Abstract
Eukaryotic translation initiation factor-4E (eIF4E) recognizes and binds the m7 guanosine nucleotide at the 5′ end of eukaryotic messenger RNAs; this protein-RNA interaction is an essential step in the initiation of protein synthesis. The structure of eIF4E from wheat (Triticum aestivum) was investigated using a combination of x-ray crystallography and nuclear magnetic resonance (NMR) methods. The overall fold of the crystallized protein was similar to eIF4E from other species, with eight β-strands, three α-helices, and three extended loops. Surprisingly, the wild-type protein did not crystallize with m7GTP in its binding site, despite the ligand being present in solution; conformational changes in the cap-binding loops created a large cavity at the usual cap-binding site. The eIF4E crystallized in a dimeric form with one of the cap-binding loops of one monomer inserted into the cavity of the other. The protein also contained an intramolecular disulfide bridge between two cysteines (Cys) that are conserved only in plants. A Cys-to-serine mutant of wheat eIF4E, which lacked the ability to form the disulfide, crystallized with m7GDP in its binding pocket, with a structure similar to that of the eIF4E-cap complex of other species. NMR spectroscopy was used to show that the Cys that form the disulfide in the crystal are reduced in solution but can be induced to form the disulfide under oxidizing conditions. The observation that the disulfide-forming Cys are conserved in plants raises the possibility that their oxidation state may have a role in regulating protein function. NMR provided evidence that in oxidized eIF4E, the loop that is open in the ligand-free crystal dimer is relatively flexible in solution. An NMR-based binding assay showed that the reduced wheat eIF4E, the oxidized form with the disulfide, and the Cys-to-serine mutant protein each bind m7GTP in a similar and labile manner, with dissociation rates in the range of 20 to 100 s−1.
Protein synthesis in eukaryotic cells is a highly regulated process that begins with the interaction of eukaryotic translation initiation factor-4E (eIF4E) with the m7GpppN cap group at the 5′ end of the mRNA (for review, see McKendrick et al., 1999; Pestova et al., 2001; Dever, 2002; Sonenberg and Dever, 2003). The eIF4E protein binds with its partner eIF4G to form the eIF4F complex (Keiper et al., 1999; Prévôt et al., 2003), which serves as a scaffold for the assembly of initiation factors eIF4A, eIF4B, eIF3, and poly(A)-binding protein. This protein-mRNA complex recruits the 40S ribosome with its attendant initiation factors (eIF1, eIF1A, eIF2, eIF5B, and eIF5) prior to scanning for the initiator AUG codon (Asano et al., 2000; Pestova et al., 2000; Sonenberg and Dever, 2003).
The role of eIF4E in translation has been extensively studied, particularly in mammalian and yeast (Saccharomyces cerevisiae) systems. In mammals, protein synthesis can be regulated by modulating the ability of eIF4E to interact with eIF4G (for review, see Richter and Sonenberg, 2005); this regulation is carried out by a group of repressor proteins called 4E-binding proteins (4E-BPs; Raught and Gingras, 1999; Pyronnet et al., 2001). The 4E-BPs function by mimicking eIF4G and are able to bind and sequester eIF4E, blocking formation of eIF4F, a complex of eIF4E and eIF4G. The 4E-BPs can be phosphorylated by the FRAP/mTOR intracellular signaling pathway (Raught and Gingras, 1999), which releases eIF4E, enabling it to form a complex with eIF4G. In mammals, eIF4E is subject to direct phosphorylation at Ser-209 by the kinases MNK1 and MNK2, which are activated by the mitogen-activated protein kinase signaling pathways (Fukunaga and Hunter, 1997; Waskiewicz et al., 1997; Raught and Gingras, 1999; Lachance et al., 2002). The C terminus of mammalian eIF4G has an MNK1-binding site that facilitates access of MNK1 to eIF4E (Pyronnet et al., 1999; Waskiewicz et al., 1999; Bellsolell et al., 2006). The effect of phosphorylation on mammalian eIF4E is still a subject of debate, with observations linking eIF4E phosphorylation to both increased and decreased protein synthesis (Pyronnet et al., 1999; Naegele and Morley, 2004; Orton et al., 2004; Worch et al., 2004; Arquier et al., 2005; Reiling et al., 2005). Phosphorylation of eIF4E does not appear to be necessary for protein synthesis in vitro or in vivo (McKendrick et al., 2001), and the mutation of the functional equivalent of Ser-209 in Drosophila melanogaster to Ala-209 is viable but does not have normal growth and development (Lachance et al., 2002). It has been reported that phosphorylation alters the ability of eIF4E to bind capped mRNAs in vitro (Scheper et al., 2002); however, phosphorylation has recently been reported to have no effect on ligand dissociation rate (koff) for cap analogs in vitro (Slepenkov et al., 2006). 5′ caps on longer (12 nucleotide) RNAs have increased affinity for both phosphorylated and unphosphorylated eIF4E, suggesting that there may be additional stabilizing interactions with eIF4E and the mRNA (Slepenkov et al., 2006).
Higher plants possess an equivalent form of eIF4E and in addition have an isozyme form of the protein, eIF(iso)4E (Allen et al., 1992; Browning et al., 1992; Browning, 1996, 2004). Plant eIF(iso)4E is approximately 50% similar in primary sequence to plant eIF4E (Allen et al., 1992; Metz et al., 1992). Furthermore, eIF(iso)4E is found in complex with an isozyme form of eIF4G, eIF(iso)4G (Lax et al., 1985; Browning et al., 1987, 1992). eIF4G and eIF(iso)4G share some similarity in their domains for the binding of additional factors eIF4A, eIF3, and poly(A)-binding protein but differ significantly in molecular mass, 180,000 versus 82,000 kD, respectively (Browning, 1996; Metz and Browning, 1996). Both the eIF4F (complex of eIF4E/eIF4G) and eIF(iso)4F [complex of eIF(iso)4E/eIF(iso)4G] have similar activities in supporting the initiation of translation in vitro (Lax et al., 1985; Browning et al., 1992). The role of the isozyme form of the eIF4E cap-binding protein in plants is not yet clear. Fluorescence quenching experiments have indicated that wheat (Triticum aestivum) eIF(iso)4F prefers hypermethylated cap structures and mRNAs with less secondary structure compared to wheat eIF4F (Balasta et al., 1993; Gallie and Browning, 2001). Plant eIF(iso)4F has also been shown to localize to maize (Zea mays) microtubules both in vitro and in vivo, suggesting that localization of eIF(iso)4F may have a role in the translation of specialized proteins within the cell (Bokros et al., 1995). Arabidopsis (Arabidopsis thaliana) eIF4E and eIF(iso)4E mRNAs are differentially expressed (Rodriguez et al., 1998), with the eIF4E mRNA being expressed in all tissue types except certain root cells, while eIF(iso)4E mRNA is particularly abundant in floral organ cells and young, developing tissues (Rodriguez et al., 1998). It was also demonstrated that Arabidopsis eIF4E could functionally complement a yeast eIF4E knockout, whereas eIF(iso)4E could not (Rodriguez et al., 1998). Resistance to some plant viruses has been shown to reside in the genes for eIF4E or eIF(iso)4E (for review, see Kang et al., 2005b; Robaglia and Caranta, 2006). These results suggest that although the biochemical properties of eIF4E and eIF(iso)4E are similar, there are differences in their function in plant cells.
The structures of eIF4E in complex with mRNA cap analog from several different organisms have previously been determined. The structure of yeast eIF4E was solved by NMR methods (Matsuo et al., 1997), and the murine eIF4E (Marcotrigiano et al., 1997) and human eIF4E (Tomoo et al., 2002, 2003) structures were solved by x-ray crystallography. These molecules were found to have identical topologies, with each protein containing a mixed eight-stranded β-sheet and three α-helices. In each case, the purine ring of the cap analog was bound to the protein using a π-π stacking interaction involving invariant Trp residues. A solution structure of apo-eIF4E from yeast was recently reported, where the protein was found to undergo changes in structure and internal motions in response to ligand binding, and a structural basis for a coupling between eIF4G and mRNA cap binding was described (Volpon et al., 2006).
In this work, we describe the first structure of an eIF4E protein from a plant, determined using a combination of x-ray crystallography, NMR, and mutational methods. These results are part of an effort to further understand the functional aspects of eIF4E that are unique to plants and provide some insight into the molecular basis of the functional differences between eIF4E and eIF(iso)4E.
RESULTS
Crystal Structure of the Wild-Type Wheat ΔN-eIF4E
The initial crystals of recombinant wheat eIF4E took over 1 year to grow. Data were collected in house and a molecular replacement solution was obtained using murine eIF4E as a test model (Marcotrigiano et al., 1997). Inspection of the electron density map and several rounds of refinement revealed that 38 amino acids at the N terminus of the protein were not visible. N-terminal sequence analysis of protein from dissolved crystals confirmed that these 38 residues had been removed during the prolonged crystallization period. These N-terminal amino acids are not well conserved between species. A similar number of amino acids were not visible at the N terminus of the murine and human forms of eIF4E (Marcotrigiano et al., 1997; Tomoo et al., 2002), and NMR results have shown that the N-terminal 35 amino acids of the yeast eIF4E are flexible in solution (Matsuo et al., 1997). To expedite additional crystallography work, the DNA encoding wheat eIF4E was truncated to produce a modified protein with 38 N-terminal residues removed; this protein is called ΔN-eIF4E. ΔN-eIF4E crystallized in about 2 weeks under the same conditions as the proteolyzed protein and had identical unit cell parameters.
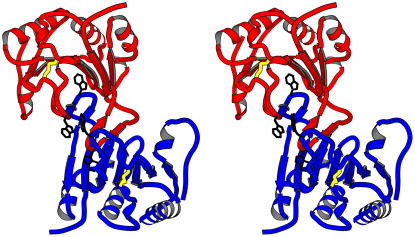
The crystals of ΔN-eIF4E belong to hexagonal space group P61 with the following cell constants: a = b = 66.3 Å, c = 180.9 Å. The crystallographic asymmetric unit of wheat ΔN-eIF4E is a dimer, exhibiting noncrystallographic symmetry along a pseudo 2-fold axis (Fig. 1). The Vm of the ΔN-eIF4E crystals is 2.8 Å3/D.
Figure 1.
