Abstract
The L to M reaction of the bacteriorhodopsin photocycle includes the crucial proton transfer from the retinal Schiff base to Asp85. In spite of the importance of the L state in deciding central issues of the transport mechanism in this pump, the serious disagreements among the three published crystallographic structures of L have remained unresolved. Here we report on the x-ray diffraction structure of the L state, to 1.53 – 1.73 Å resolutions, from replicate data sets collected from six independent crystals. Unlike in earlier studies, the partial occupancy refinement uses diffraction intensities from the same crystals before and after the illumination to produce the trapped L state. The high reproducibility of inter-atomic distances, and bond angles and torsions of the retinal, lends credibility to the structural model. The photoisomerized 13-cis retinal in L is twisted at the C13=C14 and C15=NZ double-bonds so that the Schiff base does not lose its connection to wat402 and therefore to the proton acceptor Asp85. The protonation of Asp85 by the Schiff base in the L → M reaction is likely to occur, therefore, via wat402. It is evident from the L state structure that various conformational changes involving hydrogen-bonding residues and bound water begin to propagate from the retinal to the protein at this stage already, and in both extracellular and cytoplasmic directions. Their rationales in the transport can be deduced from the way their amplitudes increase in the intermediates that follow L in the reaction cycle, and from the proton transfer reactions with which they are associated.
Keywords: bacteriorhodopsin, proton pumps, transport mechanism, membrane proteins, protein crystallography, photointermediates
Much progress has been made in recent years in describing how protons are transported in the light driven pump, bacteriorhodopsin. The structure of the protein in the non-illuminated, but light-adapted, BR-state is available to 1.4–1.6 Å resolution1, 2, and the participation of the retinal Schiff base, acidic side-chains, and numerous bound water molecules, in the internal proton transfers and in proton release and uptake at the two membrane surfaces are now understood. High-resolution x-ray structural models of nearly all of the intermediate states of the transport cycle1, 3–15 give a step by step atomic level “movie” containing details of the structural rearrangements in the protein up to the M and N states. Large-scale conformational changes in the second half of the photocycle have been described, in turn, by cryo-electron microscopy16, 17, low-resolution x-ray and neutron diffraction projection maps18–22, EPR spectroscopy with site-specific spin-labels23–27 and cysteine reactivity changes28 at the protein surface. The changes in the strength of the retinal Schiff base counter-ion, inter-atomic distances at functionally important locations, and dihedral angles in the retinal have been determined29–33 with solid-state NMR. Rotation of retinal bonds, hydrogen-out-of-plane (HOOP) motions, protonation changes in the protein, and perturbations of polar groups and the peptide backbone as well as water and a hydrogen-bonded aqueous continuum, are described34–38 by vibrational spectroscopy. Combined quantum-mechanics/molecular dynamics calculations39, 40 of the barriers to changes in local geometry, and their comparison with the measured thermodynamics of the photocycle reactions, have produced a suggested reaction mechanism for the early events in the cycle.
According to the crystal structures,1, 41 it is because the binding site does not initially accommodate the changed shape of the polyene that photoisomerization produces a twisted 13-cis,15-anti retinal. Relaxation of this high-energy state is what drives both the critical deprotonation of the Schiff base and the ensuing stepwise conformational changes of the protein that propagate toward the two membrane surfaces. There is an emerging consensus from both crystal structures and spectroscopic information on how these conformational changes allow conduction of a proton from the cytoplasmic aqueous interface to the retinal and from the retinal to the extracellular interface. However, important elements of the transport mechanism have remained controversial because there is disagreement about the structure of the L intermediate. The protonation of the anionic Asp85 by NZ+-H of Lys216 (the Schiff base connection of Lys216 to the retinal) is the crucial step in the transport, and it occurs in the L to M reaction. Its mechanism is determined by the pKa difference that develops between the proton donor to its acceptor, and by the path taken by the transferred proton. Alternatively, protonation of Asp85 might be by a water molecule that dissociates and leaves behind a hydroxyl ion, and in this case the proton pump was proposed30, 31, 33, 42–45 to be, in fact, a hydroxyl ion pump. Thus, the central issues in the transport depend on how the geometry of this region is changed from the equilibrium that exists in the BR-state to the unstable arrangement in the L state.
In the BR state wat402 receives a hydrogen-bond from the Schiff base of the all-trans retinal and donates hydrogen-bonds to the carboxylates of Asp85 and Asp212. This stable structure will be perturbed upon photoisomerization of the retinal to 13-cis,15-anti. The three crystal structures reported1, 8, 15 for the K state are in agreement with the conclusions from changes in the C=N stretch frequency that had suggested46, 47 that the Schiff base loses its hydrogen-bond. There is no such consensus for the L state. Although produced in the crystals by similar illumination protocols, for this intermediate three very different crystallographic models have been reported. They suggest different means for energy conservation in the cycle and different paths for transferring the Schiff base proton to Asp85. The crystallographic considerations in evaluating these models48 have been reviewed. In model a), from data to 2.1 Å resolution8 and a claimed 70% occupancy for L (although there is a differing opinion49), the Lys216 NZ+-H vector has turned away from wat402 that had connected it to Asp85 in the BR state, and now points into the cytoplasmic, hydrophobic region. Wat402 is missing from the model, but bending of helix C, with Asp85, toward the Schiff base site (although without a decrease in the Asp85 to Schiff base distance) was suggested to facilitate transfer of the Schiff base proton to the aspartate. The transfer is driven, presumably, by the new environment of the protonated Schiff base that is unfavorable for a charged group. In model b), from data to 1.62 Å resolution7 and 60% occupancy for L, the hydrogen-bond of the Schiff base to wat402, lost in K, is re-established. This is accomplished by twists in two of the retinal double-bonds and an increase in the bond angle at C13, so as to maintain, roughly, the contour of the all-trans retinal in spite of the C13=C14 bond rotation. Protonation of Asp85 in this model is therefore via wat402, and driven by the relaxation of the strain in the retinal permitted by loss of interaction between the Schiff base and the aspartate, as follows from a previous suggestion31, once they lose their electric charges. In model c), from data to 2.4 Å resolution12 (but in a larger unit cell than the others) and 20% occupancy for L, the Schiff base is turned to the cytoplasmic side together with wat402, maintaining the hydrogen-bond of the Schiff base. Movement of wat402 to the other side of the retinal is made possible by rotation of the Leu93 side-chain to make room for it. The proton transfer in this model is driven, presumably, by the unfavorable free energy of the uncompensated charge of the anionic Asp85 left behind, but the path of the proton will be tortuous and may have to pass via the OH group of Thr89.
Contrary to expectation, as discussed below in more detail, the various kinds of non-crystallographic information about the L state have not decided unambiguously in favor of any one these models. The FTIR O-H stretch bands in L in the wild-type protein and several mutants are consistent only with model c), because they suggest50–52 that the Schiff base becomes hydrogen-bonded to a water molecule specifically in the cytoplasmic region. The chemical shift of 15N labeled Schiff base in L by solid-state NMR30 is consistent only with models b) and c) because it indicates the existence of strong Schiff base counter-ion interaction. The NMR results suggest30 twists around the C13=C14 and C15=NZ double bonds in L, consistent with model b). Theoretical calculations40, on the other hand, preclude such twists, and are in conflict with the NMR and crystallographic data. The calculated barrier to proton transfer from the Schiff base to Asp85 agrees with the measured rates, but the predicted structure for L is not consistent with any of the three crystallographic models.
In view of this controversy, we made a new attempt to determine the structural changes in the L state. The data we report here produces an improved model, because i) crystals with higher diffraction quality were used, ii) the structure of L was determined in the partial occupancy refinement with a BR model from the same crystal, and iii) the statistical significance of the changes could be assessed from six independent determinations. The new model is consistent with model b), but with some differences.
Results and Discussion
Validity of the data and the refinement
Bacteriorhodopsin is a rigid transmembrane heptahelical bundle, and in all reports the structural differences between the BR state and the L intermediate are small. Thus, the main source of error in determining the structural changes in L will be the crystal-to-crystal variations relative to the model of the BR state used in the partial occupancy refinements. Warnings of damage from exposure to x-rays15 make the use of the same crystal for collecting data on both BR and L states seem risky, but the number of x-ray photons in our data acquisition protocol is at or below53 the “safe15” limit. In a test of radiation damage we compared models refined from two sets of data collected from the same non-illuminated crystal. Decrease of scattering from carboxyl groups is diagnostic15 of the damage. The average of the temperature factors of the carboxyl oxygen atoms of Asp85, Asp212, Glu194 and Glu204, acidic residues of various sensitivities to radiation damage in this protein, increased by 3 ± 4 Å2 between the first and second coordinate sets. The corresponding values for CA and CB of the same residues remained virtually unchanged, as did the temperature factors (and positions) of retinal atoms. Because the decrease of scattering from the carboxyl oxygen atoms seems small enough to be acceptable, and the number of unique reflections collected remains about the same, we expect that the overall quality of the data is not significantly degraded by exposure to the level of radiation used.
