Abstract
We evaluated the application of gas chromatography-mass spectrometry metabolic fingerprinting to classify forward genetic mutants with similar phenotypes. Mutations affecting distinct metabolic or signaling pathways can result in common phenotypic traits that are used to identify mutants in genetic screens. Measurement of a broad range of metabolites provides information about the underlying processes affected in such mutants. Metabolite profiles of Arabidopsis (Arabidopsis thaliana) mutants defective in starch metabolism and uncharacterized mutants displaying a starch-excess phenotype were compared. Each genotype displayed a unique fingerprint. Statistical methods grouped the mutants robustly into distinct classes. Determining the genes mutated in three uncharacterized mutants confirmed that those clustering with known mutants were genuinely defective in starch metabolism. A mutant that clustered away from the known mutants was defective in the circadian clock and had a pleiotropic starch-excess phenotype. These results indicate that metabolic fingerprinting is a powerful tool that can rapidly classify forward genetic mutants and streamline the process of gene discovery.
The discovery of gene function through classical genetics remains a powerful approach in the postgenomic era. Identification of mutants via phenotypic screens or suppressor screens retains several advantages over reverse genetic techniques. Prior knowledge of the genes involved is not needed and gene function can be affected in diverse ways through chemically induced point mutations. However, forward genetic approaches also have major limitations. First, the phenotypes used as the basis of a given screen may arise for different reasons, which may or may not relate to the topic studied. When the selected phenotype results from a secondary or pleiotropic change, mutants may be inappropriately selected for analysis, wasting time and resources. Second, bottlenecks remain in the procedures of classification and analysis of such mutants. These limitations restrict the rate of discovery of gene function. Therefore, we sought to develop a method for effective preliminary characterization of classical genetic mutants using starch metabolism in Arabidopsis (Arabidopsis thaliana) as a model system.
In Arabidopsis leaves, starch is synthesized concurrently with Suc during photosynthesis. Suc is exported to support growth and respiration, whereas starch accumulates in leaves as insoluble granules. All starch is remobilized during the following night to sustain metabolism. Reverse genetic studies have cast doubt on supposed key enzymes of starch breakdown, whereas the characterization of starch-excess (sex) mutants (plants still containing starch at the end of the night) has revealed the existence of previously unsuspected steps in the pathway (Zeeman et al., 2007). Continued characterization of novel sex mutants is likely to yield further insight, but progress is limited by the increasing number of genetic loci involved and the possibility of pleiotropic starch-excess phenotypes. We investigated whether metabolic profiling could refine the selection of sex mutants for further genetic analysis.
The use of high-throughput chromatography-linked mass spectrometry (MS) for metabolomic studies has enabled novel approaches that complement the primary technologies of functional genomics (expression arrays and proteomics; Bino et al., 2004). To date, metabolic profiling has been most effectively applied to analysis of organisms that have already been characterized genetically. There are fewer examples of metabolic profiling being used in an exploratory fashion together with classical genetics (Jander et al., 2004; Keurentjes et al., 2006; Schauer and Fernie, 2006). Here, we combine gas chromatography (GC)-MS with classical genetics and show that rapid metabolic profiling can be successfully used to predict which novel sex mutants are most likely to be genuinely affected in the pathway of starch breakdown.
RESULTS AND DISCUSSION
Selection of Mutants and Sampling
To determine the effects of deficiencies in starch metabolism on the metabolic profile of Arabidopsis, we selected two mutants defective in starch biosynthesis, phosphoglucomutase (pgm) and isoamylase2-1 (isa2-1), and four defective in starch degradation, sex1-3, maltose-excess1-1 (mex1-1), disproportionating enzyme2-4 (dpe2-4), and sex4-1. For comparison, we chose the mutant triose phosphate/phosphate translocator (tpt-1) that accumulates starch as a pleiotropic effect. These mutants are described in Supplemental Figure S1. Four uncharacterized sex mutants (Deg172, Deg263, sex3, and Ke103), in which the mutations were unknown, were also analyzed. These lines were originally selected from ethyl methanesulfonate (Deg263 and sex3), x-ray (Deg172), and fast neutron mutagenized (Ke103) populations by staining the leaves for starch using iodine at the end of a dark period. Wild-type plants contain almost no starch at this time and do not stain. Mutants contained elevated starch and stained dark brown to black (data not shown). The sex3 mutation was originally designated as TL54 (Caspar et al., 1991). All mutants are in the Columbia (Col) ecotype background, except tpt (Wassilewskija [Ws]) and Ke103 (RLD).
