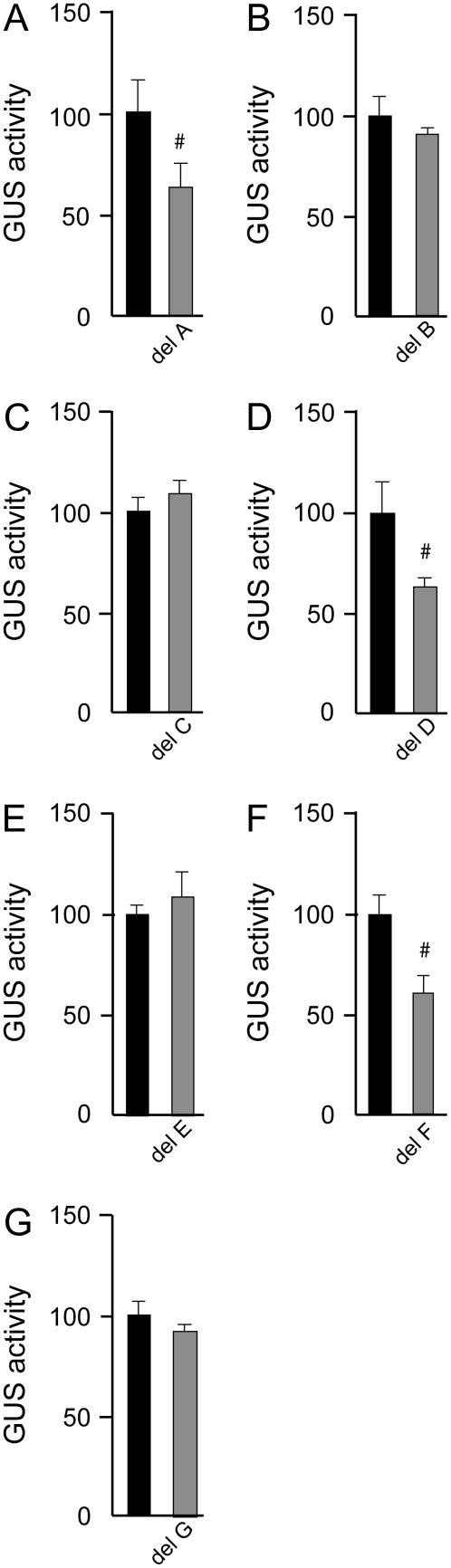
Figure 3.
Functional analysis of the AtAOX1c promoter region. Regions of the AtAOX1c promoter were used to drive the expression of GUS and activity was measured after biolistic transformation into suspension cells, and leaves from 4-week-old Col-0 and ΔAt2g19080 plants. For suspension cells and leaves from Col-0 plants, the GUS activity obtained with the −973-bp promoter construct was set to 100% (y axis) and the GUS activities supported by various deletions of this promoter were expressed in a relative manner. For ΔAt2g19080 plants, the GUS activity was 4 to 5 times greater and thus the values for these samples were normalized to the values obtained for the −973-bp construct in Col-0 leaf samples to allow the magnitude of the changes in expression to be visualized. Comparing each deletion to the preceding construct assessed the significance of changes in GUS activity. #, Significance with a P value < 0.01 using Student's t test. GUS activities for the ΔAt2g19080 leaf samples are compiled from independent analysis of both lines. A diagrammatic representation of the deletion constructs is shown in the first image (not to scale). [See online article for color version of this figure.]
Deletion analysis of the region −973 bp upstream of the transcriptional start site of the AtAOX1c promoter revealed several regulatory regions, acting in a positive or negative manner. Overall, the activity of each promoter construct was similar in each test system, although the magnitude of changes in the ΔAt2g19080 leaves was 3- to 4-fold higher than observed in Col-0 leaves and suspension cells. Deletion of bases −973 to −430 bp resulted in slightly increased GUS activity in cells and Col-0 leaf (50%) and a 400% increase in ΔAt2g19080 leaves (Fig. 3). Deletion to −293 bp identified a positive response region, and, again, the magnitude of the change in activity was greater in the ΔAt2g19080 leaf tissue compared to Col-0 and cells. Deletion of the −293-bp fragment to −253 and −187 bp revealed more negative response regions in all three test systems, except that the −253- to −187-bp response was absent in ΔAt2g19080, likely due to the larger response on deletion from −293 to −253 bp. Further deletions revealed positive response regions between −187 and −143 bp, as well as between −143 and −112 bp. Slight differences were observed between the three test systems. For instance, the pattern of GUS activity from −187 bp followed the same declining trend in all three; however, the 10-fold drop in GUS activity between −143 and −112 bp, in ΔAt2g19080, resulted in the subsequent deletions only producing relatively small decreases that were not found to be statistically significant, whereas in Col-0 leaves this decline in GUS activity was more evenly spread across the deletions.
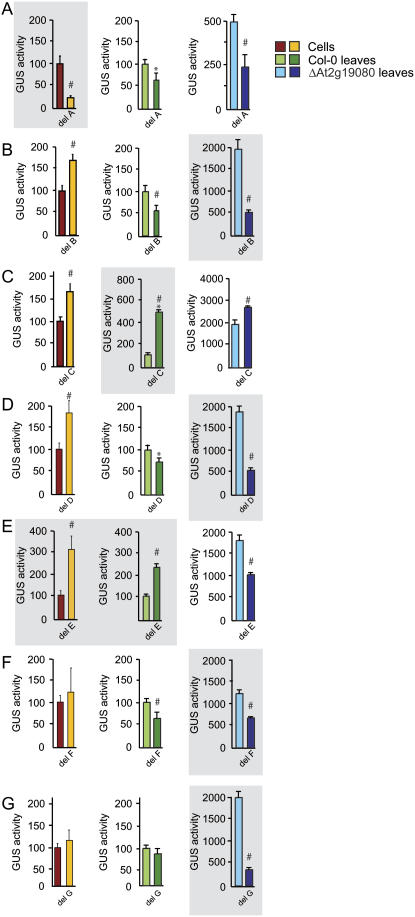
Analysis of the seven predicted elements in the three test systems revealed that elements A and C had the same effect in all of them, as positive and negative regulators, respectively (Fig. 4). Elements B, D, and F had the same effect in leaf tissue from Col-0 and ΔAt2g19080 in that they were all positive regulators, whereas in cells they were found to be negative (B and D) or to have no significant effect (F; Fig. 4). Element E displayed the same effect in cells and Col-0 leaf as a negative regulator, but was a positive regulator in ΔAt2g19080.
Figure 4.
Functional analysis of the predicted motifs in AtAOX1c. The function of the sequence elements A to G identified in the AtAOX1c promoter were tested by deleting these elements and testing their ability to drive GUS expression compared to the unmutated promoter region. Elements were deleted in the promoter regions of −973, −430, −253, and −187 bp as appropriate to the position of the element. The activity was tested in suspension cells as well as Col-0 and ΔAt2g19080 leaf tissue. For cells and Col-0 leaf tissue, normalized GUS activity driven by the unmutated promoter was set to 100% and all other activities were expressed in a relative manner. For ΔAt2g19080 leaf tissue, values were expressed against the activity of the unmutated promoter fragment in Col-0 tissue. Shading indicates the tissue in which each element had the greatest effect, as determined by the fold changes in activity resulting from the deletion. *, Significance with a P value < 0.05 using Student's t test. #, Significance with a P value < 0.01 using Student's t test. GUS activities for the ΔAt2g19080 leaf samples are compiled from independent analysis of both lines. The elements outlined in Figure 1 are designated on the left of each image.
Analysis of the functionality of each element revealed tissue-specific effects. Element A played a large role in GUS activity in cells, with a 75% decrease in activity upon its deletion, but the decrease was 50% or less in leaf tissue of Col-0 and ΔAt2g19080. Element B was shown to repress promoter activity in cells, but had a positive role in Col-0 and ΔAt2g19080 leaf tissue, with an almost 4-fold loss in activity in ΔAt2g19080 leaves, indicating that it played a large role in the up-regulation of AtAOX1c in the mutant leaf. Deletion of element C had the highest impact in Col-0 leaf, whereas deletion of element D displayed maximal effect in ΔAt2g19080, similar to element B, and deletion of element E had a greater impact in cells and Col-0 leaves. Element G was only active in ΔAt2g19080 as a positive regulator (Fig. 4).