Diffraction data were collected from six independent crystals, in each case both before and after illumination as described in Methods. The data and refinement statistics for one of these pairs is in Table 1. The data statistics were similar for the other sets (for the 12 data sets the average resolution was 1.60 ± 0.06 Å, the I/σ(I) for all reflections was 39.3 ± 3.5 and for the highest resolution shell 2.1 ± 0.1, Rmerge for all reflections was 3.7 ± 0.8% and for the highest resolution shell 60.0 ± 3.3%, and the mosaicity was 0.51 ± 0.18°).
Table 1.
X-ray data collection and refinement statistics for data sets before and after illumination of a bacteriorhodopsin crystal. The illumination, with red laser at 170K as described in Methods, converted bacteriorhodopsin to the L state with 60% occupancy.
| before illumination | after illumination | |
|---|---|---|
| Data resolution range, Å | 1.53 – 25.0 | 1.53 – 25.0 |
| Total observations | 170708 | 172256 |
| Unique reflections | 33846 | 33922 |
| Rmergea, b, % | 3.3 (621) | 2.9 (57.7) |
| Average I/σ(I)b, c | 41.7 (1.9) | 42.4 (2.1) |
| Completenessb, % | 96.9 (87.0) | 96.9 (86.5) |
| Mosaicity, ° | 0.52 | 0.35 |
| Refinement range, Å | 1.53 – 25.0 | 1.53 – 25.0 |
| Structure factors | 33815 | 33890 |
| Restraints | 7241 | 20914 |
| Parameters | 7056 | 7056 |
| Twin ratio | 48:52 | 48:52 |
| Protein atomsd | 1720 | 1720e |
| Retinal atomsd | 20 | 20e |
| Water moleculesd | 26 | 26e |
| R-factorf,g, % | 15.19 (14.13) | 15.33 (14.34) |
| Rfreeg, h, % | 18.84 (18.07) | 19.45 (18.45) |
| Average protein B, Å2 | 29.8 | 32.3e |
| Average retinal B, Å2 | 20.6 | 21.5e |
| Average water B, Å2 | 34.2 | 35.1e |
| Deviation from ideal bond lengths, Å | 0.013 | 0.011 |
| Deviation from ideal bond angle distances, Å | 0.029 | 0.026 |
Rmerge (I) = ∑hkl ∑i | Ihkl,i − 〈Ihkl〉 |/∑hkl ∑i |Ihkl,i|, where 〈Ihkl〉 is the average intensity of the multiple Ihkl,i observations for symmetry-related reflections.
values in parentheses are for the 1.53 to 1.59 Å resolution shell.
I/σ(I), average of the diffraction intensities, divided by their standard deviations.
model for the data set after illumination is refined as two partial occupancy conformations, but one is fixed as the model from the data set before illumination.
for the model of the L state only (all other parameters refer to the entire data sets).
R-factor = ∑hkl|Fobs − Fcalc|/∑hkl|Fobs|, where Fobs and Fcalc are observed and calculated structure factors, respectively.
values are for all data, those in parentheses for F > 4σ(F).
Rfree = ∑hkl ε T|Fobs − Fcalc|/∑hkl ε T|Fobs|, where a test set (5% of the data) is omitted from the refinement in such a way that all structure factors in each of several thin resolution-shells were selected to avoid bias from merohedral twinning.
The refinement will produce the coordinate set that best accounts for the diffraction intensities, but when identifying alternative conformations it is reassuring to have, additionally, visual comparison of the final model with an unbiased density map. This seems especially important when there is disagreement among reported models for the same state of a protein. Conformational changes induced in crystals were suggested to be validated54 by calculating Fobsafter− Fobsbefore difference maps between the two data sets (i.e., before and after the conformation change, and without refinement), with phase information only from the first data set. In our previous attempt7 to determine the structure of L such a map was dominated by random noise and density differences from changes in local scattering, partly obscuring the meaningful positive and negative density features at the retinal. With both data sets collected from the same crystal now, the difference maps are improved but still noisy. A light minus dark Fobs map, averaged from the six pairs of data sets, is shown in Figure 1(a). At the usual contour level of 2.5–3.0 σ for such difference maps, no difference features were seen, and the contour levels are set to ± 0.45 σ. As in other such maps of bacteriorhodopsin intermediates8–11, but at higher contour levels, the noise seems as great, or greater than, the coherent features. In order to evaluate what would be expected in this map, we prepared the idealized difference Fobs map in Figure 1(b) from the refined coordinates for L (see below) and BR, setting the temperature factors to 20.0 Å2 for the main chain and to 30 Å2 for all other atoms. This map will contain density peaks only from the atomic displacements, and some of its features are indeed readily assignable to the conformational changes. The contour levels required for this are ± 0.25 σ. If the experimental map in Figure 1(a) contained no noise, it would have density peaks of equivalent amplitude at the same contour levels. Because the amplitudes are similar at contour levels of ± 0.45 σ, other features, i.e., noise, contribute significantly to that map. In spite of the noise, however, some of the density changes of the experimental map in Figure 1(a) agree with those in the idealized map in Figure 1(b), and thus support the refined model for L. Positive and negative density peaks above and below the proximal end of the retinal chain in both maps suggest isomerization. Positive and negative features at the Lys216 side-chain, the peptide group of Ala215, and the guanidinum group of Arg82, in both maps suggest movements. Other features probably originate from noise and decrease of scattering that cover meaningful features, e.g., those expected from the displacement of water molecules in the refined model. The negative density peaks at the carboxyl oxygens of Asp85 in the experimental map must be from radiation damage because Asp85 is much more sensitive to radiation15 than Asp212, and there is no negative density associated with the latter at the contour level used. All in all, it cannot be concluded from Figure 1(a) that all of the density peaks are from distinct structural alterations, and it is not even certain that the changes are necessarily caused by the illumination. We developed therefore a different strategy for validating the data and the refined model, one suited to the specific problems we face from the small structural changes in bacteriorhodopsin, as follows.
Figure 1.
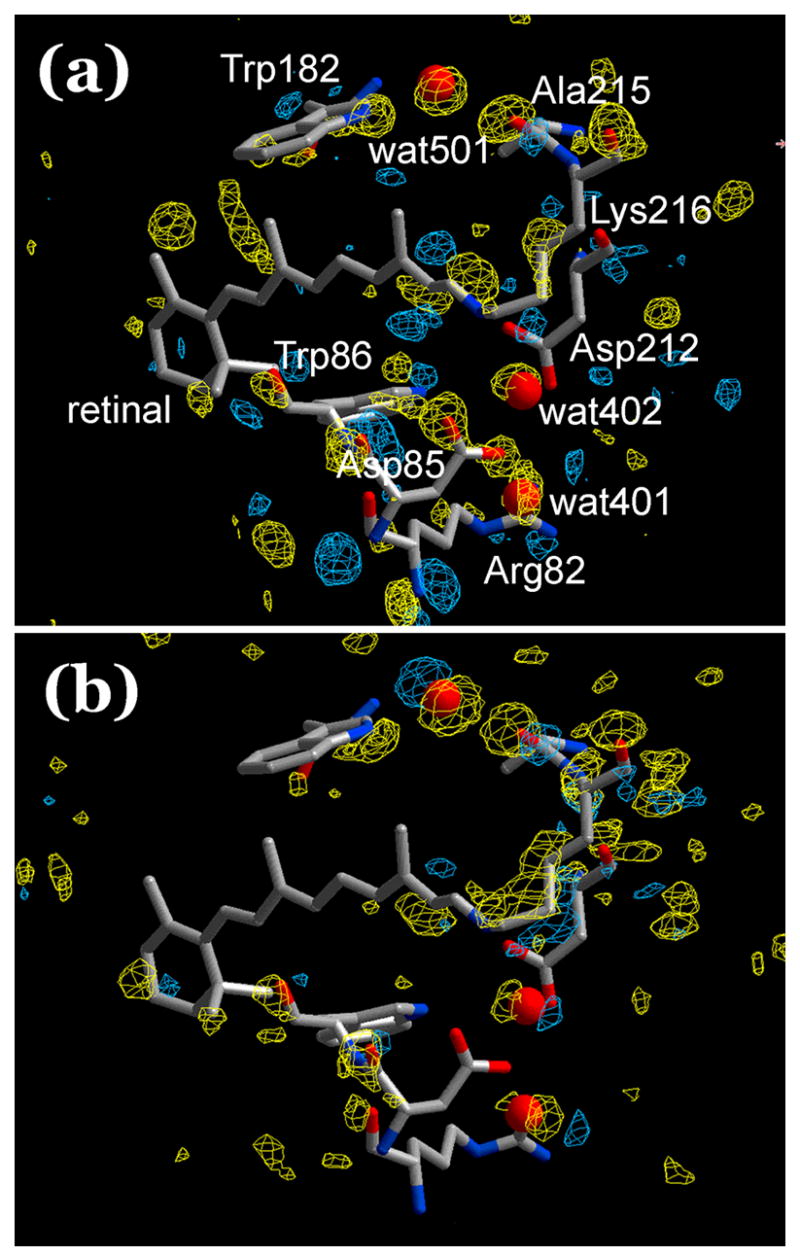
Light minus dark difference Fobs maps. Blue, positive; yellow, negative difference density. The retinal and a few residues and water from the model before illumination included. (a) Experimental difference Fobs map, using only the model before illumination as the source of phase information. Shown is the average map from six independent crystals. Contour levels ± 0.45 σ. (b) Sham Fobs difference map, prepared as (a) but from the refined models for the BR and L states. Contour levels ± 0.25 σ.