The backbone of the dimeric structure of wheat eIF4E. Molecule A of the dimer is shown in red; molecule B is shown in blue. Tryptophan side chains analogous to those previously implicated in cap binding (Trp-62 and Trp-108) are shown in black, as well as Cys-113 and -151, which form a disulfide bond, are shown in yellow. Loop 1 (residues 53–65) of each monomer extends into the neighboring molecule in the dimer, with Trp-62 occupying the center of the expected cap-binding pocket.
Three-Ångstrom electron density maps, made with diffraction data collected in house, showed a dramatic conformational change in the area of the expected cap-binding site at the dimer interface. However, due to the low resolution of the map, the peptide chain could not be unambiguously traced through this area. A crystal freezing protocol was developed that allowed 1.85 Å data to be collected at the Brookhaven synchrotron, facilitating high resolution structure determination. Crystallographic data for ΔN-eIF4E are summarized in Table I. A Ramachandran plot shows 89.6% of residues to be in the most favorable region, 9.1% in additional allowed space, and 1.3% in generously allowed space. The refined structure includes 322 solvent molecules.
Table I.
Summary of crystallographic and structure refinement statistics for the wheat eIF4E protein lacking residues 1 to 38 at its N terminus (ΔN-eIF4E) and mutant C113S of the truncated protein
| Wheat ΔN-eIF4E | Mutant C113S | |
|---|---|---|
| Space group | P61 | P21 |
| Cell constants | a = b = 66.34, c = 180.85 Å | a = 36.65, b = 58.23, c = 39.06 Å, β = 115.8° |
| Resolution (Å) (last shell) | 50.0–1.85 (1.92–1.85) | 50.0–2.25 (2.32–2.25) |
| Rmerge (%) (last shell) | 7.5 (47.7) | 6.3 (14.7) |
| <I/σI> (last shell) | 8.7 (3.9) | 16.5 (12.4) |
| Completeness (%) (last shell) | 100.0 (100.0) | 96.9 (88.9) |
| Completeness (I/σI > 3) (%) (last shell) | 81.3 (46.2) | 88.4 (72.7) |
| Unique reflections | 38,354 | 6,939 |
| Redundancy | 8.2 | 5.5 |
| Wavelength (Å) | 1.1 | 1.5418 |
| No. of residues | 354 | 177 |
| No. of atoms | 3,130 | 1,553 |
| Rworking | 0.211 | 0.200 |
| Rfree | 0.234 | 0.274 |
| rms deviation from ideality | ||
| Bonds | 0.006 | 0.007 |
| Angles | 1.180 | 1.102 |
The overall fold and the central β-sheet regions of the dimeric protein are similar to that seen in monomeric eIF4E from mammals and yeast (Marcotrigiano et al., 1997; Matsuo et al., 1997; Tomoo et al., 2002). However, the mammalian and yeast proteins differ from the wheat ΔN-eIF4E in that they contain an mRNA cap analog sandwiched between two invariant Trp rings in an aromatic stack; the wild-type wheat protein crystallized without bound cap, despite its being present in solution. The most significant differences between the protein structures occur in the region of the two loops, residues 53 to 65 (loop 1) and 103 to 116 (loop 2), which hold the cap-binding Trp; these correspond to Trp-62 and Trp-108 in wheat eIF4E. These loops were left out of early models of the wheat protein and were carefully rebuilt from a series of omit maps. In the mammalian and yeast proteins, these loops form random coils that come together with the bound cap sandwiched between the two Trp. Loop 2 is in close proximity to a short helix (residues 199–203 in the wheat protein) between β-strands 7 and 8. In the wheat eIF4E structure, these loops are extended away from the surface, opening a wide cavity between the loops and the short helix. In the cap-bound eIF4E structures, this space is occupied by the two loops and the bound cap. In the wheat protein, loop 1 is folded as a β-loop rather than random coil, and in the dimer, the loop 1 of the neighboring monomer is inserted into the cavity, forming a four-strand antiparallel β-sheet with the loop 1 of the first monomer. Trp-108 also interacts with residues of the empty cap-binding pocket of the neighboring protein in the dimer (Fig. 1). The recently reported NMR structure for the yeast apo-eIF4E indicates similar aberrant movement of the two loops that are supposed to bind the cap analog (Volpon et al., 2006).
A Novel Disulfide Bond
Wheat eIF4E contains four Cys residues. The wheat ΔN-eIF4E crystal structure showed that two of these Cys, Cys-113 and Cys-151, form a disulfide bond (Fig. 2). Interestingly, this pair of Cys is conserved in all known eIF4E proteins from higher plants, but not in animals or yeast. For example, in mouse, the homolog of Cys-151 is a Cys, but the homolog of wheat Cys-113 is Asn-107 in mouse (Fig. 3). Finding a disulfide bond in a cytoplasmic protein was somewhat surprising, because the cytoplasm of most cells is reducing. The geometry associated with the disulfide is nearly ideal. The φ and ψ angles for residues 113 and 151 are both in the allowed regions of a Ramachandran plot, and the C-S-S bond angles are 105.4° and 105.3° for residues 113 and 151, respectively. The C-S-S-C dihedral angle is −94.4°. These data suggest that the bond is not strained. From the crystallographic data alone, it was not clear whether the disulfide bond is an artifact of an aerobic purification procedure and crystallization, or if it exists under some circumstances in the cytoplasmic protein under physiological conditions.
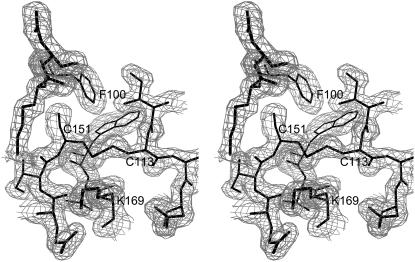
Figure 2.
A section of the 2Fo-Fc electron density map of wheat eIF4E showing the disulfide bridge between Cys-113 and Cys-151. The map is contoured at 1.0 σ.
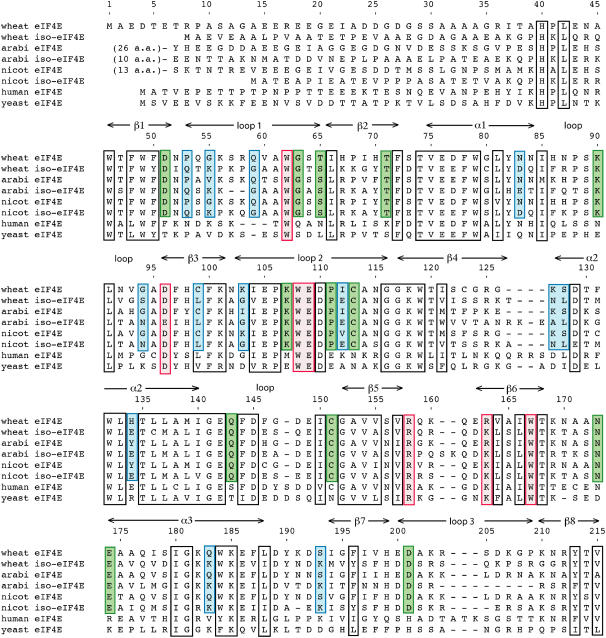
Figure 3.
Amino acid sequences of eIF4E and its isoform from higher plants aligned with human and yeast eIF4E. Sequences are shown for eIF4E and eIF(iso)4E from wheat, Arabidopsis (arabi), and Nicotiana tabacum (nicot), aligned with eIF4E from Homo sapiens (human) and yeast. Amino acids that are most directly involved in binding m7GTP are boxed in red. Amino acids that are important for stabilizing the structure of the protein are boxed in black; these are either located in the hydrophobic core or involved in other tertiary interactions and are well conserved across all eukarya. Plant-specific amino acids are boxed in green; these are conserved (>95%) in plants but differ from those in other eukarya. The plant-specific amino acids include Cys-113 and Cys-151, which form the disulfide linkage in the wheat eIF4E crystal structure, as well as several other surface residues. Blue boxes indicate amino acids that are >90% conserved in one isoform of plant eIF4E while being variable or conserved as another residue type in eIF(iso)4E and, therefore, indicative of whether the protein belongs to the eIF4E or eIF(iso)4E family. Interestingly, most of these isoform-specific residues are relatively accessible on the protein surface; hence, there is little overlap between the set of architecturally important residues (black boxes) and the isoform-specific residues (blue boxes). Although only eight sequences are shown in the figure, 55 sequences were examined in determining which residues are conserved (Joshi et al., 2005).
Crystal Structure of the C113S Mutant of Wheat ΔN-eIF4E
The Cys-to-Ser mutant C113S of wheat ΔN-eIF4E was prepared for the purpose of further investigating the role of the disulfide bond in the protein structure. We were unable to crystallize the mutant protein in the ammonium sulfate conditions that yielded wheat ΔN-eIF4E crystals. However, the mutant protein did crystallize under a different, low-salt condition. These crystals were of space group P21 with cell constants a = 36.7, b = 58.2, c = 39.1 Å, and β = 115.8°. The asymmetric unit contained a monomer, giving a Vm of 1.9 Å3/D. The 2.3-Å structure was solved by the molecular replacement method. Crystallographic data for the C113S mutant are summarized in Table I. A Ramachandran plot shows 89.0% of residues to be in the most favorable region, 10.4% in additional allowed space, and 0.6% in generously allowed space. The refined structure includes 101 solvent molecules. A ribbon drawing comparing the wild type and C113S mutant protein is shown in Figure 4.
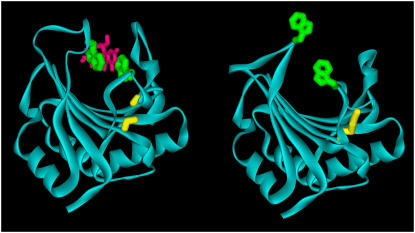
Figure 4.
Ribbon diagram comparing the structure of the wild type (right) and C113S mutant (left) of wheat eIF4E. The Trp of the cap-binding pocket are shown in green, the m7GDP is shown in magenta and the disulfide bond (right) and positions of the reduced Cys (left) are shown in yellow.