Occurrence of Elements Functional in AtAOX1c in Other AOX Genes
Elements identified by comparison of the AtAOX1c promoter and the GmAOX2b promoter were tested for function in the latter promoter using a soybean suspension cell culture (Fig. 5; (Thirkettle-Watts et al., 2003). Elements A, D, and F were found to be functional in the soybean promoter, all as positive regulators. The remaining soybean elements were found to have no effect in soybean cells. A likely reason for this is that the majority of elements tested in Arabidopsis had the greatest effect in leaf tissue and not in a cell culture system. Analysis of the upstream promoter regions of AOX genes in other plants indicated that many of the promoters contain these elements (Table I). Element B occurred in several other AOX promoter regions, whereas element G was only found in the Arabidopsis and soybean promoters.
Figure 5.
Functional analysis of the predicted motifs in GmAOX2b. The function of the sequence elements identified in GmAOX2b by comparison to AtAOX1c were tested by deleting these elements and testing the ability of the promoter to drive GUS expression compared to the unmutated soybean promoter region. Elements were deleted in the promoter regions as appropriate to the position of the element. The activity was tested in soybean suspension cells. Normalized GUS activity driven by the unmutated promoter was set to 100% and all other activities were expressed in a relative manner. #, Significance with a P value < 0.01 using Student's t test. The elements outlined in Figure 1 are designated on the left of each image.
Table I.
List of sequence elements defined as functional in the Arabidopsis AtAOX1c promoter, their occurrence in the soybean AOX2b promoter, other AOX genes, and the degenerate sequence motif defined by coexpression analysis
Sequence elements are designated motifs A to G. Numbers with each sequence refer to the position at which the motifs are present within each upstream region. Indicated in bold in the soybean sequences are bases that differ from those in AtAOX1c. Numbers with each sequence refer to the position at which the motifs are present within each upstream region. For the degenerate motifs, bases in bold indicate bases that were not included in this study. Os, Oryza sativa; Gm, Glycine max; Zm, Zea mays; At, Arabidopsis; Sg, Sauromatum guttatum; Lc, Lotus corniculatus; Car, Catharanthus roseus; Cr, Chlamydomonas reinhardtii.
| Motif | Sequence | Sequence in Soybean AOX2b | Other AOX | Degenerate Sequence |
|---|---|---|---|---|
| A | CGCCGTTA −418 to −425 bp | Reverse complement −186 to −193 bp | ZmAOX1a; OsAOX1b; AtAOX1b | ACGCCG |
| B | GTGTTC −362 to −367 bp | Reverse complement −178 to −184 bp | LcAOX2a; OsAOX1a; ZmAOX1a; CarAOX1; GmAOX1, GmAOX2a; AtAOX1a | TGTTCT |
| C | TACATAC −336 to −342 bp | Reverse complement −1,026 to −1,033 bp | CarAOX1 | ACATAC |
| D | CCCGATCC −244 to −251 bp | CCCCGACC −208 to −215 bp | OsAOX1a, OsAOX1b; LcAOX2a; SgAOX | CCCGA(T/C)C |
| E | GAAGCTAA −223 to −230 bp | Reverse complement −441 to −449 bp | CarAOX1 | GAAGCT |
| F | CTGTGTAT −146 to −153 bp | Identical −138 to −146 bp | ZmAOX1a; OsAOX1a; CarAOX1 | CACTGT |
| G | TGAAGGCCTAATGA −66 to −78 bp | TCATTAGCCGCCGTCA −1,046 to −1,062 bp | – | ATTGAANNN(NN)CTAATG |
Regulatory Context of AtAOX1c
To identify the regulatory context of AtAOX1c, we searched the Arabidopsis genome for the occurrence of the elements defined as functional in AtAOX1c. A relatively small number of genes had some of the elements occurring individually in their promoter region (Supplemental Fig. S2). This was largely due to the fact that the elements were generally longer than 6 bp. For instance, the G element of 13 bp occurred in only three other promoter regions and not in combination with any other elements. For the other elements, these were found in the promoter regions of 133 genes (element D) and up to 7,283 genes (element B). The occurrence of multiple elements was limited because no other promoter region was found to contain five or more elements and only one gene contained four elements (Supplemental Fig. S2, green shading). This gene (At2g17140) encodes a pentatricopeptide repeat-containing (PPR) protein. The family of genes encoding PPR proteins is greatly expanded in plants compared to animals and is largely located in mitochondria or plastids (Rivals et al., 2006). Analysis of coexpression of this gene with AtAOX1c revealed positive correlation, although it was not a direct relationship because the slope of the line was not 1 (Supplemental Fig. S3). This reveals that the shared elements are coregulating these genes, whereas other elements, present in AtAOX1c, but not in the promoter region of At2g17140, result in different magnitude of transcript changes in a variety of circumstances, accounting for the fact that perfect correlation is not observed. It is likely that, within the functional motifs identified, or overlapping with these sequences, are core elements that may occur more widely at the whole-genome level. Thus, an alternative approach was taken to gain further insight into the AtAOX1c regulatory context.
Genes coexpressed with AtAOX1c were identified using Botany Array Resource (BAR) Expression Angler (Toufighi et al., 2005) and Arabidopsis Coexpression Data Mining Tools (ACT; Jen et al., 2006; Supplemental Table S1, A and B). A functional classification of the genes identified using ACT, compared to the whole genome, reveals that hydrolase activity, as well as nucleotide and nucleic acid binding, are significantly overrepresented, whereas transferase activity is underrepresented (Fig. 6). Similar analysis with Expression Angler indicates that genes categorized into DNA- and RNA-binding/nucleic acid-binding functions are significantly overrepresented (Fig. 6), with an overlap of 16 genes between the two analyses. The coexpressed gene lists include homeobox genes, genes involved in chromosome maintenance, a gene related to sexual development in yeast (Saccharomyces cerevisiae), a gene encoding an Argonaute-like protein, a family of genes that play a role in regulating gene expression via RNAi, which are associated with meristem formation and organ identity (Kidner and Martienssen, 2005; Ronemus et al., 2006), and a number of genes encoding F-box proteins. F-box proteins, first identified in cell cycle mutants of yeast (Willems et al., 2004), are involved in auxin-mediated growth (Parry and Estelle, 2006) and have also been implicated in playing a role in mitochondrial segregation to daughter cells during cell division in yeast (Kondo-Okamoto et al., 2006). Almost all the other genes listed bind RNA or DNA or are involved in transcription or translation. Genes encoding proteases may be involved in growth and division in ubiquitin-mediated protein degradation where F-box proteins also play an essential role (Nemhauser and Chory, 2005).
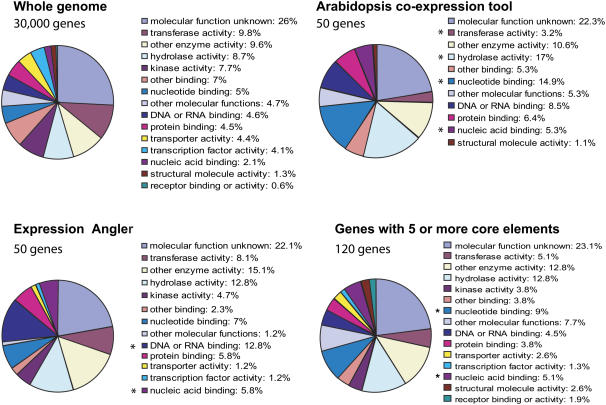
Figure 6.
Functional categorization of genes coexpressed and putatively coregulated with AtAOX1c. The 50 top hits for genes coexpressed with AtAOX1c as determined using ACT and BAR Expression Angler were classified into functional groups. Additionally, the 120 genes that share five or more core or degenerate motifs that were functional in AtAOX1c were classified into functional groups. The number of genes in each functional classification was expressed as a percentage and compared to the functional classification of the whole Arabidopsis genome. *, Functional groupings that show significant differences at a 95% confidence interval in comparison to the whole genome.
