Abstract
Diversification of transcription-associated protein (TAP) families during land plant evolution is a key process yielding increased complexity of plant life. Understanding the evolutionary relationships between these genes is crucial to gain insight into plant evolution. We have determined a substantial set of TAPs that are focused on, but not limited to, land plants using PSI-BLAST searches and subsequent filtering and clustering steps. Phylogenies were created in an automated way using a combination of distance and maximum likelihood methods. Comparison of the data to previously published work confirmed their accuracy and usefulness for the majority of gene families. Evidence is presented that the flowering plant apical stem cell regulator WUSCHEL evolved from an ancestral homeobox gene that was already present after the water-to-land transition. The presence of distinct expanded gene families, such as COP1 and HIT in moss, is discussed within the evolutionary backdrop. Comparative analyses revealed that almost all angiosperm transcription factor families were already present in the earliest land plants, whereas many are missing among unicellular algae. A global analysis not only of transcription factors but also of transcriptional regulators and novel putative families is presented. A wealth of data about plant TAP families and all data accrued throughout their automated detection and analysis are made available via the PlanTAPDB Web interface. Evolutionary relationships of these genes are readily accessible to the nonexpert at a mouse-click. Initial analyses of selected gene families revealed that PlanTAPDB can easily be exerted for knowledge discovery.
The coordinated expression control of the entirety of genes in a given cell determines its physiological state, morphology, and identity in the organism. Reprogramming the set of transcribed genes during development or physiological adaptation requires modulated activation and deactivation of regulatory factors. In eukaryotes, the transcription of protein-coding genes is controlled by complex networks of transcription-associated proteins (TAPs). Specific transcription factors (TFs) activate or repress transcription of their target genes by binding to cis-active elements. Further transcriptional regulators (TRs) include the following: (1) coactivators and corepressors, which bind and influence TFs; (2) general transcription initiation factors, which recognize core promoter elements and recruit components of the basal transcription machinery; and (3) chromatin remodeling factors, which affect the accessibility of DNA through histone modifications and DNA methylation. The modular nature of TFs, possessing DNA-binding and protein-protein interaction domains, facilitates the high diversity of transcriptional regulation.
Changes in transcriptional regulation enhance complexity at the genetic level and thus can generate novel signal transduction pathways. Such changes, mediated by recombined complexes of regulatory proteins as well as by altered regulatory sequence elements, were repeatedly proposed to be a major driving force of evolution (Doebley and Lukens, 1998; Tautz, 2000; Hsia and McGinnis, 2003; Levine and Tjian, 2003; Gutierrez et al., 2004; Carroll, 2005). Previous studies have shown that TAPs are highly specific across prokaryotic and eukaryotic lineages and that their diversity appears to be linked to their phylogenetic distance (Coulson et al., 2001; Coulson and Ouzounis, 2003). In eukaryotes, key players of the basal transcription machinery are highly conserved, whereas many families of DNA-binding TFs are taxon specific and show substantial sequence diversity (Coulson and Ouzounis, 2003). Moreover, the size and genomic fraction of TF families seem to correlate with cellular complexity (Levine and Tjian, 2003).
The evolution of eukaryotic TF genes involves the processes of specific amplification of common families through duplication and diversification, as well as the shuffling of functional domains, resulting in lineage-specific families that can facilitate novel networks of protein-protein interactions and can take over new functions. In plants, the evolution and expansion of specific gene families seem to be more pronounced than in other eukaryotes (Lespinet et al., 2002). In Arabidopsis (Arabidopsis thaliana), genes involved in transcriptional regulation were preferentially retained following whole-genome duplications (Blanc and Wolfe, 2004; Seoighe and Gehring, 2004). It could be demonstrated that TF genes show a higher duplicability as well as retention rate in seed plants compared to other crown eukaryotes and other plant genes (Shiu et al., 2005), which results in considerable lineage-specific expansion of distinct TF families in plants. Consequently, 45% of the TF genes in Arabidopsis were found to belong to families that are specific to plants (Riechmann et al., 2000). Evidence that many plant-specific proteins resemble TFs (Gutierrez et al., 2004) further supports the assumption that the increase of complexity in transcriptional regulation mechanisms has been crucial for the evolution of plants.
In recent years, much emphasis was placed on the understanding of regulatory networks controlling the transcription of genes. Genome-wide comparative analyses aid in revealing the evolution of transcriptional regulation that underlies the diversity of organisms. TAP genes and transcriptional networks have been studied extensively in unicellular organisms (e.g. Kyrpides and Ouzounis, 1999; Perez-Rueda et al., 2004; Madan Babu et al., 2006), as well as in basal metazoans (Satou and Satoh, 2005; Larroux et al., 2006) and crown eukaryotes (Messina et al., 2004; Reece-Hoyes et al., 2005). Within the plant kingdom, only two seed plants, Arabidopsis and rice (Oryza sativa), were globally investigated (for review, see Qu and Zhu, 2006) and their TAP gene families compared to those of the unicellular green alga Chlamydomonas reinhardtii, fungi, and metazoans (Riechmann et al., 2000; Shiu et al., 2005). Little is known about TAPs in nonseed plants, like the bryophyte Physcomitrella patens, and no genome-wide compendium of its TAP genes is available, as is the case for nongreen algae.
While phylogenetic studies have been carried out for single TAP families, e.g. sigma factors, LEAFY (LFY)/FLO, MADS, and AP2 (Ichikawa et al., 2004; Maizel et al., 2005; Riese et al., 2005; Shigyo et al., 2006), a large-scale phylogenetic analysis of TAP gene families from nonseed plants is still lacking. Here, we investigated and compared plant TAP gene families on a genome-wide scale across species of all three domains of life to gain insight into the evolution of transcriptional regulation in plants. We covered the whole evolutionary range from unicellular algae through bryophytes to angiosperms by including genomic-scale sequence data of the diatom Thalassiosira pseudonana, the red alga Cyanidioschyzon merolae, the green alga C. reinhardtii, the moss P. patens, the monocot rice, and the dicot Arabidopsis. The moss P. patens diverged from the ancestor of extant flowering plants at least 450 million years ago (Theissen et al., 2001; Hedges et al., 2004). It was chosen as an offset for this study because, in comparison with flowering plants, it might enable inference of the ancestral state of land plant transcriptional regulation. A comprehensive analysis of gene families can be performed using the large collection of clustered expressed sequence tag (EST) data (Rensing et al., 2002; Lang et al., 2005). Starting from the complete set of P. patens candidate TAP genes, we collected homologs using PSI-BLAST and carried out automated filtering and clustering procedures, followed by manual annotation. From the resulting ample pool of TAP genes, taxonomic distribution, lineage-specific expansion, and high-quality phylogenies were inferred.
RESULTS AND DISCUSSION
Availability: All resources are available via the PlanTAPDB Web interface (http://www.cosmoss.org/bm/plantapdb).
Compilation of the Query Dataset
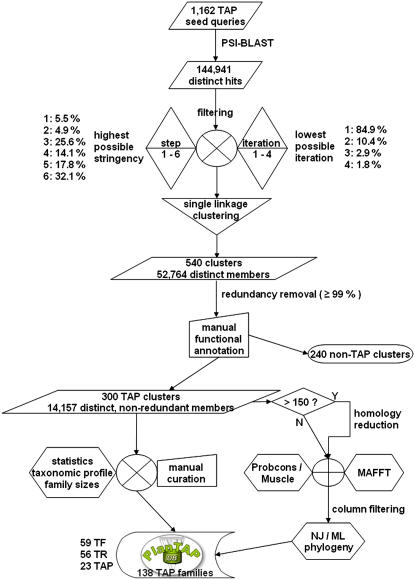
In terms of evolution, mosses are located half way between seed plants and algae and were therefore chosen as an offset for the global phylogenetic analysis of plant TAPs. In addition, mosses morphologically resemble the first plants that occupied the land (Kenrick and Crane, 1997). In the moss P. patens, a total of 1,592 putative TAPs (PTs) were identified from a comprehensive clustered and annotated EST database (Lang et al., 2005) by two strategies: (1) TBLASTN searches with plant and algae reference TAPs compiled by relaxed keyword searches, and (2) motif scans for transcription-associated domains. The resulting comprehensive set of candidate moss TAPs included nearly all TF families known from seed plants (http://arabtfdb.bio.uni-potsdam.de/v1.1/, http://ricetfdb.bio.uni-potsdam.de/v2.1/; Riechmann et al., 2000; Guo et al., 2005; Gao et al., 2006), as well as sequences putatively encoding TAPs. False-positive sequences introduced by this compilation of queries were later removed during the annotation process. To avoid potentially fragmentary virtual transcripts, we determined the full-length closest homolog for each of the moss candidate TAPs to be used subsequently as seed query sequence. For a homolog to be considered, its BLASTX match needed to be in the same frame as the original annotation of the moss candidate transcript and its predicted open reading frame (ORF). A closest homolog could be determined for about 99% of the 1,592 P. patens candidate TAPs. For 19 of the candidate sequences, no homolog was found, yet 12 of those were included into the seed query set because they contained a predicted ORF. The complete nonredundant set of closest homologs used for PSI-BLAST searches comprises 1,162 sequences (Fig. 1). This seed set contains mainly sequences of plant origin (88% Viridiplantae), around 8% of which are derived from bryophytes. Besides 5% of patented sequences, for which no taxon annotation is available, the remainder of the sequences are distributed across metazoa (3%), bacteria (2%), fungi (1%), lower eukaryotes (0.4%), viruses (0.2%), and Archaea (0.1%). The usage of PSI-BLAST enables the detection even of distant homologs (Schaffer et al., 2001), e.g. from algae, fungi, animals, Eubacteria, or Archaea.