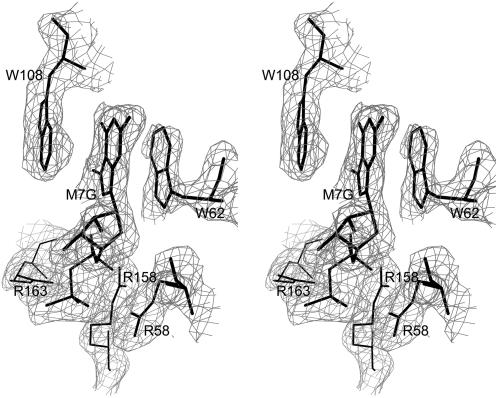
Interestingly, it was found that unlike the wild-type wheat eIF4E, which crystallized without the bound cap, the C113S mutant contained m7GDP in a binding site that is strikingly similar to that observed in crystal structures of the murine and human eIF4E (Marcotrigiano et al., 1997; Tomoo et al., 2002). As shown in Figure 5, the purine ring of the cap is sandwiched between the rings of Trp-62 and Trp-108. Conserved Glu-109 is hydrogen bonded to the methylated guanine ring. Arg-158 and Arg-163 interact with the phosphate groups of the cap; these positively charged amino acids are well conserved (as either Arg or Lys) in mammals, yeast, and plants (Fig. 3). Another remarkably conserved residue, Asp-96, forms a salt bridge with Arg-158 and is therefore likely to also be important for cap binding. The ring of conserved Trp-167 makes a van der Waals contact with the methyl group of the m7GDP ligand, providing a structural basis for the ability of the protein to discriminate between GDP and m7GDP.
Figure 5.
A section of the 2Fo-Fc electron density map of the C113S mutant of wheat eIF4E showing the bound m7GDP. The map is contoured at 0.8 σ.
Interestingly, the distance between Oγ of Ser-113 and Sγ of Cys-151 in the C113S mutant protein is only 3.7 Å. In the wild-type protein, where both of these residues are Cys, the sulfur atoms would be well placed to form a disulfide linkage with a relatively minor, but perhaps functionally significant, perturbation of the protein structure. The side chain of Ser-113 (Cys-113 in the native protein) is located relatively close to the cap-binding site; the distance between the Cβ of Ser-151 and the ring of Trp-108 is approximately 6.1 Å.
Wheat eIF4E in Solution
NMR spectroscopy is well known as a tool for providing detailed information about the structure, dynamics, and stability of proteins in solution; however, it is also an effective tool for detecting Cys that form disulfide bonds (Sharma and Rajarathnam, 2000). Specifically, the 13C NMR chemical shift of the Cβ in a reduced Cys is typically 28 to 32 ppm, while Cys that are involved in disulfides have Cβ chemical shifts near 40 ppm. An NMR method has a significant advantage over more traditional biochemical methods for disulfide detection, such as Ellman's assay (Ellman, 1959), because NMR can specifically indicate which Cys in the protein sequence are involved in a disulfide while simultaneously monitoring whether the protein has remained in a folded conformation during the oxidation attempt. In the case of wheat eIF4E, which contains four Cys residues, we found that Ellman's assay could be used to show a decrease in the number of reduced Cys under oxidizing solution conditions but did not give an unambiguous indication regarding the presence or absence of the Cys-113-Cys-151 disulfide bond.
As a first step in using NMR for disulfide bond detection, triple-resonance methods were used to assign the spectrum of 13C- and 15N-enriched wheat ΔN-eIF4E under conditions that were not intentionally oxidizing, in a solution of 20 mm potassium phosphate at pH 7, 100 mm KCl, and m7GTP that slightly exceeded the 1-mm protein concentration. The presence of m7GTP was found to significantly increase the stability of the protein in solution. The Cβ chemical shifts of all four Cys within eIF4E were identified in a three-dimensional HN(CO) CACB spectrum (Fig. 6). Cys-113 and Cys-151 were found to have Cβ chemical shifts of 29.2 and 29.1 ppm, respectively, indicating that they are not involved in a disulfide bond under these solution conditions. The other two Cys in the protein, Cys-99 and Cys-123, have Cβ chemical shifts of 29.4 and 32.1, indicating that they are also reduced. The Cys Cβ chemical shifts were unchanged in a protein sample that had been stored for 3 months at 4°C in NMR buffer without the addition of a reducing agent such as dithiothreitol (DTT), indicating that the Cys are not particularly susceptible to oxidation in solution under the conditions of the NMR experiments. The one-dimensional NMR spectrum of the protein contains resonance line widths that are typical of a 20-kD molecule, consistent with wheat ΔN-eIF4E being a monomer in solution.
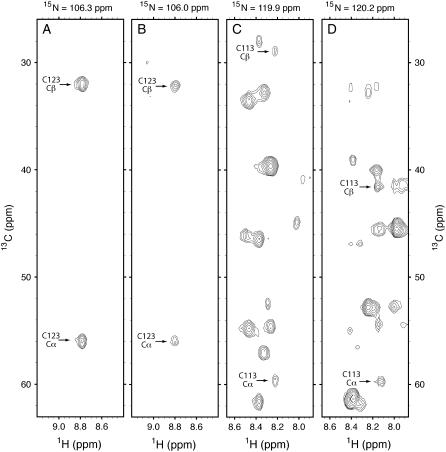
Figure 6.
Sections of three-dimensional HN(CO) CACB spectra of wheat ΔN-eIF4E showing the change in chemical shift of the Cβ of Cys-113 upon disulfide formation. Peaks indicating the 13C chemical shifts of Cα and Cβ of Cys-123 and Cys-113 are labeled. Sections A and C are from a spectrum of ΔN-eIF4E protein in 100 mm KCl, 10 mm potassium phosphate at pH 7, with m7GTP in slight excess over the protein concentration; sections B and D show the spectrum when 0.3 m ammonium sulfate and 10 mm hydrogen peroxide were added to the NMR sample to more closely mimic the conditions under which the crystals were obtained. Under these latter conditions, the 13C chemical shift of Cβ of Cys-113 changes from 29.2 to 41.4 ppm, indicative of its participation a disulfide bond, while the chemical shifts of Cβ of Cys-123 and other 1H, 13C, and 15N nuclei undergo relatively modest changes.
In an effort to determine whether the Cys-113-Cys-151 disulfide bond that was observed in the crystals of wheat ΔN-eIF4E can be formed in solution, NMR was used to study the protein under more oxidizing conditions, where 10 mm hydrogen peroxide and 0.3 m ammonium sulfate were added to the sample (the crystals were grown in ammonium sulfate). A two-dimensional 15N-1H correlated spectrum showed little or no chemical shift changes for the large majority of backbone 15N and 1H nuclei, indicating that these more oxidizing conditions did not cause the protein to denature or undergo a drastic conformational change. Resonance line widths were also not significantly changed, indicating that the protein did not dimerize in solution, as it had in the crystal. However, there was a significant decrease in intensity (or disappearance) of the NMR signals of approximately 20 backbone amide protons, mostly located in loops 1 and 2, near the cap-binding site. This decrease in NMR intensity can be attributed to an increased rate of exchange of the amide protons with the solvent and/or line resonance line broadening due to chemical exchange, either of which would be consistent with these regions of the protein becoming more flexible under the more oxidizing conditions. Most significantly, a three-dimensional HN(CO) CACB spectrum obtained under the oxidizing conditions (Fig. 6) showed a chemical shift of 41.4 ppm for Cβ of Cys-113, which is typical of a Cys involved in a disulfide bond (Sharma and Rajarathnam, 2000), presumably with Cys-151. There was also a small change of 0.2 ppm in the Cys-113 Cβ chemical shift and slight changes in the amide 15N and 1H chemical shifts of Cys-113 and adjacent amino acids. The Cβ resonance of Cys-151 was not detected in the HN(CO) CACB spectrum, due to the amide proton of Gly-152 being unobserved, likely due to its rapid exchange with protons of the solvent. The chemical shift of Cβ of Cys-123 was unchanged at 32.1 ppm under the oxidizing conditions, and the Cβ chemical shift of Cys-99 was not detected in the HN(CO) CACB spectrum due to the amide proton of residue 100 being unobserved.
In summary, the NMR data show that wheat ΔN-eIF4E does not contain a disulfide between Cys-113 and Cys-151 under solution conditions that are not intentionally oxidizing. The NMR data also provide evidence that the Cys-113-Cys-151 disulfide can be induced to form in solution under oxidizing conditions; these oxidizing conditions do not denature or induce substantial structural changes in the protein; however, there is evidence of increased flexibility in loops of the protein near the cap-binding site.
Binding of m7GTP by Wheat eIF4E in Solution
The observation that Cys-113 and Cys-151 can form a disulfide linkage without unfolding the eIF4E protein (supported by our crystallographic as well as NMR results), combined with the observation that these Cys are well conserved in plants, suggests the possibility that their oxidation state may have a role in protein function. As part of a test of this hypothesis, an investigation of the influence of disulfide formation upon the ability of the protein to bind the cap analog m7GTP was undertaken.
Biochemical methods have previously been used to study the binding of capped mRNAs and cap analogs by eIF4E from mammals and yeast in numerous studies (Carberry et al., 1990, 1991, 1992; Ueda et al., 1991; Minich et al., 1994; Niedzwiecka et al., 2002; Scheper et al., 2002), while cap binding by eIF4E from plants has been investigated to a lesser extent (Sha et al., 1995; Ren and Goss, 1996; Ruud et al., 1998; Khan and Goss, 2004; Michon et al., 2006). A fluorescence assay has been used to quantitatively assess the binding of m7GTP by the eIF4E protein, where quenching of Trp fluorescence upon stacking with the purine ring of the cap is measured as a function of protein and cap concentration (Carberry et al., 1989; Niedzwiecka et al., 2002). In the case of the wheat eIF4E and ΔN-eIF4E, the proteins were found to be unstable in the absence of ligand (as detected by NMR), thus preventing the preparation of the cap-free protein required for the fluorescence-quenching method. At low m7GTP concentrations, the protein concentration appeared to continuously decrease with time as measured by fluorescence emission or UV absorbance, perhaps due to its precipitation. Instability has also been reported for the cap-free mammalian eIF4E (Niedzwiecka et al., 2002), as well as the tendency for the protein to aggregate in the absence of cap (Niedzwiecka et al., 2005).