Using the two coexpression lists of 50 genes, we also attempted to define core or degenerate sequence elements, as outlined in “Materials and Methods.” Briefly, the two coexpression gene lists generated from BAR Expression Angler and ACT were used to predict significantly represented 6-mers compared to the whole Arabidopsis genome, using the Motif Analysis tool on The Arabidopsis Information Resource (TAIR) Web site. This sequence element prediction program, like many others, predicts hundreds of elements in a 1-kb region (Tompa et al., 2005). Thus, an additional filter was imposed on these predictions. Only predicted 6-mers that overlapped with the functional elements defined for AtAOX1c (elements A–G) were used to define putative core sequences (Table I; Supplemental Fig. S1). Elements A to G showed considerable overlap with 6-mers defined as significantly represented in the coexpression gene lists compared to the whole Arabidopsis genome. Most overlapping 6-mers occur in a large proportion of the genes in the coexpression lists (Supplemental Fig. S1, red and green bars). In addition, the two coexpression lists generated from BAR Expression Angler and ACT were combined and narrowed down to a list of genes encoding mitochondrial proteins, as described in “Materials and Methods.” This gene list was again used to predict significantly represented 6-mers overlapping with elements A to G defined in the AtAOX1c promoter region as described above. Again, considerable overlap was observed for all elements (Supplemental Fig. S1, purple bars).
These putative core sequences (outlined in Table I) were used to define a putative coregulatory environment for AtAOX1c. A list of Arabidopsis genes that contained five or more of these core sequences in their upstream regions was generated and only 120 genes met this criterion. The functional groups of nucleotide binding and nucleic acid binding were again significantly overrepresented (Fig. 6). This putative coregulatory list contains genes encoding proteins involved in cell division (At3g09840); two PPR proteins (At2g39230 and At3g22670), whose expression displayed a strong positive correlation with AtAOX1c (Supplemental Fig. S3); several proteins predicted to be targeted to mitochondria, including a subunit of cytochrome c oxidase (COX) and ATP synthase; and proteins involved in translation. Two genes encoding subunits involved in organelle biogenesis, Tic 44 and Pex 14, together with phytochrome D, also suggest that genes encoding proteins involved in organelle biogenesis are part of this putative coregulatory environment containing AtAOX1c.
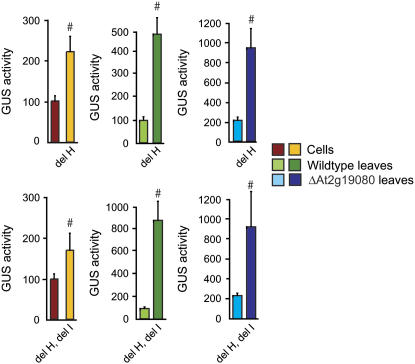
These lists of coexpressed and putatively coregulated genes suggest that AtAOX1c is regulated by growth and developmental signals. It has been reported that many nuclear-located genes encoding mitochondrial proteins contain a site II element, first identified in proliferating cell nuclear antigen (PCNA) genes (Welchen and Gonzalez, 2006). AtAOX1c is the only one of the five AOX genes in Arabidopsis that contains this element, although it appears in many genes encoding components of the cytochrome respiratory chain (Welchen and Gonzalez, 2006). To investigate the functionality of this element in the AtAOX1c promoter, we deleted both site II elements present and tested the ability of the promoter to drive the expression of GUS (Fig. 7). It was evident that deletion of only one of these site II elements at position −103 bp upstream of the transcriptional start site (element H) increased promoter activity 2-fold in cells and 4-fold in leaves from Col-0 and ΔAt2g19080 plants. The high GUS activity in ΔAt2g19080 leaf tissue was increased 4-fold, resulting in promoter activities 2- to 4-fold higher than in cells and leaf tissue from Col-0 plants. Deleting both site II elements (elements H and I) had no additional effect on promoter activity in cells, but resulted in an 8-fold increase in promoter activity in leaf tissue. Overall, it appears that AtAOX1c is coexpressed and putatively coregulated with genes playing roles in growth and division (Fig. 6; Supplemental Table S1), indicating a fundamental role for AtAOX1c in cell development and proliferation.
Figure 7.
Functional analysis of site II elements in the promoter region of AtAOX1c. Site II elements in the upstream region of AtAOX1c were deleted and the ability of these constructs to drive expression of GUS was tested in suspension cells, Col-0, and ΔAt2g19080 leaf tissue. For the suspension cells and leaf tissue from Col-0 plants, promoter activity was set to 100% for unmutated constructs, whereas GUS activity of the ΔAt2g19080 leaf tissue was normalized to leaf tissue from Col-0 plants to visualize the increase in GUS activity. #, Significance with a P value < 0.01 using Student's t test. The elements are designated as outlined in Figure 1.
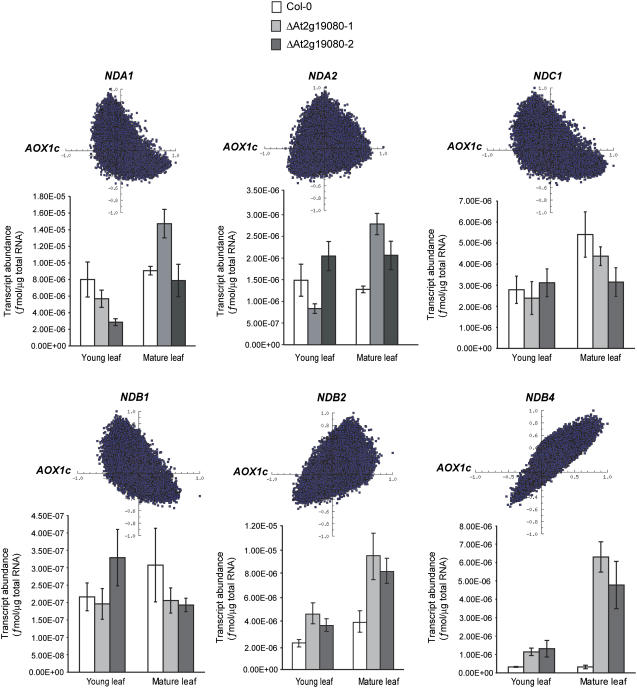
Because it has been previously reported that AtAOX1a is coexpressed with an external NAD(P)H dehydrogenase, NDB2, we investigated whether any alternative NAD(P)H dehydrogenases were coexpressed with AtAOX1c. NDB4 (At2g20800) was found in the top 20 of the 50 most correlated genes with AtAOX1c in the Expression Angler analysis. Furthermore, analysis of expression correlation plots generated using the co-correlation scatter plot function available from the ACT database (http://www.arabidopsis.leeds.ac.uk/act/coexpanalyser.php) for all seven NAD(P)H dehydrogenases revealed that NDB4 displayed the strongest correlation of coexpression with AtAOX1c (Fig. 8). To confirm this correlation, we measured transcript abundance for all seven alternative NAD(P)H dehydrogenases in Arabidopsis Col-0 and the ΔAt2g19080 lines. The expression level of NDB3 was below detection levels. It was clear that in the ΔAt2g19080 lines transcript abundance of NDB4 was up-regulated between 5- and 10-fold compared to wild-type levels, the greatest induction observed for any alternative NAD(P)H dehydrogenase gene (Fig. 8) and consistent with the increase observed in AtAOX1c (Fig. 2). Furthermore, we measured the respiratory activity of mitochondria isolated from Arabidopsis Col-0 and ΔAt2g19080 plants (Table II). Overall, it was apparent that the capacity of AOX had increased greater than 5-fold in ΔAt2g19080 mitochondria compared to Col-0. However, external NAD(P)H dehydrogenase activity was found not to be significantly different.
Figure 8.
Analysis of the coexpression of genes encoding alternative NAD(P)H dehydrogenases with AtAOX1c. Coexpression of each alternative NAD(P)H dehydrogenase with AtAOX1c was determined using the co-correlation scatter plot function on the ACT database. An output with a slope of 1 indicates perfect coexpression, whereas a slope of −1 indicates negative correlation. QRT-PCR analysis of the expression of all seven alternative NAD(P)H dehydrogenases in Col-0 and ΔAt2g19080 leaf tissue in which AtAOX1c was shown to be up-regulated was also carried out. The expression level of NDB3 was below reliable detection. [See online article for color version of this figure.]
Table II.