Figure 1.
Flowchart of TreePipe and PlanTAPDB. [See online article for color version of this figure.]
Filtering and Clustering of PSI-BLAST Results
During the PSI-BLAST searches, 369,118 hits were generated, representing a total of 144,941 distinct protein sequences (Fig. 1). To deal with the differences in degree of conservation and family size between gene families, we deployed an iterative six-step filtering scheme that optimizes the applied filtering criteria and the selected PSI-BLAST iteration for each query sequence individually. The most stringent step (6), demanding at least 45% sequence identity and 300 amino acids in alignment length, was designed to reduce domain-derived superfamilies to family or subfamily level. Smaller and more diverse superkingdom-spanning families were handled via the least stringent step (1), allowing hits from the fringe of the “twilight-zone” (Rost, 1999) with at least 25% sequence identity and 50-amino acid alignment length. The four intermediate steps (see “Materials and Methods” for details) were designed to assess conservation grades between these two extremes. In total, 115,593 hits (31%) passed the filtering procedure. The majority of sequences (90%) were filtered by steps 3 to 6 (step 3: 21%; step 4: 23%; step 5: 19%; step 6: 27%). For most of the queries (79%), results from the first PSI-BLAST iteration were preferred in order to avoid potential false-positive hits. Overlapping filtered result sets were merged to recover family relations by single linkage clustering using a stringent hit-coverage-based distance measure. The resulting 540 clusters contained 60,504 cluster members, representing 52,764 distinct sequences (Fig. 1). Because some clusters represent different yet possibly overlapping parts of one and the same gene family (see section “Cluster Annotation”), individual sequences can be part of more than one cluster, as indicated by an overlap of 12.8% among the clusters. On average, the clusters contain 112 members, with the largest cluster containing 1,182 and the smallest 21 members. The filtering and clustering procedure was developed and tested using queries derived from 93 previously determined gene families of different function covering algae, moss, and seed plants. In this test case, 91 families were recovered as expected, whereas two gene families were merged. Inspection of the merged cluster revealed that the two families are indeed part of a larger subfamily (ATPases). Therefore, the filtering and clustering procedure is able to recover family structure with good performance.
Redundancy Removal and Homology Reduction
While it greatly improves taxon sampling, the strategy to use both a huge multispecies-containing database like UniProt and the individual whole-genome protein predictions results in the detection of identical protein sequences from these overlapping databases. In addition, the same locus is often represented by more than one protein sequence due to divergent predicted gene models, splice variants, as well as sequencing and annotation errors. To cope with this problem, redundant copies of genes were eliminated prior to all functional analyses using an identity cutoff of ≥99% for sequences of the same species. The total number of cluster members was thus reduced by 30%, resulting in 42,133 total sequences, 37,247 of which are distinct.
In addition, a homology-reduced set of the 540 clusters was compiled to infer phylogenies (Fig. 1). Phylogenetic inference of large clusters is computationally costly, and the interpretation and inference of results from huge trees is difficult. As a total of 102 clusters had more than 150 members, these were condensed via stepwise homology reduction until the threshold of 150 members was reached. The homology-reduced clusters contain 29,317 cluster members in total, 26,595 of which are distinct. The average pairwise distances within clusters were found to be in the range of 12% to 95% identity with an average of 44%.
Multiple Alignments and Selection of Conserved Sites
Due to errors introduced by the alignment algorithm, a certain fraction of columns in a multiple sequence alignment (MSA) generates noise that disturbs correct inference of phylogenetic relationships (Castresana, 2000; Rosenberg, 2005). Such positions are usually removed manually in the course of a phylogenetic analysis. While current approaches to automated phylogenies (Sicheritz-Ponten and Andersson, 2001; Fuellen et al., 2003; Frickey and Lupas, 2004; Gouret et al., 2005) mostly rely on unprocessed ClustalW alignments, we placed more emphasis on the alignment quality to increase the reliability of the resulting phylogenies. Thus, we used a measure that describes evolutionary informative sites. We implemented a best-of-two approach, during which two alignments were (1) calculated using different state-of-the-art algorithms and (2) filtered using the sum-of-pairs score. In the next step, the alignment with the maximum number of remaining columns was chosen (Fig. 1). On average, the alignments consisted to 65% of gaps and were reduced to 28% of the original alignment length by applying this procedure. In 71% of the cases, the MAFFT G-INSI (Katoh et al., 2005) alignment was selected to represent the cluster, whereas ProbCons (Do et al., 2005) or Muscle (Edgar, 2004) were chosen for 29% of the clusters.
Automated Reconstruction of High-Quality Phylogenies
Many approaches to phylogenomics rely solely on a distance approach using neighbor joining (NJ; Saitou and Nei, 1987). However, NJ is known to be susceptible to noisy data, provides no confidence measures, and makes it hard to compute reliable distances for strongly divergent sequences. Probabilistic approaches, such as maximum likelihood (ML) and Bayesian methods, are known to overcome most of these problems, but both are very time consuming and thus usually not applied in large-scale phylogenomics approaches. We followed a combined approach by calculating ML consensus branch lengths using gamma-distributed rates from bootstrapped NJ topologies (Fig. 1). We compared published phylogenies of plant TAP families to those created by the approach presented here. In general, the same topology was recovered and the same conclusions could be drawn from the automatically generated phylogenies described here. For example, homologs of the floral regulator LFY, a plant-specific TF, are present in all land plants. The LFY phylogeny is characterized by two deep clefts separating (1) angiosperms from gymnosperms and ferns and (2) mosses from gymnosperms and ferns (Maizel et al., 2005). The same can be seen in the automatically generated PlanTAPDB LFY tree (TF037, accessible via the Web interface), while the increased taxonomic sampling of the cluster presented here even results in higher resolution of the phylogeny. As another example, both the automatically generated Retinoblastoma family tree, TR030, and a published phylogeny (Sabelli and Larkins, 2006) reveal lineage-specific expansion of this gene family in grasses. Phylogenetic trees of gene families can be utilized to analyze the evolution of a gene of interest, to discover orthologs, and to aid functional gene annotation. While phylogenies have been published for several plant TF families (e.g. Theissen et al., 2000; Ichikawa et al., 2004; Maizel et al., 2005; Sabelli and Larkins, 2006; Shigyo et al., 2006), this study presents phylogenies with a dense taxon resolution for plant-anchored gene families and subfamilies not only of TFs but also of TRs and PTs. These data can in turn be applied as a tool for knowledge discovery. In addition to the phylogenies described above, which are based on the homology-reduced clusters, we also calculated initial phylogenies for the full clusters prior to the homology reduction step using bootstrapped NJ.
Cluster Annotation
The functional annotation of the 540 candidate TAP clusters was inferred from identified Inter-Pro domains and associated Gene Ontology (GO) terms (Camon et al., 2004) of the cluster members after redundancy removal. A total of 482 out of 540 clusters contained one or more Inter-Pro domains with a relative occurrence of ≥80% among the nonredundant cluster members. While those were used for automated annotation, clusters with uncertain domain occurrence were manually checked and annotated. In total, only three clusters were composed of sequences from multiple unrelated TAP families. These large mixed clusters were formed due to shared DNA-binding or protein-protein interaction domains (IPR001487 Bromodomain, IPR002110 Ankyrin, IPR002713 FF domain) and were not further considered in this study. Members of 237 clusters are not directly associated with transcriptional regulation but function in related processes, such as DNA and RNA metabolism, and were also not further considered. They derive from the loose initial query selection intended to include as many as possible novel TAP families. The vast majority (94%) of the remaining 300 annotated TAP clusters (Fig. 1) contain sequences of single families or subfamilies. This confirms that the single-linkage clustering approach successfully formed clusters according to functional gene families and subfamilies. In some cases (18 clusters), closely related (sub)families are represented by a single cluster due to shared domains or conserved regions. For instance, two types of regulators of the auxin response, ARF and Aux/IAA, form one cluster (TF007) due to the shared Aux/IAA-ARF dimerization domain. Likewise, very small or orphan TAP families are sometimes completely submerged within clusters of related families (e.g. AN/TF002 and NPR1-like/TR023). Large gene families composed of several diverse subfamilies (e.g. C2C2/TF012–TF015) are sometimes represented by two types of overlapping PlanTAP clusters. They either provide a global view across the main family (C2C2 - GATA, CO-like, and Pseudo ARR-B/TF015) or exclusively span single subfamilies (C2C2 - CO-like/TF012; C2C2 - Dof/TF013; C2C2 - GATA/TF014).
TAP Gene Family Annotation
TAP clusters with the same functional annotation (main and subfamily), which had not been merged during single linkage clustering due to the stringent parameters applied there, were manually grouped, resulting in 138 families of TAPs (Supplemental Table S1). This resulted in a total number of 14,680 nonredundant TAP family members, while the remaining overlap among the families was reduced to 3.6% (14,157 distinct nonredundant family members; Fig. 1), indicating a good separation of the gene families. Fifty-four of the TAP families are represented by more than one cluster of deviating but partially overlapping composition. These multiple clusters depict the particular TAP family either from a different taxonomic perspective (e.g. restricted to the plant lineage versus covering all kingdoms) or comprise different subfamilies. Because large TAP gene families are substantially divergent outside of their conserved domains, it appears more reasonable to deduce phylogenies from subgroups to be able to utilize as much homologous sequence information as possible. The phylogenetic trees were therefore derived for each of the 300 separate TAP clusters.