In order to test the origin of the changes after the illumination, the model for the putative L state was extracted by partial occupancy refinement. First, a model for the BR state was refined from the data set before the illumination. Then, a model with two conformations was refined, the first fixed as the BR model at 40% occupancy, and the second as the putative L state with 60% occupancy (see Methods). As a control, we refined a two-conformation model for the data before the illumination also. If the changes after illumination reflect real conformational change caused by the illumination rather than noise, the second conformation in the model without illumination should be the same as the BR model. Figure 2(a) compares the second conformation (atomic colors) and BR (blue) after the illumination, superimposed on the Fobs map after illumination. In Figure 2(b) the same comparison is made for the two conformations before the illumination. It is evident from Figure 2 that after illumination the refined model contains a second conformation different from the BR state, but without illumination the two conformations virtually coincide. Thus, the changed second conformation does appear to arise as a result of illumination. The same conclusion was made in our earlier7 report on L. As a further test of the data, the second conformation after illumination should be identifiable in some way with what is expected for the L intermediate. The retinal is the most suitable part of the molecule for this. From infrared and resonance Raman studies it is widely agreed that the consequence of illumination that produces L is isomerization of the retinal. The appearance of 13-cis retinal in the second conformation for the illuminated data set is therefore strong evidence that illumination produced a meaningful change, worthy of further analysis. Reassuringly, the retinal in the second conformation in Figure 2(a) is indeed 13-cis, while without illumination the model contains only a single retinal configuration, the all-trans isomer.
Figure 2.
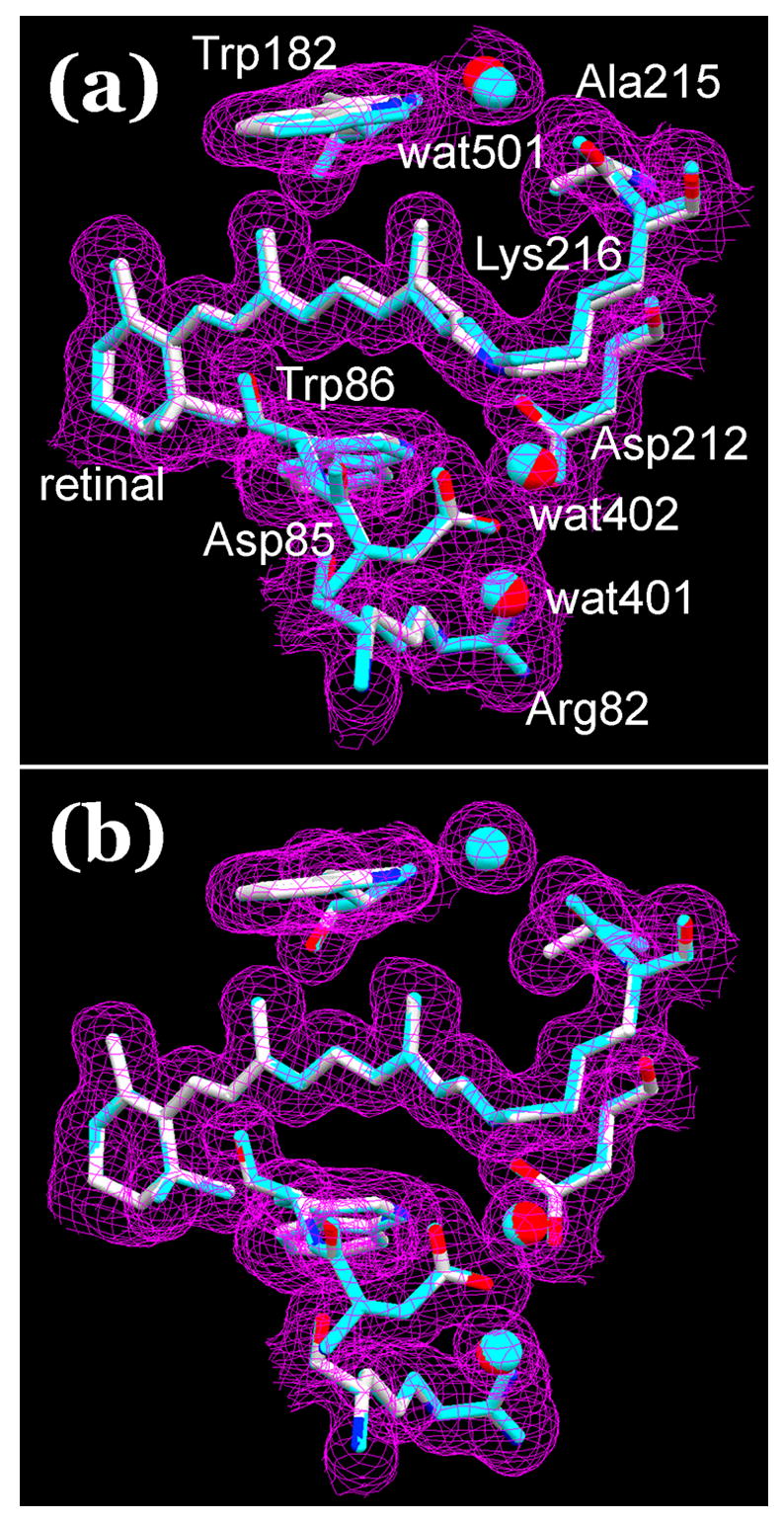
Models for data after (a) and before (b) illumination, refined as two conformations, with 40% and 60% occupancies. The first conformation (in blue) is fixed as the single conformation model before illumination (the BR state), the second (in atomic colors) is refined as the putative L state. Panel (b) is intended as a test of the idea that the second conformation in (a) is caused by illumination. The models are superimposed on Fobs maps after (b) and before (a) illumination. Contour level at 1 σ.
The Fobs maps before and after the illumination, in Figures 2(a) and (b) respectively, differ only slightly. Evidently, if L has a different conformation it will overlap the BR state considerably. Extrapolated Fobs maps contain the same information as difference Fobs maps, but easier to evaluate because they are less affected by the amplitude of noise. An extrapolated Fobs map for the second conformation (i.e., for L) was constructed by subtracting 0.40x the Fobs map of the non-illuminated crystal from the Fobs map of the illuminated crystal and dividing the result by 0.60. Figure 3 shows a stereo view of the retinal region, where there are small but consistent differences between the map calculated before the illumination and the extrapolated map (in blue and purple, respectively). The map in blue is consistent with the model for BR (also in blue), while the map in purple corresponds well to the model for L (in atomic colors). Although small, the differences in the Fobs maps justify the configurational changes of the retinal C13, C14, C15, C20, and Lys216 NZ and CE in the model (for the notation of these atoms, see below), and they are consistent also with movements of the peptide O of Ala215 and wat402 and wat501.
Figure 3.
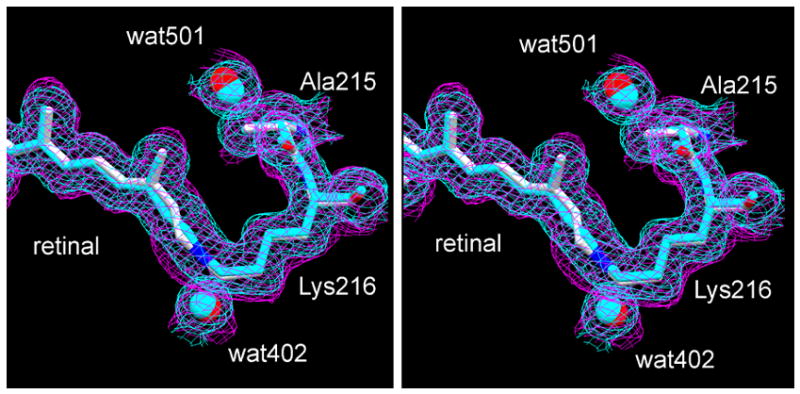
Stereo view of extrapolated Fobs maps of the retinal region. The models are the second conformations in Figure 2, in blue before illumination and in atomic colors after illumination. The extrapolated map for L (purple) was calculated from Fobs maps constructed as in Figure 2. It is overlaid on the Fobs map from data collected before illumination (blue). Contour level at 1 σ.
