Abstract
A large-scale functional genomics project was initiated to study the function of chromatin-related genes in maize (Zea mays). Transgenic lines containing short gene segments in inverted repeat orientation designed to reduce expression of target genes by RNA interference (RNAi) were isolated, propagated, and analyzed in a variety of assays. Analysis of the selectable marker expression over multiple generations revealed that most transgenes were transmitted faithfully, whereas some displayed reduced transmission or transgene silencing. A range of target-gene silencing efficiencies, from nondetectable silencing to nearly complete silencing, was revealed by semiquantitative reverse transcription-PCR analysis of transcript abundance for the target gene. In some cases, the RNAi construct was able to cause a reduction in the steady-state RNA levels of not only the target gene, but also another closely related gene. Correlation of silencing efficiency with expression level of the target gene and sequence features of the inverted repeat did not reveal any factors capable of predicting the silencing success of a particular RNAi-inducing construct. The frequencies of success of this large-scale project in maize, together with parameters for optimization at various steps, should serve as a useful framework for designing future RNAi-based functional genomics projects in crop plants.
A functional genomics approach was pursued to study the role of 130 chromatin-related maize (Zea mays) genes in controlling a range of epigenetic phenomena including paramutation, epimutation, transgene silencing, DNA methylation, and imprinting. Bioinformatics approaches were used to identify over 300 maize genes that may play a role in chromatin structure or modification (cataloged at www.ChromDB.org). In many cases, these genes belong to multigene families for which paralogous and orthologous relationships have been established (Pandey et al., 2002; Springer et al., 2003; Springer and Kaeppler, 2005). The putative proteins encoded by these genes include histone-modifying enzymes, chromatin-dependent ATPases, DNA methyltransferases, and other genes potentially involved in chromatin-level control of gene expression. This report describes the efficiencies and success of using RNA interference (RNAi) for genomic-scale analysis of gene function in maize. The results presented here are likely to be applicable to studies of other groups of maize genes.
Although there are several reverse-genetics resources available for identifying loss-of-function alleles in maize (Brutnell, 2002; May et al., 2003; Till et al., 2004), an RNAi-based approach offered us two main advantages. First, RNAi-induced mutations are dominant. Many of the assays needed to monitor the effect of reduced chromatin gene expression on epigenetic phenomena require a multiple-generation crossing scheme; dominant mutants reduce the number of crossing generations needed. The second advantage of RNAi is the potential to reduce expression of multiple, closely related genes with a single transgene locus.
RNAi can induce gene silencing at a transcriptional or posttranscriptional level depending on the sequence contained in the double-stranded RNA (dsRNA; for review, see Brodersen and Voinnet, 2006). Posttranscriptional silencing can be triggered by utilizing sequences derived from the coding region of the target gene. When abundant dsRNAs are produced by a transgene, they are processed by a dicer-like protein into small interfering RNAs (siRNAs). The siRNAs then interact with an argonaute-like protein and other associated proteins to direct degradation of mRNAs that share sequence identity (for review, see Brodersen and Voinnet, 2006; Vaucheret, 2006).
RNAi has been used successfully to silence genes in both monocots and dicots. Many studies have focused on a small number of gene targets (Chen et al., 2003; Segal et al., 2003; Travella et al., 2006). In a study that targeted a large number of genes, one of the parameters found to be important for silencing efficiency was transgene copy number (Kerschen et al., 2004). In Arabidopsis (Arabidopsis thaliana), lines bearing a single copy of an inverted repeat (IR) transgene silenced the endogenous target more efficiently than lines with multicopy insertions (Kerschen et al., 2004). This parameter, together with several others, required optimization for our large-scale study in maize.
This article describes a functional genomics effort using transgene-induced RNAi to target over 100 genes for silencing. The efficiency of this large-scale project is reported, including descriptions of the efficiency of the transformation pipeline, the frequency of reduction of target gene transcript levels, the range of stability for various transgenic events, and the spectrum of transgene behaviors.
RESULTS
Production of RNAi Lines
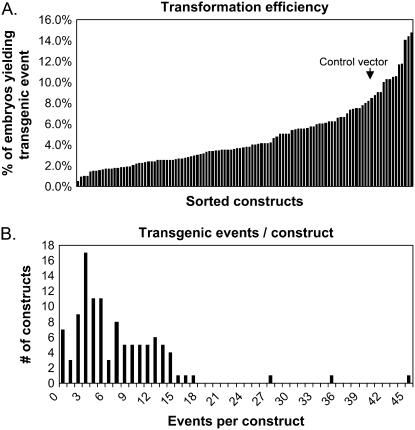
The procedures for construct design and transformation have been described previously (McGinnis et al., 2005). The relative frequencies for each step of the transformation and plant regeneration pipeline are illustrated in Table I, and a full description of the frequencies for each of the transgenic constructs is provided in Supplemental Table S1. In total, over 86,000 embryos were subjected to biolistic transformation by 106 different transgene constructs (including 104 constructs targeting chromatin genes and two control constructs), resulting in nearly 3,000 transgenic events that were identified based on their ability to grow on culture media containing bialophos. The distribution of transformation efficiencies and the number of independent events per construct are indicated in Figure 1. Only six of the 106 constructs failed to generate transgenic T0 plants. Regenerated transgenic T0 plants were crossed to the nontransgenic inbred line, B73, resulting in T1 seed stocks for 766 independent transgenic events that represent 99 of the 106 transgenic constructs used for transformation. Only about 40% of bialophos-resistant callus events yielded T0 plants. The loss of events from callus through plantlet regeneration was due in part to the high-throughput nature of the project, as slow-growing events or plants on contaminated plates were eliminated; additionally, the implementation of a Southern-blot prescreening step removed events that did not have intact transgenes. Transgenic events for two nonchromatin gene constructs, an empty vector control and a construct targeting the b1 gene (see below), showed similar rates of loss during regeneration; this argues that loss of events was not solely due to some selectively detrimental influence of reduced chromatin gene expression on plant growth or development.
Table I.
Steps and efficiencies of producing transgenic seed
| Step | Products | Total | Percent Efficiency Relative to:
|
|
|---|---|---|---|---|
| Previous Step | Callus Events | |||
| Microprojectile bombardment | Bombarded embryos | 86,675 | – | – |
| Selection | Resistant callus events after selection | 2,958 | 3 | – |
| Regeneration and transfer to greenhouse | T0 plants | 1,144 | 39 | 39 |
| Production of T1 seed | Fertile T0 plants producing T1 seed | 766 | 67 | 26 |
Figure 1.