Oxygen consumption by mitochondria isolated from Arabidopsis plants
Mitochondria were isolated from the aerial tissue of 4-week-old wild-type Col-0 and ΔAt2g19080 plants and oxygen consumption was measured using an oxygen electrode. Maximal respiratory rates using NADH or NADPH as substrates (in the presence of rotenone) were determined, as well as AOX and COX capacity. Respiration rates are presented as nmol of O2 consumed min−1 mg−1 of mitochondrial protein (mean ± se; n = 3).
| Wild-Type Col-0 | ΔAt2g19080 | |
|---|---|---|
| NADH | 85 ± 3 | 78 ± 8 |
| NADPH | 41 ± 4 | 35 ± 1 |
| AOX | 13 ± 1 | 75 ± 5 |
| COX | 203 ± 15 | 142 ± 17 |
DISCUSSION
Presently, only a few promoters of genes encoding mitochondrial proteins have been analyzed, three complex I promoters (Zabaleta et al., 1998), ThI1 (Ribeiro et al., 2005), cytochrome c (Welchen and Gonzalez, 2005), COX 5b (Welchen et al., 2004), and AtAOX1a (Dojcinovic et al., 2005). Furthermore, only four sequence motifs have been shown to be functional, two of which were identified in AtAOX1a using transient transformation approaches (Lohrmann et al., 2001; Welchen et al., 2004; Dojcinovic et al., 2005). In the case of an AOX gene like AtAOX1a, which displays tissue and developmental regulation and is stress inducible, experimental approaches cannot distinguish both sets of elements because stress induction will swamp any characterization of other control elements. Therefore, analysis of AtAOX1c, which is expressed in a variety of tissues and developmental stages, but is not induced by oxidative stress, will provide additional insight into the regulation of AOX expression. Furthermore, AtAOX1c was observed to be up-regulated approximately 10-fold in a mutant with an inactivated mitochondrial preprotein receptor, the latter inducing severe alterations in phenotype (R. Lister and J. Whelan, unpublished data). A previous analysis of cytoplasmic male sterility (cms) mutants in maize also revealed an up-regulation of different AOX isoforms, depending on the mutation that caused the cms phenotype (Karpova et al., 2002). Notably, in these plants, it was concluded that there was no increase in oxidative stress in the cms plants compared to wild type. In maize, different cms mutants induce different AOX isoforms; the NCS2 cms line exhibits induced ZmAOX2 and the NCS6 cms line exhibits induced ZmAOX3 (Karpova et al., 2002). (The terminology of AOX genes from maize follows that of Karpova et al. [2002]. As a monocot, all AOX genes in maize are an AOX1 type, but in keeping in line with the terminology of gene nomenclature for maize, the genes are labeled by successive numbers as in Karpova et al. [2002].) This indicates that different signaling pathways lead to the expression of different AOX isoforms. Additionally, for both the ΔAt2g19080 lines used in this study and the maize cms lines, the effects of the genetic lesions are developmental abnormalities (Karpova et al., 2002; R. Lister and J. Whelan, unpublished data). This links the expression of specific AOX genes to growth and development in plants rather than induction by oxidative stress.
As observed previously for AtAOX1a and NDB2, induction of AtAOX1c was accompanied by the induction of NDB4, a gene encoding an external NAD(P)H dehydrogenase (Michalecka et al., 2003; Elhafez et al., 2006). In both cases, it is not just a single terminal oxidase that is induced, but a functional alternative respiratory chain capable of oxidizing external NAD(P)H. Using the ΔAt2g19080 mutant lines, induction of AtAOX1c was shown to be accompanied by an increase in AOX capacity. Along with the increase in AtAOX1a transcript and protein, the increase in AOX capacity is a reflection of the total increase in AOX protein. Although the magnitude of induction of AtAOX1c transcript was greater than the magnitude of AtAOX1a increase (10-fold versus 2-fold), the absolute transcript abundance of AtAOX1a was still 1,000-fold greater than AtAOX1c. In contrast to the increase in AOX capacity, there was no significant increase observed in the oxidation of external NAD(P)H. The capacity of the external NAD(P)H dehydrogenase is already quite high in mitochondria from Col-0 plants and the up-regulation of NDB4 may produce a NAD(P)H dehydrogenase with altered biochemical properties (Rasmusson et al., 2004).
The demonstration that site II elements are functional in the AtAOX1c promoter is consistent with the proposal that it is regulated by cell growth and developmental signals. Site II elements were first characterized in the PCNA gene, the product of which is a cofactor of DNA polymerase involved in DNA replication and repair. Site II elements have been demonstrated to interact in vitro with proteins belonging to the Teosinte branched 1 cycloidea proliferating cell factor (TCP) family of transcription factors (Kosugi et al., 1991, 1995; Kosugi and Ohashi, 2002; Tremousaygue et al., 2003; Li et al., 2005). Within this family, class I TCP factors promote cell growth and division, whereas class II negatively regulate cell division. It was apparent that both site II elements examined in this study acted as strong negative regulators of AtAOX1c expression. This is consistent with the up-regulation of AtAOX1c in the ΔAt2g19080 lines where growth and development have been altered.
Furthermore, site II elements are proposed to function in a synergistic manner with other elements, such as the telo box domain, and multiple site II elements may bind different TCP proteins to form heterodimers and numerous combinations are possible given that 24 TCP proteins are predicted to be present in the Arabidopsis genome (Kosugi and Ohashi, 2002). It was observed that elements B, D, and E acted as either a positive or negative element, depending on the tissue. This may be explained by a sequence element binding to different transcription factors, likely to be in the same family and using the same core-binding element. Transcription factors display tissue, developmental, and stress-inducible expression patterns (Riechmann, 2002). Thus, positive or negative regulation associated with any element is dependent on the specific transcription factor present in that cell type. In addition to different classes of TCP proteins exerting positive or negative regulation, it appears that many defense-related transcription factor families have members that can activate or repress transcription (Eulgem, 2005). Thus, it is easy to visualize how a similar motif can act in a positive or negative regulatory manner, depending on the specific transcription factors that bind.
Overall, analysis of the AtAOX1c promoter revealed that regulation is complex, with positive and negative regulatory regions. This study identified nine active elements that varied in their regulatory properties, being either positive or negative and causing changes in GUS activity of different magnitudes when deleted, depending on the tissues where activity was tested. The fact that the GUS activity of the AtAOX1c promoter was 10-fold higher in leaves from ΔAt2g19080 indicates that expression of AtAOX1c is highly regulated and can be activated by various signals. All elements, except element C, functioned as positive regulators in the ΔAt2g19080 lines, whereas in suspension cells or leaves from Col-0 plants, some elements functioned as negative regulators, such as element E. The results from deleting large regions compared to that of deleting specific elements were in general agreement in that both analyses revealed positive and negative regulatory regions. For instance, deleting bases −430 to −293 bp revealed a positive regulatory region, and elements A and B in this region were positive regulators in leaf tissue. It is likely that there are several other functional elements because deletion of −973 to −430 bp revealed a negative regulatory region, yet no functional elements were defined in this region.
What is the role of an alternative respiratory pathway in cells under nonstressed conditions, especially in the context of cell growth and division? Notably, this does not just refer to AtAOX1c, but rather to AOX, in general, because a variety of studies in various plant species have indicated it is widely expressed during normal growth and development (Vanlerberghe and McIntosh, 1997; Michalecka et al., 2003; Finnegan et al., 2004; Rasmusson et al., 2004; Clifton et al., 2006; Escobar et al., 2006). One possibility is that AOX and associated NAD(P)H dehydrogenases are expressed at a basal level that leads to equilibrium for ROS production. ROS are constantly produced in a cell and are inactivated by a variety of antioxidant mechanisms (Sweetlove and Foyer, 2004). Basal-level expression of AOX may be especially important in proliferating cells where DNA is being replicated because ROS have the potential to be mutagenic and the prevention of ROS accumulation is therefore necessary to protect against an increase in mutations. The presence of AOX acts as a preoxidant defense, preventing the production of ROS and thereby protecting mitochondrial DNA from mutation. It is interesting to note that plant mitochondrial DNA has extremely low rates of mutation, albeit with rare exceptions, in comparison to their animal counterparts (Wolfe et al., 1987; Palmer and Herbon, 1988; Cho et al., 2004) and the presence of AOX in plants may play a role in maintaining these low rates of mutation. In fungal systems, expression of AOX has been directly associated with a decrease in the production of ROS, an increase in mitochondrial genome stability, restoration of fertility, and an increased lifespan (Lorin et al., 2001, 2006). Furthermore, growth and division places demands on the cell that may require the function of AOX. Growth is energy demanding in terms of ATP required for DNA replication, transcription, translation, and also in the requirements of carbon skeletons for various biosynthetic purposes, such as amino acid and nucleotide synthesis. These carbon skeletons can be supplied from the anaplerotic reactions of the TCA cycle, but this may compromise energy production. An alternative respiratory pathway, including an external NAD(P)H dehydrogenase, has the capacity to oxidize external (or cytosolic) NAD(P)H in contrast to complex I of the cytochrome chain, which oxidizes intramitochondrial NAD(P)H. In this context, it has been previously proposed that the role of AOX is to maintain growth (Parsons et al., 1999; Sieger et al., 2005). It achieves this by maintaining a supply of ATP either from cytosolic or mitochondrial generated reducing equivalents acting as a mechanism to ensure energy homeostasis (Hansen et al., 2002; Moore et al., 2002). Induction of AOX under stress conditions that inhibit the production of ATP via oxidative phosphorylation fits into this overall model, but is just one aspect of a specialized role for AOX in balancing energy and biosynthetic metabolism.