The orientation of the Schiff base NZ-H vector in the L state is of special interest (see Introduction), and we were careful to avoid being misled by a false minimum in the refinement that uses a conjugate gradient search. In our previous report7, the refinement produced a single result, but with better data this did not need to be so. When the starting coordinates for the retinal were varied to include a large number of configurations, the solutions in fact converged to two different states for the retinal. In one, from all starting configurations with the Schiff base pointing to the extracellular direction, the retinylidene nitrogen remains hydrogen-bonded to wat402 and thus connected to Asp85 and Asp212. In the other, from all starting configurations with the Schiff base pointing to the cytoplasmic direction, the nitrogen rotates away from wat402 to almost hydrogen-bonding distance of OG1 of Thr89 (3.40 Å, not shown). The global statistics of the fit, i.e., the R-factor and Rfree, could not decide between these models as they were virtually the same. This is perhaps not surprising because the differences involve very few atoms. However, there are spectroscopic and crystallographic grounds on which a decision can be made. As discussed in another section below in more detail, both NMR and FTIR spectra show47, 55 that in L there is a strong counter-ion to the Schiff base. This is contrary to the model with cytoplasmic orientation. If the Schiff base were the hydrogen-bond donor to Thr89, the N-H-O angle would be an unrealistic low 84°. With the Thr side-chain as the donor the angle would be more acceptable, but this is not possible because in L, as in the BR state, OG1 of Thr89 is the hydrogen-bond donor56 to OD1 of the anionic Asp85, at a 2.91 Å distance. Further, in this model the negative charges of Asp85 and Asp212 do not effectively shield the protonated Schiff base because the latter has moved as far as 4 Å away from the carboxylate oxygens, and is no longer connected to the wat402/Asp85/Asp212 complex. Thus, with this cytoplasmic orientation, the Schiff base is without a hydrogen-bond and a counter-ion. To evaluate the model from crystallographic criteria, we compare the two models with an extrapolated omit Fobs electron density map for L. Both Fobs maps for the extrapolation were produced with the coordinate set from the non-illuminated crystal for the phases, i.e., the BR state, as for Figure 2, but in this case with retinal atoms C13, C14, C15, C20, and Lys216 NZ and CE omitted so as not to bias the result (all other atoms virtually coincided in the two models). Figures 4(a) and (b) show the two models for the retinal, with such an extrapolated map for L. The good fit of all retinal and Lys216 atoms of the first model with NZ facing to the extracellular direction in Figure 4(a), and the poor fit of the Ret C14, C15, C20, and Lys216 CE and CD atoms of the other model in Figure 4(b), into the densities argues that the second model represents a local, and not global, minimum. We conclude that the first model is the correct structure of the retinal in the L state. However, we found that the current data are not good enough for a three-conformation refinement, i.e., to decide whether L is heterogeneous. On crystallographic grounds, at least, the presence of a small amount of the second 13-cis configuration of the retinal cannot be excluded.
Figure 4.
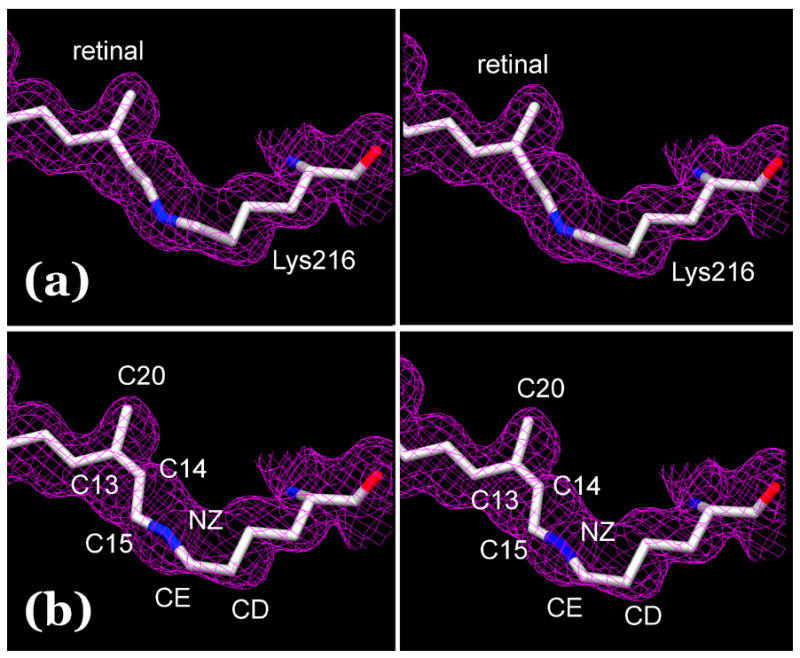
Stereo view of two alternative models for the retinal in the L state. Models (a) and (b) were refined as described in the text. They are overlaid on an extrapolated omit map for L, calculated from Fobs maps constructed without phase information for the relevant part of the retinal and Lys216 as explained in the text. Contour level at 1 σ.
Changes of the retinal in the L state
The improvements in the present work have produced a retinal configuration in L highly reproducible in six independent crystals. The retinal geometry is very different from those reported9, 10, 12 by other groups. Mainly because use of a BR model from the same crystal made the partial occupancy refinement more accurate, the geometry is somewhat different also from the one we had reported7 earlier. Figures 3, 4, and 5 illustrate the changed shape of the retinal, and Table 2 shows bond angles for the BR and L states along its single and double bonds. Except at Lys216 NZ of the L state where the standard deviation is ± 7° in the L state, they are reproducible within 1 to 3°. In a retinal free of constraints, trans-to-cis rotation around the C13=C14 bond would cause a sharp bend in the polyene chain, but in L the end-to-end distance of the retinal is hardly changed. This requires that at least one bond angle is increased7 over its value in the BR state. Two angles are seen to increase significantly, at C13 (i.e., the angle formed by C12–C13=C14) and C14, by 5 ± 2o each. Together with the double-bond torsions (see below), these will assure that the polyene chain remains extended and its overall contour resembles that of the all-trans. This was found1 also for the K state.
Figure 5.
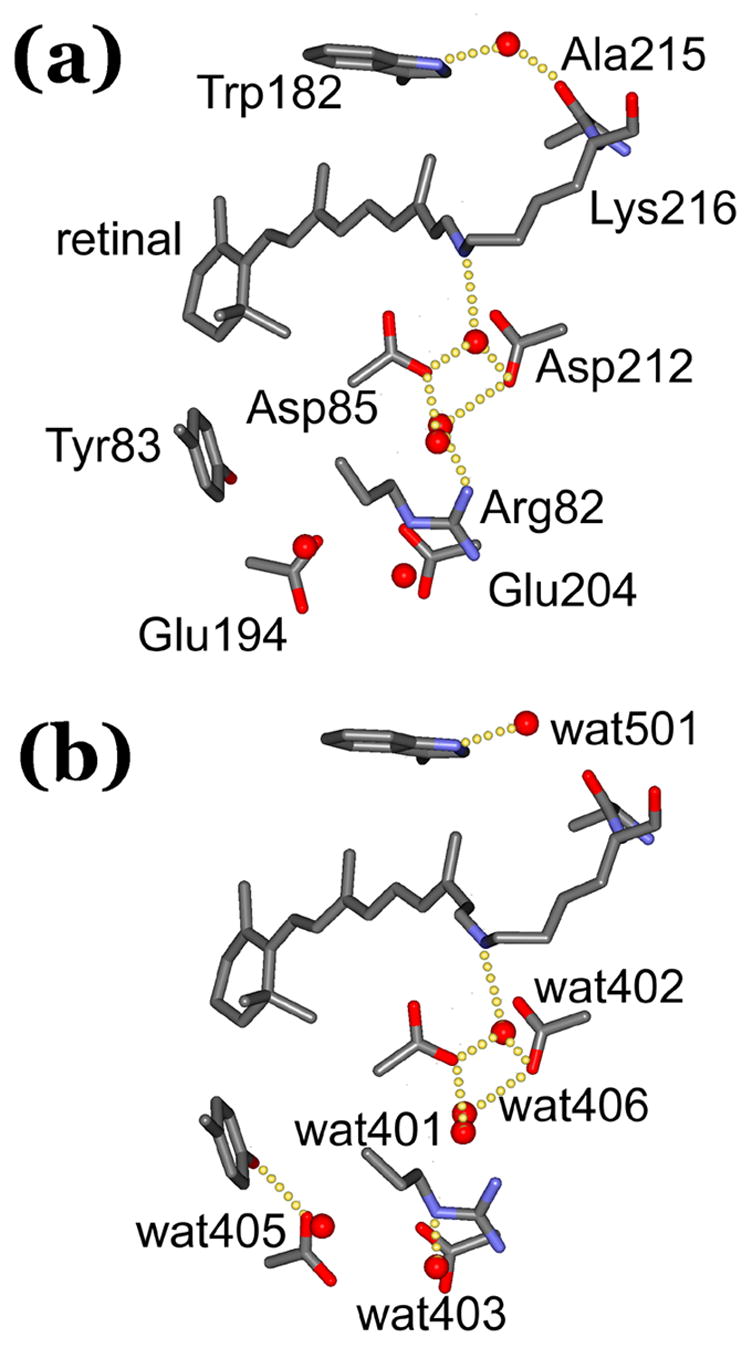
Structural differences between (a) the BR state and (b) the L state. Selected residues and water are shown to illustrate some of the changes discussed in the text and the Tables. For clarity, the residues included are labeled in (a), and the water molecules in (b). The images illustrate the deduced presence or absence of hydrogen-bonds. In some of the cases, the formation or breaking of hydrogen-bonds is exaggerated as they refer only to significant decreases or increases in inter-atomic distance, respectively (for more accurate information see Table 5).