Analysis of transformation efficiency. A, The transformation efficiency (expressed as the no. of independent transgenic events that produced T1 plants divided by the total no. of embryos targeted for transformation) is depicted for each of the 106 constructs that were used for transformation. The arrow indicates the transformation efficiency achieved using the empty control vector. B, The number of transgenic events per construct is shown.
Each transgenic line was assigned a unique numeric designation, composed of the construct number and the transgenic event from which the line was derived. For example, line 3832.001 is the first transgenic event derived from callus transformed with construct pMCG3832; this transgenic line is distinct from 3832.004, which represents a line derived from the fourth transformation event of the same construct. Multiple transgenic lines were carried forward for each construct, with the assumption that each unique transgenic event represents a transgene insertion in a distinct genomic location. By isolating multiple transgenic lines, we hoped to minimize positional effects and event quality differences when interpreting phenotypes of the transgenic plants. In addition, producing multiple lines for each construct provided the potential to recover an “allelic series” with different levels of knock-down in target gene expression (Chuang and Meyerowitz, 2000).
Proof of Concept: Silencing of an Endogenous Gene with Transgene-Induced RNAi
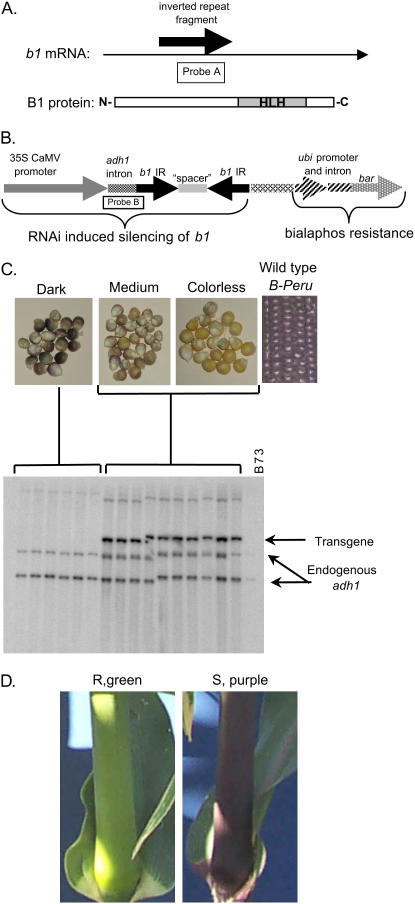
The pMCG2973 construct was designed to target the b1 gene for silencing to provide a visual assay to examine the effectiveness of the RNAi strategy and stability of the lines generated. The b1 gene codes for a transcriptional activator of the anthocyanin biosynthetic pathway (Chandler et al., 1989), and its expression results in purple pigment production. Several different alleles have been described for this gene (Styles et al., 1973; Coe, 1979; Selinger and Chandler, 1999). These alleles share sequence identity in their protein coding regions but differ substantially in their promoter sequences (Radicella et al., 1992; Selinger et al., 1998; Selinger and Chandler, 2001). The promoter differences account for the characteristic tissue-specific expression patterns of the alleles. For example, the B-Intense (B-I) allele leads to intense pigmentation in vegetative plant structures, such as leaf sheaths, whereas the B-Peru allele leads to strong pigmentation in the kernel aleurone. In designing an RNAi construct to silence b1, the IR sequence was selected from the protein coding region, such that both the B-I and the B-Peru alleles could be silenced by this transgene (Radicella et al., 1992; Selinger et al., 1998; Selinger and Chandler, 2001). Figure 2 shows the locations of the probes used in the experiments described below, the relationship of the mRNA sequence utilized in the IR construct relative to the B1 protein (Fig. 2A), and the structure of the IR transgenic construct introduced into plants (Fig. 2B). The bar gene is the selectable marker, which allows the presence of the transgene to be tracked by testing for bialophos resistance. Herein, the term “resistant” is used to describe plants that are resistant to bialophos and/or glufosinate, and the term “susceptible” is used to describe plants that are not herbicide resistant.
Figure 2.
Transgene-induced silencing of maize b1 alleles. A, Location of IR fragment and Probe A (used in this study) relative to the b1 mRNA and protein. B, Diagram of the pMCG2973 construct used to make transgenic lines to silence maize b1 alleles and the location of Probe B (used in this study). C, Seed color phenotype and Southern-blot data performed on leaf tissue from plants that germinated from seed of each color using Probe B. A band was present, indicating the presence of the 2973.004 transgene in medium and colorless kernels. D, Phenotypes of plants heterozygous for B-I with (R, green) or without (S, purple) the 2973.029 transgene. R, Resistant; S, susceptible.
Three unique insertion events for the b1:IR transgene, named 2973.004, 2973.011, and 2973.029, were evaluated at the T1 stage by crossing with B-Peru testers. The B-Peru allele is dominant over the colorless b1-B73 allele (present in the T1 plants) and is expressed in kernel aleurone, resulting in purple pigmentation in kernels that are homozygous or heterozygous for B-Peru. All progeny resulting from a cross of a hemizygous transgenic plant and a B-Peru homozygous stock will be heterozygous at the b1 locus (B-Peru/ b1-B73) and are expected to segregate 1:1 for the presence:absence of a hemizygous transgene locus if carrying a single transgenic locus. One of the three events, 2973.011, failed to show evidence of silencing in the T2 generation and was not further analyzed. The kernels derived from crosses of the other two transgenic events display three different levels of pigmentation: dark, medium, and colorless. Analysis of DNA isolated from plants produced by germinated seeds from each color class revealed that all plants resulting from dark kernels lacked the b1:IR transgene, while the plants from colorless and medium kernels had the b1:IR transgene (Fig. 2C), consistent with the IR transgene leading to silencing of B-Peru. The colorless kernels likely represent the highest efficiency of silencing of the B-Peru allele, whereas the medium-colored kernels probably represent partial silencing of the B-Peru allele.
