Abstract
Recently, the NADPH-dependent short-chain dehydrogenase/reductase (SDR) salutaridine reductase (E.C. 1.1.1.248) implicated in morphine biosynthesis was cloned from Papaver somniferum. In this report, a homology model of the Papaver bracteatum homolog was created based on the x-ray structure of human carbonyl reductase 1. The model shows the typical α/β-folding pattern of SDRs, including the four additional helices αF′-1 to αF′-4 assumed to prevent the dimerization of the monomeric short-chain dehyrogenases/reductases. Site-directed mutagenesis of asparagine-152, serine-180, tyrosine-236, and lysine-240 resulted in enzyme variants with strongly reduced performance or inactive enzymes, showing the involvement of these residues in the proton transfer system for the reduction of salutaridine. The strong preference for NADPH over NADH could be abolished by replacement of arginine residues 44 and 48 by glutamic acid, confirming the interaction between the arginines and the 2′-phosphate group. Docking of salutaridine into the active site revealed nine amino acids presumably responsible for the high substrate specificity of salutaridine reductase. Some of these residues are arranged in the right position by an additional αE′ helix, which is not present in SDRs analyzed so far. Enzyme kinetic data from mutagenic replacement emphasize the critical role of these residues in salutaridine binding and provide the first data on the molecular interaction of benzylisoquinoline alkaloids with enzymes.
The short-chain dehydrogenases/reductases (SDRs) constitute a large protein family catalyzing NAD(P)(H)-dependent oxidation/reduction reactions. At present, roughly 3,000 members are known from all living organisms and they exhibit a wide substrate spectrum, including alcohols, sugars, steroids, aromatic compounds, and xenobiotics (Kallberg et al., 2002). SDRs consist of a one-domain subunit of about 250 amino acids with the cofactor binding site in the N-terminal part and substrate binding in the C-terminal part (Jörnvall et al., 1995). The two main characteristics of this protein family are the highly conserved TGxxxGhG motif for coenzyme binding and the YxxxK motif, which, together with an upstream Ser residue, represents the catalytic center (Oppermann et al., 1997). In this catalytic triad, the Lys lowers the pKa of the Tyr hydroxyl group, which functions as the catalytic base, whereas the Ser stabilizes the substrate (Jörnvall et al., 1995). Additionally, an Asn residue has recently been proposed to stabilize the position of the catalytic side Lys, thereby forming a proton relay system involving water (Filling et al., 2002). Apart from these classical SDRs, there are the extended, divergent, intermediate, and complex SDRs, which show slightly altered amino acid sequences in the conserved regions compared to the classical ones (Kallberg et al., 2002). The classical SDRs can be further categorized according to their cofactor preference and are named either cD, in case they prefer NAD(H), or cP, if they prefer NADP(H). The same is valid for the extended SDRs, which fall either in group eD or group eP, respectively (Persson et al., 2003).
Several SDR enzymes have been shown to be involved in the biosynthesis of different secondary metabolites in plants. Thus, secoisolariciresinol dehydrogenase in podophyllotoxin biosynthesis as well as (−)-isopiperitenol dehydrogenase (ISPD), (−)-isopiperitenone reductase (ISPR), (−)-menthone:(−)-menthol (MMR) and (−)-menthol:(+)-neomenthol reductase (MNR), all acting in the monoterpenoid pathway leading to menthol, have been shown to belong to the classical SDRs (Xia et al., 2001; Ringer et al., 2003, 2005; Davis et al., 2005). The most thoroughly investigated SDRs are involved in the biosynthesis of tropane alkaloids. In this pathway, two SDRs exhibiting 64% amino acid identity with different product specificities have been identified. Tropinone reductase I (TRI) catalyzes the reduction of the keto group in tropinone to the hyoscyamine precursor tropine, and TRII reduces tropinone at the same position to Ψ-tropine leading to calystegines (Dräger, 2006). Determination of the x-ray structure and site-directed mutagenesis revealed the decisive amino acid residues conferring the product specificities of TRI and TRII (Nakajima et al., 1998, 1999). In contrast to the TRs, data describing structure and mode of substrate binding for other SDRs in plant secondary metabolism are not available.
Recently, the cDNA of salutaridine reductase (SalR), reducing the keto group of salutaridine to a hydroxyl as an intermediate step in morphine biosynthesis, was isolated and identified as a member of the classical SDRs with preference for NADPH as cofactor (Ziegler et al., 2006). The presence of two basic residues downstream of the coenzyme binding motif classifies SalR into the family cP3 according to Persson et al. (2003). The recombinant enzyme showed exclusive substrate specificity toward salutaridine as well as strict product specificity, forming only 7-(S)-salutaridinol and not its stereoisomer 7-epi-salutaridinol, which is a prerequisite for subsequent reactions in morphine biosynthesis. Whereas the TRs and ISPD have been shown to occur as homodimers in their native form (Ringer et al., 2005; Dräger, 2006), SalR as well as ISPR, MMR, and MNR are active as monomers (Ringer et al., 2003; Davis et al., 2005; Ziegler et al., 2006). Therefore, SalR showed a much higher similarity to ISPR, MMR, and MNR of 60% than to ISPD and the TRs with 20%.
In addition to SalR, cDNAs encoding seven enzymes of morphine biosynthesis have been cloned and characterized (Kutchan, 1998; Facchini, 2001; Hashimoto and Yamada, 2003; Ikezawa et al., 2003; Ounaroon et al., 2003; Samanani et al., 2004). Despite this large number of cDNAs, structural information on enzymes of benzylisoquinoline metabolism has only recently been obtained for the aldo-keto reductase codeinone reductase (Bomati et al., 2005). Based on the x-ray structure of chalcone reductase, a model of codeinone reductase was created; however, data on substrate binding have not been reported. Generally, the mode of binding to enzymes of this economically important class of compounds is not known. Knowledge about substrate binding to enzymes in benzylisoquinoline biosynthesis might help in the modification of substrate specificities in order to improve or invent new biomimetic synthetic steps for the synthesis of pharmaceutically important alkaloids.
In this study, we constructed a model of SalR by homology modeling based on its similarity to the structure of the previously crystallized SDR human carbonyl reductase 1 (HsCbr1; Tanaka et al., 2005). Furthermore, the substrate salutaridine as well as the cofactor NADPH were docked into the protein model. These data were then taken as a basis to elucidate the reaction mechanism, the cofactor preference, as well as the substrate binding by site-directed mutagenesis.
RESULTS
Comparative Modeling of SalR
Since overexpression of SalR from Papaver somniferum, described in Ziegler et al. (2006), resulted in low quantities of protein, the enzyme was cloned by reverse transcription-PCR from Papaver bracteatum and subsequently overexpressed as described for the P. somniferum cDNA. Although the P. bracteatum SalR only showed 13 amino acid substitutions compared to P. somniferum, overexpression and subsequent purification by His-tag affinity chromatography resulted in high quantities of purified protein with a yield of 10 mg/L bacterial culture. Since this facilitates the purification and characterization of the mutant proteins generated by site-directed mutagenesis, the model of the tertiary structure was created from the P. bracteatum sequence.
