Abstract
F-box proteins constitute a large family in eukaryotes and are characterized by a conserved F-box motif (approximately 40 amino acids). As components of the Skp1p-cullin-F-box complex, F-box proteins are critical for the controlled degradation of cellular proteins. We have identified 687 potential F-box proteins in rice (Oryza sativa), the model monocotyledonous plant, by a reiterative database search. Computational analysis revealed the presence of several other functional domains, including leucine-rich repeats, kelch repeats, F-box associated domain, domain of unknown function, and tubby domain in F-box proteins. Based upon their domain composition, they have been classified into 10 subfamilies. Several putative novel conserved motifs have been identified in F-box proteins, which do not contain any other known functional domain. An analysis of a complete set of F-box proteins in rice is presented, including classification, chromosomal location, conserved motifs, and phylogenetic relationship. It appears that the expansion of F-box family in rice, in large part, might have occurred due to localized gene duplications. Furthermore, comprehensive digital expression analysis of F-box protein-encoding genes has been complemented with microarray analysis. The results reveal specific and/or overlapping expression of rice F-box protein-encoding genes during floral transition as well as panicle and seed development. At least 43 F-box protein-encoding genes have been found to be differentially expressed in rice seedlings subjected to different abiotic stress conditions. The expression of several F-box protein-encoding genes is also influenced by light. The structure and function of F-box proteins in plants is discussed in light of these results and the published information. These data will be useful for prioritization of F-box proteins for functional validation in rice.
In plants, like other living organisms, protein turnover is a key regulatory mechanism in many cellular processes, including cell cycle, circadian rhythms, cell lineage specification, metabolic control, flower development, embryogenesis, stress responses, and various signal transduction pathways. The ubiquitin (Ub)/26S proteasome pathway is responsible for selective degradation of most intracellular proteins in eukaryotes (Smalle and Vierstra, 2004). Proteins targeted by this pathway are first modified by the attachment of Ub polymers. These polyubiquitinated substrates are recognized by the 26S proteasome and degraded. Ubiquitination of target protein occurs in three steps (Deshaies, 1999; Smalle and Vierstra, 2004). In first step, Ub moiety is activated by the Ub-activating enzyme (E1) and then transferred to the Ub-conjugating enzyme (E2). Finally, substrate recognition and ubiquitination occurs through Ub-protein ligases (E3), which catalyze the transfer of activated Ub to appropriate targets. Skp1p-cullin-F-box (SCF) protein is a major class of plant E3 ligases, which is composed of four subunits: cullin1/Cdc53, Rbx1/Roc1/Hrt1, Skp1, and an F-box protein. The F-box protein performs the crucial role of conferring specificity to the complex for appropriate targets.
F-box proteins contain a conserved F-box domain (40–50 amino acid) at their N terminus, which interacts with Skp1. The name F box was given because it was identified first at the N-terminal region of cyclin F (Bai et al., 1994). The C terminus of F-box proteins generally contains one or several highly variable protein-protein interaction domains, for example, Leu-rich repeat (LRR), kelch repeat, tetratricopeptide repeat (TPR), and WD40 repeat that interact with specific targets. Many reported F-box proteins have been identified as SCF components. However, F-box proteins function in non-SCF complexes as well (Clifford et al., 2000; Galan et al., 2001; Kim et al., 2002). Over the past few years, several F-box proteins have been identified in eukaryotes such as yeast (Saccharomyces cerevisiae), nematodes, flies, humans, and plants (Kipreos and Pagano, 2000; Gagne et al., 2002; Kuroda et al., 2002; Jin et al., 2004).
Regulation of various developmental processes in plants, for example photomorphogenesis, circadian clock regulation, self incompatibility, and floral meristem and floral organ identity determination, involves F-box proteins (Sullivan et al., 2003; Moon et al., 2004; Smalle and Vierstra, 2004). So far, functions of only 23 F-box proteins (18 from Arabidopsis [Arabidopsis thaliana]) have been reported from plants (Supplemental Table S1). UNUSUAL FLORAL ORGANS (UFO), the first F-box protein identified in plants, has a role in floral meristem identity and floral organ development (Levin and Meyerowitz, 1995; Samach et al., 1999; Laufs et al., 2003; Hepworth et al., 2006). LKP1/ZTL, LKP2/FKL, and FKF1 F-box proteins are involved in photomorphogenesis, circadian clock regulation, and control of flowering time (Nelson et al., 2000; Somers et al., 2000, 2004; Schultz et al., 2001; Imaizumi et al., 2003, 2005). Recently, it has been demonstrated that many F-box proteins, including TIR1, which has been known for a long time to regulate the stability of Aux/IAA proteins, act as auxin receptors in Arabidopsis (Gray et al., 2001; Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005). The role of F-box proteins in ethylene (EBF1 and EBF2) and gibberellic acid (SLY1 and SNE) signaling has also been established (Guo and Ecker, 2003; McGinnis et al., 2003; Potuschak et al., 2003; Dill et al., 2004; Gagne et al., 2004; Strader et al., 2004). The F-box protein COI1 has been implicated in jasmonate-regulated defense responses (Xie et al., 1998; Xu et al., 2002; Devoto et al., 2005). Another F-box protein, SON1, negatively regulates systemic acquired resistance and NIM1-independent defense response (Kim and Delaney, 2002). S-locus F-box genes have been identified in several plants that control pollen function of self incompatibility (Lai et al., 2002; Qiao et al., 2004a, 2004b; Ushijima et al., 2004; Wang et al., 2004; Sonneveld et al., 2005).
The Arabidopsis genome encodes a superfamily of F-box proteins (approximately 600–700 members) with diverse C-terminal domains (Gagne et al., 2002; Kuroda et al., 2002). The presence of a strikingly large number of F-box proteins implies that a plethora of SCF complexes, which recognize a wide array of substrates, are possible. Yeast two-hybrid analysis showed that several putative F-box proteins interact with one or more members of the Arabidopsis Skp1-like (ASK) family (Gagne et al., 2002; Kuroda et al., 2002), indicating that many of these proteins are components of SCF complexes. Different groups have classified Arabidopsis F-box proteins in several subfamilies or subgroups on the basis of phylogenetic relationship or unique functional domains (Gagne et al., 2002; Kuroda et al., 2002). The functions of most of the F-box proteins are yet to be studied, despite the fact that they play crucial roles in many aspects of plant growth and development. A great deal of experimental work will be required to determine the specific biological function of each of these genes comprising the F-box family. The current availability of rice (Oryza sativa) genome sequence (International Rice Genome Sequencing Project, 2005) allows the identification of F-box protein-encoding genes in this monocotyledonous species and their comparative analysis with Arabidopsis, which will be useful in terms of studying the functional and evolutionary diversity of this superfamily in plants.
To our knowledge, the function of only two F-box proteins has been reported in rice (Supplemental Table S1). Gibberellin-insensitive dwarf2 acts as a positive regulator of gibberellic acid signaling (Itoh et al., 2003; Sasaki et al., 2003; Gomi et al., 2004) and dwarf3 (D3) controls tiller bud activity (Ishikawa et al., 2005). In this study, 687 potential F-box proteins have been identified in rice genome and classified into 10 subfamilies on the basis of type of domain(s) present at their C terminus. Several putative novel conserved motifs have also been identified in proteins that do not harbor any known functional domain other than the F box. The chromosomal locations and phylogenetic relationship of F-box proteins have been reported. A comprehensive expression analysis of F-box protein-encoding genes at several stages of development of rice, including vegetative growth, floral transition, and panicle and seed development has been performed. In addition, evidence has been provided for the regulation of expression of several F-box protein-encoding genes by light and abiotic stress.
RESULTS AND DISCUSSION
Identification and Classification of F-Box Proteins
F-box proteins are represented by a large family in various organisms. There are 11 F-box protein-encoding genes predicted in yeast, 22 in Drosophila, 68 in human, 74 in mouse, and 326 in Caenorhabditis elegans (Kipreos and Pagano, 2000; Jin et al., 2004). In Arabidopsis, however, a strikingly large number of F-box protein-encoding genes (600–700) have been predicted (Gagne et al., 2002; Kuroda et al., 2002). The recent completion of genome sequencing of rice (International Rice Genome Sequencing Project, 2005) provides a powerful tool to identify genes encoding F-box proteins in this most important cereal crop. BLAST searches of the rice (subsp. japonica cv Nipponbare) genome performed using Hidden Markov Model (HMM) profile of F-box domain as query and subsequent analysis identified 687 nonredundant potential F-box proteins. The Institute of Genomic Research (TIGR) locus, open reading frame length, protein length, and chromosomal location of each of these genes are listed in Supplemental Table S2. The BLASTP search of these 687 F-box proteins in the annotated proteins of indica rice (cv 93–11) genome available at BGI-RISe Rice Genome Database (http://rise.genomics.org.cn; Yu et al., 2005) revealed that most of these proteins are conserved in both subspecies (data not shown).