NMR spectroscopy was used as an alternative to fluorescence methods for the investigation of the equilibrium and dynamics of the eIF4E-m7GTP interaction. NMR has the advantage that it can be used without preparing ligand-free samples of the protein. In addition, the characteristic 1H NMR spectrum of the protein could be monitored to maintain confidence that all of the protein was folded when ligand binding was measured. Upon introducing an excess of ordinary (12C rich) m7GTP into a sample of the protein that had been purified with 13C-labeled ligand, it was observed that the pool of free ligand reached an equilibrium mixture of 12C and 13C in less than 90 s, which is how long it took to acquire a 13C NMR spectrum after mixing. This observation immediately suggested weak ligand binding and put a lower limit on koff for the protein-ligand interaction. A two-dimensional 1H-1H NOE spectrum of a mixture of wheat ΔN-eIF4E and excess m7GTP shows that separate (and broadened) resonances are observed for the free and bound ligand (Fig. 7). Cross peaks between resonances of the free and bound m7GTP indicate that a significant fraction of the free and bound pools exchange during the 200-ms NOE mixing time (Fig. 7); this NOE spectrum is remarkably similar to that of the Trp repressor protein obtained with a similar concentration of excess Trp (Schmitt et al., 1995). The observation that the resonances of the protein and free ligand have the same sign in a two-dimensional NOE spectrum is also consistent with weak ligand binding (Post, 2003). The 1H resonances of excess free m7GTP are broadened upon the addition of a relatively small (less than stoichiometric) amount of wheat ΔN-eIF4E protein (Fig. 8). These observations indicate that the bound and free m7GTP are in slow exchange on the NMR time scale. An analysis of the NMR line widths (Lian and Roberts, 1993) was used to derive the ligand dissociation rate (koff) for samples of wild-type, mutant, oxidized, and reduced eIF4E protein.
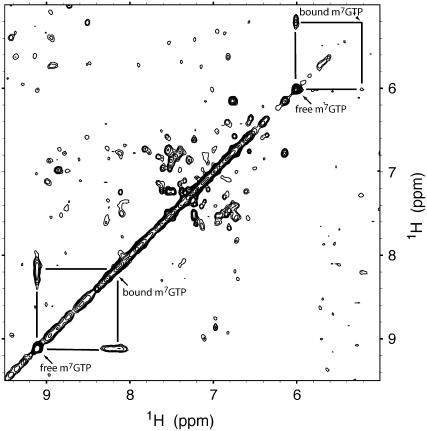
Figure 7.
A section of a two-dimensional 1H-1H NOE spectrum of 1 mm wheat ΔN-eIF4E in the presence of 5 mm m7GTP with the chemical shifts of free and bound m7GTP indicated by arrows. The spectrum was obtained at 25°C with a 200-ms NOE mixing time. The resonances of the free m7GTP have the same sign as those of the protein, consistent with weak ligand binding, and the resonances of the bound m7GTP are broadened, consistent with slow exchange on the NMR time scale. The off-diagonal exchange peaks between the free and bound m7GTP are striking, similar to those observed in the spectrum of the Trp repressor protein in the presence of excess Trp (Schmitt et al., 1995) in a similar example of weak ligand binding. This NOE spectrum was obtained using a jump-return method for suppressing the solvent water signal, which tends to attenuate the signals of peaks near 5 ppm.
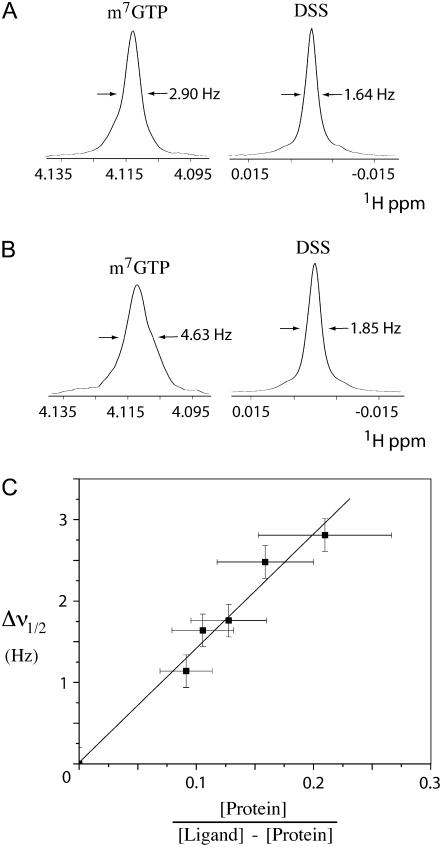
Figure 8.
A, Sections of a one-dimensional 1H NMR spectrum of a mixture of 0.2 mm m7GTP and the chemical shift reference standard DSS in the absence of protein. B, Sections of the 1H NMR spectrum of a sample containing 0.25 mm m7GTP and a less-than-stoichiometric amount (0.03 mm) of the wheat ΔN-eIF4E protein, showing that the wheat ΔN-eIF4E broadens the methyl peak of the free m7GTP, as expected in the case of slow exchange on the NMR time scale. The width of the DSS resonance shows relatively little change upon the addition of the protein. C, A representative graph showing that the width of the free m7GTP methyl resonance varies with protein and ligand concentrations, consistent with labile ligand binding and the model of slow exchange on the NMR timescale. [Protein] and [Ligand] refer to the total concentrations of wheat ΔN-eIF4E protein and m7GTP, respectively. Δν1/2 is defined as the width of the m7GTP methyl resonance at half height minus the width of the DSS resonance.
Results of the NMR-based binding assay showed that the reduced wheat ΔN-eIF4E, the oxidized form of the protein with the Cys-113-Cys-151 disulfide, and the C113S mutant protein each weakly (and similarly) bind m7GTP. For the reduced protein, koff was found to be in the range of 48 ± 15 s−1; oxidized protein with the disulfide was found to have koff of 73 ± 26 s−1. The C113S mutant that cannot form the disulfide was found to have koff in the range of 38 ± 15 s−1. The stated uncertainty in each value is indicative of the estimated uncertainties in the NMR line width measurements, protein and ligand concentrations, and the variability in data obtained using different protein preparations. It therefore appears that there is only a small (1.5×) difference in the dissociation rate for the cap analog m7GTP for the three forms of the wheat ΔN-eIF4E protein as measured by NMR. This implies that Kd for the binding of m7GTP by wheat ΔN-eIF4E is greater than or equal to approximately 10−7 m−1, which is toward the high end of the range reported for other (nonplant) forms of eIF4E, determined using fluorescence-quenching assays (Carberry et al., 1992; Niedzwiecka et al., 2002; Khan and Goss, 2004) and stopped-flow kinetic methods (Slepenkov et al., 2006). Interestingly, our NMR-based assay indicates that koff for eIF4E from yeast is 12 ± 2 s−1, which is significantly different from the reduced, oxidized, and mutant forms of the wheat protein and suggests yeast eIF4E binds m7GTP more tightly than the wheat eIF4E (data not shown).
A lower limit for Kd can be estimated by assuming that kon is no more rapid than the diffusion limit (approximately 108–109 s−1), given that Kd is defined by koff/kon. This implies that Kd for the binding of m7GTP by wheat ΔN-eIF4E is approximately 10−7 m−1 or greater, which is consistent with values determined for mammalian eIF4E using fluorescence-quenching assays (Carberry et al., 1989; Ren and Goss, 1996; Niedzwiecka et al., 2002) and stopped-flow kinetic methods (Slepenkov et al., 2006).
DISCUSSION
The results of this work provide the first description, to our knowledge, of a structure of a plant form of the translation initiation factor eIF4E. As expected, the wheat protein has much in common with the mammalian and yeast forms of eIF4E. Approximately 55 residues are conserved for clearly architectural reasons, forming the hydrophobic core of the protein and other essential tertiary interactions (boxed in black, Fig. 3). The residues that are most intimately involved in binding m7GTP are also conserved across all species (boxed in red, Fig. 3). Interestingly, there are 11 residues that are remarkably well conserved in plants yet differ in identity from the amino acids in analogous position in the mammalian and yeast forms of eIF4E; these plant-specific residues are boxed in green in Figure 3 and include Cys-113 and Cys-151, which form the disulfide linkage observed in the crystal structure of the wheat protein. Other plant-specific residues (Asp-51, Lys-90, Lys-107, Glu-143, Asn-173, Glu-174, and Asp-201) are located in relatively accessible positions on the protein surface. This combination of conservation plus surface accessibility suggests that these residues are excellent candidates for being involved in functional interactions involving eIF4E that are unique to translation initiation in plants. The roles of these plant-specific surface residues will perhaps be made clear as additional biochemical data are accumulated. The presence of plant-specific eIF4E surface features should not be too surprising, because several aspects of the initiation pathway are indeed unique to plants, including the presence of two distinct forms of eIF4E and eIF4G (Browning, 2004).
Although the roles of the two eIF4E isoforms are not yet well understood, the presence of two significantly divergent forms of the protein in plants strongly suggests some isoform-specific function. It has been reported that a knockout of the single eIF(iso)4E gene in Arabidopsis grows normally, although the amount of eIF4E is increased in the mutants (Duprat et al., 2002), suggesting that the two forms have overlapping functions. Perhaps more interestingly, mutants (both naturally occurring and induced) for both eIF4E and eIF(iso)4E are resistant to infection by a number of plant viruses, particularly the potyviruses, suggesting that these plant viruses exploit the eIF4E/eIF(iso)4E machinery to their benefit (for review, see Robaglia and Caranta, 2006). The resistance phenotype arises from disruption of the direct interaction between the viral protein (VPg) linked to the 5′ end of the potyviral RNA and eIF4E or eIF(iso)4E (Leonard et al., 2004; Michon et al., 2006). However, the viruses have countered natural resistance with mutations in the VPg (Keller et al., 1998; Redondo et al., 2001; Moury et al., 2004). Naturally occurring potyvirus resistance genes have been shown to reside in the genes for eIF4E or eIF(iso)4E in lettuce (Latuca sativa) mo1 (eIF4E; Nicaise et al., 2003); pepper (Capsicum annuum) pvr1 (eIF4E) and pvr6 [eIF(iso)4E; Ruffel et al., 2002, 2006; Kang et al., 2005a]; tomato (Lycopersicon esculentum) pot-1 (eIF4E; Ruffel et al., 2005); pea (Pisum sativum) sbm1 (eIF4E; Gao et al., 2004b) and sbm2 [eIF(iso)4E; Gao et al., 2004a]; and Arabidopsis lsm1 [eIF(iso)4E; Duprat et al., 2002; Lellis et al., 2002; Sato et al., 2005] and cum1 (eIF4E; Yoshii et al., 2004; Sato et al., 2005). The barley (Hordeum vulgare) rym4/5/6 resistance genes to various strains of the bymoviruses, Barley yellow mosaic virus and Barley mild mosaic virus, were also found to be mutations in the eIF4E gene (Kanyuka et al., 2005; Stein et al., 2005).