We divided the TAP families into three categories according to their molecular function and associated GO terms: (1) DNA-binding TFs (59), which comprise direct activators or repressors of transcription; (2) TRs (56), comprising basal TFs interacting with RNA polymerase II or the core promoter, coactivators/corepressors, and chromatin remodeling factors; and (3) proteins with unknown function and/or domains that are possibly associated with transcriptional regulation (PT, 23; Fig. 1).
Previously, plant TF gene families were globally identified in two seed plants, Arabidopsis and rice (http://arabtfdb.bio.uni-potsdam.de/v1.1/, http://ricetfdb.bio.uni-potsdam.de/v2.1/; Riechmann et al., 2000; Guo et al., 2005; Gao et al., 2006). Of the previously described TF families, just 14 are not present among the annotated families due to their absence from the P. patens candidate TAP set. However, eight of those (AS2/LOB, BES1, BZR, GeBP, GFR/ENBP, HRT-like, TCP, VOZ) could be identified in the whole-genome shotgun sequences produced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/), which became available recently, i.e. they were not covered by the clustered EST database used for query compilation. This confirms earlier estimates (Rensing et al., 2002) and shows that the EST data cover the P. patens transcriptome almost completely (in terms of TAP families, the coverage is 95%). For the other six missing TF families (C2C2-YABBY, NOZZLE [NZZ], PBF-2-like/Whirly, S1Fa-like, STERILE APETALA [SAP], ULTRAPETALA [ULT]), no homologs could be identified in the P. patens genomic traces. This might be due to the actual lack of these genes in the P. patens genome (which might also be a derived feature, i.e. secondary gene loss) or because differing rates of mutation fixation render detection using only homology searches impossible. Yet, the above-mentioned results demonstrate that using moss as an offset to identify a broad scope of plant TAPs is a valid approach, as only 4% of angiosperm TF families are unaccounted for. Furthermore, it provides evidence that the majority of flowering plant TF families can be tracked down to the basal land plant P. patens. The above-mentioned TF gene families that are absent from moss are all of small size and have specialized functions in flowering plants. They probably emerged after the evolutionary split of mosses and seed plants. The vegetative and reproductive development of flowering plants is entirely different from that of mosses, the life cycle of which is dominated by a haploid gametophytic phase. They do not possess flowers, the organs for sexual reproduction of angiosperms. While mosses do contain homologs of some angiosperm (floral) homeotic genes, like KNOX (TF031) and MIKC-type MADS box (TF041), their function remains unclear (Theissen et al., 2001). On the other hand, NZZ, SAP, and ULT all play specific roles during development of flowers (Byzova et al., 1999; Schiefthaler et al., 1999; Carles et al., 2005) and are absent from P. patens. The C2C2 zinc finger protein YABBY is expressed in a polar manner and specifies the abaxial identity of lateral organs of the Arabidopsis sporophyte (Siegfried et al., 1999), while the moss sporophyte is extensively reduced and possesses no lateral organs. Likewise, spinach (Spinacea oleracea) S1F mRNA accumulates in roots and etiolated seedlings (Zhou et al., 1995), while both tissues are not present as such in P. patens. Hence, absence of these specialized TF families from a basal land plant seems plausible.
Coverage of Known TAP Families
To analyze the level of completeness of our dataset, we compared numbers of PlanTAPDB family members with the size of well-known Arabidopsis TAP families. In Supplemental Table S2, those PlanTAP families that were previously described by Riechmann and colleagues (Riechmann et al., 2000) and/or are included in the current version (Version 2; July 2006) of DATF (Guo et al., 2005) are listed. To allow comparison of PlanTAPDB Arabidopsis members with these resources, only those member sequences corresponding to The Institute for Genomic Research (TIGR) Arabidopsis loci (loci themselves or those replaced by redundant UniProt sequences) were counted. The numbers shown were ascertained immediately after filtering and clustering, as well as after redundancy removal and homology reduction (Supplemental Table S2). Fortunately, the step of redundancy removal in no case accidentally reduced the number of detected Arabidopsis loci. As expected, the homology reduction leads to a decrease in size of large families. The coverage of a minority of Arabidopsis TAP families by PlanTAPDB differs significantly due to possible annotation errors within the different resources (e.g. the C3H family, which probably also includes RNA-binding C3H zinc fingers). Taken together, the data illustrate that the PSI-BLAST approach is able to recover most of the members for the majority of gene families. However, especially in gene families with low sequence conservation apart from functional domains (e.g. MADS, HB), a significant amount of family members might be missing. This depicts an inevitable shortcoming of this automated approach for the discovery of gene families. Nevertheless, on average the filtered and nonredundant Arabidopsis loci as present in PlanTAPDB cover 81% of the previously published gene family members.
Web Interface
The PlanTAPDB Web interface (http://www.cosmoss.org/bm/plantapdb) provides dynamic access to the results generated in this study. TAP gene families can be retrieved by their accession numbers and identifiers or queried via keyword searches among the family annotations. In addition, all 37,247 TAP cluster sequences (Fig. 1) can be queried using BLAST. The PlanTAPDB portal gives an overview of all available families of TFs, TRs, and PTs in the form of grouped lists or a clickable image map of their overall taxonomic profile (described below). Both provide access to the PlanTAPDB family entry of interest via hyperlinks. The family viewer displays the results of the comprehensive manual annotation process (main family, subfamily, consensus Inter-Pro domains), as well as literature references and the list of annotated family members (including a graphical representation of their domain structure) for each of the 138 TAP families. The extensive information available for every member, e.g. Inter-Pro domains and taxon information, is cross-linked to the primary databases. The individual taxonomic profile, as well as species names and several other parameters, can be used to filter the family member list. All member sequences can be retrieved selectively in FASTA format. The cluster(s) of which a PlanTAPDB family is composed can be accessed via links to the corresponding cluster view(s) and contain the following features: (1) the cluster's description and an optional comment that provides additional information derived from the manual annotation process; (2) the distance matrix and detailed statistics in the form of histograms and box plots, describing the cluster's sequence diversity as found in the redundancy removal and the homology reduction phase of TreePipe; (3) a graphical overview describing the distribution of the sum-of-pairs score, Shannon's entropy score, the gap ratio, and the column removal threshold along the length of the complete alignment used in the selection of conserved sites; (4) the initial alignment of all cluster members used to build the distance matrix as well as the filtered alignment, which was used to infer the phylogeny, viewable and downloadable via the Jalview applet (Clamp et al., 2004); and, finally, (5) the phylogenetic trees and various parameters describing the homology-reduced ML tree topologies, which can be viewed and downloaded in New Hampshire/eXtended (http://phylogenomics.us/forester/NHX.html) format with bootstrap values and color-coded taxon information using the ATV applet (Zmasek and Eddy, 2001). The applet also allows by-node (group) retrieval of sequences displayed using the cosmoss sequence retrieval system (Lang et al., 2005). Next to the button providing the homology-reduced topologies gained by the combined ML/NJ approach, two additional trees containing all cluster members (i.e. prior to the redundancy removal and homology reduction steps) can be viewed for each cluster of a TF or TR family. The first tree displays an unrooted NJ topology with bootstrap values and the second one a midpoint-rooted NJ topology with ML branch lengths.
Different Expansion of TAP Gene Families among Algae and Plant Lineages
Previous global comparative studies of plant TAP gene families focused mainly on the subgroup of DNA-binding TFs in seed plants (for review, see Qu and Zhu, 2006). On basis of the PlanTAPDB data, we compared characteristics of plant TAP gene families across six species, for which genome-scale databases were queried during homolog detection. These included three algae, a moss, and two flowering plants to provide a broad evolutionary perspective. The total number of distinct TFs, TRs, and PTs of these species was extracted using the taxonomic annotation of the family members. The numbers of TFs detected by the approach presented here are smaller than previously published results for Arabidopsis, rice (Xiong et al., 2005; Gao et al., 2006; Qu and Zhu, 2006), and C. reinhardtii (http://chlamytfdb.bio.uni-potsdam.de/v2.0/), which is due to the stringent filtering process applied to prevent false-positive hits.
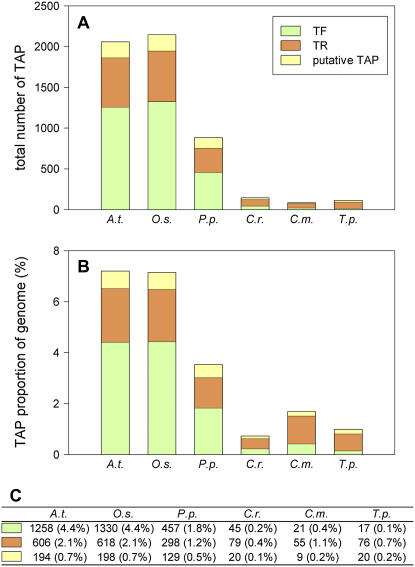
There seems to be a trend that total amounts of TAPs (Fig. 2) are associated with the number of cell types in the respective organism (there is no significant difference between Arabidopsis and rice [P = 0.84], but P. patens differs significantly from both the flowering plants and the algae in this regard [P < 0.001]). A correlation of numbers of TFs with organism complexity (which might be defined as number of cell types) has previously been described for animals (Levine and Tjian, 2003). The low amount of TF genes in the three algae as compared with the three land plants (Fig. 2A, P < 0.001) coincides with reports for basal metazoans (the demosponge Reniera, the urochordate Ciona, the worm Caenorhabditis elegans, and the fly Drosophila melanogaster), which contain a much lower amount of TFs than mammals (Riechmann et al., 2000; Reece-Hoyes et al., 2005; Satou and Satoh, 2005; Larroux et al., 2006). The fraction of TAPs per protein-coding genes in the respective genomes (Fig. 2B) depicts the same trend of association with the number of cell types.
Figure 2.