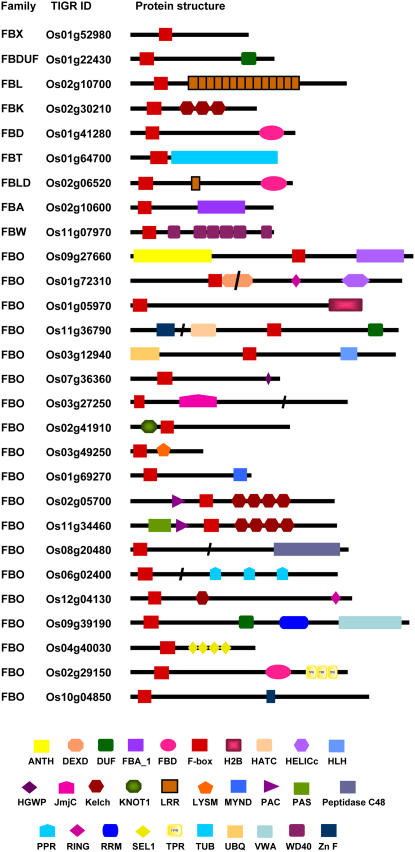
Many F-box proteins are predicted to contain various protein-protein interaction domains at their C terminus (Bai et al., 1996; Patton et al., 1998; Gagne et al., 2002; Kuroda et al., 2002) in diverse organisms. In mammals, F-box proteins have been classified into three groups, FBXW containing WD40 repeat domains, FBXL containing LRR domains, and FBXO with other domains (Jin et al., 2004). Genome-wide analyses of Arabidopsis F-box proteins revealed the presence of several domains such as LRR, kelch repeats, FBD, WD40, PAS/PAC, ring finger, tubby (TUB), and PPR (Gagne et al., 2002; Kuroda et al., 2002), allowing their classification into 19 groups (Kuroda et al., 2002). The domain search of rice F-box proteins in the SMART and PFam databases identified several other known functional domains similar to Arabidopsis. This search led us to classify the rice F-box proteins in 10 subfamilies (Table I). A large number (465) of rice F-box proteins did not show the presence of any known functional domain other than the F box and were classified as FBX subfamily members. The other 222 proteins exhibited the presence of one or more known functional domains and were classified in FBDUF (66) containing domain of unknown function (DUF), FBL (61) containing LRR domains, FBK (25) containing kelch repeats, FBD (17) containing FBD domain, FBT (14) containing TUB domain, FBLD (9) containing both LRR and FBD domains, FBA (4) containing F-box associated domain (FBA_1), FBW (2) containing WD40 repeats, and FBO (24) containing other domains, including PAS, PAC, PPR, TPR, ring finger, zinc finger, MYND, helicase, and SEL1 repeats (Table I; Fig. 1; Supplemental Fig. S1; Supplemental Table S2).
Table I.
Classification of 687 F-box proteins based on their domain architecture
| Subfamily | Domain Content | No. in Category |
|---|---|---|
| FBX | Unknown/none | 465 |
| FBDUF | DUF | 66 |
| FBL | LRR | 61 |
| FBK | Kelch | 25 |
| FBD | FBD | 17 |
| FBT | TUB | 14 |
| FBLD | LRR and FBD | 9 |
| FBA | FBA | 4 |
| FBW | WD40 | 2 |
| FBO | Others | 24 |
Figure 1.
Domain organization of representative F-box proteins from each family. Members of FBO family with different domain(s) are also shown. On the left, family name to which corresponding F-box protein belongs and TIGR locus ID are given. Domain abbreviations are: ANTH, ANTH domain; DEXD, DEAD-like helicase domain; FBA_1, F-box associated domain; FBD, domain present in F-box and BRCT domain containing proteins; H2B, histone H2B; HATC, hAT family dimerization domain; HELICc, helicase superfamily C-terminal domain; HLH, helix-loop-helix domain; HGWP, HGWP motif; JmjC, Jumonji; Kelch, kelch repeats; KNOT1, knottins; LysM, Lys motif; MYND, MYND finger; PAC, motif C terminal to PAS motifs; PAS, PAS domain; Peptidase C48, Ulp1 protease family C-terminal catalytic domain; PPR, pentatricopeptide repeats; RING, ring finger; RRM, RNA recognition motif; SEL1, sel1-like repeats; UBQ, Ub domain; VWA, von Willebrand factor type A domain; WD40, WD40 repeats; ZnF, zinc finger.
The most abundant domains containing F-box proteins are those containing DUF domain. Most of the members of FBDUF subfamily contain DUF295 domain except for Os05g18660, Os07g33400, and Os09g37590, which contain DUF1618, DUF635, and DUF246 domains, respectively. LRRs are 20 to 29 amino acid motifs with positionally conserved Leu or other aliphatic residues. Only 25 F-box proteins containing kelch repeats were identified, a number much less than that of Arabidopsis. Kelch repeats are present in a large fraction of rice and Arabidopsis F-box proteins (Gagne et al., 2002; Kuroda et al., 2002; this study) and appear to be the unique feature of plant F-box proteins, as currently no yeast or mammalian F-box protein displays the presence of kelch repeats. WD40 repeats and LRRs are the predominant domains present at the C terminus of yeast and mammalian F-box proteins (Bai et al., 1996; Cenciarelli et al., 1999; Winston et al., 1999; Jin et al., 2004). Although kelch and WD40 repeats have similar tertiary structure like a β-propeller (Smith et al., 1999; Adams et al., 2000), they have no sequence similarity. This suggests the convergent evolution of a subset of F-box proteins originating a common tertiary structure specialized in protein-protein interaction. Seventeen rice F-box proteins were predicted to contain FBD domain, a plant-specific domain of approximately 80 amino acids, which is also present in BRCT domain containing proteins. Fourteen F-box proteins exhibited the presence of C-terminal TUB domain. This 120 amino acid domain was first identified in mouse TUB1, which is involved in controlling obesity (Kleyn et al., 1996). The other nine F-box proteins harbor both the LRR repeats and FBD domains. FBA domain was predicted in four F-box proteins.
Three of the F-box proteins (Os06g47890, Os11g34460, and Os02g05700) have PAS/PAC domain in addition to kelch repeats and may function as flavin-binding photoreceptors. Similar proteins in Arabidopsis, ZTL, LKP2, and FKF1 have been implicated in control of flowering time and circadian rhythms (Nelson et al., 2000; Somers et al., 2000, 2004; Schultz et al., 2001; Imaizumi et al., 2003, 2005). JmjC domain, which is supposed to be a metal-binding site (Clissold and Ponting, 2001), was found in two F-box proteins (Os03g27250 and Os11g36450). DNA-binding domains such as zinc finger, MYND, C2H2 zinc finger, helix loop helix, and ring finger were also found in rice F-box proteins, which may be directly or indirectly involved in transcriptional regulation. In addition, PPR and TPR domains were predicted in rice F-box proteins that are responsible for processing and/or translation of organeller mRNAs (Small and Peeters, 2000).
A large fraction (465) of predicted rice F-box proteins do not harbor any known functional domain other than the F box. Therefore, the unknown putative conserved motifs in F-box proteins of FBX subfamily were investigated using the Multiple Em (Expectation Maximization) for Motif Elicitation (MEME) program (Bailey and Elkan, 1995). The statistical significance of a motif predicted by MEME is based on its log likelihood ratio, its width, and number of occurrences. The e value is an estimate of the expected number of motifs with the given log likelihood ratio (or higher) and with the same width and number of occurrences that one would find in a similarly sized set of random sequences. More than 30 putative statistically significant (e value less than e-100) motifs could be identified (Supplemental Table S3). Each of these motifs was more than 10 amino acids in length and was conserved in at least five of the predicted F-box proteins. Some of these motifs (motifs 1, 5, and 19) were conserved in as large as 138 rice F-box proteins. BLAST searches showed that most of these motifs were conserved in other plants also; however, some of them were present only in rice sequences (data not shown). These putative novel motifs may be critical for interaction of F-box proteins with other proteins, like other known domains, and may provide specific function(s) to these proteins. However, the biological significance of these motifs remains to be elucidated. Based on the BLAST searches, large-scale multiple alignments, and analysis using PFam-B databases, some new motifs have been identified in Arabidopsis F-box proteins (Gagne et al., 2002; Kuroda et al., 2002). Although the consensus sequences of these motifs have not been described, some of these motifs were found to be conserved in other plant species also (Kuroda et al., 2002). The BLAST search of some representative Arabidopsis F-box proteins harboring these motifs showed very weak similarity with rice F-box proteins (data not shown). These results suggest that rice and Arabidopsis F-box proteins contain some specific motifs, which may perform species-specific functions.
Gene Structure and Chromosomal Distribution
To study the gene structure, the number of introns present within the open reading frame of each F-box protein gene was determined by analysis of their exon-intron organization. Interestingly, 40.76% (280 of 687) of F-box protein-encoding genes are predicted to be intronless (Supplemental Table S2), which is a much higher percentage than predicted for rice genes overall (19.9%; M. Jain, P. Khurana, A.K. Tyagi, J.P. Khurana, unpublished data). Forty five of 66 members of FBDUF subfamily, 18 of 25 members of FBK subfamily, and all the four members of FBA subfamily are intronless. Such intronless gene families can evolve rapidly either by gene duplication or reverse transcription/integration (Lecharny et al., 2003; Lurin et al., 2004; Jain et al., 2006c).
