Abstract
The composition of photosystem II (PSII) in the chlorophyll (Chl) d-dominated cyanobacterium Acaryochloris marina MBIC 11017 was investigated to enhance the general understanding of the energetics of the PSII reaction center. We first purified photochemically active complexes consisting of a 47-kDa Chl protein (CP47), CP43′ (PcbC), D1, D2, cytochrome b559, PsbI, and a small polypeptide. The pigment composition per two pheophytin (Phe) a molecules was 55 ± 7 Chl d, 3.0 ± 0.4 Chl a, 17 ± 3 α-carotene, and 1.4 ± 0.2 plastoquinone-9. The special pair was detected by a reversible absorption change at 713 nm (P713) together with a cation radical band at 842 nm. FTIR difference spectra of the specific bands of a 3-formyl group allowed assignment of the special pair. The combined results indicate that the special pair comprises a Chl d homodimer. The primary electron acceptor was shown by photoaccumulation to be Phe a, and its potential was shifted to a higher value than that in the Chl a/Phe a system. The overall energetics of PSII in the Chl d system are adjusted to changes in the redox potentials, with P713 as the special pair using a lower light energy at 713 nm. Taking into account the reported downward shift in the potential of the special pair of photosystem I (P740) in A. marina, our findings lend support to the idea that changes in photosynthetic pigments combine with a modification of the redox potentials of electron transfer components to give rise to an energetic adjustment of the total reaction system.
Keywords: Acaryochloris marina, FTIR, reaction center, photosynthesis, electron transfer
Chlorophyll (Chl) a has a ubiquitous role as an electron donor in the photochemical reactions of oxygenic photosynthesis, in which two kinds of photosystem, namely photosystem I (PSI) and photosystem II (PSII), cooperatively drive photosynthetic electron flow from water to NADP+. The reduced cofactor, NADPH, is then used for CO2 fixation. Chl a is a key pigment that serves as an electron donor called the special pair in PSI and PSII. Acaryochloris spp. are unique cyanobacteria that differ from the majority of photosynthetic organisms by having Chl d (3-desvinyl-3-formyl Chl a) (1–4) as the major pigment (>95%); Chl a is a minor component but is never absent (5, 6). In photosynthetic organisms, changes in pigment composition affect both the function of pigments and their reaction environments, including the modified proteins that accommodate them. Photosynthetic pigments function in two roles: as light-harvesting components and as electron transfer components. Light harvesting is mainly governed by the orientation and energy levels of pigments, and, in this context, a particular excitation energy level is not an absolute precondition for function, but a relative one. On the other hand, electron transfer reactions are governed by an absolute redox potential, because the photosynthetic oxidation of water requires a very high potential, whereas reduction of NADP+ requires a very low potential. For this reason, it is of particular interest to understand the effects of pigment changes on the electrochemical properties of the system and not solely the light-harvesting properties.
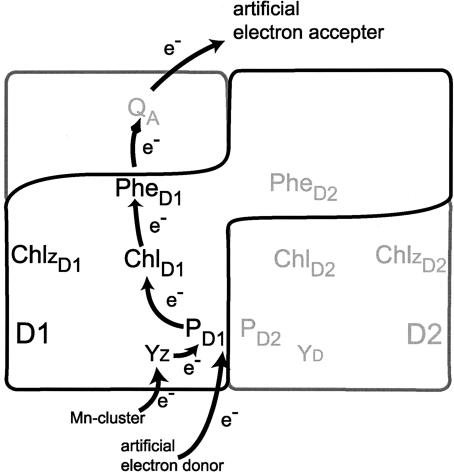
PSII is essential for the oxidation of water, and therefore a high oxidation potential and the means to realize this in the protein matrix are of primary importance. An accepted scheme for electron transfer and the organization of PSII cofactors is shown in Fig. 1. The existence of Acaryochloris spp. with a majority of Chl d pigment composition raises the inevitable question as to the constitution of PSII in these species and whether changes in the pigment species inevitably induce changes in the photosynthetic reaction system. The energy of the light absorbed by Chl d is lower than for Chl a, and yet oxygenic photosynthesis is still operative. If the special pair (PD1 and PD2) (Fig. 1) was found to comprise a Chl d homodimer or a Chl d–Chl a heterodimer, our current understanding of the overall energetics of the water oxidation system would need to be modified. This is a critical question for PSII, and Acaryochloris spp. are suitable candidates to enhance our general understanding of PSII function. We have previously proposed a Chl a homodimer as the special pair in Acaryochloris spp. based on delayed fluorescence (DF) measurements in the nanosecond time range at 77 K on intact cells (6, 7); other reports have differed in assignment of the special pair (8). The nature of the special pair in PSII is a key and fundamental question in oxygenic photosynthesis, and here we revisit this question with the hope of adding clarity to a technically difficult area.
Fig. 1.
Scheme for electron transfer and cofactor organization of PSII RC. Arrows show the direction of electron transfer. Pigments located on the D1 protein are shown with a subscript D1, and those on the D2 protein are shown with a subscript D2. PD1 and PD2, the special pair; ChlD1 and ChlD2, the accessory Chl; PheD1, the primary electron acceptor, pheophytin; PheD2, pheophytin; ChlzD1 and ChlzD2, peripheral Chl; QA, the secondary electron acceptor; YZ, secondary electron donor; YD, tyrosine on the D2 protein.
P680, the special pair of PSII in oxygenic photosynthetic organisms, was initially detected by a flash photolysis technique (9) and was later spectrally characterized in detail (10, 11). Isolation of a D1–D2–cyt b559 complex [PSII reaction center (RC)] from spinach (12) accelerated detailed characterization. The history of this field indicates that a purified sample is necessary for the reliable identification and characterization of the special pair in a given organism. We have therefore purified PSII complexes from Acaryochloris marina MBIC 11017 and have identified cofactors including the special pair using several different spectroscopic approaches. Based on new data obtained from purified samples we now identify the special pair in A. marina MBIC 11017 as a Chl d homodimer (PD1 and PD2) (Fig. 1) and the primary electron acceptor as pheophytin (Phe) a (PheD1) (Fig. 1). We also discuss overall energetics in the context of an improved general understanding of PSII reaction processes and mechanisms in oxygenic photosynthesis.