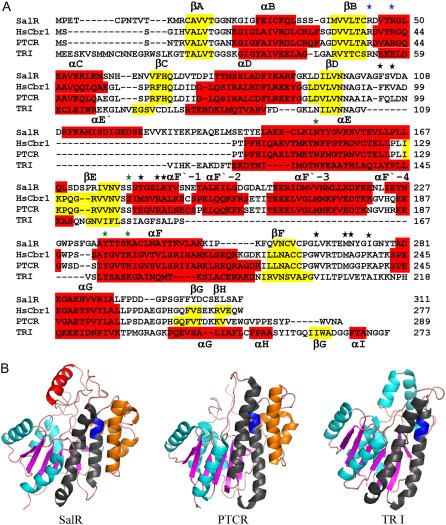
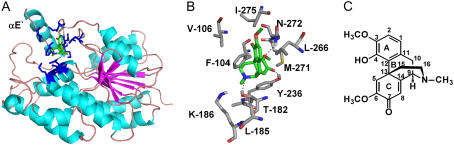
The amino acid sequences of P. bracteatum SalR showed 35% and 31% identity to the previously crystallized SDRs HsCbr1 (Tanaka et al., 2005) and porcine testicular carbonyl reductase (PTCR; Ghosh et al., 2001), respectively. Because of the higher similarity to the enzyme from humans, this was taken as the template for the SalR model. The model consists of six parallel β-sheets, which are each flanked on each site by three parallel α-helices (Fig. 1A). The βA-βF segment represents a double tortuous α/β-motif with alternating β-sheets and α-helices, and forms the classical Rossman fold for cofactor binding, which is typical for the SDRs. This pattern of secondary elements aligns almost perfectly to the one observed in HsCbr1 and PTCR. In contrast to the dimeric SDRs, such as TRI, the monomeric SalR, HsCbr1, and PTCR showed an insertion of about 45 amino acids between βE and αF, which mainly form helices denoted as αF′-1 to αF′-4 (Fig. 1A). These four helices obstruct access to the αE and αF helices, which represent the surface for the interaction of the dimeric SDRs (Ghosh et al., 2001; Fig. 1B). It is therefore assumed that these additional helices cause the monomeric nature of HsCbr1, PTCR, and SalR. Compared to the animal enzymes, there is an additional insertion of about 30 amino acids preceding αE. Since this element partially forms an α-helix with 14 amino acids, it was named αE′.
Figure 1.
A, Amino acid alignment of SalR from P. bracteatum with HsCbr1 (pdb code: 1wma), PTCR (pdb code: 1n5d), and TRI from Datura stramonium (pdb code: 1ae1). α-Helices are highlighted in red and β-sheets in yellow. The numbering of the structural elements complies with Ghosh et al. (2001). The stars indicate the residues implicated in cofactor preference (blue), catalysis (green), or that are putatively involved in salutaridine binding (black). B, Ribbon diagrams showing, from left to right, the tertiary structures of SalR, PTCR, and TRI. Turquoise: α-Helices; magenta: β-sheets; blue: the catalytic site YxxxK motif; gray: the αE and αF helices responsible for the dimerization in TRI; orange: the αF′-1 to αF′4 helices putatively preventing dimerization of SalR and PTCR; red: the αE′ helix in SalR.
Characterization of SalR from P. bracteatum
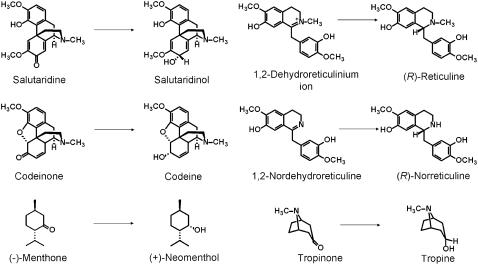
The open reading frame of the SalR cDNA from P. bracteatum consisted of 936 bp, which coded for a protein of 311 amino acids with a calculated molecular mass of 34.05 kD and an pI of 4.72. The recombinant His-tagged version of the protein exhibiting a calculated molecular mass of 36.6 kD was very efficiently produced in Escherichia coli and could, therefore, be purified to homogeneity by cobalt affinity chromatography with a high yield of 10 mg/L bacterial culture. Only a single protein band was detectable on SDS-PAGE at 36 kD, confirming the purity and intactness of the recombinant protein (Supplemental Fig. S1). Enzyme assays using salutaridine as substrate only yielded 7(S)-salutaridinol as product, not the stereoisomer 7-epi-salutaridinol. Similarly, the reverse reaction from salutaridinol to salutaridine was only performed with the 7(S)-derivative. Other compounds, such as the intermediates in morphine biosynthesis, codeinone and dehydroreticulinium ion, or the N-demethylated derivative of the latter, nordehydroreticuline, were not converted by P. bracteatum SalR. Similarly, no conversion of tropinone and (−)-menthone, both substrates for SDR enzymes in tropane alkaloid or monoterpenoid biosynthetic pathways, respectively, was detected (Fig. 2). The enzyme exhibited a broad temperature optimum around 40°C for both reactions. The pH optimum was at pH 6 for the reduction, with a steep decrease by 90% of activity within 1.5 pH units on both sides of the optimum, whereas the oxidation was most efficient at pH 9.5, with a slightly shallower decrease toward more acidic pH values showing 15% residual activity at pH 6.
Figure 2.
Structures of compounds tested for SalR specificity. The compounds and the expected reduced products are shown.
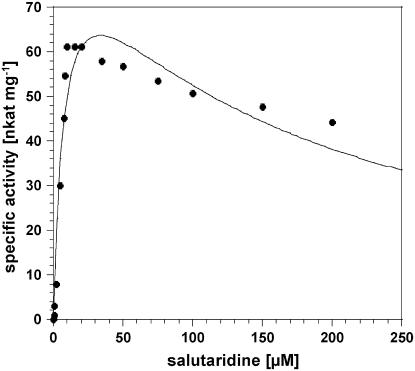
Determinations of the reaction velocity at different salutaridine concentrations showed that the enzyme does not obey Michaelis-Menten kinetics, but is instead subjected to substrate inhibition with a decrease in activity at a substrate concentration higher than 20 to 30 μm (Fig. 3). Extraction of the kinetic parameters yielded Km and Ki values for salutaridine of 7.9 μm and 140 μm, respectively, and a theoretical Vmax of 93 nkat mg−1 (Table I). Since this value can never be even approximately achieved because of strong substrate inhibition, the optimum velocity, Vopt, is taken as the measure of enzyme performance, which was calculated to 63 nkat mg−1. This value is also considered in the determination of Kcat and the catalytic efficiencies Kcat Km−1 throughout the data presented in this report. In contrast to the reduction reaction, the data for the oxidation reaction as well as for the cofactors showed the hyperbolic pattern according to Michaelis-Menten kinetics, and the Kcat and Kcat Km−1 values are based on Vmax. The kinetic data revealed a 4-fold higher affinity toward salutaridinol compared to salutaridine and a 9-fold higher reaction velocity for the reverse reaction, summing up to an almost 50-fold higher catalytic efficiency for the oxidation compared to the reduction. The higher catalytic efficiency for the oxidation reaction has already been reported for the native as well as the recombinant SalR from P. somniferum. A similar trend was observed for the cofactors with 10- and 5-fold higher Vmax and catalytic efficiency, respectively, for NADP, but a 2-fold lower affinity for NADP compared to NADPH.