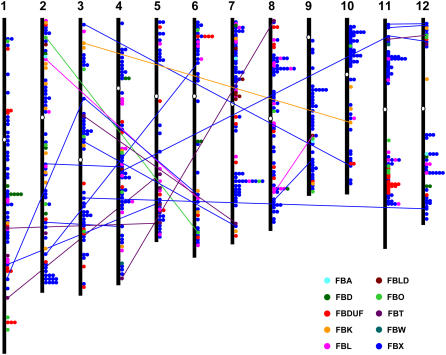
The 687 F-box protein-encoding genes were found to be distributed randomly on all the 12 rice chromosomes. The huge size of F-box family indicates that it has evolved through a large number of duplication events in rice. The rice genome has undergone genome-wide duplication events, including polyploidy, which has great impact on the amplification of members of a gene family in the genome. For fine mapping, the position (in bp) of each F-box protein gene was determined on rice chromosome pseudomolecules available at TIGR (release 4) and has been represented diagrammatically in Figure 2 (the exact position in bp and orientation of each F-box protein gene on rice chromosome pseudomolecules is given in Supplemental Table S2). Substantial clustering of F-box protein-encoding genes was evident on different chromosomes. Several F-box protein-encoding genes were arranged in tandem repeats of two to nine genes, either in the same or inverse orientation, representing localized gene duplications. Interestingly, at several positions, the F-box protein-encoding genes present in tandem belonged to different families, suggesting the diversification by domain shuffling after tandem duplications. Recently, Yu et al. (2005) presented evidence for the ongoing individual gene duplications in rice, which provide a never ending raw material for studying gene genesis and their functions. The duplication of the F-box protein-encoding genes is also associated with chromosomal block duplications in rice. Fifty F-box protein-encoding genes were found to be located on the duplicated segmental regions of rice chromosomes mapped by TIGR (Fig. 2, Supplemental Table S4). All but four of the F-box protein-encoding genes located on duplicated segments belong to same subfamily. However, four F-box protein-encoding genes present on duplicated chromosomal segments between chromosome 8 and 9 (Os08g35930 and Os09g27090) and chromosome 11 and 12 (Os11g04330 and Os12g04130) belong to different families, further supporting the idea of diversification of F-box protein-encoding genes by domain shuffling. Interestingly, eight of the 14 FBT subfamily proteins were present on duplicated chromosomal segments. Since the number of F-box protein-encoding genes present on duplicated chromosomal segments is much smaller than those present in tandem, the tandem duplications of chromosomal regions appear to have played a major role in expansion of this gene family. These results suggest that the expansion of F-box protein-encoding genes in rice is probably, in large part, due to localized gene duplications. In Arabidopsis also, tandem duplications have been implicated in the expansion of F-box protein gene family (Gagne et al., 2002).
Figure 2.
Genomic distribution of F-box protein-encoding genes on rice chromosomes. F-box protein-encoding genes classified in different families are shown in different colors. One circle represents one F-box protein gene. White ovals on the chromosomes (vertical bar) indicate the position of centromeres. Chromosome numbers are indicated at the top of each bar. The F-box protein-encoding genes present on duplicated chromosomal segments are connected by colored lines according to their families. The position (bp) and orientation of each F-box protein gene on TIGR rice chromosome pseudomolecules (release 4) is given in the Supplemental Table S2.
Phylogenetic Analysis
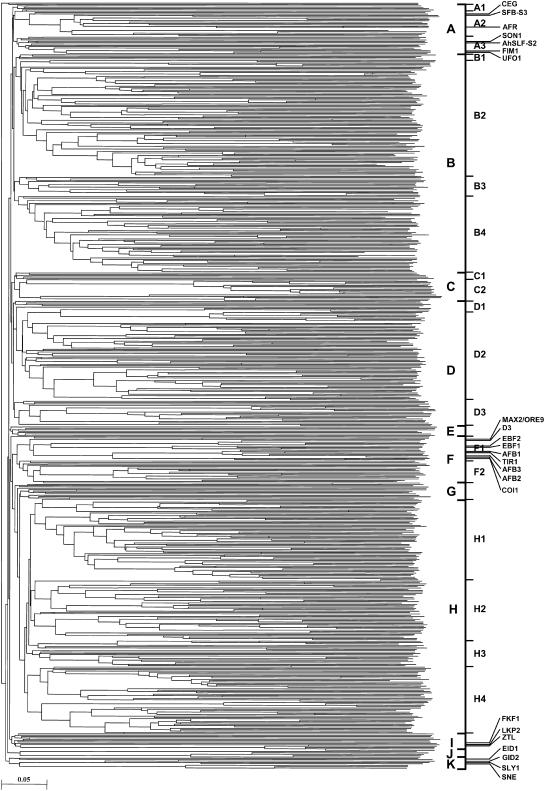
To examine the phylogenetic relationship among rice F-box proteins, an unrooted tree was constructed from alignments of their full-length protein sequences along with F-box proteins with known function in Arabidopsis and other plants (Fig. 3; Supplemental Fig. S2). Manual analysis of the phylogenetic tree revealed 11 distinct clades (A–K) of rice F-box proteins. The clades A to D, F, and H were further divided into 18 subclades, giving 23 distinct groups of proteins in total. Two proteins, Os06g49530 and Os01g14270, did not seem to belong to any of these groups. The investigation of C-terminal domains present in F-box proteins clustered together within a group revealed a striking clustering. Most members of the same subfamily were clustered together. For example, all the F-box proteins in group A2, including Arabidopsis AFR protein, belong to FBK subfamily. Similarly, clade B (groups B1–B4) mostly contains the members of FBX subfamily. All but one of the 14 members of FBT subfamily grouped together in group C2. Likewise, 56 (of 66) members of FBDUF subfamily were included in group D2. All the F-box proteins in group F1 and most of proteins in group F2 belong to FBL subfamily. Similarly, the F-box proteins with identical C-terminal domain(s) clustered together in the phylogenetic tree constructed from the alignments of their F-box domain sequences (data not shown). This correlation suggests the coevolution of F-box domain with the target C-terminal protein-protein interaction domain, corroborating the results from phylogenetic analysis of Arabidopsis F-box proteins (Gagne et al., 2002). However, some groups of F-box proteins had widely different C-terminal domains. The members of FBA, FBW, and FBO subfamilies were grouped with F-box proteins belonging to other subfamilies. Such clustering with similar F-box domain but dissimilar C-terminal domains together further supports the idea that domain shuffling had contributed to expansion of F-box protein diversity (Morgenstern and Atchley, 1999).
Figure 3.
Phylogenetic relationship among the rice F-box proteins. The unrooted tree was generated using ClustalX program by neighbor-joining method. F-box proteins grouped into 11 distinct clades (A–K). The clades A to D, F, and H were further divided into 18 subclades (A1–A3, B1–B4, C1–C2, D1–D3, F1–F2, and H1–H4). The position of F-box proteins with known function from Arabidopsis and other plants used for construction of phylogenetic tree is indicated. A larger version of the tree is available as Supplemental Figure S2.
To examine the expansion of F-box proteins in rice vis-à-vis Arabidopsis, harboring similar domains, unrooted trees were constructed from alignments of full-length protein sequences of three representative subfamilies, FBL, FBK, and FBT (Supplemental Fig. S3). This analysis revealed that most rice and Arabidopsis F-box proteins cluster in species-specific distinct clades. This result indicates that F-box proteins harboring similar domains expanded in a species-specific manner; probably only a few members originated from the common ancestral genes that existed before divergence of monocots and dicots. This type of divergence between a monocotyledonous (rice) and a dicotyledonous (Arabidopsis) species has been observed for other large gene families as well (Bai et al., 2002; Zhang et al., 2005; Jain et al., 2006a, 2006c). Furthermore, significant differences were found in the degree of expansion of F-box proteins. For example, Arabidopsis contains 100 kelch repeat proteins (Gagne et al., 2002), however, only 25 such proteins are predicted in rice. Whether the degree of expansion is species-specific and represents unique functional requirement, will be unraveled when more and more genome sequences become available and gene function analyzed. Further analysis of phylogenetic tree constructed from rice and Arabidopsis F-box proteins containing kelch repeats revealed that one major group of proteins present in Arabidopsis was completely absent in rice except for one protein (Supplemental Fig. S3B). It can be speculated that these proteins were lost in rice or evolved in Arabidopsis after divergence of monocots and dicots and may perform dicot-specific functions.
Digital Expression Analysis
Three different approaches were undertaken for expression analysis of rice F-box protein-encoding genes, which involved the use of already available information. The first approach was the survey for availability of any full-length cDNA (FL-cDNA), expressed sequence tag (EST), or peptide sequence(s) in databases corresponding to rice F-box protein-encoding genes. For this search, the gene expression evidence search page available at TIGR was used. This analysis revealed that one or more FL-cDNA, EST, and/or peptide sequence(s) were available for 368 (53.6%) of 687 F-box protein-encoding genes (Supplemental Table S5), indicating that a large percentage of these genes are expressed. However, the frequency of ESTs for F-box protein-encoding genes varied greatly from one to 62. Also, the EST sequences were derived from various rice tissue/organ libraries, indicating the differential expression of F-box protein-encoding genes. The second approach was to study organ-specific expression of F-box protein-encoding genes using microarray data from an earlier study (Ma et al., 2005), which described the expression pattern at whole genome level in several representative rice organs/tissues, including seedlings, shoots, roots, heading-stage panicles, and filling-stage panicles. Expression for a total of 361 F-box protein-encoding genes (each of them represented by a unique 70-mer oligo on the microarray) could be retrieved and among them 287 genes were expressed in at least one of the above mentioned rice organs/tissues (Supplemental Fig. S4). Several F-box protein-encoding genes exhibited tissue/organ-specific expression. There were 10, 10, 11, 21, and 13 F-box protein-encoding genes specifically expressed in seedlings, shoots, roots, heading-stage panicles, and filling-stage panicles, respectively (Supplemental Fig. S4). The specific expression of highest number of F-box protein-encoding genes in heading panicle demonstrates their predominant role in panicle development.