Results
Subunit Composition of the Purified PSII Complexes.
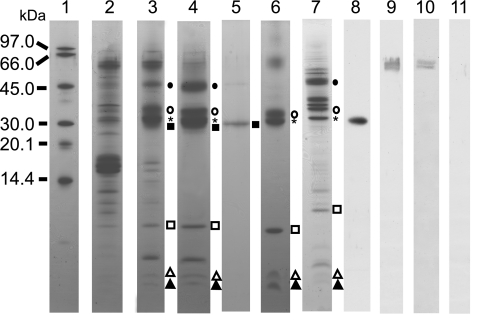
We purified dimeric PSII complexes using a combination of density gradient centrifugation and two types of column chromatography. Subunits in the PSII complexes were identified by SDS/PAGE and sequence analysis. As purification proceeded, the polypeptide pattern became simpler (Fig. 2, lanes 2–4). In thylakoid membranes, many polypeptides were present (Fig. 2, lane 2), whereas in partially purified PSII complexes several of these bands were lost, primarily phycobiliproteins (Fig. 2, lane 3). The subunit composition of the final samples (Fig. 2, lane 4) comprised 47-kDa Chl protein (CP47) (Fig. 2, filled circles), D1 (Fig. 2, asterisks), D2 (Fig. 2, open circles), cytochrome (cyt) b559 α-subunit (Fig. 2, open squares), and several small polypeptides; this composition was comparable to purified spinach PSII RC (Fig. 2, lane 6). The D1 subunit was detected specifically by Western blot analysis (Fig. 2, lane 8). The D1 band was somewhat broad and stained more heavily than the D2 band; this may be because of overlap with CP43′ (PcbC) (Fig. 2, filled squares), because the latter has very similar mobility to D1 (Fig. 2, lane 5). When a small amount of sample was loaded onto SDS/PAGE, the Coomassie brilliant blue-stained D1 band separated into two bands (data not shown), suggesting the presence of both D1 and CP43′. The presence of both components was confirmed by peptide mass fingerprinting (data not shown). The stoichiometry of the two components was estimated to be close to 1:1 based on staining intensities on SDS/PAGE. In the low-molecular-weight region three bands were discernible. Based on comparisons with spinach PSII RC (Fig. 2, lane 6) and Synechocystis sp. PCC 6803 (hereafter referred to as Synechocystis) PSII (Fig. 2, lane 7), the lower two subunits were assigned as PsbI (Fig. 2, open triangles) and cyt b559 β-subunit (PsbF; Fig. 2, filled triangles), respectively. One additional unknown colorless small polypeptide was present in our preparations; its partial sequence was obtained, but we were not able to assign it to any known polypeptide. The purified samples did not contain PSI complexes; Western blotting with anti PsaA/B antibody gave a positive signal for thylakoid membranes (Fig. 2, lane 9) and for partially purified PSII complexes (Fig. 2, lane 10), but no band for the purified PSII complexes (Fig. 2, lane 11). The results indicate that the purified PSII complexes consist of CP47, CP43′, D1, D2, cyt b559, PsbI, and one additional small polypeptide and are thus comparable to purified complexes obtained from spinach (13, 14) and Synechocystis (Fig. 2, lane 7). Compared with previous reports on isolated PSII complexes from A. marina (8, 15, 16), our sample was highly purified as judged by the subunit composition.
Fig. 2.
SDS/PAGE and Western blot analysis of PSII complexes isolated from A. marina. Lane 1, molecular marker; lane 2, thylakoid membranes; lane 3, partially purified PSII preparations after DEAE Toyopearl chromatography; lane 4, purified PSII complexes; lane 5, CP43′; lane 6, PSII RC isolated from spinach; lane 7, PSII core complex isolated from Synechocystis; lane 8, Western blot of purified PSII complex (lane 4) with anti-D1; lanes 9–11, Western blot with anti-PsaA/B; lane 9, thylakoid membranes (lane 2); lane 10, partially purified PSII (lane 3); lane 11, purified PSII complex (lane 4). Individual marks represent CP47 (filled circles), D2 (open circles), D1 (asterisks), CP43′ (filled squares), cyt b559 α-subunit (open squares), PsbI (open triangles), and cyt b559 β-subunit (filled triangles).
Photochemical Activity.
Our PSII complexes were photochemically active, showing a diphenylcarbazide (DPC)-2,6-dichlorophenol-indophenol (DCIP) photoreduction activity of 230 μmol/mg Chl/hr; isolation of photochemically active complexes from a Chl d-dominated cyanobacterium has not been reported. The observed level of activity was enough for further analysis: for comparison, the primary electron donor of PSII was initially identified by using spinach PSII RC with an activity of 170 μmol/mg Chl/hr DPC-DCIP activity (17), prepared by using the procedure of Nanba and Satoh (12).
Pigment Composition.
We identified four kinds of pigment in the purified complexes: Chl d, Chl a, Phe a, and α-carotene; Chl d′ was not detected. The quantities of individual pigments per two molecules of Phe a were 55 ± 7 Chl d, 3.0 ± 0.4 Chl a, and 17 ± 3 α-carotene (n = 4). Chl a was unambiguously detected, consistent with observations on intact cells (7). The content of Chl d was higher than expected from the crystal structure (18–20). The presence of CP43′ may be a primary reason why certain pigments were found to be present at higher levels than expected. Biochemical analyses sometimes gave rise to higher pigment contents, e.g., 40–55 Chl (21, 22) than are observable in the crystal structure; this tendency is more pronounced for lipids (23).