The two b1:IR transgenic events, 2973.004 and 2973.029, were outcrossed for multiple generations. While 2973.029 displayed evidence of silencing through the T5 generation, 2973.004 failed to show silencing beyond the T2 generation. The 2973.029 event was used in crosses to test whether it could silence another allele of b1 and whether silencing could be observed in tissues other than aleurone. T1 plants that were hemizygous for event 2973.029 were crossed with plants homozygous for the B-I allele, which is highly expressed in the sheaths and culms of homozygous and heterozygous plants. The resulting progeny included individuals with purple and green coloration (Fig. 2D). The purple plants were phenotypically identical to nontransgenic B-I plants, and, in all cases tested, the purple phenotype cosegregated with herbicide susceptibility. The green plants, which have low levels of B-I expression, contained the b1:IR transgene. This indicates that the B-Peru:IR can silence another allele, B-I, in mature plant tissues. The transgenic, green individuals were outcrossed, and silencing continued to cosegregate with herbicide resistance in all subsequent generations (to T5).
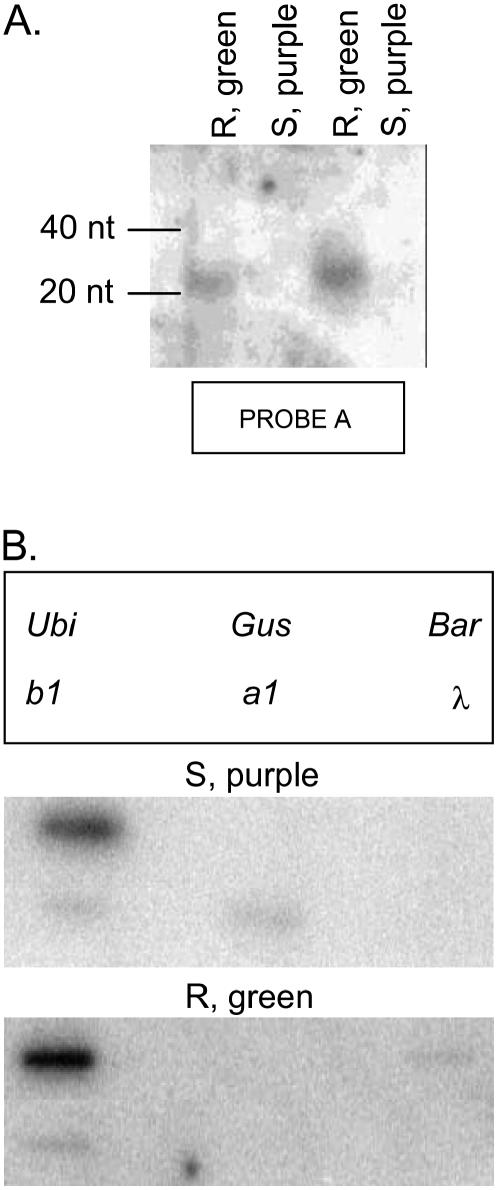
To explore the molecular nature of the silencing, tissue was harvested from the sheaths of purple susceptible progeny and green resistant progeny for analysis of small RNAs characteristic of RNA silencing. Small RNAs approximately 21 bp in length with homology to b1 could be detected in the green resistant plants, but not in the purple susceptible plants (Fig. 3A). Nuclear run-on transcription assays on nuclei prepared from sheath tissue were used to determine if the silencing induced by the IR transgene was transcriptional or posttranscriptional. The results indicated that in purple susceptible (nontransgenic) plants, both b1 and a1 (a gene involved in anthocyanin biosynthesis and responsive to transcriptional activation by b1) are transcribed. However, in the green resistant (transgenic) plants, the b1 gene was transcribed, but a1 was not (Fig. 3B). This observation suggests a posttranscriptional silencing event, whereby b1 mRNA is produced but the mRNA is degraded by transgene-induced RNA silencing before a protein can be translated.
Figure 3.
Molecular analysis of b1-IR lines. A, Northern-blot analysis of small RNA-enriched samples, hybridized with Probe A (Fig. 2). Mobility of molecular size standards is shown at left. B, Nuclear run-on analysis of sheath tissue from purple sensitive (S) and green resistant (R) plants. Section at left indicates positions of DNAs loaded on slot blots. Ubi, Ubiquitin; GUS, β-glucuronidase; Bar, bialophos resistance gene; b1 and a1, anthocyanin genes; λ, bacteriophage λ. The β-glucuronidase and λ genes were included as negative controls, as expression was not expected. Ubiquitin was included as a positive control and expected to be the same for both treatments.
To determine if the silenced phenotype was heritable in the absence of the transgene, the transgenic B-Peru or B-I plants were crossed to nontransgenic plants. The progeny without the transgene displayed the expected wild-type phenotype, indicating that silencing by the IR transgene was not heritable in the absence of the transgene (data not shown). The proof-of-concept experiments using the B-Peru RNAi construct indicated that this type of construct and system can produce loss-of-function alleles for the targeted genes. Additionally, this series of experiments provided evidence that the effectiveness and stability of transgenes can vary.
Assessment of Transgene Segregation and Complexity
The inclusion of a linked selectable marker, the bar gene, in all transgene constructs means that crossing hemizygous T0 plants with an intact transgene to nontransgenic B73 should produce T1 plants that segregate 50% nontransgenic susceptible plants and 50% hemizygous transgenic resistant plants. To test for this cosegregation in all T1 families containing one of the 104 transgene constructs designed to target a chromatin related gene, genomic DNA isolated from five or more resistant plants and from two or more susceptible plants was subjected to Southern-blot analysis using a probe (probe B; Fig. 2) derived from the transgene. A majority of the T1 families (205 out of 278 analyzed) showed cosegregation of the IR transgene with resistance. For subsequent analysis of these families, herbicide resistance was used to indicate the presence of a transgene. T1 families in which the transgene did not cosegregate with resistance were omitted from further analyses.
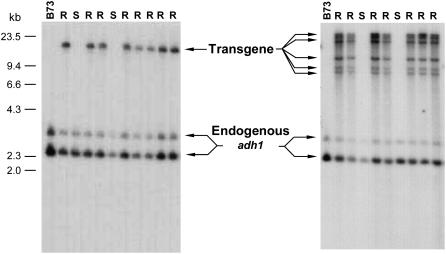
The DNA gel-blot analysis also provided a way to evaluate the complexity and approximate copy number of the inserted transgene. Biolistic transformation typically produces insertions composed of an array of multiple transgenes. We attempted to minimize the complexity of transgene insertions by using low DNA concentrations for the transformations (McGinnis et al., 2005). The screening of almost 300 independent transgenic events by DNA gel-blot analysis revealed that about one-third of the resulting transgenic lines had relatively simple banding patterns, indicating a low copy number of transgenic inserts (Fig. 4, left blot). The other approximately two-thirds of the transgenic lines yielded complex banding patterns indicative of high-copy transgenic inserts (Fig. 4, right blot).