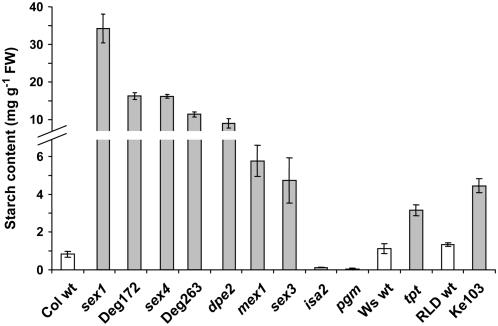
Mutants were grown in short days (8-h photoperiod). In these conditions, the fraction of photoassimilated carbon partitioned into starch is greater than in long days (Chatterton and Silvius, 1980; Gibon et al., 2004) and mutants unable to synthesize or to degrade starch are retarded in growth (Caspar et al., 1985, 1991). Leaf samples were harvested 1 h before the end of the night when the impact of altered nighttime metabolism is expected to be maximal. Wild-type plants, which normally contain around 10 mg starch/g fresh weight at the end of the day, had almost completely degraded their reserves (Fig. 1). As expected, pgm and isa2 contained almost no starch at this point. This is because pgm is unable to make any starch (Caspar et al., 1985), whereas isa2 accumulates predominantly soluble glucan (phytoglycogen), the majority of which is metabolized in the first half of the night (Zeeman et al., 1998b). The other mutants, including the four uncharacterized lines, contained higher levels of starch than the wild types.
Figure 1.
Starch content of leaves of wild-type (white bars) and mutant (gray bars) plants harvested 1 h before the end of the night. The insoluble fractions of perchloric acid extracts were assayed for starch. FW, Fresh weight. Values are the means ± se (n = 4). Note the scale change on the y axis.
Metabolite Profile Analysis
We investigated the metabolite content of four individuals of each genotype. Extracted polar metabolites were subjected to GC-MS analysis as described previously (Roessner et al., 2000, 2001). The nature of each peak in each chromatogram was assessed via its retention time and mass spectrum (e.g. Fig. 2). Over 100 metabolites were detected in each sample, but only reliably detected compounds were used for subsequent comparisons. These encompassed a range of chemical classes but comprised mainly sugars, sugar alcohols, amino acids, and organic acids. The amount of each metabolite was corrected according to an internal standard (ribitol) added to each sample during extraction to account for slight variations in subsequent sample loading.
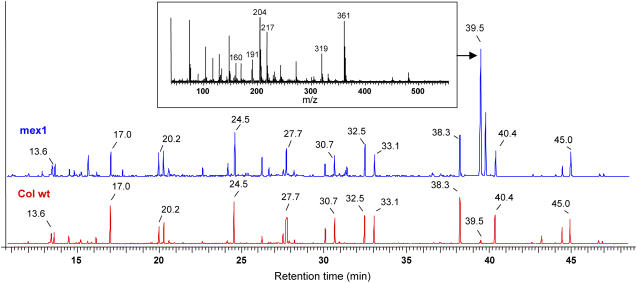
Figure 2.
Example of GC-MS results from Col wild-type (red) and mex1 (blue) extracts. The different metabolites were identified by their retention time and characteristic mass spectra. The detailed mass spectrum of a metabolite found in large amounts in mex1 and in low abundance in the wild type is inset. This peak corresponds to maltose.
Samples were analyzed in three batches. Aliquots of four technical replicate extracts (from pooled Col wild-type plants) were run in each batch, yielding similar results, confirming that the results from the three batches were comparable. The profiles from the individual wild-type and mutant plants (biological replicates) were first normalized with respect to the mean values of the technical replicates run in the respective batch (Supplemental Table S1). This normalization preserves the ecotype-specific variation in metabolites. To compare the mutants of different ecotypes, we performed a second transformation. All replicates (including those of the individual wild types) were normalized with respect to the means of the respective wild-type individuals (Supplemental Table S2).