MATERIALS AND METHODS
Cloning of Arabidopsis and Soybean Promoter Regions
Cloning of the soybean (Glycine max) GmAOX2b promoter has been previously described (Thirkettle-Watts et al., 2003). The Arabidopsis (Arabidopsis thaliana) AtAOX1c promoter region was cloned using specific primers designed to introduce SalI and NcoI on either end of the amplified fragment (AtAOX1c F, GTAGCTTTCCCGGGCTGAAC; AtAOX1c R, GTAGTGACCATGGTTCGGAT). Amplification was performed using the Expand high-fidelity PCR system (Roche Diagnostics) and visualized using agarose gel electrophoresis. Fragments of interest were subsequently purified using a PCR purification kit (Qiagen) and ligated into the pCR2.1 vector (Invitrogen), according to the manufacturer's instructions. The pCR2.1 vector was then digested using the appropriate restriction enzymes, and promoter regions were then subcloned into the pGEM-GUS vector via ligation.
Deletion constructs and motif deletions were produced using the Quikchange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. For AtAOX1c, restriction sites were introduced to the 5′-end of the promoter region to produce deletion constructs of sizes −973, −430, −293, −253, −187, −143, −112, −47, −2, and +55 bp (Fig. 1). Specific elements, labeled A to I, were mutated in the appropriate promoter construct with the numbers indicating the bases deleted: element A, −418 to −425 bp; element B, −362 to −367 bp; element C, −336 to −342 bp (deleted from −430 promoter fragment); element D, −244 to −251 bp; element E, −223 to −230 bp (deleted from −253 promoter fragment); element F, −146 to −153 bp (deleted from −187 promoter fragment); element G, −66 to −78 bp; element H, −98 to −104 bp; and element I, −84 to −88 bp (deleted from −112 promoter fragment). All numbers refer to the position relative to the transcriptional start site, with the 5′-region referred to as negative and the transcriptional start site as +1. The constructs were made as translational fusions to the GUS reporter gene, with the ATG of AtAOX1c used as the start codon. Thus, for all constructs, the 5′-untranslated region (107 bp) is included, except for the +55-bp construct.
For the GmAOX2b promoter, a deletion series of the promoter revealed positive and negative regulatory regions as reported previously (Thirkettle- Watts et al., 2003). Sequence element deletions were produced in the appropriate deletion construct as follows: element A, −186 to −193 bp (deleted from −402 promoter fragment); element B, −178 to −184 bp (deleted from −220 promoter fragment); element C, −1,026 to −1,033 bp (deleted from −1,223 promoter fragment); element D, −208 to −215 bp (deleted from −402 promoter fragment); element E, −441 to −449 bp (deleted from −655 promoter fragment); element F, −138 to −146 bp (deleted from −220 promoter fragment); and element G, −1,046 to −1,062 bp (deleted from −1,223 promoter fragment). Because the transcriptional start site for GmAOX2b is not known, all numbers refer to the translational start site, with the A of ATG denoted +1.
Suspension Cell Culture, Plant Growth, and Treatments
Suspension cells from Arabidopsis (ecotype Landsberg erecta) leaf tissue were grown in 250-mL flasks with medium rotating at 130 rpm at 22°C for 16 h at 100 μE m−2 s−1 light conditions and 8 h of dark (Gamborg, 1968; Sweetlove et al., 2002). Suspension cells from soybean leaf tissue were grown in 250-mL flasks with medium rotating at 130 rpm at 28°C, under approximately 20 μE m−2 s−1 light (Gamborg, 1968; Djajanegara et al., 2002). Cells were subcultured (1:6 [v/v] dilution) at 7-d intervals to maintain exponential cell growth. Arabidopsis plants, ecotype Col-0, were grown at 22°C under long-day conditions. Arabidopsis ΔAt2g19080 plants were germinated on Murashige and Skoog agar plates (Gamborg, 1968) and homozygotes were transplanted into soil. Plants were grown in the long-day conditions described above. For transient expression analysis, suspension cells at 7 d old (for Arabidopsis) and 6 d old (for soybean) were aseptically coated onto filter paper (55-mm diameter; Whatman no. 1) and placed onto solid medium containing the appropriate culture medium supplemented with 200 mm mannitol and incubated for 2 h at 22°C in the light (for Arabidopsis) or 28°C in the dark (for soybean). Arabidopsis leaves used for transient transformation were placed on solid medium containing Arabidopsis culture medium supplemented with 200 mm mannitol and placed under the same conditions as the cell culture.
Biolistic Transformation
For transformation of suspension cell culture, 5 μg of each GUS reporter construct and the luciferase calibration construct were precipitated onto 5 mg of 1-μm (average diameter) gold particles (Chempur) using 2.5 m CaCl2 (Sigma-Aldrich) and 100 mm spermidine (Sigma-Aldrich). For each bombardment, 0.5 mg of particles was used. Bombardment was carried out under vacuum using helium pressure of 1,400 kPa. After transformation, cells were incubated at 22°C for 16 h at approximately 100 μE m−2 s−1 light conditions and 8 h of dark for 24 h (Arabidopsis) or 48 h of dark (soybean) on the medium-impregnated filter paper discs. Transformation of leaf tissue was performed using the PDS-1000 system using the Hepta adaptor (Bio-Rad Laboratories), using 10 μg of each GUS reporter construct and the luciferase calibration construct precipitated onto 3 mg of gold microparticles with a diameter of 1 μm (INBIO). Transformation was performed using rupture discs with a 1,100 psi pressure rating (INBIO) as per the manufacturer's instructions (Bio-Rad Laboratories). After transformation, Arabidopsis leaf tissue was incubated under the same conditions as cell culture for 24 h.
Assays for Luciferase and GUS
Transformed cells and leaves were disrupted by grinding in a mortar and pestle under liquid nitrogen. Broken cells were extracted with the lysis buffer and protocol supplied with the luciferase assay system kit (Roche). Luciferase activity assays were carried out according to the manufacturer's instructions and activity was measured at 2-s intervals over 20 s, using the Polarstar Optima (BMG Labtechnologies). GUS activity was determined using the fluorimetric GUS assay (Jefferson et al., 1987). Fluorescence was measured using Polarstar Optima (excitation at 355 nm, emission at 460 nm) at 3-min intervals over 1 h. GUS activity was normalized by dividing the GUS fluorescence value by the luciferase activity for each sample, thus eliminating variation due to transformation efficiency. A minimum of three replicate bombardments was carried out per construct. GUS activities measured for each construct were expressed as a percentage of the control (largest promoter fragment or unmutated promoter), which was normalized to a value of 100%.
To determine statistical significance for the deletion series of AtAOX1c in cells, wild-type leaves, and ΔAt2g19080 leaves, two sample t tests assuming unequal variances were performed. Each deletion series transformed into different tissue types was treated independently for this analysis. Within each series, each deletion construct was compared to the construct immediately preceding it in size (e.g. the −430-bp construct was only compared to the −973-bp construct). Thus, these tests do not determine relative importance or dependence between various promoter sections, but determine changes in promoter activity between each deletion construct. For comparison of GUS activities of the motif deletions compared to that of the unmutated promoter, a two-sample t test assuming unequal variances was also performed. In all cases, a significant difference was defined as P ≤ 0.05.