Massively parallel signature sequencing (MPSS) provides a sensitive quantitative measure of gene expression for nearly all genes in the genome (Brenner et al., 2000). The third approach was to extract information about MPSS tags available from the Rice MPSS Project (http://mpss.udel.edu/rice/) for each F-box protein gene. Data from 16 MPSS libraries representing 12 different tissues/organs of rice was extracted for both 17 base and 20 base signature libraries. MPSS tags were available for 456 and 350 F-box protein-encoding genes in 17 base and 20 base signature libraries, respectively. This further strengthens the idea that most of F-box protein-encoding genes are expressed. However, significant signatures (that uniquely identify individual genes) were found for only 290 and 260 F-box protein-encoding genes in 17 base and 20 base signature libraries, respectively. Substantial differences were found in number of tags (in tpm) representing F-box protein-encoding genes, indicating from marginal (1–3 tpm) to strong (>250 tpm) expression (Supplemental Table S6). A large percentage (40%–50%) of F-box protein-encoding genes exhibited moderate expression (26–250 tpm). Only a few genes were expressed at a high level (>250 tpm). Further, we identified F-box protein-encoding genes with signatures present in only one library by comparisons between different MPSS libraries for both 17 base and 20 base signature datasets. Subsequently, the genes common in both 17 base and 20 base signature libraries were identified with tissue-specific expression (Supplemental Table S7). These F-box protein-encoding genes may play important and specific roles in the biology of these diverse tissues and their upstream regions may have experimental utility as tissue-specific promoters. Highest number of genes (7) showed specific expression in mature pollen followed by root (5). This observation is consistent with the role of F-box protein-encoding genes in controlling pollen function of self incompatibility (Lai et al., 2002; Qiao et al., 2004a, 2004b; Ushijima et al., 2004; Wang et al., 2004; Sonneveld et al., 2005).
Microarray Analysis of Expression Profiles of F-Box Protein-Encoding Genes during Panicle and Seed Development
DNA microarrays can measure the individual transcript level of tens of thousands of genes simultaneously, thus providing a high-throughput means to analyze gene expression at the whole genome level. The whole genome microarray data can also be used to analyze the expression of a subset of genes of interest. To achieve gene expression profiling of F-box protein-encoding genes, microarray analysis was performed using Affymetrix rice whole genome arrays. The rice tissues/organs and developmental stages selected for microarray analysis include seedling, seedling root, mature leaf, Y leaf (leaf subtending the shoot apical meristem [SAM]), SAM, and various stages of panicle (P1–P6) and seed (S1–S5) development. Different developmental stages of panicle and seed development have been categorized according to panicle length and days after pollination (dap), respectively, as follows: up to 0.5 mm, SAM; 0 to 3 cm, floral transition and floral organ development; 3 to 10 cm, meiotic stage; 10 to 15 cm, young microspore stage; 15 to 22 cm, vacuolated pollen stage; 22 to 30 cm, mature pollen stage; 0 to 2 dap, early globular embryo (approximately 25 cell stage); 3 to 4 dap, middle and late globular embryo (150–800 cell stage, onset of coleoptile, SAM and radicle differentiation; early milky endosperm); 5 to 10 dap, embryo morphogenesis (protrusion of leaf primordia and organ enlargement; late milky endosperm); 11 to 20 dap, embryo maturation (soft dough endosperm); 21 to 29 dap, dormancy and desiccation tolerance (hard dough endosperm). These stage specifications are approximations based on information from Itoh et al. (2005).
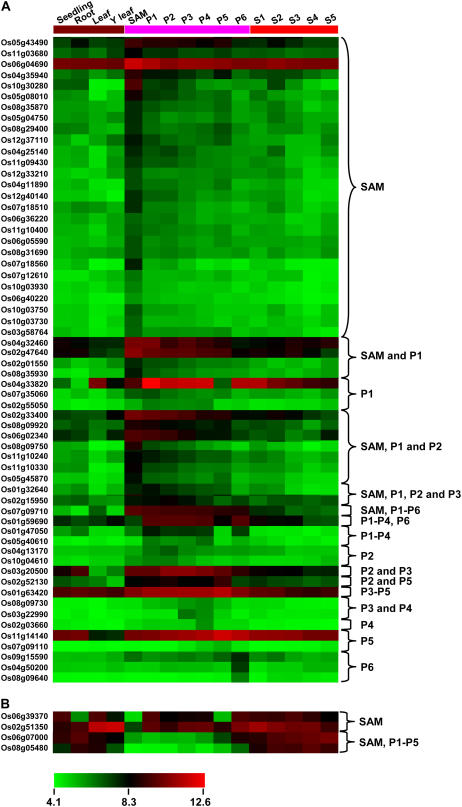
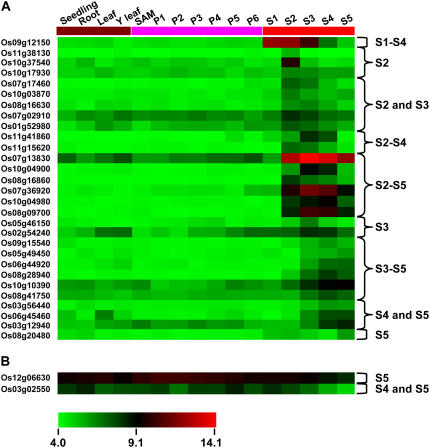
Following whole-chip data processing (quality controls, normalization, probe summarization, variance stabilization, and log transformation), the log signal values for 617 F-box protein-encoding genes represented on the array were extracted. Average log signal values for all the 617 F-box protein-encoding genes from three biological replicates of each sample are given in Supplemental Table S8. A hierarchical cluster display of average log signal values of these genes is also presented in Supplemental Figure S5. The signal values indicate that most of these F-box protein-encoding genes are expressed in at least one of the rice vegetative organs and/or stages of development analyzed. Subsequently, differential expression analysis was performed to identify F-box protein-encoding genes with most abundant expression both during panicle and seed development stage(s). We defined a gene as differentially expressed at a given stage only if the expression level of the gene at that stage was significantly higher (more than 2-fold) than the levels at all the other stages. For this, differentially expressed genes were identified for two datasets of panicle and seed development with respect to vegetative organs, including seedling, root, mature leaf, and Y leaf. This analysis revealed that a total of 125 and 81 F-box protein-encoding genes were differentially expressed in at least one of the stages of panicle and seed development, respectively, as compared to vegetative organs. Further analysis revealed that 33 genes were common among the differentially expressed genes during panicle and seed development stages as compared to vegetative organs. In the next step, the differentially expressed F-box protein-encoding genes at any stage of the panicle development as compared to seed development stages were identified. Similarly, the differentially expressed F-box protein-encoding genes at any stage of the seed development as compared to panicle development stages were identified. This analysis revealed that 64 and 31 F-box protein-encoding genes were preferentially expressed in at least one of the stages of panicle and seed development, respectively (Figs. 4 and 5). The differential expression of some representative genes in panicle and seed developmental stage(s) identified from microarray data analysis has also been confirmed by real-time PCR analysis (Supplemental Fig. S6). It is thus conceivable that these F-box protein-encoding genes may perform specific roles during different stages of development.
Figure 4.
Expression profiles of rice F-box protein-encoding genes differentially expressed during panicle development. Expression profiles of 60 up-regulated (A) and four down-regulated (B) F-box protein-encoding genes in SAM or any stage of panicle development (P1–P6) as compared to vegetative reference tissues/organs (seedling, root, mature leaf, and Y leaf) and seed developmental stages (S1–S5) are presented. The average log signal values of F-box protein-encoding genes in various tissues/organs and developmental stages (mentioned at the top of each lane) are presented by cluster display. The color scale (representing log signal values) is shown at the bottom.
Figure 5.
Expression profiles of rice F-box protein-encoding genes differentially expressed during seed development. Expression profiles of 29 up-regulated (A) and two down-regulated (B) F-box protein-encoding genes in any stage of seed development (S1–S5) as compared to vegetative reference tissues/organs (seedling, root, mature leaf, and Y leaf), SAM, and panicle developmental stages (P1–P6) are presented. The average log signal values of F-box protein-encoding genes in various tissues/organs and developmental stages (mentioned at the top of each lane) are presented by cluster display. The color scale (representing log signal values) is shown at the bottom.
SAM comprises a small group of dividing cells that give rise to aerial parts of the plant. After producing stem and leaves during vegetative growth SAM undergoes transition in its fate to reproductive development marked by formation of floral meristem followed by flowers. Molecular studies have identified several genes whose activities are induced during transition to flowering and that play a primary role in the determination of both floral meristem and floral organ identity in rice (Tang et al., 2005; Furutani et al., 2006). The role of F-box protein, UFO1, the first F-box protein identified in plants, in determination of floral meristem and floral organ identity and flower development in Arabidopsis is well established (Patton et al., 1998; Samach et al., 1999; Wang et al., 2003; Hepworth et al., 2006). Our analysis revealed a strikingly large number (36 of 64) of F-box protein-encoding genes are differentially expressed in SAM and/or early panicle development (P1) stage (Fig. 4), suggesting that F-box protein-encoding genes play crucial roles in floral transition, determination of floral meristem and floral organ identity, and early flower development in rice. As many as 28 F-box protein-encoding genes were also differentially expressed in other stages of panicle development (P2–P6), indicating their role in gametophyte development (Fig. 4). The role of several plant F-box proteins in controlling pollen function of self incompatibility has been reported in other plant species (Lai et al., 2002; Qiao et al., 2004a, 2004b; Ushijima et al., 2004; Wang et al., 2004; Sonneveld et al., 2005).