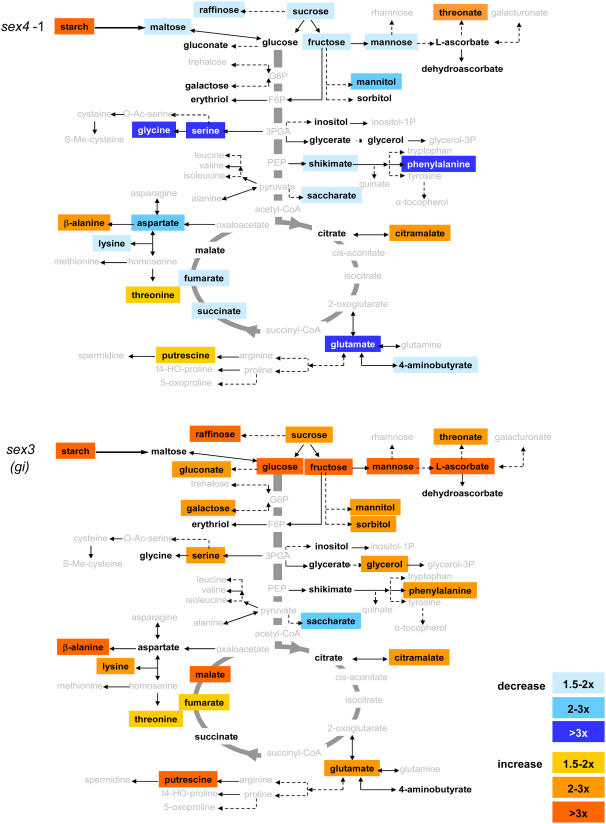
Displaying the changes in metabolite levels (relative to the respective wild type) on a metabolic map revealed distinct patterns of increases and decreases throughout central metabolism in each genotype (e.g. Fig. 3). Of the compounds measured, only Glc and maltose are intermediates of starch metabolism. A point-by-point analysis of the data is outside the scope of this study and it is important to note that these measurements are made on samples taken from a single time point. However, in general, known mutants affected in starch metabolism (e.g. sex4; Fig. 3) had lower levels of sugars that may be derived from starch during the night (e.g. Suc, Glc, Fru, Man). However, sex3, a novel sex mutant, had increased sugars relative to the wild type (Fig. 3). Appreciable variation in the levels of sugar alcohols, amino acids, and free fatty acids was also observed between the different genotypes.
Figure 3.
Metabolic maps of central metabolism displaying the changes in the mean metabolite levels in sex4 and sex3 relative to the wild type (data from Supplemental Table S2). Increases are shown in orange and decreases are shown in blue. The color intensity indicates the magnitude of the changes compared to the wild type as indicated (bottom right). Metabolites in gray were not measured. Metabolites in black were measured but unchanged. Note that not all measured metabolites are visualized on these metabolic maps.
Statistical Analysis
Given the number and complexity of the changes in metabolite levels, we applied unsupervised statistical methods (hierarchical cluster analysis [HCA] and principal component analysis [PCA]; Fiehn et al., 2000; Quackenbush, 2001; Roessner et al., 2001), and developed a supervised algorithm to analyze the data.
The metabolic profiles from individual replicate samples of each genotype (Supplemental Table S2) were analyzed by HCA (Fig. 4). In most cases, biological replicates clustered tightly together, indicating that biological variability for a given genotype was small relative to the difference between genotypes. Repeating the analysis on the raw dataset (including metabolites not reliably detected between technical replicates) or the singly normalized dataset (Supplemental Table S1) did not alter the overall groupings (data not shown). In these cases, the three wild types could be distinguished from each other, but still formed one cluster. Thus, although present, differences between the wild-type ecotypes were minor compared with the differences between the mutants.
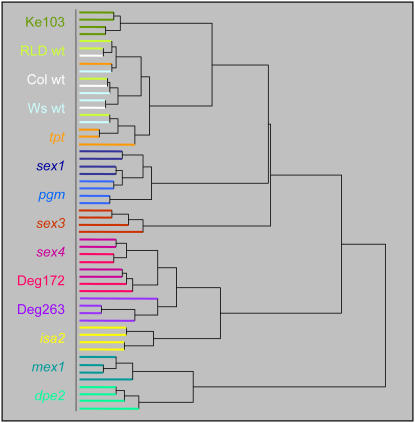
Figure 4.
HCA of metabolic profiles. Normalized metabolite data (Supplemental Table S2) were transformed by log10: analysis was performed with Euclidean distances, using an incremental linkage method to generate the clusters. Each branch represents the metabolite profile of a single biological sample. Colors indicate the genotype, given on the left.
The known starch mutants formed three clusters in HCA (Fig. 4). The first contained mex1 and dpe2, deficient in the export of maltose from the chloroplast (Niittylä et al., 2004) and its subsequent metabolism in the cytosol (Chia et al., 2004; Lu and Sharkey, 2004), respectively (see Supplemental Fig. S1). Given that these are consecutive steps in the same pathway, it is not surprising that their metabolite profiles are similar. The second group contained pgm (starchless phenotype; Caspar et al., 1985) and sex1 (severe starch-excess phenotype; Yu et al., 2001). Although these mutants are opposites in terms of their starch phenotype, the effects of the two mutations on metabolism at the end of the dark period may be similar. In pgm, sugars accumulate during the day, but are very rapidly depleted at night and, by the end of the night, the plant is starved of carbohydrates (Thimm et al., 2004; Bläsing et al., 2005). In sex1, the plant is able to make starch during the day, but cannot remobilize it at night. Thus, we would also expect sex1 plants to be carbohydrate starved at the end of the night. We predict that there would be differences in the metabolic profiles of these genotypes if samples were harvested at the end of the day rather than at the end of the night. The third group contained isa2 (reduced glucan accumulation; Delatte et al., 2005) and sex4. Although depleted in starch when sampled (Fig. 1), isa2 mutants do synthesize an appreciable amount of soluble glucans during the day, which is degraded during the first hours of the subsequent night (Zeeman et al., 1998b; Delatte et al., 2005). Thus, isa2 plants will exhaust their supplies late into the dark period. The sex4 mutant is deficient in a protein or glucan phosphatase involved in the regulation of starch degradation (Niittylä et al., 2006; Worby et al., 2006), although its phosphorylated targets are not yet known. This mutant can degrade its starch, but at a reduced rate compared with the wild type. Interestingly, the metabolic profiles of two of the uncharacterized sex lines (Deg263 and Deg172) fell into this last cluster, whereas the profiles of the other two (sex3 and Ke103) clustered separately from all other lines. The tpt mutant (which accumulates starch as a pleiotropic effect; Schneider et al., 2002) clustered with the wild types. Because starch breakdown is not affected in this mutant, it seems reasonable that the nighttime metabolite profile is normal. During the day, when tpt partitions more carbohydrate into starch, a difference from the wild type might be expected.