We determined the plastoquinone (PQ)-9 content to be 1.4 ± 0.2 per two Phe a molecules (n = 4), indicating that QA (Fig. 1) was associated with PSII. This was consistent with the observed DCIP photoreduction activity, because electron transfer to DCIP is known to arise from QA. Additionally, the light-minus-dark FTIR difference spectrum showed the appearance of a ferrocyanide band arising from electron transfer from QA (data not shown). The results clearly show that QA was active in our preparations. The content of cyt b559 was estimated at one per 55 Chl d by an oxidation–reduction difference spectrum, i.e., one cyt b559 per one PSII.
Absorption and Fluorescence Spectra.
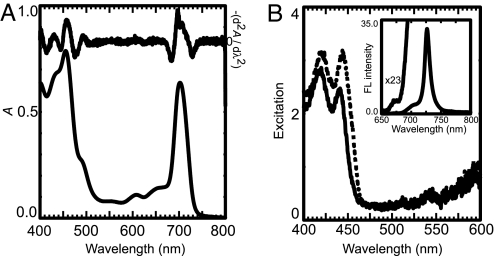
At 283 K, an absorption maximum was observed at 702 nm (Fig. 3A), significantly shorter than in previous reports (8, 15, 16). The second derivative spectrum indicated the presence of distinct bands at 697 and 708 nm, with possibly a third band present near 715 nm. In a previous report (8) a 725-nm band was observed in PSII, but we have found that this was due to contamination by PSI. In the Qy region of Chl, peaks for Phe a and Chl a were not detected because of the high content of Chl d. An α-carotene band was observed at 490 nm. The Soret bands of Phe a and Chl a were not resolved. The CD spectrum of the complexes is shown in supporting information (SI) Fig. 6.
Fig. 3.
Absorption and fluorescence spectra of purified PSII complexes. (A) Absorption spectrum at 283 K. The second derivative spectrum is shown in the upper part, after inversion. (B) Fluorescence excitation spectra at 77 K monitored at 675 nm (solid line) and 679 nm (dotted line). (Inset) Fluorescence spectrum at 77 K upon excitation at 435 nm.
Fluorescence spectra at 77 K showed three bands at 728, 700, and ≈680 nm on excitation at 435 nm (Fig. 3B Inset). The bands at 728 and 700 nm arose from Chl d, whereas the third band derived from a mixture of Chl a and Phe a, as indicated from the excitation spectrum (Fig. 3B). The presence of Chl a (443 nm) and Phe a (418 nm) was unambiguously demonstrated in fluorescence excitation spectra.
Identification of the Special Pair.
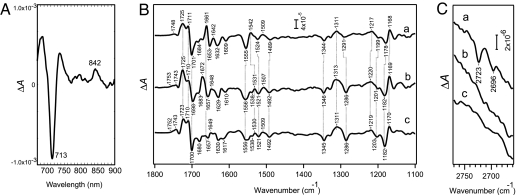
The light-induced difference spectrum of the PSII special pair was obtained at 298 K (Fig. 4A). The spectrum showed reversible changes; two kinds of difference spectra, namely during-minus-before illumination, and during-minus-after illumination, gave identical results. A negative peak was detected at 713 nm, and a positive peak was detected at 842 nm. The former corresponds to bleaching of the special pair, whereas the positive peak corresponds to a cation radical of the special pair. In the Chl a region there was no signal. This represents a spectrum for the special pair in PSII of A. marina, and it strongly suggests that the special pair consists of Chl d.
Fig. 4.
Identification of the special pair of PSII complexes. (A) Identification by the reversible absorption change of the sample in the Qy region at 298 K. Difference spectrum during blue light illumination is shown. (B and C) Identification by FTIR spectroscopy. Shown are light-induced cation-minus-neutral FTIR difference spectra of the special pair in PSII core complexes of A. marina (line a) and Synechocystis (line b) and in PSII membranes of spinach (line c). Separate overlays are shown for 1,800–1,100 cm−1, the region involving C O stretching and chlorin ring vibrations (B), and 2,770–2,660 cm−1, the region involving the CH stretching vibration of the 3-formyl group (C). Spectra were recorded at 250 K.
O stretching and chlorin ring vibrations (B), and 2,770–2,660 cm−1, the region involving the CH stretching vibration of the 3-formyl group (C). Spectra were recorded at 250 K.
Fig. 4B shows a cation-minus-neutral FTIR difference spectrum (1,800–1,100 cm−1) of the special pair (P+/P) in PSII complexes of A. marina (line a), compared with P680+/P680 in PSII core complexes of Synechocystis (line b) and PSII membranes (BBY particles) of spinach (line c). This region reflects the C O stretching and chlorin ring vibrations of Chls (24, 25). The overall spectral features were very similar among the three spectra. Prominent positive peaks at 1,725–1,723 and 1,711 cm−1, accompanied by a strong negative peak at 1,701–1,698 cm−1, are attributed to 131-keto C
O stretching and chlorin ring vibrations of Chls (24, 25). The overall spectral features were very similar among the three spectra. Prominent positive peaks at 1,725–1,723 and 1,711 cm−1, accompanied by a strong negative peak at 1,701–1,698 cm−1, are attributed to 131-keto C O vibrations, whereas strong or medium peaks at 1,680–1,620 cm−1 correspond mainly to polypeptide backbones. Bands in the region of 1,617–1,150 cm−1 are due to chlorin ring modes, which result from complex couplings among CC and CN stretching and CH bending vibrations, and hence are sensitive to differences between different Chl species.
O vibrations, whereas strong or medium peaks at 1,680–1,620 cm−1 correspond mainly to polypeptide backbones. Bands in the region of 1,617–1,150 cm−1 are due to chlorin ring modes, which result from complex couplings among CC and CN stretching and CH bending vibrations, and hence are sensitive to differences between different Chl species.