Several genes play a critical role in seed development and grain filling in crop plants. A close coordination of gene expression among many important pathways is required during this biological process of paramount importance. In rice, the expression of genes involved in different pathways has been demonstrated to be coordinately controlled in a synchronized fashion during grain filling (Anderson et al., 2003). Recently, the differential expression of a significantly large number of genes involved in various cellular processes during seed maturation in hexaploid winter wheat (Triticum aestivum cv Mercia) has been reported (Wilson et al., 2005). Because the cellular gene machinery is affected greatly during seed development and grain filling, it is expected that the expression of F-box protein-encoding genes involved in protein turnover is affected as well. In our study, 31 F-box protein-encoding genes were differentially expressed in at least one or more of the developmental stages of seed development in rice (Fig. 5), as described earlier. The transcript abundance of 29 F-box protein-encoding genes was more than 2-fold higher during various stages of seed development (Fig. 5A). Conversely, two (Os03g02550 and Os12g06630) F-box protein-encoding genes showed decreased transcript levels during later stages (S4 and/or S5) of seed development (Fig. 5B). These results suggest the involvement of F-box proteins in rice seed development.
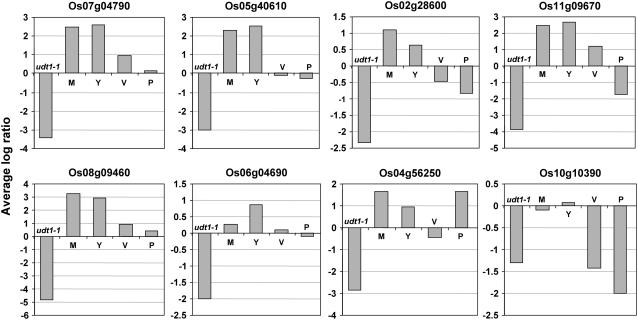
F-Box Protein-Encoding Genes in Anther Development
Recently, a rice basic helix-loop-helix transcription factor, UNDEVELOPED TAPETUM 1 (UDT1), has been identified as a major regulator of early tapetum development and pollen mother cell meiosis (Jung et al., 2005). Tapetum is the innermost sporophytic layer in the anther wall, which is thought to play a crucial role in the development and maturation of microspores. DNA microarray analysis revealed that several genes were down-regulated in udt1-1 mutant anthers (Jung et al., 2005). During the survey of these genes, we found that at least eight F-box protein-encoding genes (Os07g04790, Os05g40610, Os02g28600, Os11g09670, Os08g09460, Os06g04690, Os04g56250, and Os10g10390) reported here were significantly down-regulated (more than 2-fold) in udt1-1 anthers as compared to wild-type anthers (Fig. 6). These genes were differentially expressed at different stages of anther development, including meiosis, young microspore, vacuolated pollen, and pollen mitosis also, as compared to paleas/lemmas (Fig. 6). One of these genes, Os05g40610, was found to be up-regulated during early panicle development (P1–P4 stages) in our analysis also. The difference between our study and the Jung et al. (2005) study for other genes are most likely due to the different tissues (panicle in our study and anthers in study by Jung et al., 2005) used for the microarray analysis. Taken together, these results suggest that these F-box protein-encoding genes might be involved in rice anther development and represent the targets of UDT1 transcription factor.
Figure 6.
Expression of F-box protein-encoding genes with altered expression in udt1 mutant. Eight F-box protein-encoding genes were found to be down-regulated in udt1 mutant more than 2-fold. Average values of log ratios with base 2 for the udt1-1/wild-type anthers at meiosis to the young microspore stages (udt1-1) from three replicate experiments are shown. Average values of log ratios with base 2 for meiosis stage anthers/paleas/lemmas (M), young microspore stage/paleas/lemmas (Y), vacuolated pollen stage/paleas/lemmas (V), and pollen mitosis stage/paleas/lemmas (P) from two replicate experiments are shown.
The F-box proteins are known to interact with Skp1 protein, another component of the SCF complex. The Arabidopsis genome contains 20 Skp1-like genes called ASK, which exhibit clear tissue-specific expression during floral organ development (Marrocco et al., 2003). Several of the ASK promoters directed the postmeiotic expression of GUS in the male gametophyte (Marrocco et al., 2003). A PFam profile search for Skp1 family dimerization domain (PF01466) in the TIGR rice genome annotation database revealed the presence of at least 23 putative skp1-like genes in the rice genome, some of which are preferentially expressed in panicle development stages (data not shown). The coregulation of gene expression of F-box and Skp1-like gene family members, which interact with each other, may be crucial for the regulation of floral organ development, particularly male gametophyte by Ub degradation pathway.
Light Regulation
In addition to the direct effects, light also exerts its influence by modifying the rhythms generated by circadian clock (Somers et al., 1998; Chen et al., 2004). Although the molecular components of circadian clock in plants are not fully understood, the major control is likely to be the protein turnover mediated by Ub-proteasome pathway. The F-box proteins, LKP1/ZTL, LKP2/FKL, and FKF1, represent the important components of circadian clock. ZTL acts as a light-dependent regulator of proteolytic degradation of clock-associated proteins (Somers et al., 2000). FKF1 regulates the expression of the flowering time gene CONSTANS (CO) by degrading CYCLING DOF FACTOR 1, a repressor of CO transcription (Nelson et al., 2000; Imaizumi et al., 2003, 2005). In addition, COP9 signalosome has been known to interact with SCF complexes and act as a negative regulator of photomorphogenesis (Chamovitz et al., 1996). Also, the involvement of two F-box proteins, AFR and EID1, in phytochrome A-mediated light signaling in Arabidopsis has been demonstrated (Dieterle et al., 2001; Harmon and Kay, 2003; Marrocco et al., 2006).
Earlier, a genomic study of rice gene expression in response to light has been performed by microarray analysis, which demonstrated that the expression of a significant component of rice genome is regulated by different light qualities (Jiao et al., 2005). To investigate the light regulation of rice F-box protein-encoding genes, microarray data obtained in the study by Jiao et al. (2005) was analyzed. The expression of only 327 F-box protein-encoding genes could be retrieved under different light conditions. Examination of expression ratios between light- and dark-grown rice seedlings indicates that a significant number (54) of F-box protein-encoding genes were regulated by white light (Supplemental Table S9). We found that 28 genes were up-regulated and 26 genes were down-regulated by at least 2-fold, with a p value less than 0.05. However, under different light quality conditions, less number of genes was regulated as compared to white light (Supplemental Table S9) and most of them were regulated by specific monochromatic lights. Only three (two up-regulated and one down-regulated) genes were differentially expressed in far-red light. In red light, three F-box protein-encoding genes were up-regulated. However, four genes were up-regulated and two down-regulated in blue light. Quantitative difference was found between white light and monochromatic light regulated gene expression in rice at the whole genome level also (Jiao et al., 2005). Putative orthologs of Arabidopsis F-box proteins known to be involved in light signaling could be demarcated in rice on the basis of their phylogenetic relationship, Os06g44500 for AFR1, Os11g34460 for FKF1, Os03g27250 for EID1, and Os06g47890 and/or Os02g05700 for LKP2 and/or ZTL. Although these genes did not show any differential light regulation in the data analyzed, the possibility of their involvement in light signal transduction pathway cannot be completely ruled out. These F-box proteins containing similar functional domains may perform similar functions in rice and Arabidopsis. Taken together, these results suggest that expression of a number of F-box protein-encoding genes is regulated by light, indicating light control of protein turnover in rice, which may be critical for several light-dependent cellular processes.
Expression of F-Box Protein-Encoding Genes under Abiotic Stress Conditions
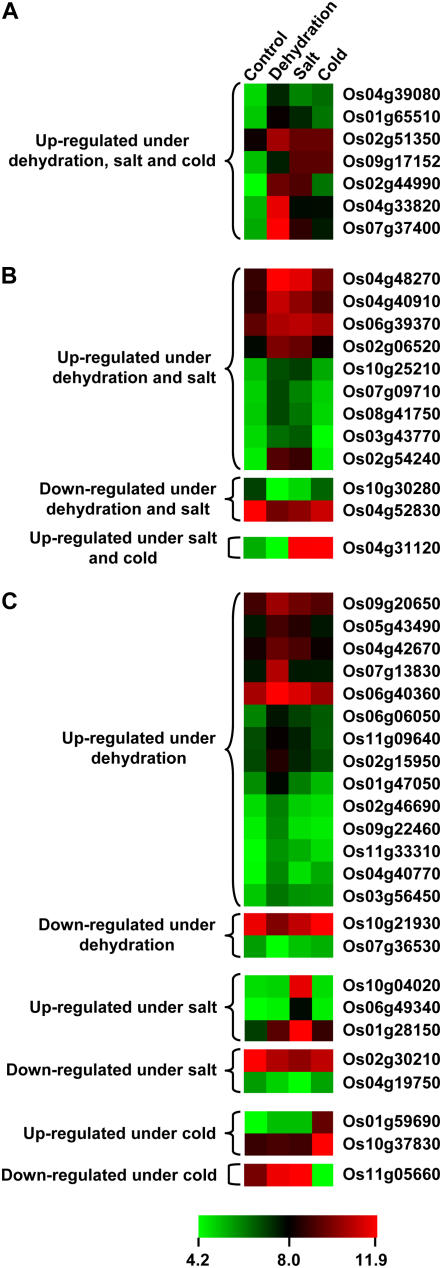
Plants respond to adverse environmental conditions by eliciting various physiological, biochemical, and molecular responses, leading to changes in gene expression. Because abiotic stresses affect the cellular gene machinery, it is quite likely that the components of protein degradation machinery, such as F-box proteins, are affected as well. To address this question, expression of rice F-box protein-encoding genes was analyzed under abiotic stress conditions by microarray analysis performed with total RNA isolated from rice seedlings subjected to salinity, desiccation, and cold treatment. We were able to identify 43 F-box protein-encoding genes that are differentially expressed (36 and 7 genes were up- and down-regulated, respectively) equal to or more than 2-fold with 95% confidence (p value less than 0.05), under one or more of the above-mentioned stress conditions (Fig. 7). Seven of these genes were up-regulated under all the abiotic stress conditions analyzed (Fig. 7A). The other 12 F-box protein-encoding genes were differentially expressed under any two stress conditions (Fig. 7B). Nine F-box protein-encoding genes were up-regulated and two were down-regulated under salt and desiccation stresses but not under cold stress. One F-box protein gene showed up-regulation under salt and cold stress. However, 24 other F-box protein-encoding genes were differentially expressed under only one stress condition; 16 under desiccation stress (14 up-regulated and 2 down-regulated), five under salt stress (3 up-regulated and 2 down-regulated), and three under cold stress (2 up-regulated and 1 down-regulated; Fig. 7C). The real-time PCR results of differential expression of some representative genes under abiotic stress condition(s) are consistent with the microarray data (Supplemental Fig. S6).