Similar groupings were obtained using PCA. Viewed in the dimensions of the first and second principal components, samples separated into three groups (Supplemental Fig. S2A). The first group contained mutants affected in starch metabolism and the two uncharacterized mutants, Deg263 and Deg172. The second group contained the wild types, tpt and Ke103. The third group contained sex3 alone. As with HCA, relationships between the metabolite profiles of mutants affected in starch metabolism (e.g. mex1 and dpe2; sex1 and pgm; isa2 and sex4) could be observed, and further resolution obtained when the data were viewed in the dimensions of the first and third principal components (Supplemental Fig. S2B).
Many of the metabolites had significant weightings in PCA (Supplemental Fig. S2, C and D), indicating that separation of the genotypes is due to changes in many metabolites. Removal of each metabolite individually, followed by reclustering by HCA did not significantly alter the groupings (data not shown), except in the case of maltose. When the maltose data were removed, mex1 and dpe2 still clustered together, but within a group that also contained sex1 and pgm (data not shown). This is consistent with the idea that, being unable to metabolize maltose, mex1 and dpe2 are depleted in metabolites that are derived from starch at night, as would be the case for sex1 and pgm.
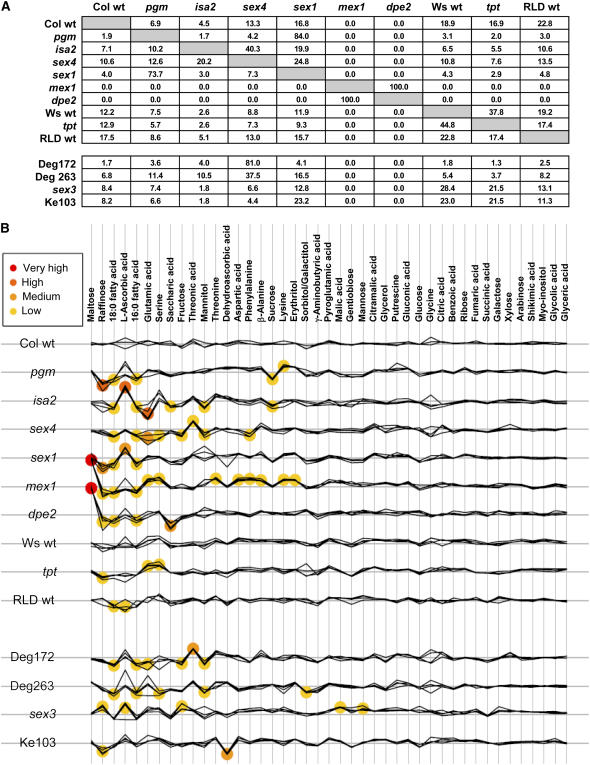
Our supervised algorithm (see “Materials and Methods” and Supplemental Methods) was used to provide independent statistical evaluation of the metabolite data. This approach uses the metabolic profiles of the known mutants and wild types to estimate the probability that each of the unknown mutants is a known type. The analysis also estimates the probability that each of the known genotypes may be classified to each of the other known genotypes, given that it is not to be classified to itself. The resulting probabilities (given as percentages; Fig. 5A) suggest that Deg172 is most likely to be sex4. Similarly, Deg263 is most likely to be sex4, although the percentage is lower. Percentages for sex3 and Ke103 are spread primarily between Ws wild type, RLD wild type, tpt, and sex1, although none is high. Thus, the metabolic profiles of sex3 and Ke103 are sufficiently different from all the others to be defined as new types. The relationships between the known genotypes inferred from the percentages reflect the groupings obtained using the unsupervised methods. Interestingly, mex1 is classified to dpe2 (and vice versa) with certainty (100%) and none of the other genotypes seems like these two. This is consistent with the fact that the mutations in these lines affect consecutive steps in the pathway of starch breakdown. Although the two metabolic profiles are not identical, the model is overconfident in its assignment due to the magnitude of the increase in maltose seen only in these two lines. Figure 5B shows the actual metabolite data, with a scaled increase in the posterior log odds that the metabolite level for a genotype-metabolite combination differs from the baseline. The metabolites are ordered according to the sums of these log odds. Although maltose has the strongest overall change, this is due entirely to the very large individual effects for mex1 and dpe2. Changes in other metabolites are more evenly distributed across types. This analysis usefully identifies the metabolites that contribute most to establishing the relationships between the profiles of the different genotypes. Indeed, using only the 21 metabolites with the highest combined log odds, identified in Figure 5B, to repeat HCA (data not shown) yielded a result very similar to that obtained using all the metabolite data (Fig. 4).