Figure 3.
Reaction velocity of SalR versus substrate concentration showing substrate inhibition at low salutaridine concentrations.
Table I.
Kinetic parameters of SalR of P. bracteatum for the forward and reverse reaction and the cofactors
| Substratesa
|
Cofactorsbc
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Km | Ki | Vmax | Vopt | Kcat | KcatKm−1 | Km | Vmax | Kcat | KcatKm−1 | |
| μm | μm | nkat mg−1 | nkat mg−1 | s−1 | s−1m−1 103 | μm | nkat mg−1 | s−1 | s−1m−1 103 | |
| Salutaridine | 7.9 ± 2.8 | 140 ± 57 | 93.8 ± 15 | 63.2 ± 8 | 2.3 ± 0.3 | 291 | 3.5 ± 0.8 | 57.5 ± 2.3 | 2.1 ± 0.1 | 600 |
| Salutaridinol | 1.5 ± 0.2 | – | 588 ± 16 | – | 21.6 ± 0.6 | 14,400 | 7.0 ± 0.9 | 598 ± 17 | 21.9 ± 0.6 | 3,128 |
NADPH as cofactor for salutaridine, NADP for salutaridinol, 250 μm each.
NADPH as cofactor for salutaridine; salutaridine concentration 10 μm.
NADP as cofactor for salutaridinol; salutaridinol concentration 50 μm.
Cofactor Preference and Catalytic Active Site of SalR
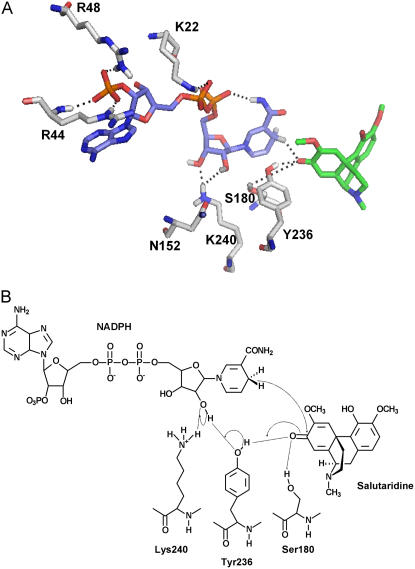
Comparing the reaction velocities for the forward reaction using either NADPH or NADH as cofactor showed an approximately 70% higher Kcat value for the nonphosphorylated cofactor. However, the affinity was more than 300-fold higher toward the phosphorylated counterpart, resulting in a 200-fold higher catalytic efficiency for NADPH compared to NADH (Table II). According to Persson et al. (2003), the preference for the phosphorylated cofactor is caused by the presence of basic residues, either Arg or Lys, either in the TGxxxGhG motif or at the C-terminal end of the second β-sheet. In the SalR model, the negatively charged 2′-phosphate group of NADPH is surrounded by two Arg residues, Arg-44 and Arg-48 (Fig. 4A). Their guanidinium groups are believed to form specific interactions, either by hydrogen bonding or by electrostatic forces, to the phosphate group. To evaluate the importance of these residues for the NADPH preference of SalR, we exchanged the Args by Glu and by a Lys residue. With a decrease in affinity by a factor of more than 2 for NADPH and an increase in affinity by 40% for NADH, the substitution of Arg-48 for Lys only resulted in subtle changes in the catalytic efficiency. However, introducing the negatively charged amino acid Glu in this position resulted in a 20-fold decrease in catalytic efficiency for NADPH, which was based on the large decrease in affinity for this cofactor. Nevertheless, a reversal in cofactor specificity was not observed since this enzyme variant only exhibited slightly increased affinity for NADH compared to the wild-type enzyme, but a reduction in Kcat by 50%. Replacing Arg-44 by Glu resulted in almost equal catalytic efficiencies for both cofactors. This was most attributable to a strongly reduced affinity and reaction velocity for NADPH, whereas the kinetic parameters for NADH only changed slightly.
Table II.
Kinetic parameters with respect to the cofactors of mutated variants of SalR designed for the investigations on cofactor preference
| Variant | Cofactor | Km | Vmax | Kcat | KcatKm−1 |
|---|---|---|---|---|---|
| μm | nkat mg−1 | s−1 | s−1m−1 103 | ||
| Wild type | NADPH | 3.5 ± 0.8a | 57.3 ± 2.3 | 2.1 ± 0.093 | 600 |
| NADH | 1,190 ± 43b | 102 ± 1.1 | 3.7 ± 0.04 | 3.1 | |
| R48K | NADPH | 9.4 ± 1.2c | 60.6 ± 1.7 | 2.2 ± 0.06 | 234 |
| NADH | 645 ± 72b | 77 ± 2.4 | 2.8 ± 0.09 | 4.3 | |
| R48E | NADPH | 104 ± 10d | 95 ± 3.3 | 3.5 ± 0.12 | 33.6 |
| NADH | 884 ± 141e | 45.8 ± 2.4 | 1.7 ± 0.08 | 1.9 | |
| R44E | NADPH | 187 ± 29d | 18.7 ± 1.02 | 0.7 ± 0.03 | 3.7 |
| NADH | 1,257 ± 91f | 159 ± 3.7 | 5.8 ± 0.14 | 4.6 |
Salutaridine concentration 10 μm.
Salutaridine concentration 100 μm.
Salutaridine concentration 20 μm.
Salutaridine concentration 50 μm.
Salutaridine concentration 150 μm.
Salutaridine concentration 250 μm.
Figure 4.
A, Amino acid residues conferring cofactor preference and participating in the catalytic cycle. The colors for the C-skeleton of each structure are: green, salutaridine; light blue, NADPH; and gray, amino acids. Hydrogen, nitrogen, oxygen, and phosphorus atoms are shown in white, blue, red, and orange, respectively. Only interacting hydrogen atoms are shown. Interactions are indicated by a dotted line. B, Schematic drawing of the catalytic cycle. Interactions are indicated by a dotted line and the proton transfer system by arrows.