Figure 7.
Expression profiles of rice F-box protein-encoding genes differentially expressed under abiotic stress conditions. Expression profiles of F-box protein-encoding genes differentially expressed under all three (A), any two (B), and specific (C) stress condition(s) as compared to the control seedlings are presented. The average log signal values of F-box protein-encoding genes under control and various stress conditions (mentioned at the top of each lane) are presented by cluster display (values are given in Supplemental Table S10). Only those genes that exhibited 2-fold or more differential expression with a p value of less than or equal to 0.05, under any of the given abiotic stress conditions are shown. The color scale (representing log signal values) is shown at the bottom.
The role of COI1, an Arabidopsis F-box protein, is well established in jasmonic acid (JA)-regulated defense responses. COI1 forms an integral part of an SCF E3 Ub ligase that is predicted to target repressors of JA signaling to the proteasome for degradation (Devoto et al., 2002; Xu et al., 2002). COI1 binds a histone deacetylase, RPD3b, and may regulate the expression of genes involved in JA response through modulating the activity of RPD3b and possibly other proteins (Devoto et al., 2002). Recently, requirement of COI1 for a significantly large number of JA- and wound-induced gene transcription has been demonstrated (Devoto et al., 2005). Another F-box protein, SON1, has also been reported to negatively regulate defense response independent of salicylic acid and systemic acquired resistance, possibly via Ub-proteasome pathway (Kim and Delaney, 2002). The F-box proteins, Os05g37690 and Os01g63420, represent the closely related orthologs of Arabidopsis COI1 and may perform similar function in rice. These genes did not show significant differential expression under abiotic stress conditions analyzed in this study. However, no closely related ortholog of SON1 could be predicted in rice. The direct role of F-box proteins in abiotic stress responses has not been demonstrated so far. Our study demonstrates that F-box proteins play a significant role in abiotic stress pathway and the corresponding genes may be a valuable resource for producing stress-tolerant transgenic crop plants.
Plant stress responses often mimic certain normal developmental processes (Cooper et al., 2003). Interaction between plant development and environmental conditions implies that some genes must be coregulated by both environmental factors and developmental cues. We found nine (Os01g47050, Os01g59690, Os02g15950, Os02g51350, Os04g33820, Os05g43490, Os06g39370, Os07g09710, and Os10g30280) and three (Os02g54240, Os07g13830, and Os08g41750) F-box protein-encoding genes that were differentially regulated under stress conditions and were specifically expressed at one or more panicle and seed developmental stage, respectively. Several examples of such interaction of developmental regulation and stress responses exist. Phospholipase D and its product phosphatidic acid that play a role in pollen germination and pollen tube growth in tobacco (Nicotiana tabacum; Potocky et al., 2003; Zonia and Munnik, 2004) are also involved in various stress responses like water deficit, salts, wounding, and elicitation (Wang, 2002). A systematic regulation of gene expression has been found to drive the developmental processes and stress responses in Arabidopsis (Chen et al., 2002), indicating that an overlap of genes occurs between the developmental processes and stress responses. Moreover, a network of rice genes that are associated with stress response and seed development has been reported (Cooper et al., 2003). Recently, microarray analysis demonstrated that a significant number of pollination/fertilization-related genes are indeed regulated by dehydration and wounding in rice (Lan et al., 2005). This suggests that the genetic programs regulating them are likely to be related. This study also provides evidence that a number of candidate F-box protein-encoding genes are likely to be involved in critical developmental processes and stress responses, but their direct relationship needs experimental validation.
CONCLUSION
The F-box protein-mediated targeted protein degradation is critical for several key cellular processes. A very large number of F-box proteins have been predicted in rice and Arabidopsis. However, the function of only a few plant F-box proteins has been established as of now. The field of F-box proteins is in fact currently emerging. This study provides insights into the functions of F-box proteins in rice. For example, several F-box protein-encoding genes displayed specific expression during various stages of panicle and seed development. In addition, F-box proteins appear to serve as the key components of the machinery involved in regulating plant growth and development throughout its life cycle and their expression is influenced by light and abiotic stresses. The leads provided here would pave the way for elucidating the precise role of individual F-box protein in rice by adopting RNAi strategy or insertion mutagenesis.
MATERIALS AND METHODS
Database Search
The BLAST search of all the annotated proteins in whole rice (Oryza sativa) genome at TIGR (release 4) was performed using the HMM profile (build 2.3.2) of F-box domain (PF00646) downloaded from PFam, as query. The HMM profile for 44 amino acid-long F-box domain has been generated by alignments of 545 seed sequences. This search resulted in the identification of 901 proteins with an e-value cutoff of 1.0. Of the 901 proteins, only 772 showed the presence of F-box domain with confidence (e value less than 1.0) by SMART/PFam, when checked individually. Among these, 72 proteins were removed because they represented different gene models present at the same locus in rice genome. Moreover, 13 proteins annotated as retrotransposons or transposable elements were removed before further analysis. For convenience, we have removed LOC suffix from all TIGR locus identifications (IDs) representing F-box proteins in this study.
Sequence Analysis
Domains in F-box proteins were identified using SMART and PFam with an e value cutoff of less than 1.0. The unknown conserved motifs were investigated by MEME (http://meme.sdsc.edu) version 3.5.2 (Bailey and Elkan, 1995). A limit of 50 motifs with other options set to default values was specified. MEME is a tool for discovering motifs in a group of related DNA or protein sequences. MEME uses statistical modeling techniques to automatically choose the best width, number of occurrences, and description for each motif. Multiple alignment analyses were performed using ClustalX (version 1.83) program (Thompson et al., 1997). The unrooted phylogenetic trees were constructed by neighbor-joining method (Saitou and Nei, 1987) and displayed using NJPLOT program (Perriere and Gouy, 1996). Exon-intron organization was determined using genome browser tool at TIGR (http://www.tigr.org/tigr-scripts/osa1_web/gbrowse/rice/). Manual curation and assessment of the gene structure of each predicted F-box protein remains to be done.
Localization of F-Box Protein-Encoding Genes on Rice Chromosomes
All the sequenced contigs of japonica cv Nipponbare have been physically constructed as pseudomolecules at TIGR (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml), representing the 12 rice chromosomes. Each of the F-box protein-encoding genes were positioned on these rice chromosome pseudomolecules by the BLASTN search. The presence of F-box protein-encoding genes on duplicated chromosomal segments was investigated by segmental genome duplication of rice available at TIGR (http://www.tigr.org/tdb/e2k1/osa1/segmental_dup/index.shtml) with the maximum length distance permitted between collinear gene pairs of 100 kb.
Plant Material and Growth Conditions
Rice (subsp. indica var IR64) seeds were obtained from the Indian Agricultural Research Institute. After disinfection with 0.1% HgCl2 for 10 min and thorough washing with reverse-osmosis (RO) water, seeds were soaked overnight in RO water. Seedlings were grown in RO water in a culture room maintained at 28°C ± 1°C with a daily photoperiodic cycle of 14-h light and 10-h dark. For salt treatment, 7-d-old light-grown rice seedlings were transferred to a beaker containing 200 mm NaCl solution for 3 h. For desiccation, 7-d-old light-grown rice seedlings were dried for 3 h between folds of tissue paper at 28°C ± 1°C, in a culture room. For cold treatment, the seedlings were kept at 4°C ± 1°C for 3 h. The seedlings kept in water for 3 h, at 28°C ± 1°C, served as control. Mature leaf, Y leaf, and different stages of panicle (up to 0.5 mm, SAM; 0–3 cm, P1; 3–5 cm, P2; 5–10 cm, P3; 10–15 cm, P4; 15–22 cm, P5; 22–30 cm, P6) and seed (0–2 dap, S1; 3–4 dap, S2; 5–10 dap, S3; 11–20 dap, S4; 21–29 dap, S5) development were harvested from rice plants grown under greenhouse or field conditions. Roots were harvested from 7-d-old light-grown seedlings grown in RO water.