Figure 5.
Probability-based comparisons of the metabolite profiles of wild types and mutants. A, Probabilities (percentages) that each metabolic profile is identified as that of a known type. In the top part of the chart, known types are compared with one another, establishing similarity-based relationships. In the bottom part, the four unknown mutants are compared with the known types. B, Visualization of metabolite changes in each of the wild types or mutants. Biological replicates are plotted together. Deviations from the baseline for each metabolite of each genotype indicate the relative increases or decreases relative to the technical replicate samples. Colored circles centered on specific metabolite-genotype combinations give an indication of its importance to the classification in A.
Mapping the Mutations in Three Novel sex Lines
Statistical analyses of the metabolite data allow us to speculate that Deg263 and Deg172 are mutants with defects in starch metabolism because their metabolite profiles are related to those of known mutants. In contrast, sex3 and Ke103 have metabolite profiles unrelated to those of known mutants, suggesting that they may have defects elsewhere in metabolism, leading to secondary effects on starch levels. Alternatively, sex3 and Ke103 may represent new classes of mutations in which the pathway of degradation is affected in a way not previously described.
We mapped the mutations in Deg172, Deg263, and sex3 to discover the affected genes. The mutation in Deg172 segregated tightly with the marker CIW4 on chromosome 3, close to the SEX4 locus. Deg172 was isolated from the same x-ray mutagenized population as sex4-1 (Zeeman et al., 1998a). Therefore, we investigated whether Deg172 carried the same deletion of the SEX4 gene as sex4-1 using PCR. The reaction with primers for the 5′ region of At3g52180 failed to amplify the same fragment in both sex4-1 and Deg172 (data not shown), consistent with a gene deletion (Niittylä et al., 2006). We crossed Deg172 with sex4-1 and observed no complementation. Thus, we conclude that Deg172 is a reisolate of sex4-1. It is interesting to note that sex4-1 has been backcrossed to the wild type three times, removing approximately 87% of the other mutations, whereas Deg172 has not. Their coclustering shows that the metabolic profile of Deg172 is due mainly or exclusively to the mutation at the SEX4 locus.
The mutation in Deg263 mapped to chromosome 4 between markers NGA8 (26.56 cM) and TL19SE (31 cM). This region contains a gene encoding a starch-debranching enzyme (isoamylase3; At4g09020), recently implicated in starch degradation (Wattebled et al., 2005; Delatte et al., 2006). We sequenced the ISA3 gene in Deg263 and identified a point mutation in exon 11, creating a stop codon. We crossed Deg263 to a T-DNA insertion line for At4g09020 (GABI_KAT_280G10; isa3-2), which also displays a starch-excess phenotype (Delatte et al., 2006). All the F1 plants were starch excess. Thus, the starch-excess phenotype in Deg263 is due to the point mutation in the ISA3 gene. We obtained the metabolic profile of isa3-2; this coclustered tightly with the profile of Deg263 (Supplemental Fig. S3). We designate Deg263 as isa3-4. The mutant with the metabolic profile most similar to these isa3 mutants is sex4 and both accumulate starch to the same degree. It is tempting to infer a functional relationship in this similarity. For example, ISA3 could be a downstream target of SEX4 regulation. This hypothesis is under investigation. However, we emphasize that the measurement of a broad set of metabolites from a range of biochemical pathways, even if performed in more depth, can provide, at best, circumstantial evidence for such a relationship. The related metabolic profiles may simply reflect the similar phenotypic severity of the two mutants. The similarity between the profile of isa2 to those of sex4 and isa3 supports this latter interpretation because isa2 has an altered-starch phenotype but not a sex phenotype.