Upon closer examination of this ring vibration region, some clear differences were observed. The peaks at 1,170, 1,182, 1,220, 1,286, 1,492, and 1,521 cm−1, which agreed within 1 cm−1 between Synechocystis and spinach, showed shifts by 2–5 cm−1 in A. marina. In addition, spectral features in the 1,560–1,530 cm−1 region were significantly different between Synechocystis and spinach on the one hand and A. marina on the other (Fig. 4B). These differences indicate that the chemical structure of the chlorin ring in the primary donor of A. marina is somewhat different from that of P680 formed by Chl a. The data strongly suggest that the special pair of PSII in A. marina consists of Chl d.
Definite evidence for Chl d should be obtainable from the vibrations of a 3-formyl group. The CH stretching vibration of a formyl (−CHO) group gives rise to a peak at ≈2,700 cm−1, which is not obscured by other CH stretching bands from methyl or methylene groups (2,800–3,000 cm−1). This peak thus serves as a good marker for the formyl group (26). Indeed, in the P740+/P740 FTIR difference spectrum of PSI complexes from A. marina (27, 28), Sivakumar et al. (28) observed 3-formyl CH bands at 2,727 and 2,715 cm−1, providing solid evidence that P740 comprises two Chl d molecules. In our P+/P difference spectra of PSII complexes from A. marina, two clear peaks are observed at 2,723 and 2,696 cm−1 (Fig. 4C, line a), whereas no peaks are observed in this region in the P680+/P680 spectra of Synechocystis and spinach (Fig. 4C, lines b and c). The negative intensities may indicate that cation radical formation perturbs these peaks mainly as an intensity decrease. The presence of the formyl CH stretch also provides strong evidence that the special pair of PSII comprises Chl d molecules.
If the special pair is formed by Chl d, the C O stretching band of the 3-formyl group should be present in the 1,670–1,640 cm−1 region of the A. marina P+/P spectrum (Fig. 4B, line a) in analogy to the 7-formyl C
O stretching band of the 3-formyl group should be present in the 1,670–1,640 cm−1 region of the A. marina P+/P spectrum (Fig. 4B, line a) in analogy to the 7-formyl C O band of Chl b (29). Indeed, the Raman spectrum of Chl d exhibits a 3-formyl C
O band of Chl b (29). Indeed, the Raman spectrum of Chl d exhibits a 3-formyl C O peak at 1,659 cm−1 (30, 31). Unfortunately, the amide I bands of proteins overlap in this region, and hence definite identification of the formyl C
O peak at 1,659 cm−1 (30, 31). Unfortunately, the amide I bands of proteins overlap in this region, and hence definite identification of the formyl C O band in the P+/P spectrum is not possible. Nevertheless, it is notable that in the spectrum of A. marina (Fig. 4B, line a) a prominent positive peak is present at 1,661 cm−1, where negative features were observed in other two species (Fig. 4B, lines b and c).
O band in the P+/P spectrum is not possible. Nevertheless, it is notable that in the spectrum of A. marina (Fig. 4B, line a) a prominent positive peak is present at 1,661 cm−1, where negative features were observed in other two species (Fig. 4B, lines b and c).
Summarizing the results from the difference absorption spectrum of the reversible change, together with the FTIR spectra, we conclude that the special pair of PSII in A. marina consists of a Chl d dimer, and we have named this P713.
Identification of the Primary Electron Acceptor.
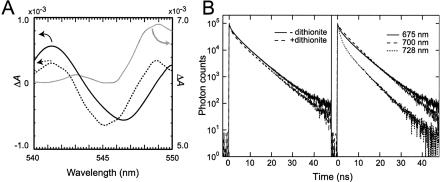
We further investigated the identity of the primary electron acceptor in PSII of A. marina. Because Phe a was present in our preparation and DPC-DCIP photoreduction was clearly demonstrated, Phe a was a probable candidate for the primary electron acceptor. Even though Phe a had been assigned as the primary electron acceptor based on an electrochromic shift (32), this assignment had not been verified by using difference absorption spectra. We have therefore determined the photoaccumulation of reduced Phe a. Usually this measurement is made in the presence of sodium dithionite (12); however, we were not able to detect any signal for Phe a in A. marina under these conditions (Fig. 5A, solid gray line; note that the scale shown on the right side applies and that the baseline is shifted). We therefore first removed QA by dithionite treatment before repeating the measurement. In the absence of QA we detected a clear signal at 547 nm, corresponding to photoaccumulation of reduced Phe a (Fig. 5A, solid black line) accompanied by changes at 515 and 418 nm (data not shown), indicating for the first time that Phe a functions as the primary electron acceptor in A. marina. In contrast, the PSII core of Synechocystis clearly showed a reversible electronic difference absorption spectrum for Phe a at 545 nm in the presence of QA and dithionite (Fig. 5A, dotted line); a similar spectrum has been reported for spinach PSII RC (12). Differences in the Phe a signal in the presence of dithionite strongly suggest that the redox potential of Phe a is shifted to a higher value in A. marina than that in Synechocystis and spinach. We investigated this point further by measuring DF for A. marina PSII complexes at 77 K. In the absence of dithionite we detected DF at 685 nm (τ = 15.1 ns, amplitude 0.12%), but this was suppressed in the presence of dithionite, as the overall fluorescence decay became faster (Fig. 5B Left). The effect of dithionite was reversible; when samples with dithionite were washed thoroughly we detected photochemical activity, i.e., P+ formation, by FTIR spectrum (data not shown). The results indicate that suppression of Phe a accumulation is probably induced by changes in the redox potential of Phe a in A. marina.
Fig. 5.