Isolation of Arabidopsis Mitochondria
Mitochondria from 5-week-old Arabidopsis Col-0 and ΔAt2g19080 mutant plants used for western-blot analysis and respiratory activity measurements were prepared by cutting plant leaf tissue into smaller segments and disrupting cells in 200 mL of grinding medium (0.5 m Suc, 25 mm tetrasodiumpyrophosphate, 1% [w/v] polyvinylpyrrolidone-40, 2 mm EDTA, 10 mm KH2PO4, and 1% [w/v] bovine serum albumin), using a mortar and pestle. Subsequent material was then filtered through Miracloth and cheesecloth. Arabidopsis mitochondria were isolated according to standard protocols as described previously (Millar et al., 2001).
Immunodetection
Western-blot analysis of mitochondrial proteins isolated from Col-0 and ΔAt2g19080 mutant Arabidopsis plants was carried out as described previously (Howell et al., 2006). Mitochondria were probed with antibodies to the outer membrane marker porin and the inner membrane protein AOX (Dr. Tom Elthon, University of Nebraska). Immunoreaction was detected using the BM luminescence western-blotting kit (Roche) and quantitative light emission was recorded using a luminescent image analyzer (LAS 1000). Western blots were analyzed using Image Gauge Version 3.0 software.
Respiratory Measurements
For respiratory measurements on isolated mitochondria, 75 to 150 μg of mitochondrial protein were added to 1 mL of reaction medium (0.3 m mannitol, 10 mm TES, 5 mm KH2PO4 10 mm NaCl, 2 mm MgSO4, 0.1% [w/v] bovine serum albumin, pH 7.5) and oxygen consumption was measured at 25°C in a Clarke-type oxygen electrode. The following reagents and inhibitors were added as described below to the reaction medium to the final concentrations indicated to examine mitochondrial function: NADH (1.5 mm), NADPH (1.5 mm), ADP (0.25 mm), CaCl2 (1 mm), rotenone (5 μm), ATP (0.3 mm), succinate (5 mm), myxothiazol (5 μm), pyruvate (5 mm), dithiothreitol (2 mm), nPG (50 μm), ascorbate (10 mm), cytochrome c (50 μm), Triton X-100 (0.05% [w/v]), and cyanide (1 mm). NADH or NADPH-dependent respiration was measured in the presence of ADP to maximize respiratory rate, CaCl2 to activate the external NAD(P)H dehydrogenases, and rotenone to ensure the respiratory rates measured were due to engagement of a rotenone-insensitive NADH dehydrogenase. AOX capacity was measured in the presence of succinate, NADH, ATP, and ADP to maximize electron flux, myxothiazol to block cytochrome pathway operation, and pyruvate and dithiothreitol to fully activate AOX. nPG was added to ensure that the oxygen consumption measured was due to AOX activity. COX capacity was measured by the solubilization of mitochondrial membranes with Triton X-100 and the provision of electrons by a reduced cytochrome c regenerating system consisting of exogenous cytochrome c and ascorbate. Cyanide was added to ensure that the oxygen consumption measured was due to COX activity.
QRT-PCR Analysis of Gene Expression
Leaf tissue was harvested from Col-0 and mutant plants at 2 weeks of age (young leaf) and at 4 weeks of age (mature leaf) and snap frozen in liquid N2. Samples were collected in triplicate. Total RNA isolation and cDNA synthesis were carried out as described previously (Lister et al., 2004). Transcript levels were assayed using the iCycler and iQ Supermix and iQ SYBR Supermix (Bio-Rad Laboratories) as described previously (Thirkettle-Watts et al., 2003; Lister et al., 2004). QRT-PCR primers (5′-3′) used for the AtAOX1b and AtAOX1d were as follows: AtAOX1b F, CACAGCCATCTTTTGATTCCTA; AtAOX1b R, CATTCAGTTCCATCTTCTTTGG; AtAOX1d F, CATCTTCGGTACAATCCTCC; and AtAOX1d R, CACTTCCAAGCTGAACCGTC.
All other assays used primers described previously (Clifton et al., 2005; Elhafez et al., 2006). Each of three independent cDNA preparations was assayed twice for each transcript analyzed. Transcript abundance was presented as absolute transcript abundance as determined by comparison to an internal standard of known concentration and ses were calculated for every data point.
Construction of Gene Lists for Experimentally Tested Elements
In this study, for each experimentally identified functional sequence element in the AtAOX1c promoter, a gene list was generated containing all Arabidopsis genes with these sequences in their upstream regions using the Patmatch function on the TAIR Web site (AGI, 2000; http://www.arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl). For each element, the nucleotide sequence was searched against Locus Upstream Sequences—1,000 bp (DNA). Elements were searched in both DNA strands (forward direction and reverse complement), with no mismatch allowed and a maximum of one hit per sequence. Once gene lists were generated, they were compiled such that the overlap (i.e. genes containing more than one element) could be identified. These data were presented in a seven-way Venn diagram, kindly provided by Frank Ruskey and Mark Weston (Department of Computer Science, University of Victoria (Ruskey and Weston, 2006).
AtAOX1c Coexpression Analysis
Arabidopsis gene lists containing the most highly correlated genes with AtAOX1c in terms of expression profiles across publicly available microarray studies were generated independently using several programs. A list of the 50 most highly correlated genes with AtAOX1c was generated using BAR Expression Angler (Toufighi et al., 2005; http://bbc.botany.utoronto.ca/ntools/cgi-bin/ntools_expression_angler.cgi) searching the NASCarrays 392 dataset with an R-value cutoff of 0.7 under default settings. An independent list of the 50 most correlated genes with AtAOX1c was generated using the Clique Finder function on the ACT database (Jen et al., 2006; http://www.arabidopsis.leeds.ac.uk/act/coexpanalyser.php) under default settings with R values ranging from 0.82 to 0.93. The overlap between the lists consisted of 16 genes.
Functional Categorization Analysis
Functional categorization using GO annotations was performed on the Arabidopsis whole-genome set, along with the independent lists of the most highly correlated genes with AtAOX1c generated in Expression Angler and ACT as described above. Functional categorizations for each gene list were obtained from TAIR using the GO annotations functional categorization function (http://www.arabidopsis.org/tools/bulk/go/index.jsp). The percentage distribution of each category for the different gene lists was compared to that of the whole genome. A T-score was calculated for each comparison using a two-sample z-statistic where T-scores over 2.0 (corresponding to a 95% confidence interval) were considered significant. A two-sample z-statistic was used to determine significant differences in proportions of functional groups. Standard normal distribution was assumed. The comparisons were performed using an online tool (http://www.marketviewresearch.com/SC/tisamples.asp).
Expression Correlation Plots of Alternative Respiratory Components
Correlation plots were generated using the co-correlation scatter plot function on the ACT database under default settings (http://www.arabidopsis.leeds.ac.uk/act/coexpanalyser.php), which plots Pearson correlation coefficients for two probes using 322 ATH1 arrays (Exp_ID: 2_1–2_41). Correlation plots were generated for AtAOX1c (At3g27620) against all the genes encoding alternative NAD(P)H dehydrogenases (At1g07180, At2g29990, At4g28220, At4g05020, At4g21490, At2g20800, and At5g08740).