Abundance of plant TAP genes. The absolute (A) and relative (B) amounts of TAP genes in six species (see Supplemental Table S1 for abbreviations) are shown as bar charts and numerical values (C). The absolute gene numbers were inferred from the NCBI taxonomy information of all TAP family members. The relative abundance of TAPs is shown in relation to the total number of predicted proteins within the respective organism. TFs are shown in green, TRs in orange, and PTs in yellow.
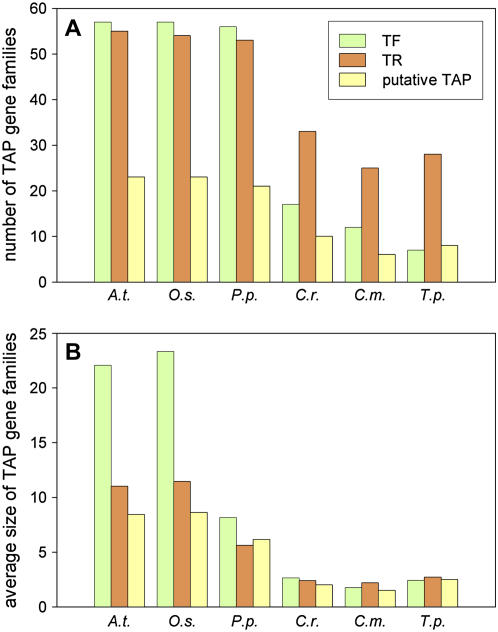
The gene family data (Fig. 3A) reveal an extensive (4.7-fold) increase in the number of different TF gene families with the transition from the three algae (average 12.0 ± 5.0 families) to the three land plants studied here (average 56.7 ± 0.6). The number of TR families exhibits the same trend (average 28.7 ± 4.0 versus 54.0 ± 1.0), but less pronounced (1.9-fold), indicating an increased importance of TF genes for the evolution of the three land plants in question. Consistent with this, components of the basal transcriptional machinery and general TFs are known to be conserved across the three domains of life, while DNA-binding TFs have been shown to evolve in a lineage-specific way in plants as well as in animals (Coulson and Ouzounis, 2003; Gutierrez et al., 2004). A relationship between the increasing number of plant TAP families and the gain in morphological complexity has been hypothesized before (Doebley and Lukens, 1998; Hsia and McGinnis, 2003; Gutierrez et al., 2004). Additionally, because basal multicellular metazoans already contain most of the TF families present in mammals (Riechmann et al., 2000; Messina et al., 2004; Reece-Hoyes et al., 2005; Larroux et al., 2006), which is not the case for the comparison of algae and land plants as shown above, this explosion of gene family number might well be related to the switch from unicellularity to multicellularity. This theory is further supported by the fact that the fraction of human TAP families present in unicellular fungi is drastically reduced as compared to metazoans (Riechmann et al., 2000; Coulson and Ouzounis, 2003; Messina et al., 2004). In concordance with this, nearly all of the different plant TAP gene families are already present in the basal land plant P. patens (Fig. 3A). However, in Arabidopsis and rice, the size (but not the number) of TAP gene families is significantly increased (Fig. 3B, P < 0.001), which might reflect the more complex body plan and specialization of the two angiosperms as compared with P. patens. However, it should be noted that the differences in average gene family size might also be due to inheritance from the respective last common ancestor and thus might not be related to morphological complexity. Yet, the role of lineage-specific expansion of gene families for the evolution of eukaryotes was studied before and predominantly occurs in plants, especially in the case of TAP gene families (Lespinet et al., 2002; Shiu et al., 2005).
Figure 3.
Number and size of plant TAP families. Number (A) and average gene family size (B) of TAP families in six species (see Supplemental Table S1 for abbreviations) are shown. The average gene family size was calculated as the ratio of absolute number of family members per number of families. TFs are shown in green, TRs in orange, and PTs in yellow.
Species-Specific Expansion of Individual TAP Families
The absolute size of the 138 annotated TAP families for the above-mentioned six species is shown in Supplemental Table S1. The size distribution of the Arabidopsis TF gene families correlates well with published results (Qu and Zhu, 2006), although the families are generally smaller due to the stringent elimination of false-positive hits applied in this study. The overall lineage-specific expansion of family number and size is evident from Figure 3, as well as from the absolute values in Supplemental Table S1. On average, TAP gene families are 2 to 3 times larger in P. patens than in the three algae, while in Arabidopsis and rice the TR and PT families show an approximately 4-fold increase and TF families a 9-fold increase as compared to the algae (Fig. 3). Consistent with this, recent comparative studies revealed that TAP family sizes in Arabidopsis and rice often expand with similar rates (Shiu et al., 2005; Xiong et al., 2005). We also determined gene families that were subject to individual expansion against the background of lineage-specific evolution, i.e. families in which above-average expansion of distinct gene families per species occurred. The data underlying Supplemental Table S1 for each of the three TAP groups (TF, TR, and PT) were normalized using the respective total amount of genes per group, and significantly deviating families were highlighted by framing. In total, 29 families exhibit species-specific expansion, two of which are present in Arabidopsis, one in rice, 10 in P. patens, four in C. reinhardtii, nine in C. merolae, and three in T. pseudonana. Moss and the algae contain more specifically expanded TAP families (e.g. HIT and CONSTITUTIVE PHOTOMORPHOGENIC1 [COP1] in P. patens, PcG and SBP in C. reinhardtii, FHA and TFb2 in C. merolae, DUF833 in T. pseudonana) than the two seed plants, which might be due to the fact that the overall expansion rate is less pronounced in the former organisms.
As an example, members of a distinct branch of the His triad family (TF033, HIT) known from animals (Kijas et al., 2006) and fungi are only present in rice and moss. Interestingly, the human HIT protein Aprataxin, which belongs to this family, has recently been shown to be involved in the protection against genotoxic stress by interaction with proteins that are involved in DNA repair (Gueven et al., 2004). Apparently, the forefather of this particular gene was already present in ancestral eukaryotes but has been lost in some plant and algal lineages. The P. patens Aprataxin-like protein might be involved as an upstream component of DNA mismatch repair (Trouiller et al., 2006) and thus might be related to the high efficiency of homologous recombination observed in the moss (Kamisugi et al., 2005).
Taxonomic Distribution of Plant TAP Families across All Domains of Life
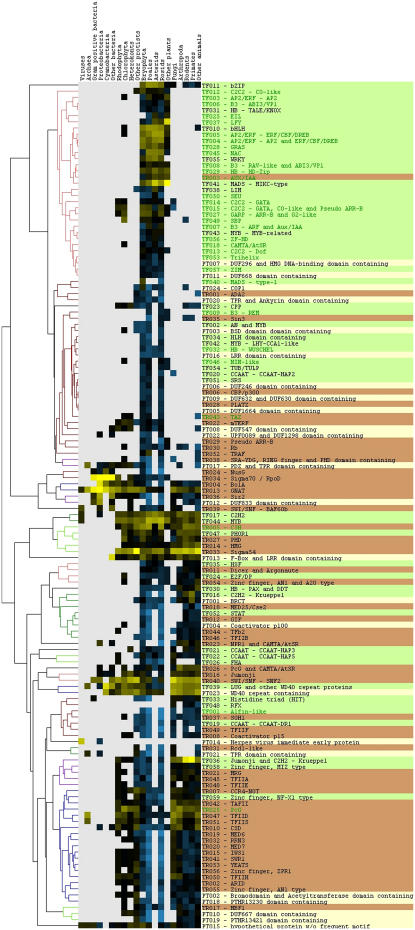
For visualization of the distribution of TAP family members across all taxonomic lineages, a taxonomic profile was created and is presented as a heat map in Figure 4. Initial tests using taxonomic resolution fixed at the kingdom or order level, respectively, were not able to resolve the expected phylogeny of the contributing taxa using columnwise clustering (data not shown). Therefore, those taxonomic groups that contributed significantly to the overall distribution were selected as columns; the remainder of the Eubacteria, protists, plants, and animals were gathered into the respective “other” columns. Thus, a nonredundant representation of the taxonomic distribution was created that is able to resolve the expected phylogeny using columnwise clustering. To overcome the sampling bias presented by fully sequenced genomes, the columns were normalized. Subsequent clustering yielded the significantly correlated groups depicted in Figure 4. The top half of the taxonomic profile contains families that are predominantly present in plants. Within these, the first significantly correlated cluster is almost completely composed of large plant TF families, most of which have been described as plant specific before (highlighted by green text color), while the second cluster contains a mixture of plant TAP families not yet discovered in Asterids. Only a few families, mostly TRs, are abundant in both prokaryotes and eukaryotes (located mainly in the middle part of the profile). The families in the second half of the profile are shared between plants and other eukaryotes and are sometimes present in Eubacteria and Archaea as well. The TR families accumulate within these clusters, especially in the lowest part. This distribution correlates very well with published data (Riechmann et al., 2000; Coulson et al., 2001; Coulson and Ouzounis, 2003) and indicates that TFs often fulfill lineage- or kingdom-specific functions, while basal components of transcriptional regulation are conserved across different eukaryotic kingdoms or sometimes even across the primary domains of life. The profile gives a good impression about the distribution of certain families or clusters of families among taxonomic groups. It can be applied to narrow down the probable function of PTs, such as PT007 (DUF296 and HMG DNA-binding domain containing), which is located in the topmost significantly correlated cluster that is mainly composed of plant-specific TFs. The hypothesis that PT007 might represent a novel TF family is fortified by the domain structure of the members, most of which contain the two PFAM (Finn et al., 2006) domains AT_hook (PF02178) and DUF296 (PF03479), which are known to be present in this particular order in a class of proteins that is thought to have DNA-binding activity. Overexpression of a protein containing DUF296 led to late flowering and modified leaf development in Arabidopsis (Weigel et al., 2000). In addition, the taxonomic profile can be employed to reveal families with biased profiles, which point at interesting evolutionary differences. As an example, among the plant-specific upper part there are clusters that, besides mosses and seed plants, contain sequences from “other,” i.e. nonphotosynthetic, protists, such as PT020 (TPR and Ankyrin domain containing; Fig. 4). A closer look reveals that the cluster contains sequences from the kinetoplastid parasites Trypanosoma cruzi and Leishmania major. While the Alveolata, such as the malaria parasite Plasmodium, harbor the remnant of a plastid, the so-called apicoplast (Waller and McFadden, 2005), this is not the case for the Kinetoplastida. Yet, they belong to the photosynthetic Euglenozoa, and, based on some plant-like nuclear genes, it has been argued that they have undergone secondary loss of a plastid (Hannaert et al., 2003), which coincides nicely with the data from cluster PT020.