The exact role of eIF4E and eIF(iso)4E in virus resistance is not clear. A number of hypotheses have been proposed for the eIF4E/eIF(iso)4E-mediated viral resistance. These include disruption of host translation initiation (Lellis et al., 2002), stabilization of the viral RNA from degradation (Lellis et al., 2002), disruption of trafficking of the viral RNA (Lellis et al., 2002), interference with viral replication (Robaglia and Caranta, 2006), sequestration of initiation factors (Kanyuka et al., 2005), and interference with cell to cell movement (Gao et al., 2004b). In the case of resistance of Pepper veinal mottle virus, both the pvr2 (eIF4E) and pvr6 [eIF(iso)4E] mutations are required for resistance (Ruffel et al., 2006). The role of the interaction of the VPg and eIF4E/eIF(iso)4E in the viral infection/replication cycle is far from clear (Robaglia and Caranta, 2006).
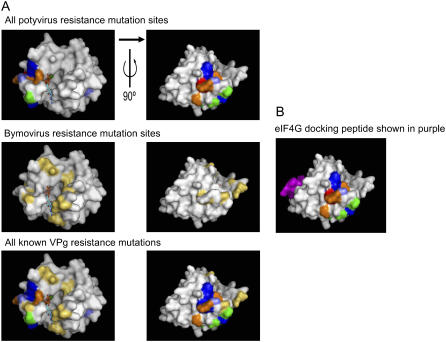
A number of structural models for plant eIF4E have been proposed based on the murine or human structure (Marcotrigiano et al., 1997; Tomoo et al., 2002) to show the positions of these naturally occurring mutations in plant eIF4E (Gao et al., 2004b; Kang et al., 2005a; Kanyuka et al., 2005; Ruffel et al., 2005; Stein et al., 2005; Albar et al., 2006; Robaglia and Caranta, 2006). These models predicted that the mutations are largely on the surface of eIF4E and some are near the cap-binding pocket. However, very few of the naturally occurring mutations have been shown to affect cap binding per se (Kang et al., 2005a). The residues known to be involved in potyvirus or bymovirus resistance are shown at the comparable position in wheat eIF4E (see Fig. 9) and are numbered to be consistent with the wheat eIF4E sequence in Figure 3. The naturally occurring resistance mutations in eIF4E for potyviruses in tomato (red), lettuce (blue), pepper (orange), and pea (green) and the bymovirus resistance mutations in eIF4E (yellow) are modeled in Figure 9. Most of the resistance genes carry more than one amino acid mutation, although the lettuce mo12 is a single mutation (S55P; Nicaise et al., 2003), and a single mutation (Q161H) generated by ethyl methanesulfonate mutagenesis confers bymovirus resistance (Kanyuka et al., 2005). These single mutations suggest that by disrupting a single contact, the VPg interaction is compromised. Generally, two regions of the eIF4E protein are targeted in virus resistance, one near the cap-binding site, but not directly part of it, and another rotated 90° from the cap-binding pocket (see Fig. 9). These two regions on different facets of the eIF4E molecule suggest that the VPg may have two binding sites for optimal interaction. The single amino acid mutations at different sites of eIF4E suggest that binding at both sites is necessary for viral infection. Only four of the mutations would appear to have any potential direct effect on cap binding. The mutation at position G94 in tomato (G94R), lettuce (G94H), and pepper (G94R) points toward the phosphate, and the more hydrophilic substitutions (Arg or His) could form hydrogen bonds with the phosphate backbone of the mRNA. Pea N156K and barley Q161K also could interact with the mRNA phosphate backbone. Only barley E109G, where the Glu forms a direct hydrogen bond with the guanine ring, would have a direct effect on cap binding. The precise contact points between the VPg and the eIF4E/eIF(iso)4E appear to be optimized for each virus/host pair by coevolution, because they occur at different positions (compare potyvirus and bymovirus in Fig. 9). A recent study of the VPg-eIF4E interaction in vitro indicated that the binding of the potyvirus lettuce mosaic virus VPg and m7GDP to eIF4E were not mutually exclusive; that is, both could be bound, but binding of one ligand did reduce the affinity for the other about 15-fold (Michon et al., 2006). In addition, VPg increased the binding of a peptide containing the eIF4E-binding domain of eIF4G to eIF4E. This result suggests that the VPg is able to manipulate the eIF4E/eIF4G interaction (Michon et al., 2006). However, the eIF4G-binding domain of plant eIF4E (Fig. 9C, shown in purple) is at a considerable distance from the resistance mutations, indicating that in the case of these particular host/virus pairs, resistance is likely not a direct result of the eIF4G interaction but more likely due to a conformational change in eIF4E upon binding of VPg. Mutations in eIF4G and eIF(iso)4G have also been reported to confer resistance to various viruses in Arabidopsis and rice (Oryza sativa), respectively (Yoshii et al., 2004; Albar et al., 2006). Thus, it appears that a variety of methods for the virus to interact with the host's translational apparatus have evolved, and the hosts have responded by coevolving mutations for resistance within these genes. The structure of a plant eIF4E will be very valuable in elucidating the mechanisms for plant virus resistance and potentially for the design of virus resistant plants.
Figure 9.
Plant virus resistance mutations modeled on the wheat eIF4E structure. A, The residues implicated in plant potyvirus resistance are shaded as follows on row 1, tomato (red; Ruffel et al., 2005), lettuce (light blue; Nicaise et al., 2003), pea (green; Gao et al., 2004b), and pepper (orange; Ruffel et al., 2002; Kang et al., 2005a). The bymovirus resistance residues are in yellow on row 2 (Stein et al., 2005; Kanyuka et al., 2005). Residues implicated in more than one species are shaded in dark blue on row 3. The view on the left shows m7GDP in the binding pocket. The view on the right is rotated 90° to the right about the vertical axis. B, The mammalian eIF4G peptide (purple; Marcotrigiano et al., 1999) is modeled on the wheat structure to show its relative position to the all potyvirus resistance mutations (shaded dark blue).
The structure of wheat eIF4E described in this work, when combined with a comparative sequence analysis, provides information that can be used to gain some insight into the features that distinguish the two isoforms that occur in plants. Thirteen residues are >90% conserved as one residue type in one isoform of the eIF4E protein, while being variable or conserved as a different residue type in the other isoform (Table II). Interestingly, most of these isoform-specific residues are located in relatively accessible positions on the protein surface. Five of these residues (53, 55, 59, 103, and 112) are located in loops relatively near the site of the bound m7GDP nucleotide, suggesting that plant eIF4E and eIF(iso)4E may differ in their interactions with the mRNA cap or a region of the mRNA near the cap. Several other isoform-specific residues (83, 127, 128, 134, 183, and 193) are also located on protein surface, though relatively far from the cap-binding site. These isoform-specific surface residues (boxed in blue in Fig. 3) are excellent candidates for being the key sites that are central to isoform-specific function. Interestingly, only two of these isoform-specific residues correspond to the virus resistance mutations (the mo12 single mutation, A55P, and one in rym6, P53S), suggesting that virus resistance and isoform-specific functions are not directly linked.
Table II.
Summary of the isoform-specific amino acids that are indicative of whether a plant protein belongs to the eIF4E or eIF(iso)4E family
These residues are >95% conserved as one residue type in one isoform while being >95% conserved as a different residue type in the other isoform of eIF4E. These isoform-specific residues are excellent candidates for being participants in intermolecular interactions that may functionally distinguish eIF4E from eIF(iso)4E. A set of 55 eIF4E and eIF(iso)4E sequences from 23 diverse plant species was examined (Joshi et al., 2005). Residues are numbered to be consistent with the wheat eIF4E sequence in Figure 3.
| Residue No. | Identity in Plant eIF4E | Identity in Plant eIF(iso)4E |
|---|---|---|
| 53 | Pro | Glu |
| 55 | Variable | Lys |
| 59 | Gln/Lys | Gly |
| 83 | Asn | Asp/Glu |
| 94 | Gly | Asn/Ser |
| 99 | Cys | Leu |
| 103 | Lys | Gly |
| 112 | Ile/Val | Glu |
| 127 | Lys | Variable |
| 128 | Ser | Leu/Phe |
| 134 | His/Tyr | Glu |
| 183 | Gln | Lys |
| 193 | Ser/Thr | Lys |
The role of the disulfide bond in plant eIF4E has yet to be fully elucidated. The redox state of the plant cell can vary in response to a number of events, including light or environmental stresses (heat, drought, pathogens, etc.), and plants use redox as a mechanism for regulation of photosynthesis, seed development, and germination (Buchanan and Balmer, 2005). The apparently strong conservation of Cys-113 and Cys-151 in plants, along with their close proximity in the protein structure and observed ability to form a disulfide linkage, together raise the possibility that disulfide formation may have some biological function, such as a regulatory role. It is very intriguing to speculate that redox may be a method unique to plants for regulating the initiation of protein synthesis.
Perhaps the simplest hypothesis would be that disulfide formation influences the ability of eIF4E to bind capped mRNA, although this is not supported by the results of this work, where modest difference (1.5×) was observed in the abilities of the reduced and oxidized wheat eIF4E to bind m7GTP. However, eIF4E in vivo is part of a larger multiprotein complex, and binding of m7GTP by eIF4E is not an ideal reporter of the ability of the eIF4E-eIF4G complex to bind a capped mRNA. As a specific example, it has been shown that eIF4E binding of 5′mRNA cap is significantly stabilized by domains of eIF4G (Von der Haar et al., 2006). So the possibility does remain that the oxidation state of these conserved Cys may modulate some aspect of eIF4E function in vivo by affecting its ability to bind capped mRNA, as well as form complexes with eIF4G, other eIF4E-binding proteins, or members of the translational machinery. Further work will be necessary to establish a role, if any, of the disulfide bond present in plant eIF4E/eIF(iso)4E in the regulation of plant protein synthesis.