Table 2.
Change of bond angles of the retinal and Lys216 (degrees), between the BR and L states (averages and standard deviations from 6 independent crystals)a
| location | BR state | L state | change |
|---|---|---|---|
| Ret C12 | 124 ± 2 | 123 ± 1 | −1 ± 1 |
| Ret C13 | 111 ± 1 | 116 ± 2 | 5 ± 2 |
| Ret C14 | 123 ± 2 | 128 ± 1 | 5 ± 2 |
| Ret C15 | 115 ± 2 | 113 ± 1 | −2 ± 2 |
| Lys216 NZ | 114 ± 3 | 116 ± 7 | 2 ± 8 |
| Lys216 CE | 109 ± 3 | 111 ± 1 | 2 ± 3 |
changes of likely significance are shown in bold
Table 3 contains torsion angles for the retinal in the BR and L states, represented as deviations from the plane of the conjugated chain. They are reproducible within 3 to 6°. The somewhat twisted C13=C14 double bond in the BR state, at −29 ± 3° out of plane, is in accord with the earlier reported crystal structures1, 2. Otherwise, in the BR state the retinal is nearly completely planar, although not the Lys216 side-chain, as indicated for example by the large out-of-plane torsion around the NZ-CE single bond. In the L state, both double-bonds between C13 and NZ become considerably twisted, in the same direction and to the same extent relative to the BR state, −54 ± 7 and −46 ± 6°, respectively. In our earlier reported structure7 the two twists were in opposite directions. However, as before, the C14–C15 single bond is not twisted. The NZ-CE single-bond, on the other hand, is rotated counter to the double-bond rotations and because its extent is 109 ± 5°. i.e., about the sum of the two double-bond rotations, the Lys216 chain beyond CE is not turned by the torsions in the retinal.
Table 3.
Change of torsion angles of the retinal and Lys216 (degrees deviation from a planar 13-cis,15-anti configuration, negative is counter-clockwise when viewed from the β–ionone ring), between the BR and L states (averages and standard deviations from 6 independent crystals)a
| bond | BR state | L state | change |
|---|---|---|---|
| Ret C13 = C14 | −29 ± 3 | −83 ± 6 | −54 ± 7 |
| Ret C14 – C15 | −5 ± 4 | −7 ± 5 | −2 ± 6 |
| Ret C15 = Lys216 NZ | −6 ± 4 | −40 ± 5 | −46 ± 6 |
| Lys216 NZ - CE | −61 ± 4 | 48 ± 3 | 109 ± 5 |
changes of likely significance are shown in bold
In L, retinal atoms C13 and C15, and especially C14, undergo considerable displacements (0.38 ± 0.06, 0.43 ± 0.06 and 0.64 ± 0.08 Å, respectively), but Lys216 NZ moves much less (Figures 3, 4, and 5, Table 4). Full rotation of the C13=C14 double-bond without other changes would turn the NZ-H vector to the cytoplasmic direction. In two other reported structures9, 10,12 for L, this is the case. Because of the two partial double-bond twists, however, in our model the NZ-H vector continues to point roughly to the extracellular direction (Figure 5).
Table 4.
Atom displacements (in Å) at selected locations between the BR and L states (averages and standard deviations from 6 independent crystals)a
| atom | displacement |
|---|---|
| Ret C13 | 0.38 ± 0.06 |
| Ret C14 | 0.64 ± 0.08 |
| Ret C15 | 0.43 ± 0.07 |
| Lys216 NZ | 0.14 ± 0.06 |
| Leu93 CD1 | 0.09 ± 0.04 |
| Trp182 NE1 | 0.27 ± 0.04 |
| Trp86NE1 | 0.22 ± 0.02 |
| Asp85 CA | 0.13 ± 0.02 |
| Asp212 CA | 0.14 ± 0.03 |
| Arg82 CZ | 0.40 ± 0.14 |
| Glu194 OE1 | 1.05 ± 0.18 |
| Glu194 OE2 | 0.48 ± 0.20 |
| Glu204 OE1 | 0.18 ± 0.06 |
| Glu204 OE2 | 0.22 ± 0.13 |
changes of likely significance are shown in bold
Changes in the protein and bound water in the L state
In L, the NZ to wat402 distance is somewhat increased (Table 5), but the hydrogen-bond between these atoms is not broken. In fact, in spite of displacement of wat402 by 0.31 ± 0.16 Å (Table 6), the characteristic trigonal geometry of Lys216 NZ, Asp85 OD2 and Asp212 OD2 around this water is unchanged (Figure 5, Table 5). The position of wat402 is less well defined than the other atoms at the active center, and it has an increased temperature factor (Table 6). In another reported structure9, 10 for L this water is missing, and in a third structure12 it has moved to the cytoplasmic side of the retinal. There is no evidence for either in the model. No space is available for a disordered water molecule on the cytoplasmic side of the Schiff base.
Table 5.
Interatomic distances at selected locations (in Å) between the BR and L states (averages and standard deviations from 6 independent crystals)a
| atoms | BR state | L state | change |
|---|---|---|---|
| Ala215 O to wat501 | 3.01 ± 0.07 | 3.68 ± 0.07 | 0.67 ± 0.10 |
| Trp182 NE1 to wat501 | 2.82 ± 0.04 | 2.77 ± 0.08 | −0.05 ± 0.09 |
| Lys216 NZ to wat402 | 2.91 ± 0.05 | 3.08 ± 0.22 | 0.17 ± 0.23 |
| Asp85 OD2 to wat402 | 2.58 ± 0.05 | 2.47 ± 0.11 | −0.11 ± 0.12 |
| Asp212 OD2 to wat402 | 3.07 ± 0.04 | 3.03 ± 0.16 | −0.04 ± 0.16 |
| Wat401 to wat406 | 2.74 ± 0.03 | 2.71 ± 0.08 | −0.03 ± 0.09 |
| Arg82 NH1 to wat406 | 2.84 ± 0.08 | 3.61 ± 0.51 | 0.77 ± 0.52 |
| Arg82 NH2 to wat403 | 3.03 ± 0.10 | 2.52 ± 0.05 | −0.51 ± 0.11 |
| Glu194 OE1 to wat405 | 3.18 ± 0.14 | 2.69 ± 0.23 | −0.49 ± 0.27 |
| Glu194 OE1 to Tyr83 OH | 3.88 ± 0.13 | 2.88 ± 0.20 | −1.00 ± 0.24 |
| Glu194 OE2 to Ser193 OD1 | 3.17 ± 0.10 | 3.45 ± 0.15 | 0.28 ± 0.18 |
changes of likely significance are shown in bold
Table 6.
Displacements (in Å) and changes in temperature factors (in Å2) of water molecules between the BR and L states (averages and standard deviations from 6 independent crystals)a
| water | displacement | B factorb | ||
|---|---|---|---|---|
| BR state | L state | change | ||
| 401 | 0.13 ± 0.07 | 23 ± 1 | 29 ± 4 | 6 ± 4 |
| 402 | 0.31 ± 0.16 | 25 ± 2 | 37 ± 8 | 12 ± 8 |
| 403 | 0.25 ± 0.19 | 43 ± 5 | 49 ± 6 | 6 ± 8 |
| 404 | 0.12 ± 0.06 | 28 ± 2 | 30 ± 2 | 2 ± 3 |
| 405 | 0.42 ± 0.22 | 41 ± 2 | 41 ± 2 | 0 ± 3 |
| 406 | 0.29 ± 0.30 | 28 ± 3 | 34 ± 4 | 6 ± 5 |
| 407 | 0.13 ± 0.05 | 24 ± 2 | 27 ± 2 | 3 ± 3 |
| 501 | 0.49 ± 0.16 | 23 ± 1 | 31 ± 4 | 8 ± 4 |
| 502 | 0.20 ± 0.08 | 35 ± 4 | 35 ± 4 | 0 ± 6 |
changes of likely significance are shown in bold
rounded to nearest integer
In the region between the Schiff base and the extracellular surface there are significant movements of side-chains known to be of functional importance. The Arg82 side-chain moves, but much less than in the M states3, 4, 11, 13, 14 reported. As a result the hydrogen-bond of Arg82 NH1 to wat406 appears to be broken and the hydrogen-bond between Arg82 NH2 and wat403 is shortened (Figure 5, Table 5). Perturbation of Arg82 spreads to the Glu194/Glu204 pair. Rotation of the Glu194 carboxyl group moves OE1 toward wat405, strengthening its existing hydrogen-bond, and toward also Tyr83 OH, forming a new hydrogen-bond (Figure 5, Table 5). As shown in Figure 5 and Table 6, wat405 moves, tracking the swivel of the Glu194 carboxyl group. Glu194 OE2, on the other hand, moves away from Ser193, breaking its hydrogen-bond (Table 5). In contrast to Glu194, Glu204 does not move significantly in L (Table 4). Some of the changes resemble3 those in M2’ although smaller in magnitude, but others are in the opposite direction. Unlike in L, in the last M state the greater displacement of the Arg82 side-chains will free it from interaction with wat403. Like in L, Arg82 loses its connection to wat406 in M2’ (in fact, wat406 is missing from that structure). In M2’ the carboxyl groups of both Glu194 and Glu204 will have undergone greater rotations than in L, breaking the hydrogen-bond of Glu194 OE1 to wat405. On the other hand, as observed in L, Ser193 is no longer hydrogen-bonded to Glu194 OE1, and Tyr83 OH retains its newly acquired hydrogen-bond to Glu194 OE1, in M2’. Thus, the motions of Arg82 and Glu194, but not Glu204, reported in M, begin in L already.