Figure 4.
Taxonomic profile of the plant TAP families. The NCBI taxonomy information for all family members was parsed per annotated TAP family. The columns represent those taxonomic groups that contributed significantly to the distribution; the remainder of the Eubacteria, protists, plants, and animals are represented as “other,” respectively. After normalization of the columns (log odds ratio), the rows were clustered and visualized as a heat map (yellow = overrepresented, blue = underrepresented, black = average representation, gray = missing). In the case of overrepresentation and underrepresentation, the color intensity increases with rising distance from zero. All clusters with a centered Pearson correlation coefficient R ≥ 0.7 are displayed in color to the left of the heat map. The PlanTAPDB family names and annotations are shown to the right (green: TF; orange: TR; yellow: PT; green text: previously described as plant specific).
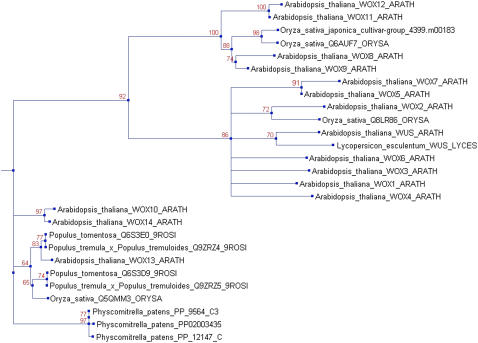
The WUSCHEL/WOX Phylogeny
The HB/WUSCHEL (WUS) family (TF032_373) exhibits a rigorous land plant-specific taxonomic profile, comprising the species Arabidopsis, tomato (Solanum lycopersicum), poplar (Populus spp.), rice, and P. patens. The consensus domains for this family are Homeobox (IPR001356), Homeodomain_like (IPR009057), and Homeodomain-rel (IPR012287). During redundancy filtering, 10 nearly identical sequences belonging to Arabidopsis, rice, and poplar were removed. The average identity between the remaining sequences is relatively low (36.26%); therefore, the alignment was reduced from an initial 950 columns to 167 columns that could be unequivocally aligned, comprising mainly the actual homeobox domain. Due to the low conservation grade of the WUS-related (WOX) gene family (e.g. 30.6% amino acid identity between Arabidopsis WOX9 and WOX14), several annotated homologs were not detected by the PSI-BLAST search and thus are missing from the above-mentioned phylogeny. To add those, all annotated Arabidopsis WUS/WOX sequences were retrieved from Swissprot. After retrieval of the remainder of the sequences using the PlanTAPDB Web interface, MSA and tree reconstruction were performed. The phylogeny is available via the Web interface as well, as an example for manually curated data to be added upon request. The resulting tree (Fig. 5) is clearly separated into two clusters, one containing Arabidopsis WUS itself as well as the majority of WOX sequences, and the other containing Arabidopsis WOX 10, 13, and 14. While WUS has been shown to be involved in shoot meristem maintenance (Mayer et al., 1998; Leibfried et al., 2005; Kieffer et al., 2006), the role of the other members of the gene family is not well defined yet, although some of the genes are involved in early embryonic cell fate decisions (Haecker et al., 2004). Given the deep cleft in the phylogenetic tree, the ancestral WUS/WOX gene probably had already acquired a paralog in the last common ancestor of all land plants. However, because P. patens homologs are exclusively present in the cluster containing the WOX 10, 13, and 14 homologs from Arabidopsis, the paralog later giving rise to WUS was probably lost from the moss lineage after the divergence from the flowering plants. While involvement in stem cell maintenance and early embryo development has been described for WUS and several WOX gene products, this is not the case for WOX 10, 13, and 14 (Haecker et al., 2004). Therefore, the function of this retained ancestral WOX homeobox TF subfamily remains enigmatic at present.
Figure 5.
Phylogeny of the WUS/WOX gene family. A manually expanded phylogeny of the PlanTAPDB WUS family (TF032_337) is presented. The ML tree is shown with quartet support values at the nodes. The protein sequences are represented by species name and accession number. [See online article for color version of this figure.]
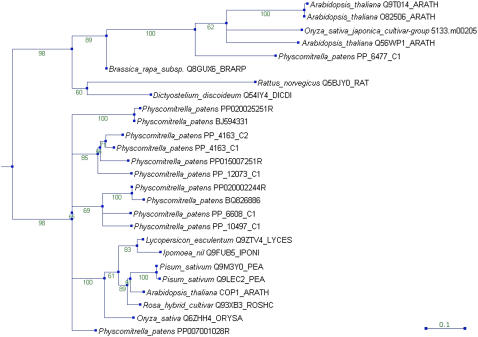
The COP1 Phylogeny
The three uppermost clusters of the taxonomic profile (Fig. 4) contain families that are generally present in plants and also appear erratically in other taxonomic groups. Among those, the PT family PT024 (COP1) can be found. It attracts attention because of the overrepresentation of moss sequences that is apparent from both the taxonomic profile (Fig. 4) and the species-specific expansion (Supplemental Table S1), which is in contrast to the generally lower amount of P. patens TAPs as compared to rice and Arabidopsis (Figs. 2 and 3). In angiosperms, the E3 Ubiquitine ligase COP1 acts as a photomorphogenesis/skotomorphogenesis switch by degradation of downstream factors in the dark, while it is inactivated by nuclear depletion in the light (Holm and Deng, 1999). In mammals, the homolog has been suggested to be involved in tumorigenesis and stress response (Yi and Deng, 2005). In Arabidopsis, a single COP1 gene is present that comprises several WD40 domains and an N-terminal RING domain. Consequently, the PT024 family was annotated using the domains WD40 (IPR001680)/WD40_like (IPR011046) and Znf_RING (IPR001841). Through redundancy removal the cluster members were reduced from 37 to 26, the redundant sequences originating from Arabidopsis, tomato, pea, and rice. The family is well conserved (average 61.39%) with even the rat homolog sharing 43.53% amino acid sequence identity with Arabidopsis COP1. The tree (Fig. 6) is clearly divided into two parts. The lower subtree contains most of the plant sequences, including Arabidopsis COP1 and several orthologs from monocots and dicots. Surprisingly, this cluster also contains a total of 11 P. patens sequences. While all the seed plant proteins in this cluster contain RING domains, this is not true for any of the moss sequences. The proteins in the upper subtree, containing some plant sequences as well as the rat and Dictyostelium homologs, do not contain RING domains, with the exception of the Brassica and Dictyostelium sequences. The Arabidopsis proteins present in this part of the phylogeny are SPA (suppressor of phytochrome A) proteins, which are dimerization partners of COP1 (Laubinger et al., 2006). Because the P. patens data are based on clustered ESTs, it is possible that too many homologs are present in the tree and that the sequences are fragmentary. Therefore, we analyzed the genomic situation by detecting and clustering all homologs within the whole-genome shotgun sequence data available via http://www.cosmoss.org. This analysis revealed that a total of nine COP1 homologs are present in the genome, all of which contain a RING domain (which was missing from the virtual transcripts because of incomplete EST data). The genomic sequences are covered by the 11 virtual transcripts present in the tree. We also detected an additional SPA homolog that lacks the RING domain (data not shown). Hence, P. patens seems to have acquired and retained several COP1 paralogs during evolution. While mosses are not able to etiolate, they certainly do possess photomorphogenesis and harbor a full complement of photoreceptors (Bierfreund et al., 2004; Ichikawa et al., 2004; Kasahara et al., 2004; Mittmann et al., 2004; Uenaka et al., 2005). Still, the expansion of this particular gene family is puzzling. It has been demonstrated recently in Arabidopsis that COP1, yet not the SPA proteins, is involved in UV-B tolerance by coordination of ELONGATED HYPOCOTYL5 (HY5) controlled as well as other pathways (Oravecz et al., 2006). The closest homologs of Arabidopsis HY5 are present in cluster TF011_518, which belongs to family TF011 (bZIP). The proteins are well conserved (identity 65.68%) and contain a single moss ortholog. Therefore, expansion of COP1 downstream factors is not apparent in moss. However, maybe the plethora of P. patens COP1 proteins aids in acquiring UV tolerance, a process that has been associated with pigment changes, e.g. in an Antarctic moss (Newsham, 2003).
Figure 6.
Phylogeny of the COP1/SPA gene family. The automatically generated PlanTAPDB phylogeny of the COP1 family (PT024) is presented. The consensus tree of 100 bootstrapped NJ trees with ML branch lengths is shown; bootstrap values are shown at the nodes. The protein sequences are represented by species name and accession number. [See online article for color version of this figure.]