Figure 4.
Southern-blot analysis of cosegregation with resistance phenotype and transgene complexity. Example blots are shown for a line with a simple, low copy number insertion (left) and a line in which the transgene contains multiple gene copies (right). For each blot, HindIII/SwaI-digested genomic DNA from the leaf of a different plant is loaded in each lane. Lanes with DNA from resistant plants are labeled R, while lanes labeled S contained DNA from sensitive plants. The probe is from the adh1 intron of maize (Probe B in Fig. 2) and recognizes the endogenous adh1 gene in addition to the IR transgene.
Assessment of Transgene Stability and Inheritance
In initial generations of outcrossing the transgenic plants, we noted that nearly one-quarter of the transgenic lines segregated less than the expected 50% resistant progeny (Supplemental Fig. S1). A series of experiments was conducted to investigate the contributions of transgene silencing and reduced transmission to the observation of reduced frequencies of resistant plants (Supplemental Text S1; Supplemental Figs. S1–S3).
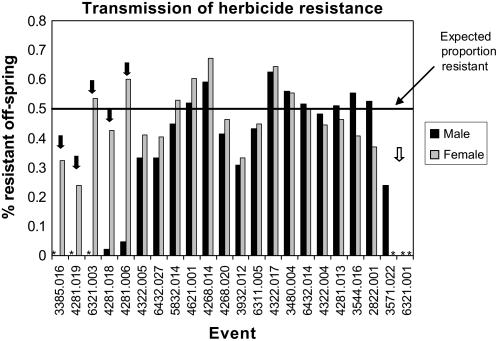
The lower-than-expected frequencies of resistant plants were more likely to be observed when the transgene was transmitted through the male parent (Fig. 5; Supplemental Table S2). The bias toward female transmission is similar to that observed for Mutator activity in maize (Walbot, 1986; Bennetzen, 1987). This bias against male transmission could be the result of reduced transmission of transgenic pollen or preferential transgene silencing in the male germline. As the male germline differentiates later than the female germline during maize development, it is possible that progressive transgene silencing during later stages of development contributes to the bias against transmission of an active transgene through the male parent. By performing the resistance screening of parental lines at multiple stages of development (including a screening at maturity), we could decrease the frequency of transgenic lines that showed reduced frequencies of resistant plants in the progeny (Supplemental Table S2). Analysis of the susceptible plants in families with reduced frequencies of resistant plants for the presence of the transgene allowed for a determination of whether the transgene was present but silent or whether the transgene was lacking in the susceptible plants (Supplemental Table S3). Of the 78 events that were tested, the reduced frequency of resistant plants was attributable to transgene silencing for 34 events and to reduced transmission for 44 events.
Figure 5.
Female and male transmission of herbicide resistance. For each transgenic event, 60 seeds derived from reciprocal crosses of a hemizygous transgenic plant with B73 were planted and scored for herbicide resistance. If stable transmission and expression of the transgene occurred, the expected frequency of herbicide-resistant progeny was 50%. For five events, the resistance phenotype was primarily transmitted through the female (black arrow). For one event, the resistance phenotype was transmitted only through the male (white arrow). The asterisks denote a case in which resistance was not transmitted through either parent.
Assessment of Transgene Stability in Self and Outcross Progeny
Based on the assumption that hemizygosity would reduce any potential trans interactions between the transgenes on homologous chromosomes that could lead to transgene silencing, our standard practice has been to maintain the transgenes in a hemizygous condition. To determine if this assumption was warranted, six individual plants, each representative of transgenic events that had demonstrated stable segregation of resistant and susceptible plants in previous generations, were outcrossed and self-pollinated. As expected, for the progeny of outcrossing, the frequency of resistant offspring was not significantly different from the expected 50% (Supplemental Table S4). However, for two of the six events, the percentage of resistant offspring from self-pollination was significantly lower than the expected 75% (P < 0.05 in χ2 test), and, for three additional events, the percentage was below 75% but not significant at P = 0.05 (Supplemental Table S4). These results suggest that outcrossing increases transgene stability.
Assessment of Reduction of Steady-State RNA Levels of Target Gene
A semiquantitative reverse transcription (RT)-PCR assay (Kerschen et al., 2004) was used to estimate the steady-state RNA level for the target gene in 230 unique transgene insertion events that represent 63 different transgenic constructs. A significant proportion of the events (79/230) showed reproducible reduction in the level of the target RNA. Nine additional events showed variable reduction in target gene RNA levels, in which some transgenic plants had reduced target gene mRNA relative to nontransgenic sibs, whereas other transgenic plants showed no reduction. The remaining 142 events did not show a detectable reduction in the amount of the target gene RNA.
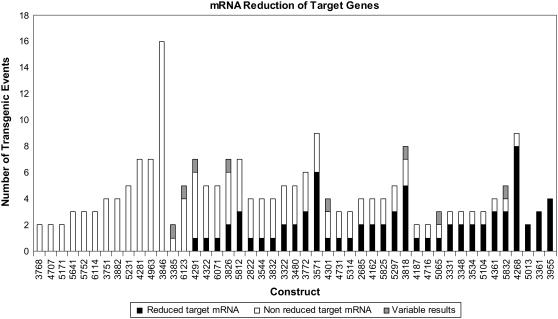
We assessed just one event for 16 of the 63 constructs. For the remaining 47 constructs, we assessed the reduction of the target RNA for multiple (2–16) events (Fig. 6). For three constructs (3361, 3955, and 5013), all events showed reduction in target gene RNA level. For 14 constructs, none of the events showed reduced target gene RNA levels. For the remaining constructs, some events had reduced RNA, but others did not.
Figure 6.
Number of independent transgenic events able to induce degradation of target gene mRNA for each construct. The construct number is indicated on the x axis. The numbers of events that showed reduced target mRNA, nonreduced target mRNA, and variable RNA reduction by RT-PCR assay are indicated for each construct.