In one of the reported structures9, 10 for L, the most important change is the bowing of helix C toward helix G. Although the unchanged distance between the Schiff base and Asp85 in the model in the same report seems to contradict it, the suggested rationale is to aid the transfer of the Schiff base proton to Asp85, as Asp85 is on helix C and Lys216 is on helix G. There is no such change of helix C in the other two reported7, 12 models for L, and lack of significant movement of the CA atoms of either Asp85 or Asp212 (Table 4) indicate that it is absent in the current model also.
On the cytoplasmic side of the retinal, the upward movement of C13 (Table 4) with C20 (Figures 3, 4, and 5) brings it into steric conflict with the Trp182 ring, which tilts away somewhat (Table 4). Trp182 NE1 retains its hydrogen-bond with wat501, in spite of displacement of the latter (Table 6) because that movement is away from Ala215 O (Table 5). The hydrogen-bond between Ala215 and wat501 is broken (Figure 5), allowing separation of helices F and G. In addition to the movement of the Trp182 indole ring, away from the retinal in response to the changed shape of the retinal polyene chain, the Trp86 indole ring on the extracellular side moves (Table 4) toward the retinal. These displacements represent the first sign of the compliance of the retinal binding site, as they are not observed1 in the K state. Other residues that form the binding site around the Ret C13 - Lys216 CD segment, i.e., Val49 and Ala5357, and Leu9358, have not moved significantly in L (not shown), and constitute continued restraints on the relaxation of the retinal at this stage of the cycle.
The most important change in the L state structure reported by another group12 is rotation of Leu93 side-chain away from the retinal, with wat402 moving into the space it vacated. However, Leu93 CD1 does not move in the current structure of the L state (Table 4).
Correspondence with structural information from non-crystallographic evidence
Solid state NMR spectra of the BR and L states to date have provided31, 33 direct information about the strength of the Schiff base/counter-ion interaction, as well as measurements and inferences about single and double-bond twists in the retinal. The counter-ion strength, from the chemical shift of 15N-labeled Lys216 NZ, is stronger in L than in the BR state30, arguing that not only has the Schiff base not lost its connection to wat402 and thus to Asp85 and Asp212, but wat402 has become more polarized. The current crystal structure contains no displacements of atoms that would explain this polarization. On the other hand, given the greater counter-ion strength in L, the extent of the blue-shift of the absorption maximum of this intermediate was argued to indicate a twist of retinal double-bonds. This is confirmed by the current crystal structure. As for single-bond twist, the only information from solid state NMR32 is on the dihedral angle (H-C-C-H) of the C14–C15 bond in the BR state. Its value of 16 ± 4° out of plane disagrees with the torsion angle (C-C-C-C) in the current structure of 5 ± 4° (Table 3). As we had pointed out before1, the two values would not be in conflict if either of the hydrogen atoms were to be somewhat out-of-plane, as suggested59 by a theoretical calculation.
The C=N stretch frequency of the protonated Schiff base of bacteriorhodopsin is coupled to N-H bend60 that shifts it from 1620 to 1644 cm−1. Although in the K state the Schiff base is still protonated, the band shifts below 1600 cm−1 and its coupling to N-H vibration is greatly decreased46, 47 suggesting strong changes in the environment and loss of the Schiff base hydrogen-bond. This is consistent with the crystallographic structures of K in which the Schiff base loses its hydrogen-bond to wat402, in one8 because wat402 has moved away, and in the two others1, 15 because rotation of the NZ+-H vector away from wat402 makes its angle unfavorable for a hydrogen-bond. However, the recovery of the C=N stretch frequency in FTIR spectra show47, 55 that in the L state the Schiff base hydrogen-bond is regained, as indicated also by the solid state NMR. This is in conflict with the L structure where wat402 is absent9, 10, but in agreement with the two others in which the hydrogen-bond with 402 is reestablished, either in the original direction7, or toward the cytoplasmic side12 where wat402 moved. It is in agreement with our current model also.
Much attention has been paid to the O-H stretch bands that appear and disappear in the L minus BR difference FTIR spectra35, because they suggest movements of water molecules to differing environments or changes of their hydrogen-bonds. Assigning the locations of these water molecules has been through the effects of single residue replacements on the frequency and amplitude of the FTIR bands, with the assumption that their effects would be local. The recent crystal structure61 of D96A indicates, however, that a single mutation can affect the hydrogen-bonding of various water molecules throughout the entire protein, making this assumption problematic. Nevertheless, from an extensive study50–52 the suggestion has been made that the cytoplasmic region contains water molecules not evident in some of the crystal structures, and that in L a new water molecule, probably wat402, appears to the cytoplasmic side of the Schiff base. This is in accord with one of the models12 for L, but not with any of the others including the current one.
A unique property of L is the appearance of a strong N-H stretch band assigned62 to Trp182 NE1. There is a specific change in the current structure that will affect the electron density at the ring NE1, i.e., loss of the hydrogen-bond of wat501, its hydrogen-bonding partner, to Ala215 O (Figure 5 and Table 5). FTIR spectra of L reveal also downshift of the C=O stretch of the protonated carboxyl of Asp115, and upshift of the C=O stretch of the protonated carboxyl of Asp96 (review35), suggesting that the hydrogen-bonding of these residues (or in the case of Asp96 perhaps even its protonated state), is altered. No changes in the hydrogen-bond lengths of Asp115 with Thr90 and Asp96 with Thr46 greater than the respective standard deviations of ± 0.03 to 0.10 Å are observed in the current model for L (not shown), as in the other models except in one9, 10 where Asp96 loses its hydrogen bond to Thr46. As at Thr46, there is no evidence in the current model for L for the structural changes at Val49 proposed63 from FTIR spectra. If the spectral changes are correctly interpreted, FTIR spectroscopy is a more sensitive method to detect small structural perturbations than crystallography at the available resolutions.
Calculation of the barrier for the Schiff base to Asp85 proton transfer was from energy-optimized structures for L, with the intent39, 40 to decide the right structure (and therefore the right reaction pathway) from agreement of the calculated and observed rates. The two alternative structures considered for L do not quite correspond to the crystallographic models. In one, Lys216 NZ+-H points to the extracellular side and is directly hydrogen-bonded to Asp212 OD1 (but not to wat402), and protonation of Asp85 involves this aspartate. In the other, NZ+-H points to the cytoplasmic side, and is within hydrogen-bonding distance of Thr89 OG1 but with an unfavorable geometry. Wat402 has remained on the extracellular side. The calculated barriers to the proton transfer to Asp85 favor the cytoplasmic orientation, the proton being conducted to Asp85 either via Thr89 OG1, or along other routes around Asp212 to the other side, facilitated40 by flexibility of protein side-chains. This conclusion is consistent with none of the crystallographic models for L or with the NMR and FTIR evidence for a strong Schiff base counter-ion. Of the two crystallographic models with a cytoplasmic orientation of Lys216 NZ+-H, in one9, 10 water402 is missing from its location, and in the other12 wat402 has moved to the cytoplasmic side.
Implications of the structure of L for the mechanism of proton transport
The current structural model for the L state supports the earlier suggestions7, 30–32, 41, 45, 53 that the energy gain from the photoisomerization of the retinal is stored in torsions of the polyene chain. The alternative hypotheses, from the two differing crystal structures for L, focused on wat402, either on its absence9, 10 in the Schiff base region, or its transport across the membrane12 in the direction opposite to the proton transport. With the current model for L, the photocycle is viewed as the progressive relaxation of the initially twisted photoisomerized retinal, and the proton transport as the consequence of the accommodation of this relaxation by the protein matrix. There are two aspects of this accommodation. First, in the L → M transition the Schiff base proton is transferred to Asp85, which converts the NZH+-wat402-OD2− dipole into a neutral assemblage from which wat402 is then lost. The deprotonated Schiff base is thereby freed up to rotate into its normal position in a 13-cis retinal, this rotation being the switch31, 64, 65 that reorients it toward the proton donor Asp96. Even before this proton transfer, however, the changes of the retinal are accompanied by perturbations that spread in the extracellular direction. Probably as the result of the movement of Arg82 and its connected water molecules wat403 and wat406, the carboxyl group of Glu194 rotates and its new position is stabilized by a hydrogen-bond to Tyr83 OH (Table 5 and Figure 5). A precise arrangement for Glu194 and Glu204 is necessary to maintain the delocalized proton in the aqueous network36 that constitutes the proton release site, because even replacement of one of the Glu residues with Asp abolishes36, 66 the observed infrared continuum. Rotation of Glu194 in L does not destroy the continuum36, but it seems relevant to proton release in the photocycle states that follow L.