Caveats
PlanTAPDB users should be aware that the automated homolog detection and clustering approach resulted in the loss of some gene families, i.e. a low percentage (approximately 4%) of plant TAP families is missing. In addition, on average 19% of the gene family members known from well-annotated genomes are lacking. To present phylogenetic trees that can be viewed on a normal computer screen, large gene families have been reduced to contain a maximum of 150 homology-condensed members. Due to the fragmentary nature of the data (incomplete genome/transcriptome data, fragmentary sequences, sampling bias), the phylogenetic analyses might be biased or flawed. Taken together, users should take appropriate caution concerning the points raised above while interpreting the data.
Potential Uses
The PlanTAPDB resource might be used as a starting point for knowledge discovery. Using the family and cluster annotation available through the Web interface, designated gene families can be located, e.g. by name or member sequence accession number. MSAs of the gene families as well as arbitrary sequence subsets can be retrieved. The taxonomic profile (Fig. 4, also available via the Web interface) and the overrepresentation analysis (Supplemental Table S1) might be employed to detect biased taxonomic distribution. Descriptive data, such as sequence conservation, gene family size, species distribution, and alignment properties, are available. Cross-links to sequence, domain, and literature databases enable simple access to related information. Finally, the phylogenetic trees offer an evolutionary vantage point for nonexperts.
CONCLUSION
So far, most comparative analyses dealing with plant TAPs have focused on TFs of Arabidopsis and rice. To broaden our evolutionary understanding of transcriptional regulation in plants, we have included three algae and a moss into the present analysis, as well as the complete UniProt database. In addition, we have analyzed both TFs and TRs, and have detected several novel PT families. Using automated methods, a stringent detection and representation of gene clusters has been established that can easily be expanded to cover more genomes in the future, while manual curation of gene clusters into families assures their quality. High-quality phylogenetic trees were created from these clusters and are available through an easy-to-use Web interface together with a multitude of accompanying data, such as alignments, domain-based family annotation, and taxonomic profiles. Instant knowledge discovery using the PlanTAPDB is straightforward, as has been demonstrated using several examples. In addition, such comparative data can be applied to aid phylogenomics.
The general expansion of both the total number of TAP genes and the amount of TAP families seems to coincide with organism complexity. A dramatic increase in the complexity of transcriptional regulation, particularly at the level of TFs, might have occurred after the development of multicellularity, respective the transition from water to land. Subsequently, during land plant evolution, the intricacy of the previously established TF families enhanced again, possibly reflecting large-scale morphological and physiological changes paralleling angiosperm radiation. Apart from these general trends, distinct TAP gene families were subject to expansion in individual species. Interesting details about the evolution of the stem cell regulator WUS, the photomorphogenesis switch COP1, and the genotoxic stress-related HIT gene family were revealed.
MATERIALS AND METHODS
Sequence Datasets
For the identification of Physcomitrella patens transcription-associated EST sequences, National Center for Biotechnology Information (NCBI) Entrez (Geer and Sayers, 2003) was utilized to query GenPept (Benton, 1990) Release 141. The Arabidopsis Information Resource (TAIR; Rhee et al., 2003) resources were searched via keyword. GenPept Release 151 and the TIGR Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) predicted proteins (see below) were used for the closest homolog determination. For the collection of homologs throughout the available protein space using PSI-BLAST, the UniProt Knowledgebase Release 7.1 (http://www.ebi.uniprot.org/database/download.shtml) was used. In addition, the following organism-specific protein databases were included. Arabidopsis: 28,952 predicted proteins, TIGR ATH1.pep 01/04 (ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/a_thaliana/annotation_dbs/ATH1.pep). Rice: 88,149 predicted proteins, TIGR OSA1.pep 04/04 (ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_2.0/). P. patens: EST were clustered and assembled according to Lang et al. (2005), http://www.cosmoss.org Release 03/04. For the resulting virtual transcripts, ORF were predicted using FrameD (Schiex et al., 2003) and ESTscan 2.0 (Iseli et al., 1999) with P. patens-specific models, yielding a total of 52,458 ORFs. Thalassiosira pseudonana: 11,397 predicted proteins from Release 1.0, Department of Energy Joint Genome Institute (http://genome.jgi-psf.org/thaps1/thaps1.download.ftp). Cyanidioschyzon merolae: 5,013 translated mRNAs, Release 11/04 (http://merolae.biol.s.u-tokyo.ac.jp/download/cds_nt.fasta). Chlamydomonas reinhardtii: 19,832 predicted proteins from Release 2.0, Department of Energy Joint Genome Institute (http://genome.jgi-psf.org/chlre2/chlre2.download.ftp). For the calculation of Figure 2B, the recently corrected number of protein-coding genes for rice, 30,000, has been used (Itoh et al., 2007), and the estimated number of 25,000 protein-coding genes for P. patens (Rensing et al., 2002; Lang et al., 2005).
Software
The results and resources presented here were generated using an automated phylogeny pipeline that utilizes BLAST and PSI-BLAST (Altschul et al., 1997), Inter-ProScan 4.2 (Quevillon et al., 2005), EMBOSS 3.0.0 (Rice et al., 2000), MAFFT 5.8 (Katoh et al., 2005), ProbCons 1.1 (Do et al., 2005), Muscle 3.52 (Edgar, 2004), Phylip 3.65 (Felsenstein, 1989), Tree-Puzzle 5.2 (Schmidt et al., 2002), a modified version of the puzzleboot script (http://www.tree-puzzle.de/puzzlebootREADME.txt), and the PostgreSQL 8.0.8 (http://www.postgresql.org) relational database. This so-called TreePipe is able to construct phylogenetic trees for large datasets without manual interference and is implemented with Perl 5.8.7 (http://www.perl.com), SQL, and shell scripts, making use of the Bioperl CVS “live” branch (Stajich et al., 2002) and the Bio∷Phylo Version 0.09 (http://search.cpan.org/∼rvosa/Bio-Phylo-0.09/) packages. The extensive data that are collected throughout the pipeline are stored in a relational database schema developed for this project, called TreePipeDB. The PlanTAPDB Web interface is implemented using mod_perl 2.0 (http://perl.apache.org/) and Javascript with the TreePipeDB as backend. For the interactive exploration of MSA and phylogenetic trees, we integrated the Jalview multiple alignment editor 2.08.1 (http://www.jalview.org/) and ATV phylogenetic tree viewer 2.0 BETA (http://www.phylogenomics.us/atv/) java applets.
Identification of the TAP Query Set
NCBI GenBank was queried using the keywords “transcription factor,” “transcription activator,” “transcription repressor,” and “transcription regulator,” as well as taxon IDs of Viridiplantae and nongreen algae (txids 33090, 136419, 3027, 33682, 38254, 2830, 2763, 33634). Additionally, Arabidopsis loci were extracted from TAIR matching the keyword “transcription factor.” With this reference set of 7,476 TAPs, the clustered P. patens EST sequences were searched by TBLASTN. A total of 286 PFAM HMM profiles and 67 PROSITE patterns of transcription-associated domains without taxonomic restriction were used for motif searches in the same database. A total of 1,592 nonredundant P. patens candidate TAP sequences were identified. Full-length closest homologs of the 1,592 moss candidate TAP transcripts were determined via BLASTX (Altschul et al., 1997) with an E-value cutoff of 1E-3 against GenPept and the TIGR Arabidopsis and rice predicted protein databases. The resulting hits were filtered using an alignment length and percent identity threshold of 50 amino acids and 25%, respectively.
PSI-BLAST Searches and Filtering of the Results
PSI-BLAST searches were performed against the UniProt Knowledgebase, all available whole-genome predicted protein databases of plants and algae, and the predicted ORF of the P. patens virtual transcripts using an E-value threshold of 1E-4, a profile inclusion threshold of 1E-5, and four iterations. Up to 500 results per query were considered and parsed into the TreePipeDB. Each result set (composed of one query and its hits after one of the four PSI-BLAST iterations) was run through a series of six filter steps with increasing stringency concerning the length and percent identity of the PSI-BLAST matches (step 1: 25% identity/50-amino acid alignment length; step 2: 30%/60 amino acids; step 3: 35%/80 amino acids; step 4: 45%/100 amino acids; step 5: 45%/150 amino acids; step 6: 45%/300-amino acid length). For each query and iteration, the filtering process determines the first filtering step that reduces the result set to ≤50 and ≥5 members. Afterward, the optimal iteration (plus determined filtering step) is chosen for each query, using a set of sequentially applied criteria: (1) the most stringent possible filtering step, (2) the maximal number of remaining sequences, and (3) the lowest iteration step (in order to select result sets with low amounts of false-positive hits).
Clustering of the Filtered Result Sets
Single-linkage clustering using a stringent hit-coverage-based distance measure was implemented in Perl and the TreePipeDB backend. Result sets of two queries were merged if they shared at least one hit covering the same region of this hit sequence. The length of the region to be shared depends on the previously selected filter step, namely, the most stringent filter step possible (e.g. result set A overlaps with B on hit X). A was filtered using step 6 and B using step 5. Hence, A and B can only then be merged into a cluster if they overlap to at least 300 amino acids (step 6 criteria) on sequence X. Result sets without any significant overlaps were added as single-query clusters. For all cluster members, the corresponding NCBI taxonomy annotation was retrieved and stored in TreePipeDB.