Changes in the redox potential of Phe a in A. marina. (A) Light-induced absorption difference spectra in the Qx region of Phe a. Shown are QA-depleted PSII complexes from A. marina (solid black line) and PSII complexes with sodium dithionite (solid gray line). The dotted line shows the PSII core from Synechocystis. For the solid black line and the dotted line, the scale on the left applies. (B Left) Suppression of DF in the presence of sodium dithionite. The excitation wavelength was 400 nm. Fluorescence decay at 685 nm was measured at 77 K in the absence (solid line) or presence (dashed line) of 4 mM sodium dithionite. (B Right) Wavelength-dependent fluorescence decay curves of DF at 77 K in the absence of dithionite.
Identification of Other Cofactors.
DF in the nanosecond time region at 77 K was observed only in the Chl a region for isolated PSII complexes (Fig. 5B Right), consistent with observations on intact cells (6, 7). This represents direct evidence of the involvement of Chl a in the electron transfer pathway of PSII. Two molecules of ChlZ (Fig. 1) were identified as Chl d by using FTIR difference spectra and UV/Vis absorption difference spectra measured on the same sample under the same conditions (SI Fig. 7).
Discussion
Purity of Samples.
Our PSII complexes consist of CP47, CP43′, D1, D2, cyt b559, PsbI, and a small polypeptide. Although the last component could not be assigned to a known subunit of PSII, it was clear that all of the essential subunits required for photochemical reactions were present in our preparation. We tested a wide range of fractions from column chromatography for PSI contamination using antibodies. Partially purified PSII complexes (Fig. 2, lane 3) still contained PSI (Fig. 2, lane 10), indicating that it is difficult to remove PSI during purification of PSII. By comparing the SDS/PAGE pattern of our sample with purified PSII samples in published reports (8, 15, 16), we concluded that the methods previously reported were not adequate to fully purify PSII complexes from A. marina.
Our pigment analysis showed that Chl a was definitely a component of PSII complexes in A. marina. A low Chl a content in the PSII complexes has been reported previously (8), but this could be due to contamination with PSI, based on comparison of our SDS/PAGE pattern with the reported results. At the same time, the high content of Chl d in our PSII complexes may possibly be attributed to the presence of CP43′; this Chl protein is predominantly associated with Chl d (15, 16). It is not clear at present whether the binding of CP43′ reflects a native property or is an artificial replacement of CP43 during the isolation procedures.
Identification of the Special Pair (PD1 and PD2).
The electronic absorption difference spectrum of the special pair showed bleaching at 713 nm and a positive band at 842 nm (Fig. 4A); we have thus designated the special pair P713. The peaks were red-shifted by 33 and 22 nm, respectively, compared with P680 and the 820-nm cation radical band. The FTIR difference spectrum of P+/P indicates that the Chl species responsible for this reaction was Chl d. Solid evidence was provided by the presence of stretching bands at 2,723 and 2,696 cm−1 corresponding to 3-formyl CH (Fig. 4C, line a); the presence of a strong positive peak at 1,661 cm−1 as a candidate for the 3-formyl C O vibration (Fig. 4B, line a) supports this conclusion.
O vibration (Fig. 4B, line a) supports this conclusion.
The appearance of two 3-formyl CH peaks at 2,723 and 2,696 cm−1 indicates that the 3-formyl groups of the two Chl d of the special pair differed in molecular environments in the RC proteins. The stronger intensity in the former peak may indicate that the positive charge is distributed mainly on the Chl d molecule with the 2,723-cm−1 formyl CH band. The charge distribution in P+ of A. marina appears to be similar to that in P680+, as shown by the similar shape of the 131-keto C O doublets at ≈1,725 and ≈1,711 cm−1 (Fig. 4B). This observation also supports the assignment of a Chl d homodimer as the special pair in A. marina, because a Chl d–Chl a heterodimer would be expected to give a significantly altered charge distribution in P+.
O doublets at ≈1,725 and ≈1,711 cm−1 (Fig. 4B). This observation also supports the assignment of a Chl d homodimer as the special pair in A. marina, because a Chl d–Chl a heterodimer would be expected to give a significantly altered charge distribution in P+.
We had previously assigned indirectly PD1 and PD2 as Chl a (Fig. 1) based on measurements of DF on intact cells in the nanosecond time region at 77 K (6, 7). This observation was confirmed in our PSII complexes (Fig. 5B Right). The direct assignment of a homodimer of Chl d to the special pair now also sheds new light on the origin of DF. In the case of conventional PSII systems containing only Chl a and Phe a, DF originates from Chl a, but its assignment has been controversial (33). It could be either P680 or ChlD1 (Fig. 1). Our current results may suggest that the origin of DF is ChlD1. This point will need further confirmation, as well as identification of the charge separation site in PSII. There are two possibilities for the charge separation site; one is on the special pair, (34) and the other is on ChlD1 (35, 36).
Another group has reported the spectral properties of the special pair using a PSII fraction that lacked oxygen evolution activity.‡‡ The authors reported a broad difference spectrum in the 660- to 740-nm region at 288 K and attributed the 725-nm peak to PSII RC Chl. The spectrum reported was very different from our observations (Fig. 4A). Although part of the spectrum contained a signal from PSII RC Chl (P713), the spectrum may also contain an additional unknown signal. This may arise from a lower purity of the samples used for analysis.
Function of Phe a and Changes in Its Potential.
It is generally thought that, in PSII of oxygenic photosynthetic systems comprising Chl a/Phe a, the potential of P680 is between +1.1 and +1.2 V and that of P680* is between −0.74 and −0.64 V, with Phe a being at −0.61 ± 0.03 V (37). Relative to the Chl a/Phe a systems, the redox potentials in A. marina are modified. When the special pair of PSII in A. marina is assigned as P713, the energy gained by light absorption at 713 nm can be estimated as 1.75 V. This is lower by 0.09 V than the energy gained from light of 680 nm. If the redox potential of the special pair in A. marina is unchanged from that of P680, as suggested by Shevela et al. (38), the expected potential of P713* should be between −0.65 and −0.55 V, and that of Phe a in A. marina should be a little higher. The potential of sodium dithionite is −0.53 V. Phe a in A. marina was actually reduced in the presence of dithionite (Fig. 5A), and therefore the in vivo potential of Phe a is probably comparable to, or higher than, that of dithionite. When the amino acid sequences of D1 and D2 proteins were compared, a significant difference was observed close to PheD1 (unpublished data). The change in the redox potential is attributed to an adjustment induced by the change in pigment species in A. marina. The most important function of the PSII reaction system is water oxidation, and all systems are subject to adjustment to this end.