Second, as the bend of the retinal chain at C13 becomes more and more acute, Ret C20 comes into steric conflict with the Trp182 ring. The C20 to Trp182NE1 distance first decreases, then increases7, 29, in the first half of the cycle as the indole ring is pushed and then yields to move further away in response to the pressure. The deflection of Trp182 in L breaks the hydrogen-bond of the connected wat501 to Ala215 O on the cytoplasmic side (Figure 5 and Table 5) that connects helices F and G. Loss of the wat501 to Ala215 O hydrogen-bond is also observed in the M states3, 4 (although in one of the studies the BR state11 used to refine the M model contains no water equivalent to wat501, and in another13 wat501 loses its other hydrogen-bond, to Trp182, also). Interestingly, breaking of the Ala215-wat501 hydrogen-bond occurs also in the non-illuminated D96A mutant61, where the perturbation of Ala215 originates not from the retinal but from loss of the Asp96-Thr46 hydrogen bond, and propagated via Thr46 O, wat502, and Lys216 O. The deduced long-range coupling between the retinal and Asp96 over a > 10 Å distance via alternating covalent and hydrogen-bonds was suggested to separate the cytoplasmic ends of helices B and C, and thereby develop cavities near Asp96 that will recruit water4–6 and lower its pKa later in the photocycle. The separation of helices F and G, on the other hand, might be to trigger the outward tilt of helix G that aids the creation of a hydrogen-bonded chain of four water molecules6 to conduct a proton from Asp96 to the unprotonated Schiff base in the N state.
Thus, the structure of L contains hints at the causes of all of the structural changes that occur later in the photocycle. This is not unexpected if these can be traced back to the transformations of the retinal that begin in the K state already1. Only in the N state does the retinal binding site conform fully to the changed, 13-cis,15-anti configuration70, and thus the earlier intermediates, like L, should be viewed as quasi-stable states along this equilibration coordinate.
Methods
Data collection
The crystals, grown in cubic lipid phase as described before68, were thin hexagonal plates about 120 μm x 120 μm x 10–15 μm. Pieces of the cubic phase with encased crystals were soaked overnight in the mother liqueur plus 0.5% octylglucoside, and the crystals were extracted and mounted by mechanical manipulation with a nylon loop. Most of the crystals diffracted only to about 2 Å, and only a very small fraction were of the quality required for the present work.
Diffraction data were collected at 100K before illumination, and then from the same crystals after illumination at 170K and rapid re-cooling with a nitrogen-stream to 100K. Warming to 170K was in ca. 10 mins, cooling back to 100K in ca. 20 mins. Illumination was with a He-Ne laser at 628 nm and 5 mW, parallel with the c-axis in the crystals. The photostationary state, whose composition at this temperature depends only on the relative forward and back-reactions between the BR and L states,7, 49 will contain 60% of the L state. Illumination was for 10 mins on one side followed by 180° turn of the crystal and another 10 mins on the other side to ensure that amount of the L state in the photostationary state would reach the limiting value7, 49 determined before. There was no decrease in the amount of L state upon cooling after the illumination (our unpublished experiments). The measurements were at beamline BL 11-1 of SSRL (Stanford, California), using a 3 x 3 array CCD detector (ADSC, San Diego). For each data set 90 images with 1.0° oscillation angle were collected, integrated and scaled69 with HKL2000, in the P63 space group, with minor variations from a = 61 Å, b = 61 Å, c= 110 Å, α = 90°, β = 90°, γ = 120°.
Refinement
Refinement of models70 was with SHELXL-97, which handles the merohedral twinning71 of the crystals by comparing a twinned model with the twinned data, and re-twinning the model at each cycle of refinement. For each crystal a model for the BR state was obtained from the data collected before illumination, by 20-cycle refinement of a single conformation. Input model was 1C3W.pdb, but without including lipid atoms. Then, two-conformation models were refined using the data collected after illumination. The first conformation was fixed, with 40% occupancy, as the BR state model of the same crystal, and the second conformation refined, with 60% occupancy, as the putative model for the L state. The Fobs electron density maps shown were manipulated72 with Mapman, and drawn73 with Setor. The atom maps for Figure 1(b) were constructed74 with the SFALL program in the CCP4 suite.
Protein Data Bank accession codes
The coordinates and diffraction amplitudes for the structures of the BR state (before illumination), and the BR and L states (after illumination), from one of the crystals (Table 1) were deposited in the Protein Data Bank under entry codes 2NTU and 2NTW, respectively. Diffraction data from the other five crystals, measured for each before and after illumination, are available on ScienceDirect as supplementary material.
Supplementary Material
Acknowledgments
The authors are grateful to the beam staff at SSRL for essential assistance. The work was supported in part by grants to J.K.L. from NIH (R01-GM29498) and from DOE (DEFG03-86ER13525).
Abbreviations used
- BR state
unphotolyzed but light-adapted bacteriorhodopsin
- K,L,M,N states
intermediates of the bacteriorhodopsin photocycle
- octylglucoside
n-octyl-β-D-glucopyranoside
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schobert B, Cupp-Vickery J, Hornak V, Smith S, Lanyi J. Crystallographic structure of the K intermediate of bacteriorhodopsin: conservation of free energy after photoisomerization of the retinal. J Mol Biol. 2002;321:715–726. doi: 10.1016/s0022-2836(02)00681-2. [DOI] [PubMed] [Google Scholar]
- 2.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 Å resolution. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 3.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structural changes in bacteriorhodopsin during ion transport at 2 Angstrom resolution. Science. 1999;286:255–261. doi: 10.1126/science.286.5438.255. [DOI] [PubMed] [Google Scholar]
- 4.Luecke H, Schobert B, Richter HT, Cartailler JP, Rosengarth A, Needleman R, Lanyi JK. Coupling photoisomerization of the retinal in bacteriorhodopsin to directional transport. J Mol Biol. 2000;300:1237–1255. doi: 10.1006/jmbi.2000.3884. [DOI] [PubMed] [Google Scholar]
- 5.Lanyi JK, Schobert B. Crystallographic structure of the retinal and the protein after deprotonation of the Schiff base: the switch in the bacteriorhodopsin photocycle. J Mol Biol. 2002;321:727–737. doi: 10.1016/s0022-2836(02)00682-4. [DOI] [PubMed] [Google Scholar]
- 6.Schobert B, Brown LS, Lanyi JK. Crystallographic structures of the M and N intermediates of bacteriorhodopsin: assembly of a hydrogen-bonded chain of water molecules between Asp96 and the retinal Schiff base. J Mol Biol. 2003;330:553–570. doi: 10.1016/s0022-2836(03)00576-x. [DOI] [PubMed] [Google Scholar]
- 7.Lanyi JK, Schobert B. Mechanism of proton transport in bacteriorhodopsin from crystallographic structures of the K, L, M1, M2, and M2’ intermediates of the photocycle. Journal of Molecular Biology. 2003;328:439–450. doi: 10.1016/s0022-2836(03)00263-8. [DOI] [PubMed] [Google Scholar]
- 8.Edman K, Nollert P, Royant A, Belrhali H, Pebay-Peyroula E, Hajdu J, Neutze R, Landau EM. High-resolution X-ray structure of an early intermediate in the bacteriorhodopsin photocycle. Nature. 1999;401:822–826. doi: 10.1038/44623. [DOI] [PubMed] [Google Scholar]
- 9.Royant A, Edman K, Ursby T, Pebay-Peyroula E, Landau EM, Neutze R. Helix deformation is coupled to vectorial proton transport in the photocycle of bacteriorhodopsin. Nature. 2000;406:645–648. doi: 10.1038/35020599. [DOI] [PubMed] [Google Scholar]
- 10.Edman K, Royant A, Larsson G, Jacobson F, Taylor T, Van Der SD, Landau EM, Pebay-Peyroula E, Neutze R. Deformation of helix C in the low temperature L-intermediate of bacteriorhodopsin. J Biol Chem. 2004;279:2147–2158. doi: 10.1074/jbc.M300709200. [DOI] [PubMed] [Google Scholar]