Up to now, the catalytic mechanism of the SDRs was exclusively investigated in mammalian, insect, or bacterial systems showing the involvement of a catalytic triad consisting of Ser, Lys and Tyr in the transfer of hydrogen from NAD(P)H to the substrate (Obeid and White, 1992; Chen et al., 1993; Oppermann et al., 1997). Recently, a conserved Asn residue in the αE helix was elucidated to interact with active site residues in a bacterial steroid dehydrogenase (Filling et al., 2002). Based on the sequence alignment, the catalytic motif YxxxK in SalR can be localized to the αF helix with the Tyr at position 236 (Fig. 1A). However, at the presumed position for the Ser residues, three Sers at positions 179 to 181 are present in SalR. The homology model shows the close proximity of Tyr-236 and Lys-240 to the substrate and the cofactor (Fig. 4A). Of the three Ser residues, only the side chain of Ser-180 points to the catalytic center, whereas the hydroxyl groups of the other Sers face away from the substrate and the cofactor (data not shown). Furthermore, the carbonyl group of the side chain of Asn-152 forms a hydrogen bond to the side chain of Lys-240. Asn-152 was a likely candidate to participate in the catalytic cycle as proposed by Filling et al. (2002). These residues were chosen for site-directed mutagenesis to evaluate their importance in the catalytic cycle of SalR. Substitution of Tyr-236 for Phe and of Lys-240 for Glu resulted in complete loss of enzyme activity (Table III). The enzyme variant with an Ala exchange for Ser-180 still showed activity, but the catalytic efficiency dropped to 0.3% of the original level. This was mainly due to the dramatic decrease in the catalytic constant from 2.3 s−1 to 0.0025 s−1, but also the interaction of salutaridine with the enzyme was weakened by a factor of 3. In contrast, the Km for the substrate remained unchanged in the N152A variant, and only the reaction velocity decreased by a factor of 6, resulting in 15% of the catalytic efficiency compared to the wild-type SalR. The kinetic parameters for the cofactor showed a similar trend with a decrease in affinity by a factor of 3 and a strongly reduced Vmax value of 0.091 s−1 for the S180A variant. Unlike for the substrate, the N152A substitution resulted, in addition to a similar decrease in Vmax, in a reduced affinity toward the cofactor by a factor of 3 (data not shown).
Table III.
Kinetic parameters with respect to the salutaridine of mutated variants of SalR designed for the investigations on catalytic site residues and substrate binding
NADPH concentration 250 μm.
| Variant | Km | Ki | Vopt | Kcat | KcatKm−1 |
|---|---|---|---|---|---|
| μm | μm | nkat mg−1 | s−1 | s−1m−1 103 | |
| Wild type | 7.9 ± 2.8 | 140 ± 57 | 63.2 ± 8 | 2.3 ± 0.3 | 291 |
| N152A | 7.9 ± 2.6 | 229 ± 81 | 9.8 ± 1.3 | 0.36 ± 0.05 | 46 |
| S180A | 25.3 ± 3.6 | 179 ± 19 | 0.068 ± 0.01 | 0.0025 ± 0.0002 | 0.099 |
| K240E | Inactive | ||||
| Y236F | Inactive | ||||
| V106A | 9.4 ± 1.1 | 168 ± 21 | 121 ± 0.9 | 4.5 ± 0.3 | 479 |
| F104A | 52.5 ± 6.2 | 203 ± 33 | 123 ± 10 | 4.5 ± 0.3 | 86 |
| L266A | 96 ± 6.9 | 840 ± 82 | 34.2 ± 1.6 | 1.3 ± 0.06 | 13 |
| L266S | 98.3 ± 6.3 | 1,356 ± 143 | 30.0 ± 1.1 | 1.1 ± 0.05 | 11 |
| L266V | 44.5 ± 3.8 | 1,423 ± 197 | 50.3 ± 2.3 | 1.8 ± 0.09 | 40 |
| M271T | Inactive | ||||
| N272T | Inactive | ||||
Mode of Substrate Binding of SalR
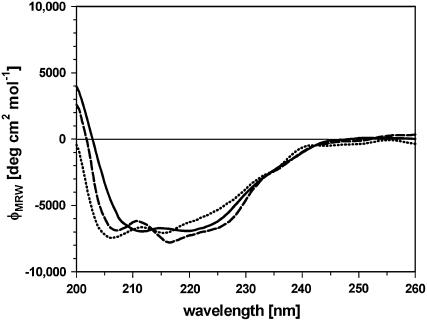
The docking arrangement of salutaridine in the active site of SalR shows that rings B and C of the substrate are completely buried in the enzyme, whereas ring A is pointing to the periphery (Fig. 5A). The docking revealed that the binding site is formed by amino acids residing in the loops between βD and αE′, and βF and αG, as well as in the αF′-1 helix. Most important, the carbonyl group of salutaridine forms a hydrogen bond to the hydroxyl group of the catalytic active Tyr-236 and the carbonyl carbon is in close proximity of 3.4 Å to the transferred hydrogen of the cofactors. Further recognition sites are Leu-266 forming strong hydrophobic interactions with the aromatic ring system A, the cyclohexane ring B, and the cyclohexadiene ring C containing the carbonyl group to be reduced (Fig. 5B). Weaker hydrophobic interactions can be observed between the side chain Phe-104 and ring C, the methoxy group at position 6, and carbons 15 and 16 of the piperidine ring. Further hydrophobic interactions are imaginable between Leu-185 and the N-methyl group, and Met-271 and Ile-275 and the methoxy groups at positions 6 and 3, respectively, as well as between Val-106 and carbon 15. Furthermore, a hydrogen bond is formed between the side chain of Asn-272 and the methoxy oxygen atom at position 6 and between Thr-182 and the nitrogen atom of the substrate. An interaction with the nitrogen atom also exists with Lys-186. Based on the interactions proposed by the model, several amino acids were exchanged to reveal their importance for substrate binding experimentally. Whereas the substitution of Val-106 for Ala resulted in an increased catalytic efficiency, which was mainly based on the higher turnover number, the F104A variant exhibited a 3-fold lower efficiency (Table III). This was mainly due to the 6-fold lower affinity toward the substrate, despite the doubling of the catalytic constant. The efficiency of the enzyme was also decreased by substitutions at position 266. Shortening the hydrophobic side chain of Leu by replacement with Ala, or introducing a hydrophilic group by a Ser residue, increased the Km more than 10-fold and decreased the turnover rate by 50%, resulting in catalytic efficiency to 5% of the wild-type enzyme. Interestingly, when Leu was replaced by an amino acid with a longer hydrophobic side chain than Ala, the efficiency increased by a factor of 4 compared to the L266A substitution. However, whereas the catalytic constant approached wild-type levels, the L266V variant exhibited a more than 5-fold lower affinity for salutaridine. In most of the variants, a decrease in affinity toward the substrate was accompanied by an increase in the Ki value for the substrate inhibition. The kinetic parameters for the cofactor showed increases in Km values for NADPH by factors of 2 and 3 in the cases of the Leu and Phe substitutions, respectively. The Vmax for NADPH was roughly doubled in the Val-106 and Phe-104 variants, whereas it was decreased by 50% in L266A and L266S (data not shown). Substitutions of Met-271 and Asn-272 for Thr rendered the enzyme inactive. However, CD spectra indicate an overall change in the structure of both mutated proteins compared to the wild-type enzyme (Fig. 6). Whereas the two minima at 212 and 220 represent the dominating proportion of α-helical elements in the wild-type protein, the minimum at 205 nm for M271T and N272T indicated a shift toward more random coil elements. However, the replacement of Met-271 for Ala or Val, or Asn-272 for Ala, also resulted in an active enzyme (data not shown). All enzyme variants were investigated for a possible change in substrate specificity and stereospecificity. None of the proteins showed any conversion of salutaridine to 7-epi-salutaridinol. Similarly, all SalR variants kept the substrate specificity observed for the wild type, in that no conversion of codeine, dehydroreticulinium ion, nordehydroreticuline, tropinone, or menthone took place (data not shown).
Figure 5.