We are interested in identifying the attributes that lead to a successful reduction of the RNA level of the target gene. Therefore, we attempted to correlate silencing success with several features, including endogenous expression level of the target gene, region of the gene targeted, or size of the target gene. Endogenous expression levels of the target genes in nontransgenic lines (based on Affymetrix analysis of B73 seedlings) were approximated from published microarray data (Stupar and Springer, 2006). The expression level of the target gene for constructs that provided silencing for all events (or the majority of events) was compared with the expression level for the target gene of constructs that rarely, or never, resulted in reduced expression of the target gene. We did not find any significant difference in the average expression level or distribution of expression levels for these genes. The size of the target genes and the position of the targeted region (first one-third, middle, or last one-third) within the target gene were also compared for constructs with consistent silencing versus constructs with rare silencing of the endogenous target gene (data not shown), but no correlation was detected.
A Single Transgene Can Target Multiple Endogenous Genes
Many of the chromatin genes analyzed in this project belong to families of genes that share sequence similarity. Because RNAi is a homology-mediated process, an IR sequence designed to target one gene might also silence a homolog. To identify such potential secondary targets, we searched for sequence matches of at least 21 bp (size of many typical siRNAs; Vaucheret, 2006) between primary IR segments and closely related gene family members. Thus, we judged a gene to be a potential target for silencing by an IR transgene if it shared a 21-bp stretch of identity with the IR sequence.
For 11 events that resulted in reduction of the RNA levels for their primary targets, semiquantitative RT-PCR was used to assay the effect of the transgene on expression level of potential secondary targets. The overall identity between the IR sequence and the secondary target genes included in these experiments ranged from 85% to 93% (Table II), and the number of perfect 21-bp matches ranged from one to 11. Seven of 10 constructs evaluated for their ability to reduce transcript abundance for multiple genes were able to reduce steady-state RNA levels for the secondary targets. For the remaining three constructs, steady-state RNA abundance was not reduced for the secondary target, even though these targets shared 90% to 91% identity with their respective IR sequences and had at least three 21-bp regions of perfect identity with the IR. This observation suggests that sequence identity is not the sole determinant of whether an IR sequence can induce degradation of target gene mRNA.
Table II.
Silencing of multiple targets with IR constructs
| Percent Identity between Secondary Target and IR Sequence | Secondary Target Semiquantitative RT-PCR Result | No. of ≥21-bp Stretches of Perfect Match between Secondary Target and IR Sequence | Gene Pair (Primary Target/Secondary Target) |
|---|---|---|---|
| 85 | Reduced | 1 | vef101/vef102 |
| 86 | Reduced | 2 | brd101/brd104 |
| 86 | Reduced | 3 | hdt103/hdt102 |
| 89 | Reduced | 5 | mbd108/mbd111 |
| 90 | Not reduced | 3 | dmt103/dmt107 |
| 90 | Reduced | 5 | hdt101/hdt104 |
| 90 | Not reduced | 3 | mbd101/mbd120 |
| 91 | Not reduced | 6 | mbd109/mbd115 |
| 92 | Reduced | 11 | dmt102/dmt105 |
| 93 | Reduced | 3 | hda109/hda115 |
Analysis of IR Sequence Context and Silencing Frequency
Transcription of an IR sequence is thought to result in a dsRNA that is recognized and processed by dicer-like proteins. Therefore, a given IR sequence will be processed to generate a population of siRNAs, with the number of siRNAs directly related to the length of the IR fragment. These siRNAs interact with argonaute-like proteins (for review, see Vaucheret, 2006), leading to degradation of target gene mRNA. A variety of features have been shown to be important predictors of an siRNA's silencing ability in human embryonic kidney cell culture (Reynolds et al., 2004). These include GC content, Tm, and presence of adenine at position 3 or 19, guanine at position 13, and uridine at positions 10 and 19 for each predicted siRNA (positions numbered consecutively with 1 indicating the 5′ most end of the siRNA relative to the presumed direction of transcription). To determine if the specific sequences of the component siRNAs would correlate with silencing ability of maize IR sequences, the Dice-o-matic computer program was used to generate a list of all possible 21-bp siRNAs for each IR sequence, and each siRNA was evaluated for the characteristics listed above.
Multiple regression analyses were performed to determine if a relationship existed between the ability of an IR sequence to silence its target and the variables listed above. At the time of this analysis, RT-PCR data on the expression level of the endogenous target gene was available for transgenic lines harboring 48 different IR transgenes. The values for every predicted siRNA for each of the 48 IR sequences were averaged and subjected to multiple regression analysis. In no case was there a predictive relationship found between any of the tested variables and a given IR sequence's ability to silence its target.
This analysis included seven IR sequences predicted to target two endogenous genes. Five of these IR constructs led to reduced expression of both target genes, while two constructs reduced expression of only the primary gene. The latter two constructs produced nine and 18 predicted siRNAs that would share 100% identity with their respective secondary target genes. The five constructs that did effectively reduce secondary target mRNA were predicted to produce a range of 11 to 81 siRNAs with 100% identity to the secondary targets. For each of these seven constructs, the subset of siRNAs that were predicted to target both primary and secondary target genes was compared to the entire population of siRNAs produced by this IR. No single siRNA characteristic or set of characteristics was predictive of an IR construct's ability to silence a secondary target gene (data not shown). The two unsilenced secondary targets included in this analysis were expressed in the same tissues as the primary target that was successfully silenced (data not shown). As such, tissue-specific expression of the secondary target gene also did not correlate with secondary target silencing.
DISCUSSION
Transgene-induced RNAi was used to trigger specific silencing of chromatin genes in maize. In some cases, this technique was highly effective, resulting in a properly transmitted transgene that led to thorough reduction of target gene mRNA. In other cases, effectiveness was compromised by reduced transmission, transgene silencing, or failure of the IR construct to silence its gene targets. These inconsistencies in transgene transmission and silencing ability might result from the innate variability of RNAi as a biological system, from stochastic silencing or spontaneous rearrangement of the bombarded transgenes, or from inadvertently biased selection during tissue culture.
In maize, the use of RNAi involves transformation and tissue culture. As with any deleterious or lethal mutation, RNAi constructs that hinder plant growth or development are likely to be selected against during callus growth and plant regeneration. This can result in failure to recover transgenic events for a particular construct or recovery of only nonfunctional transgenic events. The use of biolistic transformation, which frequently results in complex transgene loci, might also explain some of the transgene behaviors we observed, including changes in transgene activity throughout development, biased transmission of the transgene through male and female parents, and inconsistent transgene activity over multiple generations of outcrossing. It is likely that such variation is inherent to transgenic lines per se. Others have reported variation in levels of transgene expression and frequency of transgene silencing (De Wilde et al., 2000; Vain et al., 2002; Meng et al., 2006), which in our specific case might influence maintenance of silencing of the target gene. However, we found that maintaining transgenic stocks by outcrossing rather than self-pollination and by applying herbicide selection late in development, but prior to crossing, improves the likelihood of developing stable lines that faithfully transmit active transgenes.