Defining Core Elements
For the purpose of defining a coregulatory environment of AtAOX1c, the relatively large sequence elements experimentally tested in this study were broken down further to identify a possible core sequence that may be more widely represented in coregulated genes. The top 10 coexpressed genes with AtAOX1c as defined by ACT and Expression Angler were used to generate lists of 6-mers significantly represented in these promoter regions (defined as 1,000 bp upstream of the transcriptional start site in this analysis) using the Motif Analysis function in the TAIR database (http://www.arabidopsis.org/tools/bulk/motiffinder/index.jsp). This function compares the frequency of 6-mer words in the query set to that of the whole genome (31,407 sequences) and calculates statistically significant represented motifs in the query set. The 6-mer words identified in the promoters of these two lists were then filtered to reveal 6-mers that overlapped with the functional motifs defined in this study, with a minimum overlap of 2 or more bases. Each 6-mer identified was categorized as to the number of promoter sequences from the coexpression lists in which it was present (from the original lists of 50 of coexpressed genes; i.e. 25% to 50% indicates that the motif is present 25% to 50% of the time in the promoters in this list; Supplemental Fig. S1). In addition, we combined the lists of the 50 most correlated genes generated via the two different programs and analyzed for genes encoding mitochondrial proteins (defined in the mitochondrial proteome or predicted to be mitochondrial (Heazlewood et al., 2005). This list of 13 genes was again used as the query set in the Motif Analysis function on TAIR database under the same conditions as described earlier, and the 6-mers identified were categorized as to their percentage presence in the promoter list. Because the elements predicted in this study were originally predicted via comparison with the promoter region of GmAOX2b, the corresponding elements in the GmAOX2b promoter were searched to see whether there were degenerate bases between GmAOX2b and AtAOX1c elements and therefore further define a motif core. These analyses were plotted against the elements identified experimentally and highly conserved regions were identified. These regions were then defined as possible core sequences (Table I) and were then utilized in further analyses. For each of the core elements defined, sequences were again searched against the upstream regions of all Arabidopsis genes using TAIR Patmatch function (http://www.arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl). The core sequences searched were as described in Table I, and searches were carried out as described for experimentally tested element sequences. Once gene lists were generated, they were compiled such that the overlap (i.e. genes containing more than one element) could be identified. In addition, a functional categorization using GO annotations was performed on the Arabidopsis whole-genome set and the list of genes containing five or more of the defined core elements, as described earlier, and T-scores were calculated for percentile distribution comparisons between the whole-genome categorization and the gene list generated in this study.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY303971 (GmAOX2a), S81466 (GmAOX1), U87907 (GmAOX2b), NM113135 (AtAOX1a), and NM113678 (AtAOX1c).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The putative promoter region of AtAOX1c.
Supplemental Figure S2. Representation of genes that contain at least one of the sequence elements identified as functional in AtAOX1c.
Supplemental Figure S3. Analysis of coexpressed genes encoding PTPs with AtAOX1c.
Supplemental Table S1. Genes whose expression correlated with AtAOX1c as defined by various coexpression analysis tools.
Supplementary Material
This work was supported by the Australian Research Council Centre of Excellence in Plant Energy Biology.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: James Whelan (seamus@cyllene.uwa.edu.au).
Some figures in this article are displayed in color online but in blank and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Borecky J, Nogueira FT, de Oliveira KA, Maia IG, Vercesi AE, Arruda P (2006) The plant energy-dissipating mitochondrial systems: depicting the genomic structure and the expression profiles of the gene families of uncoupling protein and alternative oxidase in monocots and dicots. J Exp Bot 57 849–864 [DOI] [PubMed] [Google Scholar]
- Cho Y, Mower JP, Qiu YL, Palmer JD (2004) Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc Natl Acad Sci USA 101 17741–17746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton R, Lister R, Parker KL, Sappl PG, Elhafez D, Millar AH, Day DA, Whelan J (2005) Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol Biol 58 193–212 [DOI] [PubMed] [Google Scholar]
- Clifton R, Millar AH, Whelan J (2006) Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim Biophys Acta 1757 730–741 [DOI] [PubMed] [Google Scholar]
- Considine MJ, Holtzapffel RC, Day DA, Whelan J, Millar AH (2002) Molecular distinction between alternative oxidase from monocots and dicots. Plant Physiol 129 949–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa JH, Jolivet Y, Hasenfratz-Sauder MP, Orellano EG, da Guia Silva Lima M, Dizengremel P, Fernandes de Melo D (2007) Alternative oxidase regulation in roots of Vigna unguiculata cultivars differing in drought/salt tolerance. J Plant Physiol doi/10.1016/j.jplph.2006.04.001 [DOI] [PubMed]
- Djajanegara I, Finnegan PM, Mathieu C, McCabe T, Whelan J, Day DA (2002) Regulation of alternative oxidase gene expression in soybean. Plant Mol Biol 50 735–742 [DOI] [PubMed] [Google Scholar]
- Dojcinovic D, Krosting J, Harris AJ, Wagner DJ, Rhoads DM (2005) Identification of a region of the Arabidopsis AtAOX1a promoter necessary for mitochondrial retrograde regulation of expression. Plant Mol Biol 58 159–175 [DOI] [PubMed] [Google Scholar]
- Elhafez D, Murcha MW, Clifton R, Soole KL, Day DA, Whelan J (2006) Characterisation of mitochondrial alternative NAD(P)H dehydrogenases in Arabidopsis: intraorganelle location and expression. Plant Cell Physiol 47 43–54 [DOI] [PubMed] [Google Scholar]
- Escobar MA, Geisler DA, Rasmusson AG (2006) Reorganization of the alternative pathways of the Arabidopsis respiratory chain by nitrogen supply: opposing effects of ammonium and nitrate. Plant J 45 775–788 [DOI] [PubMed] [Google Scholar]
- Eulgem T (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 10 71–78 [DOI] [PubMed] [Google Scholar]
- Finnegan PF, Soole KL, Umbach AL (2004) Alternative mitochondrial electron transport proteins in higher plants. In DA Day, H Millar, J Whelan, eds, Plant Mitochondria: From Genome to Function, Vol 17. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 163–230
- Finnegan PM, Whelan J, Millar AH, Zhang Q, Smith MK, Wiskich JT, Day DA (1997) Differential expression of the multigene family encoding the soybean mitochondrial alternative oxidase. Plant Physiol 114 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorani F, Umbach AL, Siedow JN (2005) The alternative oxidase of plant mitochondria is involved in the acclimation of shoot growth at low temperature. A study of Arabidopsis AOX1a transgenic plants. Plant Physiol 139 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamborg O (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50 151–158 [DOI] [PubMed] [Google Scholar]
- Hansen LD, Churchb JN, Mathesona S, McCarliea VW, Thygersonc T, Criddlea RS, Smith BN (2002) Kinetics of plant growth and metabolism. Thermochim Acta 388 415–425 [Google Scholar]
- Heazlewood JL, Tonti-Filippini J, Verboon RE, Millar AH (2005) Combining experimental and predicted datasets for determination of the subcellular localisation of proteins in Arabidopsis. Plant Physiol 139 598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzapffel RC, Castelli J, Finnegan PM, Millar AH, Whelan J, Day DA (2003) A tomato alternative oxidase protein with altered regulatory properties. Biochim Biophys Acta 1606 153–162 [DOI] [PubMed] [Google Scholar]
- Howell KA, Millar AH, Whelan J (2006) Ordered assembly of mitochondria during rice germination begins with pro-mitochondrial structures rich in components of the protein import apparatus. Plant Mol Biol 60 201–223 [DOI] [PubMed] [Google Scholar]
- Ito Y, Saisho D, Nakazono M, Tsutsumi N, Hirai A (1997) Transcript levels of tandem-arranged alternative oxidase genes in rice are increased by low temperature. Gene 203 121–129 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen CH, Manfield IW, Michalopoulos I, Pinney JW, Willats WG, Gilmartin PM, Westhead DR (2006) The Arabidopsis co-expression tool (ACT): a WWW-based tool and database for microarray-based gene expression analysis. Plant J 46 336–348 [DOI] [PubMed] [Google Scholar]
- Karpova OV, Kuzmin EV, Elthon TE, Newton KJ (2002) Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 14 3271–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA (2005) The role of ARGONAUTE1 (AGO1) in meristem formation and identity. Dev Biol 280 504–517 [DOI] [PubMed] [Google Scholar]