Overall Energetics of PSII in A. marina.
In PSII of A. marina, the special pair (PD1 and PD2) (Fig. 1) is assigned as a Chl d homodimer, and the primary electron acceptor (PheD1) (Fig. 1) is identified as Phe a. If the redox potential of the former is the same as in the Chl a/Phe a system, the potential of the latter will be lowered in accordance with a lower gain of light energy for P713. Adjustment of the potential is inevitably accompanied by modification of the protein matrix, i.e., amino acid sequences. This is consistent with the changes in potential observed for the special pair in PSI in A. marina (39); the potential of P740 has been estimated to be +0.335 V, lower by ≈0.1 V than that of P700. Nevertheless, reduction of PSI was maintained at the same level as in other oxygenic photosynthetic organisms, even after a lower gain of light energy. Water oxidation and NADP+ reduction are prerequisites for construction of an oxygenic photosynthetic system, and there is limited flexibility in these two reactions. At the same time, other components are capable of modification. This is a clear generalization of the principles of the reaction system in oxygenic photosynthesis.
There remain several unanswered questions regarding the electron transfer system in A. marina: the role of Chl a in PSII RC; redox potentials and the mechanism of control of the intermediary components from Phe a to plastocyanin; and a structure of RC indicating the alterations responsible for changes in the redox potentials. Resolution of these questions may allow A. marina to open new horizons toward understanding the reaction processes and reaction mechanisms in the electron transfer systems of oxygenic photosynthesis.
Materials and Methods
Isolation of PSII Complexes.
A. marina MBIC 11017 was photosynthetically cultured in IMK medium under continuous illumination from an incandescent light (15 μmol photons per m2·s) at 298 K. Isolation was conducted at 277 K unless otherwise indicated. Cells were suspended in a buffer solution (50 mM Mes, pH 6.5) containing 25% (wt/vol) glycerol, 10 mM MgCl2, and 5 mM CaCl2 and were passed twice through a French press at a pressure of 150 MPa. Cell debris was removed by centrifugation (2,000 × g for 5 min). Thylakoid membranes were recovered by centrifugation (37,000 × g for 20 min) and stored at 193 K until use. PSII complexes were solubilized from thylakoid membranes by gentle stirring with 1% dodecyl-β-d-maltoside for 20 min in the dark. Solubilized PSII complexes were recovered by centrifugation (37,000 × g for 30 min), and the supernatant was subjected to purification. The first step was purification on a DEAE-Toyopearl 650S column, followed by passage over a UnoQ column. Finally, fractions containing purified PSII complexes were subjected to sucrose density gradient centrifugation to remove contaminating free CP43′. From these steps we obtained a dimer form of PSII complexes. Subunit compositions were analyzed by SDS/PAGE following Ikeuchi et al. (40); the stacking gel was 4.5%, and the running gel was 16–22%. Gels were stained with Coomassie brilliant blue R-250. Antibody raised against a peptide conserved in all known D1 proteins (AgriSera, Vännäs, Sweden) was used for Western blot analysis of D1 protein. A few subunits were identified by peptide mass fingerprinting.
PSII RC was prepared from spinach as described by Nanba and Satoh (12), with slight modifications (41). PSII core from Synechocystis was purified by Ni(II)-chelate affinity chromatography after introducing a 6× His tag to the C terminus of CP47 by site-directed mutagenesis (33, 42). PSII membranes (BBY particles) were prepared by a procedure described elsewhere (43).
Absorption and Fluorescence Spectra.
Absorption and fluorescence spectra were measured by the procedures described elsewhere (7). The spectral sensitivity of the fluorometer was corrected.
Determination of Pigment Contents.
Pigments were analyzed by HPLC (GULLIVER series; Jasco, Tokyo, Japan) using procedures described elsewhere (41, 44), and were detected by a photodiode array detector (MD-915; Jasco). A standard solution containing known amounts of authentic α-carotene, Chl a, Chl d, and Phe a was used for quantitative calibration. We used published molar extinction coefficients for the individual pigments (45–47). The PQ-9 content was measured by reverse-phase HPLC as described in detail elsewhere (21).
Measurements of Absorption Difference Spectra of P713.
Light-induced difference spectra of P713 were measured with a Hitachi U-0080D photodiode array spectrophotometer at 298 K (48). To samples containing 5.3 μg/ml Chl d, 1 mM potassium ferricyanide and 100 μM silicomolybdate were added. After passing through a Corning 4-96 filter, the samples were illuminated with blue light with an intensity of 70 μmol photons per m2·s. Absorption difference spectra in the red region were obtained by subtraction of the spectra of samples under illumination from the spectra of control samples; a red filter (R-65; Toshiba, Tokyo, Japan) was used for protection from actinic light.
Measurements of Photochemically Active Phe a.
Reversible absorption changes in Phe a of the PSII core of Synechocystis were measured in the presence of 15 mM sodium dithionite and 1 mM DPC by using a Hitachi U-0080D photodiode array spectrophotometer at 298 K. Before illumination samples were held in the dark for 30 min at 277 K. Absorption changes were measured under a cross-illumination system (12) with red actinic light (250 μmol photons per m2·s); appropriate filters were used [a red cutoff filter (R-67) and a heat- and UV-absorbing filter (HA-50; Hoya, Tokyo, Japan) for the actinic beam, and a Corning 4-96 filter for the measuring beam]. The Chl concentration was 10 μg/ml.