Varying frequencies for silencing of an endogenous target have been reported in many different RNAi studies (Chuang and Meyerowitz, 2000; Fuhrmann et al., 2001; Cutter et al., 2003; Kamath et al., 2003; Waterhouse and Helliwell, 2003; Kerschen et al., 2004; Wang et al., 2005). One important note is that in many of these published studies the silencing efficiency was evaluated based on the severity of an anticipated phenotype. However, for a functional genomics project such as ours in which the gene function is unknown, it is difficult to anticipate an appropriate phenotype or set of phenotypes with which to identify the most robust lines. In the absence of such criteria, we chose to assess the success of the RNAi-silencing lines based on reduced mRNA levels for the target genes. Using a semiquantitative RT-PCR assay to determine the steady-state level for a particular region of a transcript could be applied in a high-throughput manner and allowed for identification of lines that induced mRNA degradation. Using this method, we found approximately 35% of our lines were effective at silencing their intended targets. This success rate for the production of lines with reduction of the steady-state level of the target gene is lower than that in several previous reports (Chuang and Meyerowitz, 2000; Fuhrmann et al., 2001; Kerschen et al., 2004). There are several potential explanations for the apparent discrepancy in silencing efficiency between our project and previous reports. It is possible that the previous reports are biased toward higher frequencies due to the use of genes with observable phenotypes or analysis in early transgenic generations. In addition, negative results are less frequently published, and there may be numerous examples of genes that were targeted by RNAi but were recalcitrant to this approach. Alternatively, it is possible that our success rate is abnormally low. Perhaps the lower efficiencies we observed were a factor of the large number of genes that we attempted to silence and that any project that involved this many genes would observe similar efficiency rates. Another factor might be the nature and function of the genes we targeted. Of all the previous reports of transgene-induced RNAi in plants, our efficiencies are most similar to those reported by researchers investigating a subset of Arabidopsis genes similar in number and predicted function (Kerschen et al., 2004). Genes related to chromatin structure and modification may be more difficult to silence using RNAi than other classes of genes.
In our analysis of b1 silencing by an IR transgene, two transgenic events showed silencing in initial generations, but silencing did not persist. This was true for other lines as well (data not shown). Because our RT-PCR evaluations of silencing were performed at the T2 generation, it is possible that some of the events that did not show silencing at T2 may have shown silencing in earlier generations. Evaluating silencing at a later generation may increase the likelihood of identifying the most stable transgenic events in which silencing will persist for multiple generations.
Other groups have noted that there is apparent variation in the susceptibility of different genes to silencing by RNAi (Cutter et al., 2003; Kerschen et al., 2004). To induce successful RNAi using an IR transgene, at least three distinct steps must occur: production of the dsRNA from the transgenic locus, processing of the dsRNA to form siRNAs, and degradation of the target RNA guided by the siRNAs. Our experiments tested for the outcome of the final step, a reduction in the steady-state transcript level for the target gene. Therefore, we were unable to determine whether the transgenic events that failed to reduce the target gene were the result of a failure to produce dsRNA or siRNA or to degrade the target gene. Additional work will be required to understand at what stage silencing failed in the unsuccessful lines. It is possible that the IR portion of the transgene was silenced in spite of bar gene activity, resulting in a loss of siRNAs. Another possibility is that the IR remained active and the siRNAs were generated, but for some reason posttranscriptional transcript degradation did not occur. A combination of these and likely other unidentified factors contribute to the inconsistent or lack of silencing by some constructs.
Although RNAi is a widely used technology, relatively little is known about the optimal application of this technique in plants. There are many potential factors that can contribute to a given sequence's ability to cause RNAi-induced silencing of an endogenous target. The “21-bp rule” is commonly used to predict whether a given IR will target a given gene, meaning that 21 bp of 100% identity should be enough to trigger silencing. Our data suggest that this is an oversimplification and that other factors play a role. Additional analysis will be required to determine what characteristics can be used to model and predict the silencing effectiveness of a given IR sequence in plants.
Overall, this study led to the production of a large number of stable lines that transmitted and expressed the transgene faithfully from generation to generation and that successfully induced target gene silencing. This suggests that it is possible to use an RNAi approach to generate positive results in spite of a significant degree of inherent variability. This variability should be accounted for as much as possible when analyzing RNAi-induced phenotypes and when proposing future large-scale RNAi projects in crop plants.
MATERIALS AND METHODS
Genetic Stocks
Embryos were isolated from HiII A × B F1 hybrids, as described previously (Armstrong et al., 1991; McGinnis et al., 2005). Following transformation by biolistic particle bombardment and regeneration, T0 plants were outcrossed with the maize (Zea mays) inbred line B73. In subsequent generations, stocks were typically maintained by outcrossing with B73 to maintain the transgene in a hemizygous condition. For the b1 silencing assays, T1 plants that had been outcrossed for one generation with the inbred line B73 were crossed with B-I/B-I tester, B-Peru/B-Peru tester, or B-I/B-Peru heterozygous tester. The B-I and B-Peru stocks have been described previously (Dorweiler et al., 2000). Transgenic stocks that have been tested to verify the correct transgene and reduction for the steady-state level of the target mRNA are available through the Maize Genetics Stock Center.
Transformation Protocols
Transformation protocols have been described previously in more detail (McGinnis et al., 2005). Briefly, microprojectile biolistic delivery of DNA was used to transform immature embryos. Bombardments were carried out with a low concentration of plasmid DNA (0.4 μg plasmid DNA/1 mg microcarriers) so as to achieve lower copy number transgene insertions.
siRNA Northern Blots
Tissue was harvested and RNA extracted from the sheaths of individual plants for analysis of siRNAs, with an additional step to enrich for siRNA molecules by polyethylene glycol precipitation (McGinnis et al., 2006). Polyacrylamide gel electrophoresis was used to size fractionate RNAs prior to transfer to membrane. Blotting and hybridization of northern blots were performed as described by Rudenko et al. (2003). The b1 probe utilized for this work was amplified by PCR and corresponds to the positions 422 to 1,081 in GenBank accession X57276.