Substrate binding of SalR. A, Ribbon diagrams showing the tertiary structure of SalR indicating the position of the substrate binding side. Residues shown in B are highlighted in blue. B, Close-up view of salutaridine binding site. The colors for the C-skeleton of each structure are green for salutaridine and gray for amino acids. Hydrogen, nitrogen, oxygen, and sulfur atoms are shown in white, blue, red, and yellow, respectively. Only interacting hydrogen atoms are shown. Interactions are indicated by dotted lines. C, Structure of salutaridine indicating carbon and ring numbering.
Figure 6.
CD spectra of wild-type SalR (solid line), M271T (dashed line), and N272T (dotted line).
DISCUSSION
In this work, a model of the tertiary structure of the monomeric SalR was created based on the homology to mammalian SDRs of experimentally resolved structure. The model shows the typical alternating α-helix/β-sheet pattern of the SDRs with the four helices αF′-1 to αF′-4 specific for monomeric SDR like PTCR and HsCbr1 (Ghosh et al., 2001; Tanaka et al., 2005). These helices are believed to prevent the interaction of the αD helix of the Drosophila melanogaster-related oligomeric SDRs. Attempts to substantiate this suggestion experimentally failed since purification of the protein without this amino acid stretch could not be achieved. The presence of all amino acids in favored or allowed regions, respectively, indicates the high quality of the model. The correctness of the cofactor and substrate docking is proven by site-directed mutagenesis of presumed catalytic site residues. The keto group to be reduced is located in close proximity to Tyr-236 and the hydrogen of the nicotinamide ring (Fig. 4A). Furthermore, the alignment of NADPH suggests the transfer of the proton from the B site of the nicotinamide ring. The selective transfer of this hydridion had been previously shown by Gerardy and Zenk (1993) by incubation of partially purified SalR from P. somniferum extracts with tritiated cofactor. The correct spatial arrangement of other active site residues is underlined by inactivation or strong reduction of SalR activity after S180A, K240E, and Y236F substitutions, amino acid residues well established to form the catalytic triad of SDRs (Oppermann et al., 1997). The residual activity of the S180A mutation, which led to complete loss of activity in other SDRs, suggests that the stabilization of the salutaridine intermediate can be fulfilled by the neighboring hydroxylated amino acid residues Ser-179, Ser-181, and Thr-182. Based on the distance to the keto group, Ser-181 is the most likely candidate for the formation of a hydrogen bridge to the substrate, although the distance with 5.1 Å is quite large. The 7-fold-reduced catalytic efficiency of the N152A variant is an indication of the participation of this conserved residue within the αE helix in the proton transfer system involving water molecules, Lys-240, the 2′-hydroxy group of the Rib moiety of NADPH, and Tyr-236 also in plant SDRs, as has been proposed for bacterial hydroxysteroid reductase by Filling et al. (2002). However, whereas substitution for Leu in the bacterial enzyme resulted in complete loss of activity, the N152A variant of SalR was still active. Since the side chain of Ala does not form hydrogen bridges, the involvement of the carbonyl oxygen of the peptide bond at position 152 in the establishment of the proton transfer system is possible. Alternatively, the side chain of Asn-152 might interact via a water molecule with the side chain of Lys-240, thereby supporting the positioning of this active site residue. This could explain the decreased activity of N152A. The amino group of Lys-240 is located in a distance of 2.2 Å to both hydroxy groups of the Rib moiety of NADPH, indicating their participation in the proton transfer system (Fig. 4A). However, the partial negative charge of the reduced carbonyl group of the substrate after hydride transfer might lead to a slight torsion of the Lys-240 side chain, so that also a direct interaction of this residue with salutaridine is conceivable.
SalR shows strong preference for NADPH compared to NADH and can be grouped into the class cP3 according to Persson et al. (2003). This classification is based on conserved basic residues among NADPH preferring SDRs in the TGxxxGhG motif or/and at the end of the second β-sheet. Many residues in this region have been shown to be involved in cofactor binding and discrimination. The importance of basic residues at the end of βB has been shown by site-directed mutagenesis for the NADH preferring D. melanogaster alcohol dehydrogenase, where the exchange of an Asp for an Asn and of an Ala for an Arg led to an increase in NADPH specificity (Chen et al., 1991, 1994). The reverse approach, i.e. increasing NADH specificity in an NADPH preferring SDR, has been performed by replacement of an uncharged residue by a negatively charged residue (Huang et al., 2001). However, the effect on removal of positive charges at the end of βB has not been investigated to our knowledge. In the SalR structure, Arg-44 and Arg-48, showing the closest proximity to the phosphate group, are likely candidates conferring cofactor selectivity. The importance of these residues is evident by the strong decrease in affinity to NADPH when both residues are substituted with residues of opposite charges. The most dramatic effect was observed by the R44E variant, which may be due to the repulsion between the negatively charged Glu and the phosphate group. Although, according to the model, the amino group of the Arg-44 peptide bond might also interact with the phosphate group, this is obviously not sufficient to compensate for the change in charge inferred by Glu. The less pronounced effect of R48E can be explained by the larger distance of the guanidinium group to the cofactor. Only the R44E variant showed marginally higher catalytic efficiency for NADH compared to NADPH, whereas the affinities for NADH only increased in the R48K mutation. Provided no change in the spatial arrangement of the backbone, it is conceivable that an interaction of the carboxyl groups with the 2′-hydroxy of NADH cannot take place because of the shorter length of the Glu side chain. Although our results show that Arg at position 44 is very critical for NADPH binding, other residues have been reported to interact with the cofactor and specifically with NADPH (Ghosh et al., 2001; Huang et al., 2001). These are basic residues in the TGxxxGhG motif, which are occupied by Asn-21 and Lys-22 in SalR. Further experiments might show whether substitutions of several basic charges for negative ones in the same protein are necessary to reverse the cofactor specificity of SalR.