The sex3 mutation mapped to chromosome 1 between markers CAT3a (29.9 cM) and M235 (34.01 cM). We identified GIGANTEA (GI; At1g22770) as a candidate gene in this region. GI is a component of the circadian clock and gi mutants have a number of pleiotropic phenotypes, including excess leaf starch (Eimert et al., 1995) and very late flowering (Koornneef et al., 1991). Although mutants in starch metabolism exhibit slightly delayed flowering (e.g. sex1; Eimert et al., 1995), sex3 resembles gi in that it flowers very late. We sequenced the GI gene in sex3 and found a point mutation in exon 8 causing a stop codon. We obtained the gi-3 mutant (a point mutation causes a stop codon in exon 10; Fowler et al., 1999) and confirmed its starch-excess phenotype. In the complementation test between sex3 and gi-3, all the F1 plants were starch-excess. The reason for the starch-excess phenotype of gi mutants is not known. The amount of starch made in leaves depends on the photoperiod (Chatterton and Silvius, 1980; Gibon et al., 2004) and several studies have reported that genes involved in starch metabolism show clock-dependent diurnal fluctuations in expression (Harmer et al., 2000; Smith et al., 2004; Lu et al., 2005). Thus, it is not surprising that altered starch levels are seen in mutants affected in circadian clock function. Interestingly, the levels of sugars were also increased in sex3, suggesting that starch can still be converted to sugars at night. Because the circadian clock influences many metabolic processes, the utilization of sugars may be uncoupled from their supply in sex3. This may result in sugar accumulation in the leaf, which might feed back onto starch breakdown, causing the observed starch-excess phenotype.
CONCLUSION
Identification of the three genes mutated in the novel starch-excess lines indicates that metabolic profiling, coupled with statistical analysis, can be used to classify forward genetic mutants with similar phenotypes. In this study, sample preparation and data acquisition for the metabolite profiles was rapid (2–3 weeks) compared with the time taken to map even a single mutation. Manually checking the metabolite assignments in our CG-MS chromatograms was time consuming, but is a process that can be automated. Refinement of the approach could be achieved with the selection of a second sample set. For example, samples harvested during the day could have provided a useful comparison to the samples harvested during the night in this study.
We propose high-throughput metabolite profiling as a powerful and rapid strategy to group other forward genetic mutants that have already been selected on the basis of strong common phenotypes (e.g. plants with leaves that are paler/darker green than normal or that display altered growth rates). Profile comparisons could determine which lines are allelic or carry mutations in genes with related functions. This technique could also be effectively used to classify second-site mutations (again identified through forward genetic screens) that suppress or modify a known mutant phenotype. In this case, the metabolic profiles of the suppressor mutants could be compared with one another to establish groups and contrasted with the parental mutant line rather than with the wild type. This streamlining could be especially advantageous because genetic mapping of second-site mutations can be more time consuming than mapping of single-site mutants.
MATERIALS AND METHODS
Plant Growth and Harvest
Arabidopsis (Arabidopsis thaliana) ecotypes Col-0, Ws, Landsberg erecta, and RLD and their mutants were grown in a climate-controlled chamber set to short-day conditions (8-h light at 20°C/16-h dark at 16°C, 180 μmol photons m−2 s−1 fluorescent light, 60%–75% relative humidity). Plants were grown individually in soil in 150-cm3 pots. Plants of each genotype were randomized within the growth chamber. Replicate samples comprising four to six mature leaves of an individual plant (mature rosette stage, preflowering) were harvested 1 h before the end of the night and immediately frozen in liquid N2.
Extraction and Analyses of Arabidopsis Leaf Metabolites
Leaves were ground to a powder while frozen using a mixer mill (Rentsch) with precooled adapters. For starch measurement, 50 mg of leaf powder was extracted using perchloric acid (Delatte et al., 2005). Starch in the insoluble fraction was measured as described previously (Hargreaves and ap Rees, 1988).
To extract soluble metabolites for GC-MS analysis, leaf material (40–60 mg) was extracted in 1.4 mL of 100% methanol together with 60 μL of an internal standard (0.2% [w/v] ribitol in water). The mixture was heated at 70°C for 15 min with vigorous mixing. Insoluble material was removed by centrifugation. Chloroform (0.75 mL) and water (1.5 mL) were added to the supernatant. The mixture was mixed vigorously for 30 s and the phases separated by centrifugation. Aliquots of the methanol-water phase containing the polar metabolites were taken and reduced to dryness in a SpeedVac concentrator. Samples were dissolved in 40 μL of 20 mg/mL methoxyamine hydrochloride in pyridine for 2 h at 37°C to protect the carbonyl moieties. Ten microliters of a retention time standard mixture (0.029% [v/v] n-dodecane, n-pentadecane, n-nonadecane, n-docosane, n-octacosane, n-dotracontane, and n-hexatriacontane dissolved in pyridine) were added. Acidic protons were derivatized by treatment with 70 μL N-methyl-N-[trimethylsil] trifluoroacetamide for 30 min at 37°C.