MATERIALS AND METHODS
Preparation of Full-Length Wheat eIF4E
A cDNA expression clone based on Z12616 (Metz et al., 1992) for wheat (Triticum aestivum) eIF4E was constructed by DNA amplification and inserted into plasmid pET22b (Novagen). This clone differed from Z12616 in that the DNA sequence in the first approximately 60 nucleotides was modified to reduce the high GC content, but maintained the correct amino acid sequence. The expression clone was transformed into BL21pLysS (Novagen) cells for expression. Procedures for expression and purification of wheat eIF4E for crystallization were similar to those described (Van Heerden and Browning, 1994; Mayberry, et al., 2007). Briefly, a 2.5-L culture was grown to an O.D. of approximately 0.8 at 37°C. Expression of eIF4E was induced by the addition of 0.64 mm isopropylthio-β-galactoside and grown for an additional 2 h at 37°C. The cells were harvested and resuspended in 20 mL of buffer (20 mm HEPES-KOH, pH 7.6, 0.1 mm EDTA, 2.0 mm DTT, 10% glycerol, 50 mm KCl, 100 μm GTP) containing two dissolved Mini-Protease Inhibitor tablets (Boerhinger-Mannheim). The cells were disrupted by sonication and centrifuged for 1 h in a 60Ti rotor at 47,500 rpm. The supernatant was applied to a 3-mL m7GTP-Sepharose (Pharmacia) column equilibrated in 20 mm HEPES-KOH, pH 7.6, 0.1 mm EDTA, 2.0 mm DTT, 10% glycerol, and 50 mm KCl. The column was washed with equilibration buffer until the O.D. dropped to baseline. The column was then washed with 25 mL of 20 mm HEPES-KOH, pH 7.6, 2.0 mm DTT, and 0.2 mm GTP. The GTP helps removes a protein contaminant that copurifies with eIF4E (identified as DNA J by protein sequence analysis; data not shown). The eIF4E protein was eluted from the affinity column with 25 mL of 20 mm HEPES-KOH, pH 7.6, 2.0 mm DTT, and 0.2 mm m7GTP or m7GDP. Fractions containing the highest amount of protein were pooled and concentrated to approximately 1 to 2 mg/mL for crystallization trials.
N-terminal protein sequence analysis of the crystallized eIF4E was done at the Protein Microanalysis Facility at the University of Texas at Austin.
Preparation of Truncated and Mutant Wheat eIF4E
A cDNA expression clone for wheat eIF4E truncating amino acids 1 to 38 (referred to as ΔN-eIF4E) was prepared by DNA amplification and placed in plasmid pET15b (Novagen); the C113S variant of ΔN-eIF4E was also prepared using DNA amplification. The ΔN-eIF4E protein was expressed and purified essentially the same as the full-length protein. The purification of the C113S mutant ΔN-eIF4E protein used 20 mm HEPES-KOH, pH 7.6, 0.1 mm EDTA, 2.0 mm DTT, 10% glycerol, and 50 mm KCl for washing of m7GTP-Sepharose column, and the protein was eluted with the same buffer containing 0.2 mm m7GDP. The eluted protein was concentrated to approximately 1 to 5 mg/mL for crystallization. Proteins used in NMR experiments were prepared by washing the affinity column with a buffer of 10 mm phosphate, pH 7, and 100 mm KCl prior to elution with 0.2 mm m7GTP in the same phosphate buffer. Protein samples enriched in 15N and/or 13C were prepared as above, but with M9 minimal medium containing 0.5 g/L 15N ammonium chloride and/or 2 g/L 13C Glc (Cambridge Isotope Laboratories) as the sources of nitrogen and/or carbon.
Crystallization and Data Collection
Wheat wild-type eIF4E, in elution buffer containing 0.2 mm m7GTP, was crystallized by the hanging drop method at 4°C from 1.8 to 2.4 m ammonium sulfate in 0.1 m HEPES or Tris-HCl buffer, pH 7.5 to 8.0. Prior to collecting cold stream data, a wild-type crystal was dipped for 1 to 5 s into a solution of 50% saturated sorbitol in artificial mother liquor (2 m ammonium sulfate, 25 mm Tris-HCl, pH 8.0).
C113S mutant ΔN-eIF4E in elution buffer containing 0.2 mm m7GDP was crystallized at 4°C using the hanging drop method from 28% (w/v) PEG4000, 0.1 m HEPES, pH 7.0, 20 mm phenol. Prior to data collection, the crystal was treated with cryoprotectant by transferring to artificial mother liquor (35% PEG4000, 0.1 m HEPES, pH 7.0) for 1 to 5 s. Following treatment with cryoprotectant, a wild-type or mutant crystal mounted in a cryoloop (Hampton Research) was then frozen by dipping into liquid nitrogen and placed in the cold stream on the goniostat.
Data from a C113S mutant crystal and preliminary wild-type crystal were collected in-house at 100 K on a Rigaku RAXIS IV image plate detector with a Rigaku RU-H3R rotating copper anode generator (Rigaku/MSC) operated at 50 kV and 100 mA. Data from a ΔN-eIF4E crystal were collected at 100 K on an ADSC Quantum 4 CCD detector at beamline X12-B of the National Synchrotron Light Source, Brookhaven National Laboratory. Raw data were reduced using the programs DENZO and SCALEPACK from the HKL suite (Otwinowski and Minor, 1997).
Crystal Structure Determination and Analysis
Molecular replacement with both wild-type and mutant data was carried out using murine eIF4E (Protein Data Bank ID 1EJ1; Marcotrigiano et al., 1997) as a model; the wheat and murine proteins have approximately 40% sequence identity. The molecular replacement and initial refinement with approximately 3 Å wild-type data collected in house were carried out using the Evolutionary Protein Molecular Replacement program (Kissinger et al., 1999) and X-PLOR (Brunger, 1992). Initial phasing and electron density map improvement with high resolution ΔN-eIF4E data were generated from the molecular replacement solution using the program ARP/wARP (Lamzin and Wilson, 1993; Perrakis et al., 1997). Molecular replacement with the C113S data was done with the program MOLREP (Vagin and Teplyakov, 1997).
Following manual adjustment using the program O (Jones et al., 1991), the model was refined with the Crystallography and NMR system (CNS, version 1.0) suite using the slow-cooling protocol (Brunger et al., 1998). There were several rounds of refinement followed by rebuilding of the model. To facilitate manual rebuilding of the model, a difference map and a 2Fo-Fc map, SIGMAA weighted to eliminate bias from the model (Read, 1986), were prepared. Five percent of the diffraction data were set aside throughout refinement for cross validation (Brunger, 1993). Geometry and stereochemistry of the models were analyzed throughout building and refinement using PROCHECK (Laskowski, 1993). For the purpose of building bound waters into the model, CNS was used to select solvent peaks. Candidates were positive peaks on a difference map 3.5 sds above the mean and within 3.5 Å of a protein nitrogen or oxygen atom. O was used to manually view and accept water sites. Computations were done on a Gateway Select SB computer. Model visualization and rebuilding was done on a Silicon Graphics Indy computer.
Pictures and drawings were constructed using MOLSCRIPT (Kraulis, 1991), BOBSCRIPT (Esnouf, 1999), and PyMOL (Delano Scientific). Atomic coordinates for the dimeric wheat ΔN-eIF4E (2IDR) and its monomeric C113S mutant (2IDV) have been deposited in the Protein Data Bank.
NMR Spectroscopy
NMR spectra were recorded at 25°C using a 500-MHz Varian Inova spectrometer equipped with a triple-resonance cryogenic probe and z axis pulsed field gradient. NMR samples typically contained 1.0 mm of ΔN-eIF4E protein in 90% water/10% D2O plus 100 mm KCl, a buffer of 10 mm potassium phosphate at pH 7, and m7GTP in slight excess over the protein concentration. Backbone 1H, 13C, and 15N resonance assignments were obtained using three-dimensional HNCA, HNCACB, HN(CO) CACB, HNCO, and HACACBCO spectra (Grzesiek et al., 1992; Kay, 1993; Muhandiram and Kay, 1994). Side chain resonance assignments were obtained using three-dimensional 15N-1H-1H HSQC-TOCSY and 13C-1H-1H HCCH-TOCSY spectra (Kay et al., 1993). Data were processed using NMR-Pipe (Delaglio et al., 1995) and visualized using Sparky (Goddard and Kneller, 2006). 1H, 15N, and 13C chemical shifts were referenced as recommended (Wishart et al., 1995), with proton chemical shifts relative to internal 2,2-dimethyl-2-silapentane-5-sulfonate (DSS), and 13C and 15N reference frequencies determined by multiplying the 1H reference frequency by 0.251449530 and 0.101329118, respectively.
The off rate for the interaction between wheat ΔN-eIF4E and m7GTP was derived from the width of the m7GTP methyl resonance measured at various protein and ligand concentrations. Data were obtained in NMR buffer at 25°C and fit to the equation: πΔυ − πΔυo = koff ([P]/[L] − [P]), where Δυ is the width at half height of the methyl resonance of m7GTP, Δυo is the width in the absence of protein, [P] is the total protein concentration, and [L] is the total ligand concentration (Lian and Roberts, 1993); this analysis assumes slow exchange on the NMR time scale and free ligand concentrations that significantly exceed Kd. In mixtures used for NMR line width measurements, protein concentrations were typically 0.02 to 0.06 mm, as measured by Bradford assay (Bradford, 1976), and ligand concentrations were typically in the range of 0.2 to 1.0 mm; under these conditions, broadening of the 1H resonances of the free m7GTP was typically in the range of 1 to 10 Hz due to chemical exchange. The 0.0 ppm resonance of DSS was used as an internal NMR line width standard to control for variations in shimming. NMR-based ligand binding assays were carried out in (at least) triplicate.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number Z12616.
Acknowledgments
We thank Kelley Ruud Nitka and Daniel Hirschhorn for technical assistance in the initial wheat eIF4E preparation and crystallization, and Patricia Murphy for technical assistance in the preparation of wheat ΔN-eIF4E. We thank Alexander Taylor and John Hart for data collection at beamline X12-B at the National Synchrotron Light Source, Brookhaven National Laboratory; support for the beamline comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the U.S. Department of Energy and from the National Center for Research Resources of the National Institutes of Health.
This work was supported by the National Institutes of Health (grant no. GM 63593 to J.D.R.), by the National Science Foundation (grant no. MCB–0214996 to K.S.B.), by the Welch Foundation (grant nos. F–1225, F–1353, and F–1333 to J.D.R., D.W.H., and K.S.B., respectively), and by the Center for Structural Biology from the College of Natural Sciences.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Karen S. Browning (kbrowning@mail.utexas.edu).
Open Access articles can be viewed online without a subscription.