Microarray Hybridization and Data Analysis
Isolation of total RNA and quality controls were done as described previously (Jain et al., 2006b). The microarray analysis was performed using one-cycle target labeling and control reagents (Affymetrix) using 5 μg of a high quality total RNA (Jain et al., 2006b) as starting material for each sample. Affymetrix GeneChip Rice Genome Arrays (Gene Expression Omnibus platform accession no. GPL2025) were used for microarray analysis. Target preparation, hybridization to arrays, washing, staining, and scanning were carried out according to manufacturer's instructions (Affymetrix). Affymetrix GeneChip Operating Software 1.2.1 was used for washing and scanning in Fluidics Station 450 (Affymetrix) and Scanner 3300 (Affymetrix), respectively. Sample quality was assessed by examination of 3′ to 5′ intensity ratios of Poly-A controls, hybridization controls, and housekeeping genes.
For further data analysis, the probe intensity (.cel) files were imported into Avadis prophetic (version 4.2) software (Strandgenomics). The normalization and probe summarization was performed by probe logarithmic intensity error method followed by variance stabilization of 16. The variance stabilization step stabilizes the variance across the entire range of expression including the genes with low expression. Three biological replicates of each sample with an overall correlation coefficient value of more than 0.95 were selected for final analysis. Any data set that did not meet this cutoff value was discarded. The data from 57 hybridizations was included in final analysis. Two projects were created, one for developmental series including data from 48 chips (three biological replicates for each of seedling, root, mature leaf, Y leaf, SAM, P1, P2, P3, P4, P5, P6, S1, S2, S3, S4, and S5) and other for stress series including data from 12 chips (three biological replicates for each of control seedlings and seedlings subjected to salt, dehydration, and cold stress).
To extract the IDs of probe sets present on the Affymetrix rice genome array representing the F-box protein-encoding genes, Rice Multi-Platform Microarray Search (http://www.ricearray.org/matrix.search.shtml) tool available at National Science Foundation Rice Oligonucleotide Array Project was used. Probe sets with the entire set of 11 probes (8–10 in some cases) present on the array aligned with 100% identity over the entire length with corresponding F-box protein genes were considered to be significant. The data for only one probe set for each F-box protein gene was used for expression analysis. In this way, the probes for 617 (out of 687) F-box protein-encoding genes could be identified that were represented on the Affymetrix rice genome array (probe set IDs are given in Supplemental Table S8). After normalization, variance stabilization, and log transformation of data for all the rice genes present on the chip, the log signal intensity values for rice probe IDs corresponding to F-box protein-encoding genes were extracted as a subset. All the subsequent analyses were done on this subset only. To identify differentially expressed genes, a Student's t test was performed. The genes that are up- or down-regulated equal to or more than 2-fold were considered to be differentially expressed significantly. The average of three biological replicates for each sample was used for analysis. We defined a gene as specifically enriched in a given organ only if the expression level of the gene in the organ was significantly higher (more than 2-fold) than the levels in all the other organs.
Real-Time PCR Analysis
To confirm the differential expression of representative F-box protein-encoding genes in various rice tissues/developmental stages and stress treatments identified by microarray data analysis, real-time PCR analysis was performed using gene-specific primers as described earlier (Jain et al., 2006b). The primer sequences are listed in Supplemental Table S11. At least two biological replicates of each sample and three technical replicates of each biological replicate were used for real-time PCR analysis.
FL-cDNA, EST, and MPSS Data Analysis (Digital Expression Analysis)
Gene expression evidence search page (http://www.tigr.org/tdb/e2k1/osa1/locus_expression_evidence.shtml) available at TIGR rice genome annotation was used for digital expression analysis. Each of the TIGR locus IDs corresponding to all F-box protein-encoding genes was searched to find availability of corresponding FL-cDNA, EST, and peptide sequence. Expression evidence from FL-cDNA and EST sequences was determined by minimal alignment over 90% length of the transcript with 95% identity. Peptide sequences from the Koller et al. (2002) publication were mapped to the predicted rice proteome using BLAST and those matching uniquely at 100% identity and 100% of the length of the peptide were utilized.
Expression evidence from MPSS tags was determined from the rice MPSS project (http://mpss.udel.edu/rice/) mapped to TIGR rice gene models. The signature was considered to be significant if it uniquely identifies an individual gene and shows perfect match (100% identity over 100% of the length of the tag). The normalized abundance (tags per million) of these signatures for a given gene in a given library represents a quantitative estimate of expression of that gene. MPSS expression data for 17- and 20-base signatures from 16 libraries representing 12 different tissues/organs of rice were used for the analysis. The description of these libraries is: NCA, 35 d callus; NGD, 10 d germinating seedlings grown in dark; NGS, 3 d germinating seed; NIP, 90 d immature panicle; NML, 60 d mature leaves (representing an average of four replicates; A, B, C, and D); NME, 60 d meristematic tissue; NOS, ovary and mature stigma; NPO, mature pollen; NMR, 60 d mature roots (representing an average of two replicates; A and B); NST, 60 d stem; NYL, 14 d young leaves; NYR, 14 d young roots.
Microarray data from this article have been deposited in the Gene Expression Omnibus database at the National Center for Biotechnology Information under the series accession numbers GSE6893 and GSE6901.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Distribution of rice F-box proteins in different subfamilies.
Supplemental Figure S2. Phylogenetic relationship among the rice F-box proteins.
Supplemental Figure S3. Phylogenetic relationship of rice and Arabidopsis F-box proteins containing LRR domains (A), kelch repeats (B), and TUB domain (C) at their C terminus.
Supplemental Figure S4. Organ-specific expression of F-box protein-encoding genes based on the microarray analysis from an earlier study (Ma et al., 2005).
Supplemental Figure S5. Hierarchial clustering display of all the 617 F-box protein-encoding genes represented on Affymetrix rice genome array in various rice organs and developmental stages (mentioned at the top of each lane).
Supplemental Figure S6. Real-time PCR analysis of representative F-box protein-encoding genes differentially expressed in various tissues/developmental stages and stress treatments.
Supplemental Table S1. F-box proteins with known function in plants.
Supplemental Table S2. F-box protein encoding genes in rice.
Supplemental Table S3. Putative motifs predicted in F-box proteins of FBX subfamily by MEME with an e-value less than e-100.
Supplemental Table S4. F-box protein-encoding genes present on duplicated chromosomal segments of rice.
Supplemental Table S5. Availability of FL-cDNA, EST, and/or peptide sequence(s) corresponding to rice F-box protein-encoding genes.
Supplemental Table S6. MPSS data showing variable tissue-specific abundance of rice F-box protein-encoding genes.
Supplemental Table S7. Rice F-box protein-encoding genes with tissue-specific expression as revealed from MPSS data.
Supplemental Table S8. Average log signal values of 617 F-box protein-encoding genes from three biological replicates of each sample.
Supplemental Table S9. F-box protein-encoding genes differentially expressed under different light conditions.
Supplemental Table S10. Average log signal values of 43 F-box protein-encoding genes differentially expressed under various stress conditions.
Supplemental Table S11. Primer sequences used for real-time PCR analysis.