Nuclear Run-On Analysis
Nuclear run-on analysis was performed on nuclei isolated from sheaths of individual plants, as described previously (Dorweiler et al., 2000). Briefly, nuclei from 10 to 15 g of sheath or husk tissue from plants at anthesis were prepared using a modified chromatin isolation protocol. Denatured PCR product was slot blotted onto nitrocellulose filter membranes; the sequences of the probes used herein have been described previously (McGinnis et al., 2006).
DNA Gel-Blot Analysis
DNA extractions and Southern-blot analyses were performed as described previously (McGinnis et al., 2006). Genomic DNA was extracted from healthy leaf tissue and digested with restriction enzymes predicted to isolate the IR cassette from genomic DNA (typically HindIII and SwaI). Agarose gel electrophoresis and blotting of DNA to membrane was performed according to standard protocols (Sambrook et al., 1989), and blots were hybridized with a probe specific for the adh1 intron that is included in the IR transgene. This probe was generated by PCR, resulting in a product that corresponds to positions 4,837 to 5,402 in GenBank AY572837.
Herbicide Testing
Testing for resistance to glufosinate was performed as described previously (McGinnis et al., 2005). Briefly, a herbicide solution (1:100 dilution of a stock containing 200 mg/mL glufosinate, 0.01% Tween 20) was painted on a fully emerged leaf that had been marked with black permanent marker so the application site could be identified later. After 3 to 5 d, herbicide resistance or susceptibility was scored by visual observation. A susceptible plant exhibited a yellowing or browning of the painted leaf. A resistant leaf retained a healthy green appearance. For the experiments that involved herbicide-resistant testing at multiple stages, we applied the herbicide solution to the actively growing portions of the plants using a backpack sprayer.
PCR Analysis of Transgene Presence
PCR amplification was used to test for the presence of the transgene in DNA isolated from resistant and sensitive plants. Leaf tissue was harvested and used for cetyl trimethyl ammonium bromide DNA extractions (Stupar and Springer, 2006). Two separate PCR reactions that test for the presence of two different regions of the transgene were used to test for presence of the transgene. The primers used were BstBarF1 (5′-GCC AAA TGT TGA ACG ATC TGC AGG-3′) with BstBarR1 (5′-GCA CCA TCG TCA ACC ACT ACA TCG-3′); or WaxyintronF1 (5′-GCC ACC CAA GAA ACT GCT CCT TAA G-3′) with OCS3'R1 (5′-CGG CGG TAA GGA TCT GAG CTA CAC-3′). PCR reactions were performed in a 15-μL total volume containing approximately 20 ng of DNA, 2.5 pmol of each primer, 0.065 units of HotStarTaq polymerase (Qiagen), 1.5 μL of 10× reaction buffer, and 0.1 μL of 25 mm dNTPs. Conditions of the PCR were as follows: 94°C for 15', 35 cycles of 94°C for 30″, 62°C for 30″, 72° for 1', followed by 72°C for 10'.
RT-PCR Assays
Analysis of target gene mRNA abundance was performed using RT-PCR techniques described previously (McGinnis et al., 2005).
Analysis of siRNA Sequences
Dice-o-matic (whose source code is available upon request) was implemented in the Ruby programming language. Dice-o-matic accepts as its input a sequence 21 bp or longer, which is then partitioned into consecutive 21-bp windows. Each of these 21-bp subsequences is then subjected to the eight tests identified as significant in previous studies (Reynolds et al., 2004): GC content, number of A/U bp in positions 1 to 5 and 15 to 19 of the sense strand, Tm, a lack of G and C at position 19 and the presence of adenine at position 3 or 19, guanine at position 13, and uridine at positions 10 and 19 for each predicted siRNA (positions numbered consecutively with 1 indicating the 5′ most end of the siRNA relative to the presumed direction of transcription). We found the published method for calculating Tm to be ambiguous; thus, we used an alternate formulation. Rather than a cutoff of 20°C, the temperature at which 30% of the siRNAs generated by all constructs melted (as determined using the nearest-neighbor method; SantaLucia, 1998), 56.3°C, was used. The results of each test, as well as a summary of the input sequence's performance, were output in CSV format.
After running Dice-o-matic for each of the constructs, a suite of custom Perl scripts was prepared for an initial evaluation of the output. Additionally, R was used to perform multiple regression analysis of the results to determine if there was a correlation between silencing success and the eight variables described above (R Development Core Team, 2005). First, the ability of each construct to induce silencing was quantified on the basis of the number of events that induced silencing of the endogenous target gene based on RT-PCR data. For each construct, the mean silencing value was then used for multiple regression analysis in combination with the aggregate data for each of the eight properties of all predicted siRNAs for that construct. In the cases of GC content and Tm, the actual values for each siRNA were averaged to give a mean value for each construct. For the properties that tested the presence of specific base pairs in specific locations, the percentage of siRNAs that met the condition of having the designated base pair in the designated location was used for regression analysis.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Text S1. Transgene heritability and stability.
Supplemental Figure S1. Variation for transgene stability.
Supplemental Figure S2. Male and female transmission of transgene.
Supplemental Figure S3. Seedling transgene stability.
Supplemental Table S1. Transformation efficiencies for all constructs.
Supplemental Table S2. Parent-of-origin effects on transgene inheritance.
Supplemental Table S3. Analysis of transgene silencing and reduced transmission.
Supplemental Table S4. Effect of self-pollination on transgene stability.
Supplementary Material
Acknowledgments
We are grateful to Vicki Chandler for assistance in the design and interpretation of the experiments as well as her valuable feedback on the manuscript. Anna Howell, Arthur Kerschen, Robert Sandoval, Carolyn Napoli, Rich Jorgensen, Lyudmila Sidorenko, Annie Bergmark, Anna Bredsten, Robert Stupar, Jill Mahoy, Laura Schmitt-Brunold, Dean Bergstrom, Miriam Hankins, and Barbara Sonderman all contributed to the data generation for this project, and we are very grateful for their assistance.