References
- Albar L, Bangratz-Reyser M, Hebrard E, Ndjiondjop MN, Jones M, Ghesquiere A (2006) Mutations in the eIF(iso)4G translation initiation factor confer high resistance of rice to rice yellow mottle virus. Plant J 47 417–426 [DOI] [PubMed] [Google Scholar]
- Allen ML, Metz AM, Timmer RT, Rhoads RE, Browning KS (1992) Isolation and sequence of the cDNAs encoding the subunits of the isozyme form of wheat protein synthesis initiation factor 4F. J Biol Chem 267 23232–23236 [PubMed] [Google Scholar]
- Arquier N, Bourouis M, Colombani J, Leopold P (2005) Drosophila Lk6 kinase controls phosphorylation of eukaryotic translation initiation factor 4E and promotes normal growth and development. Curr Biol 15 19–23 [DOI] [PubMed] [Google Scholar]
- Asano K, Clayton J, Shalev A, Hinnebusch AG (2000) A multifactor complex of eukaryotic initiation factors, eIE1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev 14 2534–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasta ML, Carberry SE, Friedland DE, Perez RA, Goss DJ (1993) Characterization of the ATP-dependent binding of wheat germ protein synthesis initiation factors eIF-(iso)4F and eIF-4A to mRNA. J Biol Chem 268 18599–18603 [PubMed] [Google Scholar]
- Bellsolell L, Cho-Park PF, Poulin F, Sonenberg N, Burley SK (2006) Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1. Structure 14 913–923 [DOI] [PubMed] [Google Scholar]
- Bokros CL, Hugdahl JD, Kim H-H, Hanesworth VR, Van Heerden A, Browning KS, Morejohn LC (1995) Function of the p86 subunit of eukaryotic initiation factor (iso)4F as a microtubule-associated protein in plant cells. Proc Natl Acad Sci USA 92 7120–7124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Browning KS (1996) The plant translational apparatus. Plant Mol Biol 32 107–144 [DOI] [PubMed] [Google Scholar]
- Browning KS (2004) Plant translation initiation factors: it is not easy to be green. Biochem Soc Trans 32 589–591 [DOI] [PubMed] [Google Scholar]
- Browning KS, Lax SR, Ravel JM (1987) Identification of two messenger RNA cap binding proteins in wheat germ: evidence that the 28-kDa subunit of eIF-4B and the 26-kDa subunit of eIF-4F are antigenically distinct polypeptides. J Biol Chem 262 11228–11232 [PubMed] [Google Scholar]
- Browning KS, Webster C, Roberts JKM, Ravel JM (1992) Identification of an isozyme form of protein synthesis initiation factor 4f in plants. J Biol Chem 267 10096–10100 [PubMed] [Google Scholar]
- Brunger AT (1992) X-PLOR Version 3.1: A System for X-Ray Crystallography and NMR. Yale University Press, New Haven, CT
- Brunger AT (1993) Assessment of phase accuracy by cross validation: the free R value. Methods and applications. Acta Crystallogr D Biol Crystallogr 49 24–36 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54 905–921 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56 187–220 [DOI] [PubMed] [Google Scholar]
- Carberry SE, Darzynkiewicz E, Goss DJ (1991) A comparison of the binding of methylated cap analogues to wheat germ protein synthesis initiation factors 4F and (iso)4F. Biochemistry 30 1624–1627 [DOI] [PubMed] [Google Scholar]
- Carberry SE, Darzynkiewicz E, Stepinski J, Tahara SM, Rhoads RE, Goss DJ (1990) A spectroscopic study of the binding of N-7-substituted cap analogues to human protein synthesis initiation factor 4E. Biochemistry 29 3337–3341 [DOI] [PubMed] [Google Scholar]
- Carberry SE, Friedland DE, Rhoads RE, Goss DJ (1992) Binding of protein synthesis initiation factor 4E to oligoribonucleotides: effects of cap accessibility and secondary structure. Biochemistry 31 1427–1432 [DOI] [PubMed] [Google Scholar]
- Carberry SE, Rhoads RE, Goss DJ (1989) A spectroscopic study of the binding of m7GTP and m7GpppG to human protein synthesis initiation factor 4E. Biochemistry 28 8078–8083 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6 277–293 [DOI] [PubMed] [Google Scholar]
- Dever TE (2002) Gene-specific regulation by general translation factors. Cell 108 545–556 [DOI] [PubMed] [Google Scholar]
- Duprat A, Caranta C, Revers F, Menand B, Browning KS, Robaglia C (2002) The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J 32 927–934 [DOI] [PubMed] [Google Scholar]
- Ellman GL (1959) Tissue sulfhydryl groups. Arch Biochem Biophys 82 70–77 [DOI] [PubMed] [Google Scholar]
- Esnouf RM (1999) Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr D Biol Crystallogr 55 938–940 [DOI] [PubMed] [Google Scholar]
- Fukunaga R, Hunter T (1997) MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J 16 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Browning KS (2001) eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J Biol Chem 276 36951–36960 [DOI] [PubMed] [Google Scholar]
- Gao Z, Eyers S, Thomas C, Ellis N, Maule AJ (2004. a) Identification of markers tightly linked to sbm recessive genes for resistance to Pea seed-borne mosaic virus. Theor Appl Genet 109 488–494 [DOI] [PubMed] [Google Scholar]
- Gao Z, Johansen E, Eyers S, Thomas CL, Noel Ellis TH, Maule AJ (2004. b) The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J 40 376–385 [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG (2006) SPARKY 3. University of California, San Francisco. http://www.cgl.ucsf.edu/home/sparky/
- Grzesiek S, Dobeli H, Gentz R, Garotta G, Labhardt AM, Bax A (1992) 1H, 13C, and 15N NMR backbone assignments and secondary structure of human interferon-gamma. Biochemistry 31 8180–8190 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 110–119 [DOI] [PubMed] [Google Scholar]
- Joshi B, Lee K, Maeder DL, Jagus R (2005) Phylogenetic analysis of eIF4E-family members 2. BMC Evol Biol 5 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BC, Yeam I, Frantz JD, Murphy JF, Jahn MM (2005. a) The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with tobacco etch virus VPg. Plant J 42 392–405 [DOI] [PubMed] [Google Scholar]
- Kang BC, Yeam I, Jahn MM (2005. b) Genetics of plant virus resistance. Annu Rev Phytopathol 43 581–621 [DOI] [PubMed] [Google Scholar]
- Kanyuka K, Druka A, Caldwell DG, Tymon A, McCallum N, Waugh R, Adams MJ (2005) Evidence that the recessive bymovirus resistance locus rym4 in barley corresponds to the eukaryotic translation initiation factor 4E gene. Mol Plant Pathol 6 449–458 [DOI] [PubMed] [Google Scholar]
- Kay LE (1993) Pulsed-field gradient-enhanced three-dimensional NMR experiment for correlating 13Ca./b, 13C', and 1Ha chemical shifts in uniformly 13C-labeled proteins dissolved in water. J Am Chem Soc 115 2055–2057 [Google Scholar]
- Kay LE, Xu GY, Singer AU, Muhandiram DR, Forman-Kay JD (1993) A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlations in H2O samples of proteins. J Magn Reson B 101 333–337 [Google Scholar]
- Keiper BD, Gan WN, Rhoads RE (1999) Protein synthesis initiation factor 4G. Int J Biochem Cell Biol 31 37–41 [DOI] [PubMed] [Google Scholar]
- Keller KE, Johansen IE, Martin RR, Hampton RO (1998) Potyvirus genome-linked protein (VPg) determines pea seed-borne mosaic virus pathotype-specific virulence in Pisum sativum. Mol Plant Microbe Interact 11 124–130 [DOI] [PubMed] [Google Scholar]
- Khan MA, Goss DJ (2004) Phosphorylation states of translational initiation factors affect mRNA cap binding in wheat. Biochemistry 43 9092–9097 [DOI] [PubMed] [Google Scholar]
- Kissinger CR, Gehlhaar DK, Fogel DB (1999) Rapid automated molecular replacement by evolutionary search. Acta Crystallogr D Biol Crystallogr 55 484–491 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24 946–950 [Google Scholar]
- Lachance PED, Miron M, Raught B, Sonenberg N, Lasko P (2002) Phosphorylation of eukaryotic translation initiation factor 4E is critical for growth. Mol Cell Biol 22 1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamzin VS, Wilson KS (1993) Automated refinement of protein models. Acta Crystallogr D Biol Crystallogr 49 129–147 [DOI] [PubMed] [Google Scholar]
- Laskowski RA (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26 283–291 [Google Scholar]
- Lax S, Fritz W, Browning K, Ravel J (1985) Isolation and characterization of factors from wheat germ that exhibit eukaryotic initiation factor 4b activity and overcome 7-methylguanosine 5′triphosphate inhibition of polypeptide synthesis. Proc Natl Acad Sci USA 82 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lellis AD, Kasschau KD, Whitham SA, Carrington JC (2002) Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr Biol 12 1046–1051 [DOI] [PubMed] [Google Scholar]
- Leonard S, Viel C, Beauchemin C, Daigneault N, Fortin MG, Laliberte JF (2004) Interaction of VPg-Pro of turnip mosaic virus with the translation initiation factor 4E and the poly(A)-binding protein in planta. J Gen Virol 85 1055–1063 [DOI] [PubMed] [Google Scholar]
- Lian L-Y, Roberts GCK (1993) NMR of Macromolecules: A Practical Approach. Oxford University Press, New York, pp 153–182
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK (1997) Co-crystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89 951–961 [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK (1999) Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of elF4G. Mol Cell 3 707–716 [DOI] [PubMed] [Google Scholar]
- Matsuo H, Li HJ, McGuire AM, Fletcher CM, Gingras AC, Sonenberg N, Wagner G (1997) Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol 4 717–724 [DOI] [PubMed] [Google Scholar]
- Mayberry LK, Dennis MD, Allen ML, Nitka KA, Murphy PA, Campbell L, Browning KS (2007) Expression and purification of recombinant wheat translation initiation factors eIF1, eIF1A, eIF4A, eIF4B, eIF4F, eIF(iso)4F and eIF5. Methods Enzymol (in press) [DOI] [PubMed]
- McKendrick L, Morley SJ, Pain VM, Jagus R, Joshi B (2001) Phosphorylation of eukaryotic initiation factor 4E (eIF4E) at Ser209 is not required for protein synthesis in vitro and in vivo. Eur J Biochem 268 5375–5385 [DOI] [PubMed] [Google Scholar]
- McKendrick L, Pain VM, Morley SJ (1999) Translation initiation factor 4E. Int J Biochem Cell Biol 31 31–35 [DOI] [PubMed] [Google Scholar]
- Metz AM, Browning KS (1996) Mutational analysis of the functional domains of the large subunit of the isozyme form of wheat initiation factor eIF4F. J Biol Chem 271 31033–31036 [DOI] [PubMed] [Google Scholar]