Compared to the rather unspecific hydroxysteroid reductases PTCR and HsCbr (Tanaka et al., 1992; Nakajin et al., 1997; Ghosh et al., 2001), SalR contains an additional helix between the fourth β-sheet and α-helix (Fig. 1A). The arrangement of this αE′ helix could be due to Ser-105 and Glu-130 side chains preceding and following the helix, which are in close proximity of 2.5 Å to each other and might form hydrogen bonds. Although no direct interaction between amino acids residing in the αE′ helix and salutaridine takes place, its arrangement positions the stretch between βD and αE′ in close proximity to the substrate. On this stretch, Phe-104 could be shown to exert hydrophobic interaction with ring C, the methoxy group at position 6, and carbons 15 and 16 of the piperidine ring of salutaridine (Table III; Fig. 5). The decreased substrate affinity after replacement of this residue with Ala underscores the importance of this interaction. The increased catalytic constant of this variant might be due to a less restricted access of the substrate and a faster release of the product from the active site. On the opposite side, the substrate is bound by strong hydrophobic interactions between Leu-266 and the aromatic ring system A (distance between one Cδ-atom of Leu-266 and the centroid of ring A, 3.4 Å), the cyclohexane ring B, and the cyclohexadiene ring C of salutaridine. According to this suggestion, the weakening of the hydrophobic interaction by introduction of shorter residues decreased the affinity. It is imaginable that the introduction of an Ile at that position would result in slightly lower affinity to the substrate because it should only interact with ring A of the substrate. On the other hand, the substitution for Phe should completely prevent the access of salutaridine to the active site, resulting in an enzyme incapable of converting the substrate. The participation of Met-271 and Asn-272 in substrate binding could not be fully established since substitution of both residues for Thr resulted in inactive enzymes and altered CD spectra, indicating major structural changes in the protein. Although the involvement of both residues in salutaridine binding is conceivable based on the distance of both residues to the methoxy group at position 6 (2.2 Å for Asn-272 and 4.0 Å for Met-271), they most likely exert additional interactions to preserve the structure. According to our model, Met-271 could interact hydrophobically with the Tyr-236 and thereby be responsible for the proper positioning of Leu-266. Similarly, Asn-272 could serve the same purpose by interaction with the backbone hydrogen of Gly-276 (distance: 3.7 Å). Therefore, alteration of these residues could result in a change in protein structure. Alternatively, the substitution for Thr could result in a strong change in polarity and changes in loop conformations. Especially, newly formed interaction of the Thrs with Lys-268 and Glu-270 might have inferred the observed structural changes. Further experiments are necessary to confirm the contribution of Met-271 and Asn-272 to the preservation of the structure of the enzyme and their specificity of substrate binding. Similarly, the substrate binding properties of Thr-182, Leu-185, Lys-186 (forming the bottom of the substrate binding pocket), and Leu-275, presumably interacting with the methoxy group at position 3 of the substrate, deserve closer investigations by site-directed mutagenesis. Nevertheless, the results obtained so far describe and identify for the first time, to our knowledge, critical residues forming the binding pocket for an enzyme involved in benzylisoquinoline metabolism. To our knowledge, the TRs from tropane alkaloid biosynthesis are the only enzymes in plant alkaloid biosynthesis whose structures were analyzed and the substrate binding identified by site-directed mutagenesis (Nakajima et al., 1998; Nakajima et al., 1999). In these studies, critical residues for the stereospecificity of TRI and TRII could be identified, and the specific exchange of amino acids led to the adoption of TRI activity in the TRII protein and vice versa. It was shown that the spatial orientation of the substrate tropinone in the active site pocket was critical for stereospecificity. In SalR, the substrate is tightly packed between Phe-104 and Leu-266. A turn by 180° around the vertical axis of salutaridine would lead to strong spatial interference of the piperidine ring with Leu-266, which would either lead to no substrate binding at all or would dislocate the keto group to be reduced to a distance, where a hydrogen transfer from NADPH would not be possible. Thus, the exclusive stereospecificity of SalR is most likely due to the inability of salutaridine to slide into the active site in another orientation. However, not only Phe-104 and Leu-266 are responsible for the stereospecificity since their replacements by small residues did not result in the generation of 7-epi-salutaridinol. It will be interesting to evaluate the influence of the other amino acids in the binding pocket by site-directed mutagenesis or of the exchange of several residues in one protein.
Additionally, the substrate specificity was unchanged in all the mutations we performed. Although Phe-104 as well as Leu-266 mutations drastically decrease the catalytic efficiency toward salutaridine, presumably by widening the binding pocket, the access of the other tested compounds to the catalytic center might still be prevented. Possibly, the binding pocket is still not large enough after substitutions of single amino acids to accommodate larger molecules, such as codeinone possessing an oxide bridge between carbon 5 of ring C and carbon 4 of ring A (Fig. 2). Additionally, with the keto group at position 6 of ring C in codeinone or with an isopropyl group in ortho position to the keto group in (−)-menthone, it is imaginable that both compounds have to slide much deeper into the binding pocket to approach the keto group to the catalytic Tyr residue for efficient hydrogen transfer as compared to salutaridine. This might be prevented by Thr-182, Leu-185, and Lys-186, forming the bottom of the binding pocket. It will therefore be interesting to see whether substitutions of these amino acids as well as multiple amino acid exchanges will lead to different or broader substrate specificities of the SalR.
MATERIALS AND METHODS
Plant Material
Papaver bracteatum plants were grown outdoors in Saxony-Anhalt, Germany, and stem sections were harvested in early June after petals had dropped.
Cloning and Expression of P. bracteatum SalR
Total RNA was isolated from stems with TRIzol according to the manufacturer's protocol (Invitrogen) and reverse transcribed with an oligo(dT) primer. The open reading frame of SalR was amplified using primers 16B1-5-2 (5′-ATGCCTGAAACATGTCC-3′) and 16B1-3-1 (5′-ATAACTCAAAATGCAGATAGTTCTG-3′) using PCR (30 s 94°C, 35 cycles 30 s 94°C, 30 s 54°C, 2 min 72°C, final elongation for 10 min at 72°C) using Taq-Polymerase (Promega). The resulting band of 1,000 bp was gel purified and cloned into pGEMT (Promega). For overexpression, the cDNA was excised from pGEMT using SphI and PstI and cloned into pQE-31 (Qiagen), sequenced using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems), and transformed into Escherichia coli strain SG13009. Overexpression and extraction of the recombinant protein was performed as described (Ziegler et al., 2006). For characterization of the protein, the fraction eluting between 30 mm and 60 mm imidazole from the cobalt affinity column (Talon; CLONTECH) was used after buffer exchange to 10 mm HEPES, pH 7.5. Purity of the enzymes was checked by SDS-PAGE (12% polyacrylamide) according to Laemmli (1970). Since the protein was purified to homogeneity according to SDS-PAGE, the concentration was determined at 280 nm using the molar absorption coefficient determined on the basis of the amino acid sequence.
Site-Directed Mutagenesis of SalR
The following primer pairs were used to introduce point mutations into the SalR open reading frame: R48K, R48E, R44E: 16B1bracMutf1 5′-GTTTTAACTTGTRRAGATGTAACTRRAGGTCTTGAAGC-3′, 16B1bracMutr1 5′-GCTTCAAGACCTYYAGTTACATCTYYACAAGTTAAAAC; F104A, V106A: 16B1bracMutf2 5′-GGGGTTGCAGGTKYTTCAGHTGATGCTGATCG-3′, 16B1bracMutr2 5′-CGATCAGCATCADCTGAARMACCTGCAACCCC-3′; S180A: 16B1Mutf3 5′-GAATTGTTAATGTTKCAKCATCCACGGGAAGCCTC-3′, 16B1Mutr3 5′-GAGGCTTCCCGTGGATGMTGMAACATTAACAATTC-3′; Y236F, K240A: 16B1Mutf4 5′-CGGAGCTGCATWCACAACATCARAAGCATGTTTG-3′, 16B1Mutr4 5′-CAAACATGCTTYTGATGTTGTGWATGCAGCTCCG-3′; M271T, N272T: 16B1bracMutf4 5′-GTTAAAACAGAARYGRMCTACGGCATTGG-3′, 16B1bracMutr4 5′-CCAATGCCGTAGKYCRYTTCTGTTTTAAC-3′; L266A, L266S, L266V: 16B1bracMutf5 5′-GTGAATTGTGTTTRTCCTGBTKYGGTTAAAACAG-3′, 16B1Mutr5 5′-CTGTTTTAACCRMAVCAGGAYAAACACAATTCAC-3′; and N152A: 16B1bracMutf6 5′-GAATGTCTCAAAATAVHTTACTACGGTG-3′, 16B1bracMutr6 5′-CACCGTAGTAADBTATTTTGAGACATTC-3′ (B = C/G/T, D = A/G/T, H = A/C/T, K = G/T, M = A/C, R = A/G, V = A/C/G, Y = C/T). These primers were used together with the pQE31-SalR plasmid and the proofreading Pfu DNA polymerase (Fermentas) for PCR (94°C for 30 s, 12 cycles 94°C for 30 s, 55°C for 1 min, 68°C for 5 min). Subsequently, the PCR was digested with DpnI for 2 h at 37°C, and 1 μL thereof used to transform the E. coli strain XL1BlueMRF′. Plasmids were purified, sequenced, and introduced for protein overexpression into SG13009 strains. Induction of overexpression, extraction, purification, and quantification of proteins was performed as described for wild-type SalR (Supplemental Fig. S1).