The GC-MS profiling method was largely as described previously (Roessner et al., 2000, 2001). Samples in each batch were analyzed in random order. GC-MS spectra were obtained with an AS 2000 autosampler, a GC8000 gas chromatograph coupled to a Voyager quadrupol-type mass spectrometer (ThermoFinnigan), operated by MassLab software (ThermoQuest). The mass spectrometer was calibrated according to the manufacturer's recommendations using Tris-(perfluorobutyl)-amine (CF43) as a reference gas. CG-MS hardware and settings were as described elsewhere (Roessner et al., 2000, 2001) with the following minor modifications: Samples were injected in a split-less mode; use of a 5°C min−1 temperature ramp with final temperature set to 320°C on a 30 m × 0.25 mm i.d. Rtx-5Sil MS capillary column with an integrated guard column (Restek GmbH); and use of the n-alkane mixture (see above) as retention time standards. Mass spectra were recorded at two scans/s with a mass-to-charge ratio of 50 to 600 scanning range. Samples in each batch were analyzed in random order. Metabolite assignments in each chromatogram were evaluated manually.
Statistical Analysis
PCA and HCA pattern recognition was performed using Pirouette software (Infometrix) with log10 data transformation. Different kinds of preprocessing tested yielded similar results. HCA was performed using Euclidean distances with incremental linkage. Because PCA and HCA are exploratory methods with no explicit probabilistic basis, they provide neither an assessment of the uncertainty of their output nor an ordering of the importance of the variables for pattern recognition. To obtain these and check the reliability of PCA and HCA output, we developed an approach based on normal mixture modeling of the metabolite profiles after log transformation. The interbatch variation was removed as for PCA and HCA. The supervised classification tool is a hierarchical model having three levels of variation: variation among replicates from the same genotype, around an experimental profile for that genotype; variation among experimental profiles for the same genotype; and variation in the underlying profiles for the different genotypes. The last is modeled using a mixture of point mass and normal distribution independently for each metabolite. Point mass corresponds to no change from the background level, whereas the normal distribution gives the profile for those genotype-metabolite combinations whose background level is different from zero; this occurs with a probability P. The result of incorporating the three levels of variation is a normal mixture model whose hyperparameters, including P, may be estimated by empirical Bayes. Given the estimated values of the hyperparameters, one can produce Bayesian estimates of the underlying metabolic profiles and of the probabilities for classification of each of the mutants to the known types and assess the importance of each metabolite to the empirical classification. The details are outlined in Supplemental Methods and will be described more fully by A.C. Davison and V. Partovi Nia (unpublished data).
Positional Identification of the Mutations in Deg172, Deg263, and sex3
Mutants were crossed to the Landsber erecta ecotype. F2 plants screened for the starch-excess phenotype (65 plants for Deg172, 40 for Deg263, and 70 for sex3), which were genotyped using simple sequence length polymorphisms and single nucleotide polymorphisms available on The Arabidopsis Information Resource database Web site (www.arabidopsis.org).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The pathways of starch synthesis and degradation in the chloroplasts of Arabidopsis leaves.
Supplemental Figure S2. PCA of metabolite profiles.
Supplemental Figure S3. HCA of metabolite profiles.
Supplemental Methods. Bayesian analysis of metabolite profiles.
Supplemental Table S1. Metabolite data normalized for batch variation.
Supplemental Table S2. Metabolite data renormalized for ecotype variation.
Supplementary Material
Acknowledgments
We thank Tim Caspar for sex3 seeds, Louisa Wright and George Coupland for gi3 seeds, Anja Schneider for tpt seeds, Joachim Kopka for GC-MS access time, Christopher Ball and Rebecca Alder for assistance growing plants, and Hans-Rudolf Roth, Cris Kuhlemeier, Philip Zimmermann, and Alison Smith for useful discussions.