Supplementary Material
This work was supported by the Department of Biotechnology, Government of India, the Council of Scientific and Industrial Research (research fellowship to M.J., A.N., R.A., S.R., and P.S.), and the University Grants Commission, New Delhi (research fellowship to P.A.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jitendra P. Khurana (khuranaj@genomeindia.org).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adams J, Kelso R, Cooley L (2000) The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol 10 17–24 [DOI] [PubMed] [Google Scholar]
- Anderson A, Hudson M, Chen W, Zhu T (2003) Identification of nutrient partitioning genes participating in rice grain filling by singular value decomposition (SVD) of genome expression data. BMC Genomics 4 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Richman R, Elledge SJ (1994) Human cyclin F. EMBO J 13 6087–6098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86 263–274 [DOI] [PubMed] [Google Scholar]
- Bai J, Pennill LA, Ning J, Lee SW, Ramalingam J, Webb CA, Zhao B, Sun Q, Nelson JC, Leach JE, et al (2002) Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res 12 1871–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1995) The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3 21–29 [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18 630–634 [DOI] [PubMed] [Google Scholar]
- Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M (1999) Identification of a family of human F-box proteins. Curr Biol 9 1177–1179 [DOI] [PubMed] [Google Scholar]
- Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, Deng XW (1996) The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86 115–121 [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38 87–117 [DOI] [PubMed] [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, et al (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford R, Lee MH, Nayak S, Ohmachi M, Giorgini F, Schedl T (2000) FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development 127 5265–5276 [DOI] [PubMed] [Google Scholar]
- Clissold PM, Ponting CP (2001) JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2beta. Trends Biochem Sci 26 7–9 [DOI] [PubMed] [Google Scholar]
- Cooper B, Clarke JD, Budworth P, Kreps J, Hutchison D, Park S, Guimil S, Dunn M, Luginbuhl P, Ellero C, et al (2003) A network of rice genes associated with stress response and seed development. Proc Natl Acad Sci USA 100 4945–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ (1999) SCF and cullin/ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol 15 435–467 [DOI] [PubMed] [Google Scholar]
- Devoto A, Ellis C, Magusin A, Chang HS, Chilcott C, Zhu T, Turner JG (2005) Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol 58 497–513 [DOI] [PubMed] [Google Scholar]
- Devoto A, Nieto-Rostro M, Xie D, Ellis C, Harmston R, Patrick E, Davis J, Sherratt L, Coleman M, Turner JG (2002) COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J 32 457–466 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005. a) The F-box protein TIR1 is an auxin receptor. Nature 435 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M (2005. b) Plant development is regulated by a family of auxin receptor F-box proteins. Dev Cell 9 109–119 [DOI] [PubMed] [Google Scholar]
- Dieterle M, Zhou YC, Schafer E, Funk M, Kretsch T (2001) EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev 15 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani I, Sukegawa S, Kyozuka J (2006) Genome-wide analysis of spatial and temporal gene expression in rice panicle development. Plant J 46 503–511 [DOI] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo SD, Yanagisawa S, Vierstra RD (2004) Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA 101 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JM, Wiederkehr A, Seol JH, Haguenauer-Tsapis R, Deshaies RJ, Riezman H, Peter M (2001) Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol Cell Biol 21 3105–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi K, Sasaki A, Itoh H, Ueguchi-Tanaka M, Ashikari M, Kitano H, Matsuoka M (2004) GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J 37 626–634 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414 271–276 [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115 667–677 [DOI] [PubMed] [Google Scholar]
- Harmon FG, Kay SA (2003) The F-box protein AFR is a positive regulator of phytochrome A-mediated light signaling. Curr Biol 13 2091–2096 [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Klenz JE, Haughn GW (2006) UFO in the Arabidopsis inflorescence apex is required for floral-meristem identity and bract suppression. Planta 223 769–778 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309 293–297 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA (2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426 302–306 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436 793–800 [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46 79–86 [DOI] [PubMed] [Google Scholar]
- Itoh H, Matsuoka M, Steber CM (2003) A role for the ubiquitin-26S-proteasome pathway in gibberellin signaling. Trends Plant Sci 8 492–497 [DOI] [PubMed] [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46 23–47 [DOI] [PubMed] [Google Scholar]
- Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP (2006. a) Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6 47–59 [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006. b) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345 646–651 [DOI] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP (2006. c) Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88 360–371 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Ma L, Strickland E, Deng XW (2005) Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 17 3239–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW (2004) Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev 18 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G (2005) Rice undeveloped tapetum1 is a major regulator of early tapetum development. Plant Cell 17 2705–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451 [DOI] [PubMed] [Google Scholar]
- Kim HS, Delaney TP (2002) Arabidopsis SON1 is an F-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell 14 1469–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim JH, Lee SH, Kim DH, Kang HY, Bae SH, Pan ZQ, Seo YS (2002) The novel human DNA helicase hFBH1 is an F-box protein. J Biol Chem 277 24530–24537 [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Pagano M (2000) The F-box protein family. Genome Biol 1 REVIEWS3002 [DOI] [PMC free article] [PubMed]
- Kleyn PW, Fan W, Kovats SG, Lee JJ, Pulido JC, Wu Y, Berkemeier LR, Misumi DJ, Holmgren L, Charlat O, et al (1996) Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell 85 281–290 [DOI] [PubMed] [Google Scholar]
- Koller A, Washburn MP, Lange BM, Andon NL, Deciu C, Haynes PA, Hays L, Schieltz D, Ulaszek R, Wei J, et al (2002) Proteomic survey of metabolic pathways in rice. Proc Natl Acad Sci USA 99 11969–11974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Takahashi N, Shimada H, Seki M, Shinozaki K, Matsui M (2002) Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol 43 1073–1085 [DOI] [PubMed] [Google Scholar]
- Lai Z, Ma W, Han B, Liang L, Zhang Y, Hong G, Xue Y (2002) An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol 50 29–42 [DOI] [PubMed] [Google Scholar]
- Lan L, Li M, Lai Y, Xu W, Kong Z, Ying K, Han B, Xue Y (2005) Microarray analysis reveals similarities and variations in genetic programs controlling pollination/fertilization and stress responses in rice (Oryza sativa L.). Plant Mol Biol 59 151–164 [DOI] [PubMed] [Google Scholar]
- Laufs P, Coen E, Kronenberger J, Traas J, Doonan J (2003) Separable roles of UFO during floral development revealed by conditional restoration of gene function. Development 130 785–796 [DOI] [PubMed] [Google Scholar]
- Lecharny A, Boudet N, Gy I, Aubourg S, Kreis M (2003) Introns in, introns out in plant gene families: a genomic approach of the dynamics of gene structure. J Struct Funct Genomics 3 111–116 [PubMed] [Google Scholar]
- Levin JZ, Meyerowitz EM (1995) UFO: an Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell 7 529–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Chen C, Liu X, Jiao Y, Su N, Li L, Wang X, Cao M, Sun N, Zhang X, et al (2005) A microarray analysis of the rice transcriptome and its comparison to Arabidopsis. Genome Res 15 1274–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrocco K, Lecureuil A, Nicolas P, Guerche P (2003) The Arabidopsis SKP1-like genes present a spectrum of expression profiles. Plant Mol Biol 52 715–727 [DOI] [PubMed] [Google Scholar]
- Marrocco K, Zhou Y, Bury E, Dieterle M, Funk M, Genschik P, Krenz M, Stolpe T, Kretsch T (2006) Functional analysis of EID1, an F-box protein involved in phytochrome A-dependent light signal transduction. Plant J 45 423–438 [DOI] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern B, Atchley WR (1999) Evolution of bHLH transcription factors: modular evolution by domain shuffling? Mol Biol Evol 16 1654–1663 [DOI] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101 331–340 [DOI] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M (1998) Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet 14 236–243 [DOI] [PubMed] [Google Scholar]
- Perriere G, Gouy M (1996) WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78 364–369 [DOI] [PubMed] [Google Scholar]
- Potocky M, Elias M, Profotova B, Novotna Z, Valentova O, Zarsky V (2003) Phosphatidic acid produced by phospholipase D is required for tobacco pollen tube growth. Planta 217 122–130 [DOI] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115 679–689 [DOI] [PubMed] [Google Scholar]
- Qiao H, Wang H, Zhao L, Zhou J, Huang J, Zhang Y, Xue Y (2004. a) The F-box protein AhSLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell 16 582–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Wang F, Zhao L, Zhou J, Lai Z, Zhang Y, Robbins TP, Xue Y (2004. b) The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell 16 2307–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4 406–425 [DOI] [PubMed] [Google Scholar]
- Samach A, Klenz JE, Kohalmi SE, Risseeuw E, Haughn GW, Crosby WL (1999) The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J 20 433–445 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, et al (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299 1896–1898 [DOI] [PubMed] [Google Scholar]
- Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA (2001) A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ID, Peeters N (2000) The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25 46–47 [DOI] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55 555–590 [DOI] [PubMed] [Google Scholar]
- Smith TF, Gaitatzes C, Saxena K, Neer EJ (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem Sci 24 181–185 [DOI] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282 1488–1490 [DOI] [PubMed] [Google Scholar]
- Somers DE, Kim WY, Geng R (2004) The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101 319–329 [DOI] [PubMed] [Google Scholar]
- Sonneveld T, Tobutt KR, Vaughan SP, Robbins TP (2005) Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. Plant Cell 17 37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Ritchie S, Soule JD, McGinnis KM, Steber CM (2004) Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proc Natl Acad Sci USA 101 12771–12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Shirasu K, Deng XW (2003) The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat Rev Genet 4 948–958 [DOI] [PubMed] [Google Scholar]
- Tang J, Xia H, Li D, Cao M, Tao Y, Tong W, Zhang X, Hu S, Wang J, Yu J, et al (2005) Gene expression profiling in rice young panicle and vegetative organs and identification of panicle-specific genes through known gene functions. Mol Genet Genomics 274 467–476 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Yamane H, Watari A, Kakehi E, Ikeda K, Hauck NR, Iezzoni AF, Tao R (2004) The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J 39 573–586 [DOI] [PubMed] [Google Scholar]
- Wang L, Dong L, Zhang Y, Wu W, Deng X, Xue Y (2004) Genome-wide analysis of S-locus F-box-like genes in Arabidopsis thaliana. Plant Mol Biol 56 929–945 [DOI] [PubMed] [Google Scholar]
- Wang X (2002) Phospholipase D in hormonal and stress signaling. Curr Opin Plant Biol 5 408–414 [DOI] [PubMed] [Google Scholar]
- Wang X, Feng S, Nakayama N, Crosby WL, Irish V, Deng XW, Wei N (2003) The COP9 signalosome interacts with SCF UFO and participates in Arabidopsis flower development. Plant Cell 15 1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ID, Barker GL, Lu C, Coghill JA, Beswick RW, Lenton JR, Edwards KJ (2005) Alteration of the embryo transcriptome of hexaploid winter wheat (Triticum aestivum cv. Mercia) during maturation and germination. Funct Integr Genomics 5 144–154 [DOI] [PubMed] [Google Scholar]
- Winston JT, Koepp DM, Zhu C, Elledge SJ, Harper JW (1999) A family of mammalian F-box proteins. Curr Biol 9 1180–1182 [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D (2002) The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, et al (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3 e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chen C, Li L, Meng L, Singh J, Jiang N, Deng XW, He ZH, Lemaux PG (2005) Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol 139 1107–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonia L, Munnik T (2004) Osmotically induced cell swelling versus cell shrinking elicits specific changes in phospholipid signals in tobacco pollen tubes. Plant Physiol 134 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.