Enzyme Assays and Enzyme Characterizations
The enzyme assays and the reaction product identifications and quantifications were performed as described (Ziegler et al., 2006). The data for the calculation of the kinetic parameters were collected during the initial linear phase of the enzyme reaction, which was achieved by adjusting the incubation times or the amount of enzyme. Typically, the enzyme assays contained 0.2 to 1.4 μg of purified protein and were incubated at 30°C between 20 s and 3 min. Each measurement was conducted at least three times. The kinetic data for the cofactors and the oxidation reaction were calculated using the Michaelis-Menten equation v = Vmax × S/(S + Km) or, for salutaridine because of substrate inhibition, v = Vmax/(1 + Km/S + S/Ki), where S denotes the substrate or cofactor concentration. The optimal velocity was determined by Vopt = Vmax/(1 + 2(Km/Ki)1/2). All calculations were performed using KaleidaGraph software (version 3.6; Synergy Software).
Homology Modeling of SalR
The prerequisite for homology modeling of proteins is the identification of homologous proteins with at least 30% homology for which the x-ray structure was already elucidated. A BLASTP search (Altschul et al., 1990) was performed for the SalR amino acid sequence from P. bracteatum. Except for the first nine residues, for which no alignment could be obtained, the most promising homolog was HsCbr1 (pdb code: 1wma; Tanaka et al., 2005) with 34.9% homology. Using the molecular modeling program MOE (molecular operating environment; Chemical Computing Group), the template sequence was aligned with the x-ray structure of 1wma using the BLOSOM35-substitution matrix (Henikoff and Henikoff, 1992, 1993). Ten structures were generated with subsequent energy minimization using Charmm22 (MacKerell et al., 1998) and Born-Sovation (Pellegrini and Field, 2002). All structures were checked with respect to stereochemical quality with PROCHECK (Laskowski et al., 1993) and for native folding with PROSA II (Sippl, 1990). Some outliers appearing in disallowed regions of the Ramachandran plot were either manually modified or moved to allow regions by short molecular dynamics simulations (10ps, 500 K) and subsequent minimization. Finally, all parameters for a good stereochemical quality of the structures were fulfilled by PROCHECK (84.8% in most favored region, 12.6% in additionally allowed regions, and 2.6% in generously allowed regions; no outliers). Except for a small loop of some 20 residues, the PROSA analysis showed all residues in negative energy area and a combined energy z-score of −9.19, which is in the expected range for a protein with 302 amino acid residues and native fold. The docking arrangement of NADPH was taken over from the template protein by superposition of the backbone structures of the SalR model and of the 1wma x-ray structure, followed by merging the cosubstrate to the model and subsequent energy optimization. The substrate salutaridine was docked to the active site using the automatic docking program GOLD (Genetic Optimized Ligand Docking; Cambridge Crystallographic Data Centre; Jones et al., 1995, 1997; Nissink et al., 2002; Verdonk et al., 2003) with standard values given by the program. From the resulting 30 docking arrangements, the one with expected orientation of the carbonyl group close to NADPH and the active site Tyr (see “Results”) was used for further energy refinement to capture slight induced fit modifications of the protein active site (all amino acid residues of the protein are fixed during the docking procedure).
CD Spectroscopy
CD spectra were recorded at a temperature of 30°C and with protein content between 25 μg mL−1 and 50 μg mL−1 in 10 mm HEPES, pH 7.5, using a J-810 spectral polarimeter (Jasco). All spectra consist of an accumulation of 15 single measurements per sample.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under the accession number EF184229 for P. bracteatum SalR.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. SDS-PAGE of SalR mutant proteins.
Supplementary Material
Acknowledgments
We thank Silvia Wegener for excellent technical assistance. We gratefully acknowledge the help of Dr. Ralf Globik, University of Halle, for his support with CD spectroscopy.
This work was supported by the Deutsche Forschungsgemeinschaft, Bonn (SPP 1152, Priority Program “Evolution of Metabolic Diversity”).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jörg Ziegler (joerg.ziegler@ucalgary.ca).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215 403–410 [DOI] [PubMed] [Google Scholar]
- Bomati EK, Austin MB, Bowman ME, Dixon RA, Noel JP (2005) Structural elucidation of chalcone reductase and implications for deoxychalcone biosynthesis. J Biol Chem 280 30496–30503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Jiang JC, Lin ZG, Lee WR, Baker ME, Chang SH (1993) Site-specific mutagenesis of Drosophila alcohol dehydrogenase: evidence for involvement of tyrosine-152 and lysine-156 in catalysis. Biochemistry 32 3342–3346 [DOI] [PubMed] [Google Scholar]
- Chen Z, Lee W, Chang S (1991) Role of aspartic acid 38 in the cofactor specificity of Drosophila alcohol dehydrogenase. Eur J Biochem 202 263–267 [DOI] [PubMed] [Google Scholar]
- Chen Z, Tsigelny I, Lee WR, Baker ME, Chang SH (1994) Adding a positive charge at residue 46 of Drosophila alcohol dehydrogenase increases cofactor specificity for NADP+. FEBS Lett 356 81–85 [DOI] [PubMed] [Google Scholar]
- Davis EM, Ringer KL, McConkey ME, Croteau R (2005) Monoterpene metabolism. Cloning, expression, and characterization of menthone reductases from peppermint. Plant Physiol 137 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dräger B (2006) Tropinone reductases, enzymes at the branch point of tropane alkaloid metabolism. Phytochemistry 67 327–337 [DOI] [PubMed] [Google Scholar]
- Facchini PJ (2001) Alkaloid biosynthesis in plants: biochemistry, cell biology, molecular regulation, and metabolic engineering applications. Annu Rev Plant Physiol Plant Mol Biol 52 29–66 [DOI] [PubMed] [Google Scholar]
- Filling C, Berndt KD, Benach J, Knapp S, Prozorovski T, Nordling E, Ladenstein R, Jörnvall H, Oppermann U (2002) Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J Biol Chem 277 25677–25684 [DOI] [PubMed] [Google Scholar]
- Gerardy R, Zenk MH (1993) Purification and characterization of salutaridine: NADPH 7-oxidoreductase from Papaver somniferum. Phytochemistry 34 125–132 [Google Scholar]