- Kondo-Okamoto N, Ohkuni K, Kitagawa K, McCaffery JM, Shaw JM, Okamoto K (2006) The novel F-box protein Mfb1p regulates mitochondrial connectivity and exhibits asymmetric localization in yeast. Mol Biol Cell 17 3756–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30 337–348 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Suzuka I, Ohashi Y (1995) Two of three promoter elements identified in a rice gene for proliferating cell nuclear antigen are essential for meristematic tissue-specific expression. Plant J 7 877–886 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Suzuka I, Ohashi Y, Murakami T, Arai Y (1991) Upstream sequences of rice proliferating cell nuclear antigen (PCNA) gene mediate expression of PCNA-GUS chimeric gene in meristems of transgenic tobacco plants. Nucleic Acids Res 19 1571–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Potuschak T, Colon-Carmona A, Gutierrez RA, Doerner P (2005) Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA 102 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Chew O, Lee MN, Heazlewood JL, Clifton R, Parker KL, Millar AH, Whelan J (2004) A transcriptomic and proteomic characterization of the Arabidopsis mitochondrial protein import apparatus and its response to mitochondrial dysfunction. Plant Physiol 134 777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrmann J, Sweere U, Zabaleta E, Baurle I, Keitel C, Kozma-Bognar L, Brennicke A, Schafer E, Kudla J, Harter K (2001) The response regulator ARR2: a pollen-specific transcription factor involved in the expression of nuclear genes for components of mitochondrial complex I in Arabidopsis. Mol Genet Genomics 265 2–13 [DOI] [PubMed] [Google Scholar]
- Lorin S, Dufour E, Boulay J, Begel O, Marsy S, Sainsard-Chanet A (2001) Overexpression of the alternative oxidase restores senescence and fertility in a long-lived respiration-deficient mutant of Podospora anserina. Mol Microbiol 42 1259–1267 [DOI] [PubMed] [Google Scholar]
- Lorin S, Dufour E, Sainsard-Chanet A (2006) Mitochondrial metabolism and aging in the filamentous fungus Podospora anserina. Biochim Biophys Acta 1757 601–610 [DOI] [PubMed] [Google Scholar]
- McCabe TC, Finnegan PM, Harvey Millar A, Day DA, Whelan J (1998) Differential expression of alternative oxidase genes in soybean cotyledons during postgerminative development. Plant Physiol 118 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalecka AM, Svensson AS, Johansson FI, Agius SC, Johanson U, Brennicke A, Binder S, Rasmusson AG (2003) Arabidopsis genes encoding mitochondrial type II NAD(P)H dehydrogenases have different evolutionary origin and show distinct responses to light. Plant Physiol 133 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Liddell A, Leaver CJ (2001) Isolation and sub-fractionation of mitochondria from plants. In E Schon, ed, Methods in Cell Biology on Mitochondria. Academic Press, New York, pp 53–74 [DOI] [PubMed]
- Moore AL, Albury MS, Crichton PG, Affourtit C (2002) Function of the alternative oxidase: Is it still a scavenger? Trends Plant Sci 7 478–481 [DOI] [PubMed] [Google Scholar]
- Navet R, Jarmuszkiewicz W, Almeida AM, Sluse-Goffart C, Sluse FE (2003) Energy conservation and dissipation in mitochondria isolated from developing tomato fruit of ethylene-defective mutants failing normal ripening: the effect of ethephon, a chemical precursor of ethylene. J Bioenerg Biomembr 35 157–168 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Chory J (2005) A new FronTIR in targeted protein degradation and plant development. Cell 121 970–972 [DOI] [PubMed] [Google Scholar]
- Palmer JD, Herbon LA (1988) Plant mitochondrial DNA evolves rapidly in structure, but slowly in sequence. J Mol Evol 28 87–97 [DOI] [PubMed] [Google Scholar]
- Parry G, Estelle M (2006) Auxin receptors: a new role for F-box proteins. Curr Opin Cell Biol 18 152–156 [DOI] [PubMed] [Google Scholar]
- Parsons HL, Yip JY, Vanlerberghe GC (1999) Increased respiratory restriction during phosphate-limited growth in transgenic tobacco cells lacking alternative oxidase. Plant Physiol 121 1309–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson AG, Soole KL, Elthon TE (2004) Alternative NAD(P)H dehydrogenases of plant mitochondria. Annu Rev Plant Biol 55 23–39 [DOI] [PubMed] [Google Scholar]
- Rhoads DM, Umbach AL, Subbaiah CC, Siedow JN (2006) Mitochondrial reactive oxygen species. Contribution to oxidative stress and interorganellar signaling. Plant Physiol 141 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads DM, Vanlerberghe GC (2004) Mitochondria-nucleus interactions: evidence for mitochondrial retrograde communication in plant cells. In DA Day, H Millar, J Whelan, eds, Plant Mitochondria: From Genome to Function, Vol 17. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 83–106
- Ribeiro DT, Farias LP, de Almeida JD, Kashiwabara PM, Ribeiro AF, Silva-Filho MC, Menck CF, Van Sluys MA (2005) Functional characterization of the thi1 promoter region from Arabidopsis thaliana. J Exp Bot 56 1797–1804 [DOI] [PubMed] [Google Scholar]
- Riechmann JL (2002) Transcriptional regulation: a genomic overview. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, pp 1–46 [DOI] [PMC free article] [PubMed]
- Rivals E, Bruyere C, Toffano-Nioche C, Lecharny A (2006) Formation of the Arabidopsis pentatricopeptide repeat family. Plant Physiol 141 825–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus M, Vaughn MW, Martienssen RA (2006) MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by Argonaute, Dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell 18 1559–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskey F, Weston M (2006) More fun with symmetric Venn diagrams. Theory Comput Syst 39 413–423 [Google Scholar]
- Saisho D, Nambara E, Naito S, Tsutsumi N, Hirai A, Nakazono M (1997) Characterization of the gene family for alternative oxidase from Arabidopsis thaliana. Plant Mol Biol 35 585–596 [DOI] [PubMed] [Google Scholar]
- Sieger SM, Kristensen BK, Robson CA, Amirsadeghi S, Eng EW, Abdel-Mesih A, Moller IM, Vanlerberghe GC (2005) The role of alternative oxidase in modulating carbon use efficiency and growth during macronutrient stress in tobacco cells. J Exp Bot 56 1499–1515 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Foyer CH (2004) Roles for reactive oxygen species and antioxidants in plant mitochondria. In DA Day, H Millar, J Whelan, eds, Plant Mitochondria: From Genome to Function, Vol 17. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 307–320
- Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J 32 891–904 [DOI] [PubMed] [Google Scholar]
- Thirkettle-Watts D, McCabe TC, Clifton R, Moore C, Finnegan PM, Day DA, Whelan J (2003) Analysis of the alternative oxidase promoters from soybean. Plant Physiol 133 1158–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, et al (2005) Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol 23 137–144 [DOI] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43 153–163 [DOI] [PubMed] [Google Scholar]
- Tremousaygue D, Garnier L, Bardet C, Dabos P, Herve C, Lescure B (2003) Internal telomeric repeats and ‘TCP domain’ protein-binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. Plant J 33 957–966 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Fiorani F, Siedow JN (2005) Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol 139 1806–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach AL, Ng VS, Siedow JN (2006) Regulation of plant alternative oxidase activity: a tale of two cysteines. Biochim Biophys Acta 1757 135–142 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L (1996) Signals regulating the expression of the nuclear gene encoding alternative oxidase of plant mitochondria. Plant Physiol 111 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L (1997) ALTERNATIVE OXIDASE: from gene to function. Annu Rev Plant Physiol Plant Mol Biol 48 703–734 [DOI] [PubMed] [Google Scholar]
- Welchen E, Chan RL, Gonzalez DH (2004) The promoter of the Arabidopsis nuclear gene COX5b-1, encoding subunit 5b of the mitochondrial cytochrome c oxidase, directs tissue-specific expression by a combination of positive and negative regulatory elements. J Exp Bot 55 1997–2004 [DOI] [PubMed] [Google Scholar]
- Welchen E, Gonzalez DH (2005) Differential expression of the Arabidopsis cytochrome c genes Cytc-1 and Cytc-2. Evidence for the involvement of TCP-domain protein-binding elements in anther- and meristem-specific expression of the Cytc-1 gene. Plant Physiol 139 88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E, Gonzalez DH (2006) Overrepresentation of elements recognized by TCP-domain transcription factors in the upstream regions of nuclear genes encoding components of the mitochondrial oxidative phosphorylation machinery. Plant Physiol 141 540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan J, Millar AH, Day DA (1996) The alternative oxidase is encoded in a multigene family in soybean. Planta 198 197–201 [DOI] [PubMed] [Google Scholar]
- Willems AR, Schwab M, Tyers M (2004) A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta 1695 133–170 [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84 9054–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabaleta E, Heiser V, Grohmann L, Brennicke A (1998) Promoters of nuclear-encoded respiratory chain complex I genes from Arabidopsis thaliana contain a region essential for anther/pollen-specific expression. Plant J 15 49–59 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.