- Metz AM, Timmer RT, Browning KS (1992) Isolation and sequence of a cDNA encoding the cap binding protein of wheat eukaryotic protein synthesis initiation factor 4F. Nucleic Acids Res 20 4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michon T, Estevez Y, Walter J, German-Retana S, Le Gall O (2006) The potyviral virus genome-linked protein VPg forms a ternary complex with the eukaryotic initiation factors eIF4E and eIF4G and reduces eIF4E affinity for a mRNA cap analogue 38. FEBS J 273 1312–1322 [DOI] [PubMed] [Google Scholar]
- Minich WB, Balasta ML, Goss DJ, Rhoads RE (1994) Chromatographic resolution of in vivo phosphorylated and nonphosphorylated eukaryotic translation initiation factor eIF-4E: increased cap affinity of the phosphorylated form. Proc Natl Acad Sci USA 91 7668–7672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moury B, Morel C, Johansen E, Guilbaud L, Souche S, Ayme V, Caranta C, Palloix A, Jacquemond M (2004) Mutations in potato virus Y genome-linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum. Mol Plant Microbe Interact 17 322–329 [DOI] [PubMed] [Google Scholar]
- Muhandiram DR, Kay LE (1994) Gradient-enhanced triple-resonance three-dimensional nmr experiments with improved sensitivity. J Magn Reson B 103 203–216 [Google Scholar]
- Naegele S, Morley SJ (2004) Molecular cross-talk between MEK1/2 and mTOR signaling during recovery of 293 cells from hypertonic stress. J Biol Chem 279 46023–46034 [DOI] [PubMed] [Google Scholar]
- Nicaise V, German-Retana S, Sanjuan R, Dubrana MP, Mazier M, Maisonneuve B, Candresse T, Caranta C, LeGall O (2003) The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus. Plant Physiol 132 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecka A, Darzynkiewicz E, Stolarski R (2005) Deaggregation of eIF4E induced by mRNA 5′ cap binding. Nucleosides Nucleotides Nucleic Acids 24 507–511 [DOI] [PubMed] [Google Scholar]
- Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, Gingras AC, Mak P, Darzynkiewicz E, Sonenberg N, et al (2002) Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J Mol Biol 319 615–635 [DOI] [PubMed] [Google Scholar]
- Orton KC, Ling J, Waskiewicz AJ, Cooper JA, Merrick WC, Korneeva NL, Rhoads RE, Sonenberg N, Traugh JA (2004) Phosphorylation of Mnk1 by caspase-activated Pak2/gamma-PAK inhibits phosphorylation and interaction of eIF4G with Mnk. J Biol Chem 279 38649–38657 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276 307–326 [DOI] [PubMed] [Google Scholar]
- Perrakis A, Sixma TK, Wilson KS, Lamzin VS (1997) wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr D Biol Crystallogr 53 448–455 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CUT (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA 98 7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CUT (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403 332–335 [DOI] [PubMed] [Google Scholar]
- Post CB (2003) Exchange-transferred NOE spectroscopy and bound ligand structure determination. Curr Opin Struct Biol 13 581–588 [DOI] [PubMed] [Google Scholar]
- Prévôt D, Darlix JL, Ohlmann T (2003) Conducting the initiation of protein synthesis: the role of eIF4G. Biol Cell 95 141–156 [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Dostie J, Sonenberg N (2001) Suppression of cap-dependent translation in mitosis. Genes Dev 15 2083–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N (1999) Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J 18 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Gingras AC (1999) eIF4E activity is regulated at multiple levels. Int J Biochem Cell Biol 31 43–57 [DOI] [PubMed] [Google Scholar]
- Read RJ (1986) Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr A 42 140–149 [Google Scholar]
- Redondo E, Krause-Sakate R, Yang SJ, Lot H, Le GO, Candresse T (2001) Lettuce mosaic virus pathogenicity determinants in susceptible and tolerant lettuce cultivars map to different regions of the viral genome. Mol Plant Microbe Interact 14 804–810 [DOI] [PubMed] [Google Scholar]
- Reiling JH, Doepfner KT, Hafen E, Stocker H (2005) Diet-dependent effects of the Drosophila Mnk1/Mnk2 homolog Lk6 on growth via eIF4E. Curr Biol 15 24–30 [DOI] [PubMed] [Google Scholar]
- Ren JH, Goss DJ (1996) Synthesis of a fluorescent 7-methylguanosine analog and a fluorescence spectroscopic study of its reaction with wheat germ cap binding proteins. Nucleic Acids Res 24 3629–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N (2005) Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433 477–480 [DOI] [PubMed] [Google Scholar]
- Robaglia C, Caranta C (2006) Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci 11 40–45 [DOI] [PubMed] [Google Scholar]
- Rodriguez CM, Freire MA, Camilleri C, Robaglia C (1998) The Arabidopsis thaliana cDNAs coding for eIF4E and eIF(iso)4E are not functionally equivalent for yeast complementation and are differentially expressed during plant development. Plant J 13 465–473 [DOI] [PubMed] [Google Scholar]
- Ruffel S, Dussault MH, Palloix A, Moury B, Bendahmane A, Robaglia C, Caranta C (2002) A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J 32 1067–1075 [DOI] [PubMed] [Google Scholar]
- Ruffel S, Gallois JL, Lesage ML, Caranta C (2005) The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol Genet Genomics 274 346–353 [DOI] [PubMed] [Google Scholar]
- Ruffel S, Gallois JL, Moury B, Robaglia C, Palloix A, Caranta C (2006) Simultaneous mutations in translation initiation factors eIF4E and eIF(iso)4E are required to prevent pepper veinal mottle virus infection of pepper. J Gen Virol 87 2089–2098 [DOI] [PubMed] [Google Scholar]
- Ruud KA, Kuhlow C, Goss DJ, Browning KS (1998) Identification and characterization of a novel cap-binding protein from Arabidopsis thaliana. J Biol Chem 273 10325–10330 [DOI] [PubMed] [Google Scholar]
- Sato M, Nakahara K, Yoshii M, Ishikawa M, Uyeda I (2005) Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. FEBS Lett 579 1167–1171 [DOI] [PubMed] [Google Scholar]
- Scheper GC, Van Kollenburg B, Hu JZ, Luo YJ, Goss DJ, Proud CG (2002) Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J Biol Chem 277 3303–3309 [DOI] [PubMed] [Google Scholar]
- Schmitt TH, Zheng Z, Jardetzky O (1995) Dynamics of tryptophan binding to Escherichia coli Trp repressor wild type and AV77 mutant: an NMR study. Biochemistry 34 13183–13189 [DOI] [PubMed] [Google Scholar]
- Sha M, Wang YH, Xiang T, Van Heerden A, Browning KS, Goss DJ (1995) Interaction of wheat germ protein synthesis initiation factor eIF-(iso)4F and its subunits p28 and p86 with m7GTP and mRNA analogues. J Biol Chem 270 29904–29909 [DOI] [PubMed] [Google Scholar]
- Sharma D, Rajarathnam K (2000) 13C NMR chemical shifts can predict disulfide bond formation. J Biomol NMR 18 165–171 [DOI] [PubMed] [Google Scholar]
- Slepenkov SV, Darzynkiewicz E, Rhoads RE (2006) Stopped-flow kinetic analysis of eIF4E and phosphorylated eIF4E binding to cap analogs and capped oligoribonucleotides: evidence for a one-step binding mechanism. J Biol Chem 281 14927–14938 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Dever TE (2003) Eukaryotic translation initiation factors and regulators. Curr Opin Struct Biol 13 56–63 [DOI] [PubMed] [Google Scholar]
- Stein N, Perovic D, Kumlehn J, Pellio B, Stracke S, Streng S, Ordon F, Graner A (2005) The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J 42 912–922 [DOI] [PubMed] [Google Scholar]
- Tomoo K, Shen X, Okabe K, Nozoe Y, Fukuhara S, Morino S, Ishida T, Taniguchi T, Hasegawa H, Terashima A, et al (2002) Crystal structures of 7-methylguanosine 5′-triphosphate (m7GTP)- and P1-7-methylguanosine-P3-adenosine-5′,5′- triphosphate (m7GpppA)-bound human full-length eukaryotic initiation factor 4E: biological importance of the C-terminal flexible region. Biochem J 362 539–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoo K, Shen X, Okabe K, Nozoe Y, Fukuhara S, Morino S, Sasaki M, Taniguchi T, Miyagawa H, Kitamura K, et al (2003) Structural features of human initiation factor 4E, studied by x-ray crystal analyses and molecular dynamics simulations. J Mol Biol 328 365–383 [DOI] [PubMed] [Google Scholar]
- Ueda H, Maruyama H, Doi M, Inoue M, Ishida T, Morioka H, Tanaka T, Nishikawa S, Uesugi S (1991) Expression of a synthetic gene for human cap binding protein (human IF-4E) in Escherichia coli and fluorescence studies on interaction with mRNA cap structure analogues. J Biochem (Tokyo) 109 882–889 [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30 1022–1025 [Google Scholar]
- Van Heerden A, Browning KS (1994) Expression in Escherichia coli of the two subunits of the isozyme form of wheat germ protein synthesis initiation factor 4F: purification of the subunits and formation of an enzymatically active complex. J Biol Chem 269 17454–17457 [PubMed] [Google Scholar]
- Volpon L, Osborne MJ, Topisirovic I, Siddiqui N, Borden KL (2006) Cap-free structure of eIF4E suggests a basis for conformational regulation by its ligands. EMBO J 25 5138–5149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von der Haar T, Oku Y, Ptushkina M, Moerke N, Wagner G, Gross JD, McCarthy JE (2006) Folding transitions during assembly of the eukaryotic mRNA cap-binding complex. J Mol Biol 356 982–992 [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Flynn A, Proud CG, Cooper JA (1997) Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J 16 1909–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz AJ, Johnson JC, Penn B, Mahalingam M, Kimball SR, Cooper JA (1999) Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol 19 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD (1995) 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR 6 135–140 [DOI] [PubMed] [Google Scholar]
- Worch J, Tickenbrock L, Schwable J, Steffen B, Cauvet T, Mlody B, Buerger H, Koeffler HP, Berdel WE, Serve H, et al (2004) The serine-threonine kinase MNK1 is post-translationally stabilized by PML-RARalpha and regulates differentiation of hematopoietic cells. Oncogene 23 9162–9172 [DOI] [PubMed] [Google Scholar]
- Yoshii M, Nishikiori M, Tomita K, Yoshioka N, Kozuka R, Naito S, Ishikawa M (2004) The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J Virol 78 6102–6111 [DOI] [PMC free article] [PubMed] [Google Scholar]