Redundancy Removal and Homology Reduction
For the removal of redundant sequences, a MSA was performed using MAFFT FFT-NS-2 and pairwise distances were calculated using the EMBOSS distmat program. This alignment was used to infer initial phylogenies of the complete clusters. The resulting matrix was scanned for sequence pairs from the same species with a distance ≤1 substitutions per 100 amino acids. For each pair, one representative was selected based on the originating database (UniProt sequences were preferred), sequence length, and lexical sort order of the accession number. The procedure was implemented in Perl using several Bioperl modules, including a modified version of the Bio∷Tools∷Run∷Alignment∷MAFFT module. For the parsing of the distmat distance matrices, an object-oriented Bioperl module (Bio∷Matrix∷IO∷distmat) was written. Homology reduction was implemented in the same program but follows a different strategy. Beginning with 1 substitution per 100 amino acids and heuristically increasing this distance threshold, the distance matrix is iteratively scanned for sequence pairs with the respective distance, regardless of their species. The iteration stops when the remaining representative cluster members reach a given limit (150 sequences).
Multiple Alignments and Selection of Informative Sites
Multiple alignments for a given cluster were performed using MAFFT G-INSI and ProbCons (clusters ≤150) or Muscle (clusters >150). Subsequently, sum-of-pairs scores using the BLOSUM62 substitution matrix, gap ratios, and Shannon's entropy scores (Valdar, 2002) were calculated and recorded columnwise in the TreePipeDB. Finally, columns below a sum-of-pairs score of −2 were excised from the alignment. The procedure was implemented in a Perl program, which, besides the filtering of a given MSA, also produced overview graphics of the different scores along the overall alignment.
Reconstruction of Phylogenies of the Representative Cluster Members
Phylogenies for the representative cluster members were inferred using a Perl program on all clusters. After generation of 100 bootstrapped alignments using seqboot from the PHYLIP package, ML distance matrices were computed for these alignments using puzzleboot as implemented in Tree-Puzzle. These distance matrices were then used to infer topologies by applying the NJ algorithm as implemented in PHYLIP's neighbor program. Afterward, the resulting 100 trees were used to create a ML consensus topology using Tree-Puzzle. For the two steps where Tree-Puzzle was used to compute maximum likelihoods, eight gamma-distributed rates were used to model mutation rate heterogeneity and full (exact) ML parameter estimation was performed for each gene family. Manual ML trees were created using the same parameter settings. The WAG (Whelan and Goldman, 2001) evolutionary model of sequence evolution, which is derived from a database of globular proteins, was used. The resulting phylogenetic tree offered both an overall confidence value, i.e. the ML of the tree, and confidence values for every branch in the form of bootstrap values. Finally, the trees were parsed and midpoint-rooted via an additional Perl program that also collects a large variety of parameters from the tree topologies using both Bioperl and the Bio∷Phylo modules (e.g. the longest internal branch, the Fiala stemminess [Fiala and Sokal, 1985], and the resolution) and writes them into the TreePipeDB. The initial phylogenies for the complete clusters were inferred in analogy to the procedure described above, using a Perl wrapper combining the PHYLIP tools seqboot and neighbor. However, in this case JTT distances (Jones et al., 1992) were calculated with PHYLIP's protdist and consensus trees with consense to cope with the runtime demands of clusters up to 1,182 members. Finally, the consensus topologies were used to estimate ML branch lengths with the user-tree option of Tree-Puzzle, using uniform rates and exact parameter estimation.
Cluster and Gene Family Annotation
The nonredundant cluster member sequences were annotated using Inter-ProScan 4.2 with all available databases of the Inter-Pro Release 12.1. The annotated domains and associated GO terms were stored in the TreePipeDB. Inter-ProScan searches (Quevillon et al., 2005) were performed for the 37,247 distinct cluster members after redundancy removal. A total of 99.8% of the sequences could be annotated with Inter-Pro domains. Sixty-two percent of the domains found were from the PANTHER (Mi et al., 2005), PFAM (Finn et al., 2006), and PROSITE (Hulo et al., 2006) databases. Manual curation was performed by inspection of the description lines of the enclosed UniProt sequences and by inferring the classification of Arabidopsis cluster members from DATF (Guo et al., 2005) and ArabTFDB (http://arabtfdb.bio.uni-potsdam.de/v1.1/). To further assign thus far undetected TAP families, their corresponding Arabidopsis and rice members collected from DATF (Guo et al., 2005), ArabTFDB (http://arabtfdb.bio.uni-potsdam.de/v1.1/), DRTF (Gao et al., 2006), and RiceTFDB (http://ricetfdb.bio.uni-potsdam.de/v2.1/) were used to screen the nonredundant cluster members for homologs by BLASTP.
Species-Specific Expansion, Taxonomic Profiling, and Statistical Tests
The PlanTAPDB family sizes in six genera, Arabidopsis, rice, P. patens, C. reinhardtii, C. merolae, and T. pseudonana, were inferred using the NCBI taxonomy information of the nonredundant list of family members. These values were normalized using the total amount of members per group (TF, TR, or PT) in order to account for the general differences in TAP family sizes. If the fraction of family members in a given species deviated from the arithmetic average of the group with a z score of ≥1.8, it was marked as expanded (no gene family was significantly reduced according to this criterion). The cutoff was chosen based on a distribution plot of all z scores (data not shown).
For visualization of the taxonomic composition of the TAP families (taxonomic profile), all taxa were allocated into 20 nonredundant taxonomic groups that were chosen because they contributed significantly to the distribution of NCBI taxonomy strings. After normalization for taxonomic group size (columnwise log ratio per average), the rows were used for average-linkage clustering with a centered Pearson-correlation distance and heat map visualization using Cluster 3.0 and JavaTreeview 1.0.12 (Eisen et al., 1998).
Hypothesized differences in the size distribution of TAP gene families between organisms (Fig. 3B) were tested using two-sided t tests assuming unequal variances. Fisher's exact test was used to test for hypothesized differences between total number of genes of the six organisms (Fig. 2A). The resulting P values were adjusted for multiple testing by calculating the false discovery rate (Benjamini and Hochberg, 1995).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Plant TAP family sizes in algae, moss, and flowering plants.
Supplemental Table S2. Coverage of known TAP families through PlanTAPDB.
Supplementary Material
Acknowledgments
We thank T. Kretsch, T. Laux, and M. Woriedh for helpful discussions, A.K. Prowse for critically reading the manuscript, and several anonymous reviewers for helpful comments.
This work was supported by the German Research Foundation (grant nos. Re 837/7–3 and Re 837/10–1 to R.R.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Stefan A. Rensing (stefan.rensing@biologie.uni-freiburg.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57 289–300 [Google Scholar]
- Benton D (1990) Recent changes in the GenBank On-line Service. Nucleic Acids Res 18 1517–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierfreund NM, Tintelnot S, Reski R, Decker EL (2004) Loss of GH3 function does not affect phytochrome-mediated development in a moss, Physcomitrella patens. J Plant Physiol 161 823–835 [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byzova MV, Franken J, Aarts MG, de Almeida-Engler J, Engler G, Mariani C, Van Lookeren Campagne MM, Angenent GC (1999) Arabidopsis STERILE APETALA, a multifunctional gene regulating inflorescence, flower, and ovule development. Genes Dev 13 1002–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J, Binns D, Harte N, Lopez R, Apweiler R (2004) The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res (Database issue) 32 D262–D266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles CC, Choffnes-Inada D, Reville K, Lertpiriyapong K, Fletcher JC (2005) ULTRAPETALA1 encodes a SAND domain putative transcriptional regulator that controls shoot and floral meristem activity in Arabidopsis. Development 132 897–911 [DOI] [PubMed] [Google Scholar]
- Carroll SB (2005) Evolution at two levels: on genes and form. PLoS Biol 3 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17 540–552 [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20 426–427 [DOI] [PubMed] [Google Scholar]
- Coulson RM, Enright AJ, Ouzounis CA (2001) Transcription-associated protein families are primarily taxon-specific. Bioinformatics 17 95–97 [DOI] [PubMed] [Google Scholar]
- Coulson RMR, Ouzounis CA (2003) The phylogenetic diversity of eukaryotic transcription. Nucleic Acids Res 31 653–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do CB, Mahabhashyam MS, Brudno M, Batzoglou S (2005) ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res 15 330–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Lukens L (1998) Transcriptional regulators and the evolution of plant form. Plant Cell 10 1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (1989) PHYLIP: Phylogeny Inference Package (Version 3.2). Cladistics 5 164–166 [Google Scholar]
- Fiala KI, Sokal RR (1985) Factors determining the accuracy of cladogram estimation: evaluation using computer-simulation. Evolution Int J Org Evolution 39 609–622 [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al (2006) Pfam: clans, web tools and services. Nucleic Acids Res (Database issue) 34 D247–D251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickey T, Lupas AN (2004) PhyloGenie: automated phylome generation and analysis. Nucleic Acids Res 32 5231–5238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuellen G, Spitzer M, Cullen P, Lorkowski S (2003) BLASTing proteomes, yielding phylogenies. In Silico Biol 3 313–319 [PubMed] [Google Scholar]
- Gao G, Zhong Y, Guo A, Zhu Q, Tang W, Zheng W, Gu X, Wei L, Luo J (2006) DRTF: a database of rice transcription factors. Bioinformatics 22 1286–1287 [DOI] [PubMed] [Google Scholar]
- Geer RC, Sayers EW (2003) Entrez: making use of its power. Brief Bioinform 4 179–184 [DOI] [PubMed] [Google Scholar]
- Gouret P, Vitiello V, Balandraud N, Gilles A, Pontarotti P, Danchin EG (2005) FIGENIX: intelligent automation of genomic annotation: expertise integration in a new software platform. BMC Bioinformatics 6 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueven N, Becherel OJ, Kijas AW, Chen P, Howe O, Rudolph JH, Gatti R, Date H, Onodera O, Taucher-Scholz G, et al (2004) Aprataxin, a novel protein that protects against genotoxic stress. Hum Mol Genet 13 1081–1093 [DOI] [PubMed] [Google Scholar]