Photoaccumulation of Phe a of A. marina PSII was measured after extraction of PQ by dithionite treatment. Samples were incubated with sodium dithionite (100 mM) and 0.1 mM benzyl viologen for >240 min at 277 K and were then washed. This treatment was repeated three times. To the PQ-depleted samples, DPC (2 mM) was added, and red-light illumination (250 μmol photons per m2·s) was then applied. The absorption difference spectrum was measured in the Qx region of Phe a with a Hitachi U-0080D photodiode array spectrophotometer at 298 K.
Measurements of FTIR Difference Spectra.
FTIR difference spectra of the special pair (P680 in typical PSII systems) upon cation radical formation were recorded on a Bruker IFS-66/S spectrophotometer equipped with an MCT detector (D313-L) by using the method described previously (49). Experimental details are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. A. Tanaka (Hokkaido University) for providing an antibody raised against PsaA/B. This study was supported by a Grant-in-Aid for Creative Scientific Research (Grant 17GS0314) from the Japan Society for the Promotion of Science (to M.M. and T.N.); by Scientific Research on Priority Areas “Comparative Genomics” Grants 17018022 and 18017016 from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to M.M.); and by Grants-in-Aid for Scientific Research (Grant 18350067 to S.A. and Grant 18570145 to T.N.).
Abbreviations
- Chl
chlorophyll
- CP47
47-kDa Chl protein
- cyt
cytochrome
- DCIP
2,6-dichlorophenol-indophenol
- DF
delayed fluorescence
- DPC
diphenylcarbazide
- Phe
pheophytin
- PQ
plastoquinone
- PSI
photosystem I
- PSII
photosystem II
- RC
reaction center.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701847104/DC1.
‡‡Itoh, S., Iwaki, M., Noguti, T., Kawamori, A., Mino, H., Hu, Q., Iwasaki, I., Miyashita, H., Kurano, N., Miyachi, S., et al., 12th International Congress on Photosynthesis, August 18–23, 2001, Brisbane, Australia, abstr. S6-028.
References
- 1.Miyashita H, Ikemoto H, Kurano N, Adachi K, Chihara M, Miyachi S. Nature. 1996;383:402. [Google Scholar]
- 2.Murakami A, Miyashita H, Iseki M, Adachi K, Mimuro M. Science. 2004;303:1633. doi: 10.1126/science.1095459. [DOI] [PubMed] [Google Scholar]
- 3.Miller SR, Augustine S, Olson TL, Blankenship RE, Selker J, Wood AM. Proc Natl Acad Sci USA. 2005;102:850–855. doi: 10.1073/pnas.0405667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kühl M, Chen M, Ralph PJ, Schreiber U, Larkum AWD. Nature. 2005;433:820. doi: 10.1038/433820a. [DOI] [PubMed] [Google Scholar]
- 5.Miyashita H, Adachi K, Kurano N, Ikemoto H, Chihara M, Miyachi S. Plant Cell Physiol. 1997;38:274–281. [Google Scholar]
- 6.Mimuro M, Akimoto S, Gotoh T, Yokono M, Akiyama M, Tsuchiya T, Miyashita H, Kobayashi M, Yamazaki I. FEBS Lett. 2004;556:95–98. doi: 10.1016/s0014-5793(03)01383-8. [DOI] [PubMed] [Google Scholar]
- 7.Mimuro M, Akimoto S, Yamazaki I, Miyashita H, Miyachi S. Biochim Biophys Acta. 1999;1412:37–46. doi: 10.1016/s0005-2728(99)00048-1. [DOI] [PubMed] [Google Scholar]
- 8.Chen M, Telfer A, Lin S, Pascal A, Larkum AWD, Barber J, Blankenship RE. Photochem Photobiol Sci. 2005;4:1060–1064. doi: 10.1039/b507057k. [DOI] [PubMed] [Google Scholar]
- 9.Döring G, Stiehl HH, Witt HT. Z Naturforsch B. 1967;22:639–644. doi: 10.1515/znb-1967-0614. [DOI] [PubMed] [Google Scholar]
- 10.van Gorkom HJ, Tamminga JJ, Haveman J. Biochim Biophys Acta. 1974;347:417–438. doi: 10.1016/0005-2728(74)90080-2. [DOI] [PubMed] [Google Scholar]
- 11.Davis MS, Forman A, Fajer J. Proc Natl Acad Sci USA. 1979;76:4170–4174. doi: 10.1073/pnas.76.9.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nanba O, Satoh K. Proc Natl Acad Sci USA. 1987;84:109–112. doi: 10.1073/pnas.84.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghanotakis DF, de Paula JC, Demetriou DM, Bowlby NR, Petersen J, Babcock GT, Yocum CF. Biochim Biophys Acta. 1989;974:44–53. doi: 10.1016/s0005-2728(89)80164-1. [DOI] [PubMed] [Google Scholar]
- 14.Zheleva D, Sharma J, Panico M, Morris HR, Barber J. J Biol Chem. 1998;273:16122–16127. doi: 10.1074/jbc.273.26.16122. [DOI] [PubMed] [Google Scholar]
- 15.Chen M, Quinnell RG, Larkum AWD. FEBS Lett. 2002;514:149–152. doi: 10.1016/s0014-5793(02)02315-3. [DOI] [PubMed] [Google Scholar]