- Ghosh D, Sawicki M, Pletnev V, Erman M, Ohno S, Nakajin S, Duax WL (2001) Porcine carbonyl reductase. Structural basis for a functional monomer in short chain dehydrogenases/reductases. J Biol Chem 276 18457–18463 [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Yamada Y (2003) New genes in alkaloid metabolism and transport. Curr Opin Biotechnol 14 163–168 [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG (1992) Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA 89 10915–10919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG (1993) Performance evaluation of amino acid substitution matrices. Proteins 17 49–61 [DOI] [PubMed] [Google Scholar]
- Huang Y-W, Pineau I, Chang H-J, Azzi A, Bellemare V, Laberge S, Lin S-X (2001) Critical residues for the specificity of cofactors and substrates in human estrogenic 17β-hydroxysteroid dehydrogenase 1: variants designed from the three-dimensional structure of the enzyme. Mol Endocrinol 15 2010–2020 [DOI] [PubMed] [Google Scholar]
- Ikezawa N, Tanaka M, Nagayoshi M, Shinkyo R, Sakaki T, Inouye K, Sato F (2003) Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel P450 family, from cultured Coptis japonica cells. J Biol Chem 278 38557–38565 [DOI] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC (1995) Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol 245 43–53 [DOI] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC, Leach AR, Taylor R (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267 727–748 [DOI] [PubMed] [Google Scholar]
- Jörnvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry 34 6003–6013 [DOI] [PubMed] [Google Scholar]
- Kallberg Y, Oppermann U, Jörnvall H, Persson B (2002) Short-chain dehydrogenases/reductases (SDRs). Eur J Biochem 269 4409–4417 [DOI] [PubMed] [Google Scholar]
- Kutchan TM (1998).Molecular genetics of plant alkaloid biosynthesis. In G Cordell, ed, The Alkaloids, Vol 50. Academic Press, San Diego, pp 257–316
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26 283–291 [Google Scholar]
- MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102 3586–3616 [DOI] [PubMed] [Google Scholar]
- Nakajima K, Kato H, Oda J, Yamada Y, Hashimoto T (1999) Site-directed mutagenesis of putative substrate-binding residues reveals a mechanism controlling the different stereospecificities of two tropinone reductases. J Biol Chem 274 16563–16568 [DOI] [PubMed] [Google Scholar]
- Nakajima K, Yamashita A, Akama H, Nakatsu T, Kato H, Hashimoto T, Oda J, Yamada Y (1998) Crystal structures of two tropinone reductases: different reaction stereospecificities in the same protein fold. Proc Natl Acad Sci USA 95 4876–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajin S, Tamura F, Takase N, Toyoshima S (1997) Carbonyl reductase activity exhibited by pig testicular 20 β-hydroxysteroid dehydrogenase. Biol Pharm Bull 20 1215–1218 [DOI] [PubMed] [Google Scholar]
- Nissink JW, Murray C, Hartshorn M, Verdonk ML, Cole JC, Taylor R (2002) A new test set for validating predictions of protein-ligand interaction. Proteins 49 457–471 [DOI] [PubMed] [Google Scholar]
- Obeid J, White PC (1992) Tyr-179 and Lys-183 are essential for enzymatic activity of 11 β-hydroxysteroid dehydrogenase. Biochem Biophys Res Commun 188 222–227 [DOI] [PubMed] [Google Scholar]
- Oppermann UC, Filling C, Berndt KD, Persson B, Benach J, Ladenstein R, Jörnvall H (1997) Active site directed mutagenesis of 3β/17β-hydroxysteroid dehydrogenase establishes differential effects on short-chain dehydrogenase/reductase reactions. Biochemistry 36 34–40 [DOI] [PubMed] [Google Scholar]
- Ounaroon A, Decker G, Schmidt J, Lottspeich F, Kutchan TM (2003) (R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum: cDNA cloning and characterization of methyl transfer enzymes of alkaloid biosynthesis in opium poppy. Plant J 36 808–819 [DOI] [PubMed] [Google Scholar]
- Pellegrini E, Field MJ (2002) A generalized-born solvation model for macromolecular hybrid-potential calculations. J Phys Chem A 106 1316–1326 [Google Scholar]
- Persson B, Kallberg Y, Oppermann U, Jörnvall H (2003) Coenzyme-based functional assignments of short-chain dehydrogenases/reductases (SDRs). Chem Biol Interact 143–144 271–278 [DOI] [PubMed] [Google Scholar]
- Ringer KL, Davis EM, Croteau R (2005) Monoterpene metabolism. Cloning, expression, and characterization of (-)-isopiperitenol/(-)-carveol dehydrogenase of peppermint and spearmint. Plant Physiol 137 863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringer KL, McConkey ME, Davis EM, Rushing GW, Croteau R (2003) Monoterpene double-bond reductases of the (-)-menthol biosynthetic pathway: isolation and characterization of cDNAs encoding (-)-isopiperitenone reductase and (+)-pulegone reductase of peppermint. Arch Biochem Biophys 418 80–92 [DOI] [PubMed] [Google Scholar]
- Samanani N, Liscombe DK, Facchini PJ (2004) Molecular cloning and characterization of norcoclaurine synthase, an enzyme catalyzing the first committed step in benzylisoquinoline alkaloid biosynthesis. Plant J 40 302–313 [DOI] [PubMed] [Google Scholar]
- Sippl MJ (1990) Calculation of conformational ensembles from potentials of mean force. An approach to the knowledge-based prediction of local structures in globular proteins. J Mol Biol 213 859–883 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Bateman R, Rauh D, Vaisberg E, Ramachandani S, Zhang C, Hansen KC, Burlingame AL, Trautman JK, Shokat KM, et al (2005) An unbiased cell morphology-based screen for new, biologically active small molecules. PLoS Biol 3: e128 [DOI] [PMC free article] [PubMed]
- Tanaka M, Ohno S, Adachi S, Nakajin S, Shinoda M, Nagahama Y (1992) Pig testicular 20 β-hydroxysteroid dehydrogenase exhibits carbonyl reductase-like structure and activity. cDNA cloning of pig testicular 20 β-hydroxysteroid dehydrogenase. J Biol Chem 267 13451–13455 [PubMed] [Google Scholar]
- Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD (2003) Improved protein-ligand docking using GOLD. Proteins 52 609–623 [DOI] [PubMed] [Google Scholar]
- Xia ZQ, Costa MA, Pelissier HC, Davin LB, Lewis NG (2001) Secoisolariciresinol dehydrogenase purification, cloning, and functional expression. Implications for human health protection. J Biol Chem 276 12614–12623 [DOI] [PubMed] [Google Scholar]
- Ziegler J, Voigtländer S, Schmidt J, Kramell R, Miersch O, Ammer C, Gesell A, Kutchan TM (2006) Comparative transcript and alkaloid profiling in Papaver species identifies a short chain dehydrogenase/reductase involved in morphine biosynthesis. Plant J 48 177–192 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.