- Guo A, He K, Liu D, Bai S, Gu X, Wei L, Luo J (2005) DATF: a database of Arabidopsis transcription factors. Bioinformatics 21 2568–2569 [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, Green PJ, Keegstra K, Ohlrogge JB (2004) Phylogenetic profiling of the Arabidopsis thaliana proteome: What proteins distinguish plants from other organisms? Genome Biol 5 R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131 657–668 [DOI] [PubMed] [Google Scholar]
- Hannaert V, Saavedra E, Duffieux F, Szikora JP, Rigden DJ, Michels PA, Opperdoes FR (2003) Plant-like traits associated with metabolism of Trypanosoma parasites. Proc Natl Acad Sci USA 100 1067–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Blair JE, Venturi ML, Shoe JL (2004) A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol 4 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Deng XW (1999) Structural organization and interactions of COP1, a light-regulated developmental switch. Plant Mol Biol 41 151–158 [DOI] [PubMed] [Google Scholar]
- Hsia CC, McGinnis W (2003) Evolution of transcription factor function. Curr Opin Genet Dev 13 199–206 [DOI] [PubMed] [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJ (2006) The PROSITE database. Nucleic Acids Res (Database issue) 34 D227–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa K, Sugita M, Imaizumi T, Wada M, Aoki S (2004) Differential expression on a daily basis of plastid sigma factor genes from the moss Physcomitrella patens. Regulatory interactions among PpSig5, the circadian clock, and blue light signaling mediated by cryptochromes. Plant Physiol 136 4285–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseli C, Jongeneel CV, Bucher P (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. In Proceedings of International Conference on Intelligent Systems for Molecular Biology. American Association for Artificial Intelligence, Menlo Park, CA, pp 138–148 [PubMed]
- Itoh T, Tanaka T, Barrero RA, Yamasaki C, Fujii Y, Hilton PB, Antonio BA, Aono H, Apweiler R, Bruskiewich R, et al (2007) Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res 17 175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8 275–282 [DOI] [PubMed] [Google Scholar]
- Kamisugi Y, Cuming AC, Cove DJ (2005) Parameters determining the efficiency of gene targeting in the moss Physcomitrella patens. Nucleic Acids Res 33 e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, Kagawa T, Sato Y, Kiyosue T, Wada M (2004) Phototropins mediate blue and red light-induced chloroplast movements in Physcomitrella patens. Plant Physiol 135 1388–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P, Crane PR (1997) The origin and early evolution of plants on land. Nature 389 33–39 [Google Scholar]
- Kieffer M, Stern Y, Cook H, Clerici E, Maulbetsch C, Laux T, Davies B (2006) Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 18 560–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijas AW, Harris JL, Harris JM, Lavin MF (2006) Aprataxin forms a discrete branch in the HIT (histidine triad) superfamily of proteins with both DNA/RNA binding and nucleotide hydrolase activities. J Biol Chem 281 13939–13948 [DOI] [PubMed] [Google Scholar]
- Kyrpides NC, Ouzounis CA (1999) Transcription in archaea. Proc Natl Acad Sci USA 96 8545–8550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Eisinger J, Reski R, Rensing SA (2005) Representation and high-quality annotation of the Physcomitrella patens transcriptome demonstrates a high proportion of proteins involved in metabolism in mosses. Plant Biol 7 238–250 [DOI] [PubMed] [Google Scholar]
- Larroux C, Fahey B, Liubicich D, Hinman VF, Gauthier M, Gongora M, Green K, Worheide G, Leys SP, Degnan BM (2006) Developmental expression of transcription factor genes in a demosponge: insights into the origin of metazoan multicellularity. Evol Dev 8 150–173 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Marchal V, Gentilhomme J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133 3213–3222 [DOI] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438 1172–1175 [DOI] [PubMed] [Google Scholar]
- Lespinet O, Wolf YI, Koonin EV, Aravind L (2002) The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res 12 1048–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Tjian R (2003) Transcription regulation and animal diversity. Nature 424 147–151 [DOI] [PubMed] [Google Scholar]
- Madan Babu M, Teichmann SA, Aravind L (2006) Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J Mol Biol 358 614–633 [DOI] [PubMed] [Google Scholar]
- Maizel A, Busch MA, Tanahashi T, Perkovic J, Kato M, Hasebe M, Weigel D (2005) The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308 260–263 [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815 [DOI] [PubMed] [Google Scholar]
- Messina DN, Glasscock J, Gish W, Lovett M (2004) An ORFeome-based analysis of human transcription factor genes and the construction of a microarray to interrogate their expression. Genome Res 14 2041–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al (2005) The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res (Database issue) 33 D284–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittmann F, Brucker G, Zeidler M, Repp A, Abts T, Hartmann E, Hughes J (2004) Targeted knockout in Physcomitrella reveals direct actions of phytochrome in the cytoplasm. Proc Natl Acad Sci USA 101 13939–13944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsham KK (2003) UV-B radiation arising from stratospheric ozone depletion influences the pigmentation of the Antarctic moss Andreaea regularis. Oecologia 135 327–331 [DOI] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Mate Z, Brzezinska A, Molinier J, Oakeley EJ, Adam E, Schafer E, Nagy F, Ulm R (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Rueda E, Collado-Vides J, Segovia L (2004) Phylogenetic distribution of DNA-binding transcription factors in bacteria and archaea. Comput Biol Chem 28 341–350 [DOI] [PubMed] [Google Scholar]
- Qu LJ, Zhu YX (2006) Transcription factor families in Arabidopsis: major progress and outstanding issues for future research. Curr Opin Plant Biol 9 544–549 [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33 W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Deplancke B, Shingles J, Grove CA, Hope IA, Walhout AJ (2005) A compendium of Caenorhabditis elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol 6 R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Rombauts S, Van de Peer Y, Reski R (2002) Moss transcriptome and beyond. Trends Plant Sci 7 535–538 [DOI] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16 276–277 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290 2105–2110 [DOI] [PubMed] [Google Scholar]
- Riese M, Faigl W, Quodt V, Verelst W, Matthes A, Saedler H, Munster T (2005) Isolation and characterization of new MIKC*-Type MADS-box genes from the moss Physcomitrella patens. Plant Biol (Stuttg) 7 307–314 [DOI] [PubMed] [Google Scholar]
- Rosenberg MS (2005) Evolutionary distance estimation and fidelity of pair wise sequence alignment. BMC Bioinformatics 6 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B (1999) Twilight zone of protein sequence alignments. Protein Eng 12 85–94 [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA (2006) Grasses like mammals? Redundancy and compensatory regulation within the retinoblastoma protein family. Cell Cycle 5 352–355 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4 406–425 [DOI] [PubMed] [Google Scholar]
- Satou Y, Satoh N (2005) Cataloging transcription factor and major signaling molecule genes for functional genomic studies in Ciona intestinalis. Dev Genes Evol 215 580–596 [DOI] [PubMed] [Google Scholar]
- Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF (2001) Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res 29 2994–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K (1999) Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96 11664–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiex T, Gouzy J, Moisan A, de Oliveira Y (2003) FrameD: a flexible program for quality check and gene prediction in prokaryotic genomes and noisy matured eukaryotic sequences. Nucleic Acids Res 31 3738–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A (2002) TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18 502–504 [DOI] [PubMed] [Google Scholar]
- Seoighe C, Gehring C (2004) Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet 20 461–464 [DOI] [PubMed] [Google Scholar]
- Shigyo M, Hasebe M, Ito M (2006) Molecular evolution of the AP2 subfamily. Gene 366 256–265 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Shih MC, Li WH (2005) Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol 139 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicheritz-Ponten T, Andersson SG (2001) A phylogenomic approach to microbial evolution. Nucleic Acids Res 29 545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL (1999) Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126 4117–4128 [DOI] [PubMed] [Google Scholar]
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al (2002) The Bioperl toolkit: Perl modules for the life sciences. Genome Res 12 1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D (2000) Evolution of transcriptional regulation. Curr Opin Genet Dev 10 575–579 [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Munster T, Winter KU, Saedler H (2000) A short history of MADS-box genes in plants. Plant Mol Biol 42 115–149 [PubMed] [Google Scholar]
- Theissen G, Münster T, Henschel K (2001) Why don't mosses flower? New Phytol 150 1–8 [Google Scholar]
- Trouiller B, Schaefer DG, Charlot F, Nogue F (2006) MSH2 is essential for the preservation of genome integrity and prevents homeologous recombination in the moss Physcomitrella patens. Nucleic Acids Res 34 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uenaka H, Wada M, Kadota A (2005) Four distinct photoreceptors contribute to light-induced side branch formation in the moss Physcomitrella patens. Planta 222 623–631 [DOI] [PubMed] [Google Scholar]
- Valdar WS (2002) Scoring residue conservation. Proteins 48 227–241 [DOI] [PubMed] [Google Scholar]
- Waller RF, McFadden GI (2005) The apicoplast: a review of the derived plastid of apicomplexan parasites. Curr Issues Mol Biol 7 57–79 [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18 691–699 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Liu T, Tian C, Sun S, Li J, Chen M (2005) Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol Biol 59 191–203 [DOI] [PubMed] [Google Scholar]
- Yi C, Deng XW (2005) COP1: from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15 618–625 [DOI] [PubMed] [Google Scholar]
- Zhou DX, Bisanz-Seyer C, Mache R (1995) Molecular cloning of a small DNA binding protein with specificity for a tissue-specific negative element within the rps1 promoter. Nucleic Acids Res 23 1165–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek CM, Eddy SR (2001) ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics 17 383–384 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.