This work was supported by the Roche Research Foundation and the Swiss National Science Foundation (National Centre of Competence in Research–Plant Survival) and by the Deutsche Forschungsgemeinschaft (to P.G. and A.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Samuel C. Zeeman (szeeman@ethz.ch).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bino RJ, Hall RD, Fiehn O, Kopka J, Saito K, Draper J, Nikolau BJ, Mendes P, Roessner-Tunali U, Beale MH, et al (2004) Potential of metabolomics as a functional genomics tool. Trends Plant Sci 9 418–425 [DOI] [PubMed] [Google Scholar]
- Bläsing OE, Gibon Y, Günther M, Höhne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville C (1985) Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol 79 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Lin TP, Kakefuda G, Benbow L, Preiss J, Somerville C (1991) Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol 95 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterton NJ, Silvius JE (1980) Photosynthate partitioning into leaf starch as affected by daily photosynthetic period duration in six species. Physiol Plant 49 141–144 [Google Scholar]
- Chia T, Thorneycroft D, Chapple A, Messerli G, Chen J, Zeeman SC, Smith SM, Smith AM (2004) A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant J 37 853–863 [DOI] [PubMed] [Google Scholar]
- Delatte T, Trevisan M, Parker ML, Zeeman SC (2005) Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J 41 815–830 [DOI] [PubMed] [Google Scholar]
- Delatte T, Umhang M, Trevisan M, Eicke S, Thorneycroft D, Smith SM, Zeeman SC (2006) Evidence for distinct mechanisms of starch granule breakdown in plants. J Biol Chem 281 12050–12059 [DOI] [PubMed] [Google Scholar]
- Eimert K, Wang SM, Lue WL, Chen J (1995) Monogenic recessive mutations causing both late floral initiation and excess starch accumulation in Arabidopsis. Plant Cell 7 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L (2000) Metabolic profiling represents a novel and powerful approach for plant functional genomics. Nat Biotechnol 18 1157–1161 [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Blasing OE, Palacios-Rojas N, Pankovic D, Hendriks JH, Fisahn J, Hohne M, Gunther M, Stitt M (2004) Adjustment of diurnal starch turnover to short days: depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J 39 847–862 [DOI] [PubMed] [Google Scholar]
- Hargreaves JA, ap Rees T (1988) Turnover of starch and sucrose in the roots of Pisum sativum. Phytochem 27 1627–1629 [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290 2110–2113 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Joshi V, Fraga M, Rugg A, Yu S, Li L, Last RL (2004) Application of a high-throughput HPLC-MS/MS assay to Arabidopsis mutant screening; evidence that threonine aldolase plays a role in seed nutritional quality. Plant J 39 465–475 [DOI] [PubMed] [Google Scholar]
- Keurentjes JJB, Fu JY, de Vos CHR, Lommen A, Hall RD, Bino RJ, van der Plas LHW, Jansen RC, Vreugdenhil D, Koornneef M (2006) The genetics of plant metabolism. Nat Genet 38 842–849 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Vanderveen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229 57–66 [DOI] [PubMed] [Google Scholar]
- Lu Y, Gehan JP, Sharkey TD (2005) Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol 134 2280–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Sharkey TD (2004) The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta 218 466–473 [DOI] [PubMed] [Google Scholar]
- Niittylä T, Comparot-Moss S, Lue WL, Messerli G, Trevisan M, Seymour MD, Gatehouse JA, Villadsen D, Smith SM, Chen J, et al (2006) Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem 281 11815–11818 [DOI] [PubMed] [Google Scholar]
- Niittylä T, Messerli G, Trevisan M, Chen J, Smith AM, Zeeman SC (2004) A previously unknown maltose transporter essential for starch degradation in leaves. Science 303 87–89 [DOI] [PubMed] [Google Scholar]
- Quackenbush J (2001) Computational analysis of microarray data. Nat Rev Genet 5 418–427 [DOI] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L (2000) Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23 131–142 [DOI] [PubMed] [Google Scholar]
- Schauer N, Fernie AR (2006) Plant metabolomics: towards biological function and mechanism. Trends Plant Sci 11 508–516 [DOI] [PubMed] [Google Scholar]
- Schneider A, Häusler RE, Kolukisaoglu Ü, Kunze R, van der Graaff E, Schwacke R, Catoni E, Desimone M, Flügge UI (2002) An Arabidopsis thaliana knock-out mutant of the chloroplast triose phosphate/phosphate translocator is severely compromised only when starch synthesis, but not starch mobilisation is abolished. Plant J 32 685–699 [DOI] [PubMed] [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM (2004) Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol 136 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37 914–939 [DOI] [PubMed] [Google Scholar]
- Wattebled F, Dong Y, Dumez S, Delvallé D, Planchot V, Berbezy P, Vyas D, Colonna P, Chatterjee M, Ball S, et al (2005) Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol 138 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby CA, Gentry MS, Dixon JE (2006) Laforin: a dual specificity phosphatase that dephosphorylates complex carbohydrates. J Biol Chem 281 30412–30418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TS, Kofler H, Häusler RE, Hille D, Flügge UI, Zeeman SC, Smith AM, Kossmann J, Lloyd J, Ritte G, et al (2001) The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell 13 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Northrop F, Smith AM, ap Rees T (1998. a) A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolyzing enzyme. Plant J 15 357–365 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM (2007) The diurnal metabolism of leaf starch. Biochem J 401 13–28 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Umemoto T, Lue W-L, Au-Yeung P, Martin C, Smith AM, Chen J (1998. b) A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell 10 1699–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.