- 16.Chen M, Bibby TS, Nield J, Larkum AWD, Barber J. FEBS Lett. 2005;579:1306–1310. doi: 10.1016/j.febslet.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 17.Allakhverdiev SI, Hayashi H, Nishiyama Y, Ivanov AG, Aliev JA, Klimov VV, Murata N, Carpentier R. J Plant Physiol. 2003;160:41–49. doi: 10.1078/0176-1617-00845. [DOI] [PubMed] [Google Scholar]
- 18.Kamiya N, Shen J-R. Proc Natl Acad Sci USA. 2003;100:98–103. doi: 10.1073/pnas.0135651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Science. 2004;303:1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- 20.Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Nature. 2005;438:1040–1044. doi: 10.1038/nature04224. [DOI] [PubMed] [Google Scholar]
- 21.Patzlaff JS, Barry BA. Biochemistry. 1996;35:7802–7811. doi: 10.1021/bi960056z. [DOI] [PubMed] [Google Scholar]
- 22.Rögner M, Nixon PJ, Diner BA. J Biol Chem. 1990;265:6189–6196. [PubMed] [Google Scholar]
- 23.Sakurai I, Shen J-R, Leng J, Ohashi S, Kobayashi M, Wada H. J Biochem. 2006;140:201–209. doi: 10.1093/jb/mvj141. [DOI] [PubMed] [Google Scholar]
- 24.Lutz M, Mäntele W. In: Chlorophylls. Scheer H, editor. Boca Raton, FL: CRC Press; 1991. pp. 855–902. [Google Scholar]
- 25.Noguchi T, Tomo T, Inoue Y. Biochemistry. 1998;37:13614–13625. doi: 10.1021/bi9812975. [DOI] [PubMed] [Google Scholar]
- 26.Socrates G. Infrared Characteristic Group Frequencies. 2nd Ed. New York: Wiley; 1994. pp. 88–89. [Google Scholar]
- 27.Hastings G. Appl Spectrosc. 2001;55:894–900. [Google Scholar]
- 28.Sivakumar V, Wang R, Hastings G. Biophys J. 2003;85:3162–3172. doi: 10.1016/S0006-3495(03)74734-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujiwara M, Tasumi M. J Phys Chem. 1986;90:250–255. [Google Scholar]
- 30.Cai Z-L, Zeng H, Chen M, Larkum AWD. Biochim Biophys Acta. 2002;1556:89–91. doi: 10.1016/s0005-2728(02)00357-2. [DOI] [PubMed] [Google Scholar]
- 31.Chen M, Zeng H, Larkum AWD, Cai Z-L. Spectrochim Acta A. 2004;60:527–534. doi: 10.1016/s1386-1425(03)00258-0. [DOI] [PubMed] [Google Scholar]
- 32.Razeghifard MR, Chen M, Hughes JL, Freeman J, Krausz E, Wydrzynski T. Biochemistry. 2005;44:11178–11187. doi: 10.1021/bi048314c. [DOI] [PubMed] [Google Scholar]
- 33.Mimuro M, Akimoto S, Tomo T, Yokono M, Miyashita H, Tsuchiya T. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbabio.2007.02.012. in press. [DOI] [PubMed] [Google Scholar]
- 34.Seibert M. In: The Photosynthetic Reaction Center. Deisenhofer J, Norris JR, editors. Vol 1. London: Academic; 1993. pp. 319–356. [Google Scholar]
- 35.Frese RN, Germano M, de Weerd FL, van Stokkum IHM, Shkuropatov AY, Shuvalov VA, van Gorkom HJ, van Grondelle R, Dekker JP. Biochemistry. 2003;42:9205–9213. doi: 10.1021/bi0273516. [DOI] [PubMed] [Google Scholar]
- 36.Prokhorenko VI, Holzwarth AR. J Phys Chem B. 2000;104:11563–11578. [Google Scholar]
- 37.Klimov VV, Allakhverdiev SI, Demeter S, Krasnovskii AA. Dokl Akad Nauk SSSR. 1979;249:227–230. [Google Scholar]
- 38.Shevela D, Nöring B, Eckert H-J, Messinger J, Renger G. Phys Chem Chem Phys. 2006;8:3460–3466. doi: 10.1039/b604389e. [DOI] [PubMed] [Google Scholar]
- 39.Hu Q, Miyashita H, Iwasaki I, Kurano N, Miyachi S, Iwaki M, Itoh S. Proc Natl Acad Sci USA. 1998;95:13319–13323. doi: 10.1073/pnas.95.22.13319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ikeuchi M, Inoue Y. Plant Cell Physiol. 1988;29:1233–1239. [Google Scholar]
- 41.Tomo T, Mimuro M, Iwaki M, Kobayashi M, Itoh S, Satoh K. Biochim Biophys Acta. 1997;1321:21–30. [Google Scholar]
- 42.Bricker TM, Morvant J, Masri N, Sutton HM, Frankel LK. Biochim Biophys Acta. 1998;1409:50–57. doi: 10.1016/s0005-2728(98)00148-0. [DOI] [PubMed] [Google Scholar]
- 43.Berthold DA, Babcock GT, Yocum CF. FEBS Lett. 1981;134:231–234. [Google Scholar]
- 44.Akiyama M, Miyashita H, Kise H, Watanabe T, Miyachi S, Kobayashi M. Anal Sci. 2001;17:205–208. doi: 10.2116/analsci.17.205. [DOI] [PubMed] [Google Scholar]
- 45.Watanabe T, Hongu A, Honda K. Anal Chem. 1984;56:251–256. [Google Scholar]
- 46.French CS. In: Handbuch der Pflanzenphysiologie. Pirson A, editor. Vol 5. Heidelberg: Springer; 1960. pp. 252–272. [Google Scholar]
- 47.Britton G. In: Carotenoids. Britton G, Liaaen-Jensen S, Pfander H, editors. Vol 1B. Basel: Birkhäuser; 1995. pp. 13–62. [Google Scholar]
- 48.van Gorkom HJ, Pulles MPJ, Wessels JSC. Biochim Biophys Acta. 1975;408:331–339. doi: 10.1016/0005-2728(75)90134-6. [DOI] [PubMed] [Google Scholar]
- 49.Sugiura M, Rappaport F, Brettel K, Noguchi T, Rutherford AW, Boussac A. Biochemistry. 2004;43:13549–13563. doi: 10.1021/bi048732h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.