This work was supported by the National Science Foundation (grant no. DBI–0421619 to K.C., H.K., S.K., K.M., and N.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Nathan M. Springer (springer@umn.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Armstrong C, Green C, Phillips R (1991) Development and availability of germplasm with high Type II culture formation response. Maize Genet Coop News Lett 65 92–93 [Google Scholar]
- Bennetzen JL (1987) Covalent DNA modification and the regulation of Mutator element transposition in maize. Mol Gen Genet 208 45–51 [Google Scholar]
- Brodersen P, Voinnet O (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22 268–280 [DOI] [PubMed] [Google Scholar]
- Brutnell TP (2002) Transposon tagging in maize. Funct Integr Genomics 2 4–12 [DOI] [PubMed] [Google Scholar]
- Chandler VL, Radicella JP, Robbins TP, Chen J, Turks D (1989) Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences. Plant Cell 1 1175–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Tang WH, Hong MM, Wang ZY (2003) OsBP-73, a rice gene, encodes a novel DNA-binding protein with a SAP-like domain and its genetic interference by double-stranded RNA inhibits rice growth. Plant Mol Biol 52 579–590 [DOI] [PubMed] [Google Scholar]
- Chuang CF, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA 97 4985–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe EJ (1979) Specification of the anthocyanin biosynthetic function by B and R in maize. Maydica 24 49–58 [Google Scholar]
- Cutter AD, Payseur BA, Salcedo T, Estes AM, Good JM, Wood E, Hartl T, Maughan H, Strempel J, Wang B, et al (2003) Molecular correlates of genes exhibiting RNAi phenotypes in Caenorhabditis elegans. Genome Res 13 2651–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wilde C, Van Houdt H, De Buck S, Angenon G, De Jaeger G, Depicker A (2000) Plants as bioreactors for protein production: avoiding the problem of transgene silencing. Plant Mol Biol 43 347–359 [DOI] [PubMed] [Google Scholar]
- Dorweiler JE, Carey CC, Kubo KM, Hollick JB, Kermicle JL, Chandler VL (2000) Mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12 2101–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann M, Stahlberg A, Govorunova E, Rank S, Hegemann P (2001) The abundant retinal protein of the Chlamydomonas eye is not the photoreceptor for phototaxis and photophobic responses. J Cell Sci 114 3857–3863 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 231–237 [DOI] [PubMed] [Google Scholar]
- Kerschen A, Napoli CA, Jorgensen RA, Muller AE (2004) Effectiveness of RNA interference in transgenic plants. FEBS Lett 566 223–228 [DOI] [PubMed] [Google Scholar]
- May BP, Liu H, Vollbrecht E, Senior L, Rabinowicz PD, Roh D, Pan X, Stein L, Freeling M, Alexander D, et al (2003) Maize-targeted mutagenesis: a knockout resource for maize. Proc Natl Acad Sci USA 100 11541–11546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis K, Chandler V, Cone K, Kaeppler H, Kaeppler S, Kerschen A, Pikaard C, Richards E, Sidorenko L, Smith T, et al (2005) Transgene-induced RNA interference as a tool for plant functional genomics. Methods Enzymol 392 1–24 [DOI] [PubMed] [Google Scholar]
- McGinnis KM, Springer C, Lin Y, Carey CC, Chandler VL (2006) Transcriptionally silenced transgenes in maize are activated by three mutations defective in paramutation. Genetics 173 1637–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Ziv M, Lemaux PG (2006) Nature of stress and transgene locus influences transgene expression stability in barley. Plant Mol Biol 62 15–28 [DOI] [PubMed] [Google Scholar]
- Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30 5036–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2005) R: a language and environment for statistical computing. R Foundation for Statistical Computing. http//www.R-project.org (June 20, 2006)
- Radicella JP, Brown D, Tolar LA, Chandler VL (1992) Allelic diversity of the maize B regulatory gene: different leader and promoter sequences of two B alleles determine distinct tissue specificities of anthocyanin production. Genes Dev 6 2152–2164 [DOI] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A (2004) Rational siRNA design for RNA interference. Nat Biotechnol 22 326–330 [DOI] [PubMed] [Google Scholar]
- Rudenko GN, Ono A, Walbot V (2003) Initiation of silencing of maize MuDR/Mu transposable elements. Plant J 33 1013–1025 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- SantaLucia J Jr (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci USA 95 1460–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Song R, Messing J (2003) A new opaque variant of maize by a single dominant RNA-interference-inducing transgene. Genetics 165 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger DA, Chandler VL (1999) Major recent and independent changes in levels and patterns of expression have occurred at the b gene, a regulatory locus in maize. Proc Natl Acad Sci USA 96 15007–15012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger DA, Chandler VL (2001) B-Bolivia, an allele of the maize b1 gene with variable expression, contains a high copy retrotransposon-related sequence immediately upstream. Plant Physiol 125 1363–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger DA, Lisch D, Chandler VL (1998) The maize regulatory gene B-Peru contains a DNA rearrangement that specifies tissue-specific expression through both positive and negative promoter elements. Genetics 149 1125–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Kaeppler SM (2005) Evolutionary divergence of monocot and dicot methyl-CpG-binding domain proteins. Plant Physiol 138 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Napoli CA, Selinger DA, Pandey R, Cone KC, Chandler VL, Kaeppler HF, Kaeppler SM (2003) Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol 132 907–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Springer NM (2006) Cis-transcriptional variation in maize inbred lines B73 and Mo17 lead to additive expression patterns in the F1 hybrid. Genetics 173 2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles E, Ceska O, Seah K (1973) Developmental differences in the action of r and b alleles in maize. Can J Genet Cytol 15 59–72 [Google Scholar]
- Till BJ, Reynolds SH, Weil C, Springer N, Burtner C, Young K, Bowers E, Codomo CA, Enns LC, Odden AR, et al (2004) Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol 4 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travella S, Klimm TE, Keller B (2006) RNAi based gene silencing as an efficient tool for functional genomics in hexaploid bread wheat (Triticum aestivum L.). Plant Physiol 142 6–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vain P, James A, Worland B, Snape W (2002) Transgene behaviour across two generations in a large random population of transgenic rice plants produced by particle bombardment. Theor Appl Genet 105 878–889 [DOI] [PubMed] [Google Scholar]
- Vaucheret H (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20 759–771 [DOI] [PubMed] [Google Scholar]
- Walbot V (1986) Inheritance of Mutator activity in Zea mays as assayed by somatic instability of the bz2-mul allele. Genetics 114 1293–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Iyer LM, Pancholy R, Shi X, Hall TC (2005) Assessment of penetrance and expressivity of RNAi-mediated silencing of the Arabidopsis phytoene desaturase gene. New Phytol 167 751–760 [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Helliwell CA (2003) Exploring plant genomes by RNA-induced gene silencing. Nat Rev Genet 4 29–38 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.