- 11.Sass HJ, Buldt G, Gessenich R, Hehn D, Neff D, Schlesinger R, Berendzen J, Ormos P. Structural alterations for proton translocation in the M state of wild- type bacteriorhodopsin. Nature. 2000;406:649–653. doi: 10.1038/35020607. [DOI] [PubMed] [Google Scholar]
- 12.Kouyama T, Nishikawa T, Tokuhisa T, Okumura H. Crystal structure of the L intermediate of bacteriorhodopsin: evidence for vertical translocation of a water molecule during the proton pumping cycle. J Mol Biol. 2004;335:531–546. doi: 10.1016/j.jmb.2003.10.068. [DOI] [PubMed] [Google Scholar]
- 13.Takeda K, Matsui Y, Kamiya N, Adachi S, Okumura H, Kouyama T. Crystal structure of the M intermediate of bacteriorhodopsin: allosteric structural changes mediated by sliding movement of a transmembrane helix. J Mol Biol. 2004;341:1023–1037. doi: 10.1016/j.jmb.2004.06.080. [DOI] [PubMed] [Google Scholar]
- 14.Facciotti MT, Rouhani S, Burkard FT, Betancourt FM, Downing KH, Rose RB, McDermott G, Glaeser RM. Structure of an early intermediate in the M-state phase of the bacteriorhodopsin photocycle. Biophys J. 2001;81:3442–3455. doi: 10.1016/S0006-3495(01)75976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsui Y, Sakai K, Murakami M, Shiro Y, Adachi S, Okumura H, Kouyama T. Specific damage induced by X-ray radiation and structural changes in the primary photoreaction of bacteriorhodopsin. J Mol Biol. 2002;324:469–481. doi: 10.1016/s0022-2836(02)01110-5. [DOI] [PubMed] [Google Scholar]
- 16.Subramaniam S, Gerstein M, Oesterhelt D, Henderson R. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 1993;12:1–8. doi: 10.1002/j.1460-2075.1993.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramaniam S, Henderson R. Crystallographic analysis of protein conformational changes in the bacteriorhodopsin photocycle. Biochim Biophys Acta. 2000;1460:157–165. doi: 10.1016/s0005-2728(00)00136-5. [DOI] [PubMed] [Google Scholar]
- 18.Kataoka M, Kamikubo H, Tokunaga F, Brown LS, Yamazaki Y, Maeda A, Sheves M, Needleman R, Lanyi JK. Energy coupling in an ion pump. The reprotonation switch of bacteriorhodopsin. J Mol Biol. 1994;243:621–638. doi: 10.1016/0022-2836(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 19.Kamikubo H, Kataoka M, Varo G, Oka T, Tokunaga F, Needleman R, Lanyi JK. Structure of the N intermediate of bacteriorhodopsin revealed by x-ray diffraction. Proc Natl Acad Sci U S A. 1996;93:1386–1390. doi: 10.1073/pnas.93.4.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamikubo H, Oka T, Imamoto Y, Tokunaga F, Lanyi JK, Kataoka M. The last phase of the reprotonation switch in bacteriorhodopsin: the transition between the M-type and the N-type protein conformation depends on hydration. Biochemistry. 1997;36:12282–12287. doi: 10.1021/bi9712302. [DOI] [PubMed] [Google Scholar]
- 21.Oka T, Yagi N, Fujisawa T, Kamikubo H, Tokunaga F, Kataoka M. Time-resolved x-ray diffraction reveals multiple conformations in the M- N transition of the bacteriorhodopsin photocycle. Proc Natl Acad Sci U S A. 2000;97:14278–14282. doi: 10.1073/pnas.260504897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dencher NA, Dresselhaus D, Zaccai G, Bueldt G. Structural changes in bacteriorhodopsin during proton translocation revealed by neutron diffraction. Proc Natl Acad Sci U S A. 1989;86:7876–7879. doi: 10.1073/pnas.86.20.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rink T, Riesle J, Oesterhelt D, Gerwert K, Steinhoff HJ. Spin-labeling studies of the conformational changes in the vicinity of D36, D38, T46, and E161 of bacteriorhodopsin during the photocycle. Biophys J. 1997;73:983–993. doi: 10.1016/S0006-3495(97)78131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rink T, Pfeiffer M, Oesterhelt D, Gerwert K, Steinhoff HJ. Unraveling photoexcited conformational changes of bacteriorhodopsin by time resolved electron paramagnetic resonance spectroscopy. Biophys J. 2000;78:1519–1530. doi: 10.1016/S0006-3495(00)76704-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mollaaghababa R, Steinhoff HJ, Hubbell WL, Khorana HG. Time-resolved site-directed spin-labeling studies of bacteriorhodopsin: loop-specific conformational changes in M. Biochemistry. 2000;39:1120–1127. doi: 10.1021/bi991963h. [DOI] [PubMed] [Google Scholar]
- 26.Thorgeirsson TE, Xiao W, Brown LS, Needleman R, Lanyi JK, Shin YK. Transient channel-opening in bacteriorhodopsin: an EPR study. J Mol Biol. 1997;273:951–957. doi: 10.1006/jmbi.1997.1362. [DOI] [PubMed] [Google Scholar]
- 27.Xiao W, Brown LS, Needleman R, Lanyi JK, Shin YK. Light-induced rotation of a transmembrane alpha-helix in bacteriorhodopsin. J Mol Biol. 2000;304:715–721. doi: 10.1006/jmbi.2000.4255. [DOI] [PubMed] [Google Scholar]
- 28.Brown LS, Needleman R, Lanyi JK. Conformational change of the E-F interhelical loop in the M photointermediate of bacteriorhodopsin. J Mol Biol. 2002;317:471–478. doi: 10.1006/jmbi.2002.5428. [DOI] [PubMed] [Google Scholar]
- 29.Griffiths JM, Bennett AE, Engelhard M, Siebert F, Raap J, Lugtenburg J, Herzfeld J, Griffin RG. Structural investigation of the active site in bacteriorhodopsin: geometric constraints on the roles of Asp-85 and Asp-212 in the proton- pumping mechanism from solid state NMR. Biochemistry. 2000;39:362–371. doi: 10.1021/bi991106d. [DOI] [PubMed] [Google Scholar]
- 30.Hu JG, Sun BQ, Petkova AT, Griffin RG, Herzfeld J. The predischarge chromophore in bacteriorhodopsin: a 15N solid-state NMR study of the L photointermediate. Biochemistry. 1997;36:9316–9322. doi: 10.1021/bi970416y. [DOI] [PubMed] [Google Scholar]
- 31.Herzfeld J, Tounge B. NMR probes of vectoriality in the proton-motive photocycle of bacteriorhodopsin: evidence for an ‘electrostatic steering’ mechanism. Biochim Biophys Acta. 2000;1460:95–105. doi: 10.1016/s0005-2728(00)00132-8. [DOI] [PubMed] [Google Scholar]
- 32.Lansing JC, Hohwy M, Jaroniec CP, Creemers AF, Lugtenburg J, Herzfeld J, Griffin RG. Chromophore distortions in the bacteriorhodopsin photocycle: evolution of the H-C14-C15-H dihedral angle measured by solid-state NMR. Biochemistry. 2002;41:431–438. doi: 10.1021/bi011529r. [DOI] [PubMed] [Google Scholar]
- 33.Herzfeld J, Lansing JC. Magnetic resonance studies of the bacteriorhodopsin pump cycle. Annu Rev Biophys Biomol Struct. 2002;31:73–95. doi: 10.1146/annurev.biophys.31.082901.134233. [DOI] [PubMed] [Google Scholar]
- 34.Maeda A, Kandori H, Yamazaki Y, Nishimura S, Hatanaka M, Chon YS, Sasaki J, Needleman R, Lanyi JK. Intramembrane signaling mediated by hydrogen-bonding of water and carboxyl groups in bacteriorhodopsin and rhodopsin. J Biochem (Tokyo) 1997;121:399–406. doi: 10.1093/oxfordjournals.jbchem.a021602. [DOI] [PubMed] [Google Scholar]
- 35.Maeda A. Internal water molecules as mobile polar groups for light-induced proton translocation in bacteriorhodopsin and rhodopsin as studied by difference FTIR spectroscopy. Biochemistry (Mosc ) 2001;66:1256–1268. doi: 10.1023/a:1013183302690. [DOI] [PubMed] [Google Scholar]
- 36.Garczarek F, Brown LS, Lanyi JK, Gerwert K. Proton binding within a membrane protein by a protonated water cluster. Proc Natl Acad Sci U S A. 2005;102:3633–3638. doi: 10.1073/pnas.0500421102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kandori H. Role of internal water molecules in bacteriorhodopsin. Biochim Biophys Acta. 2000;1460:177–191. doi: 10.1016/s0005-2728(00)00138-9. [DOI] [PubMed] [Google Scholar]
- 38.Tanimoto T, Furutani Y, Kandori H. Structural changes of water in the Schiff base region of bacteriorhodopsin: proposal of a hydration switch model. Biochemistry. 2003;42:2300–2306. doi: 10.1021/bi026990d. [DOI] [PubMed] [Google Scholar]
- 39.Bondar AN, Elstner M, Suhai S, Smith JC, Fischer S. Mechanism of primary proton transfer in bacteriorhodopsin. Structure. 2004;12:1281–1288. doi: 10.1016/j.str.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 40.Bondar AN, Fischer S, Smith JC, Elstner M, Suhai S. Key role of electrostatic interactions in bacteriorhodopsin proton transfer. J Am Chem Soc. 2004;126:14668–14677. doi: 10.1021/ja047982i. [DOI] [PubMed] [Google Scholar]
- 41.Lanyi JK, Schobert B. Local-global conformational coupling in a heptahelical membrane protein: transport mechanism from crystal structures of the nine states in the bacteriorhodopsin photocycle. Biochemistry. 2004;43:3–8. doi: 10.1021/bi035843s. [DOI] [PubMed] [Google Scholar]
- 42.Luecke H. Atomic resolution structures of bacteriorhodopsin photocycle intermediates: the role of discrete water molecules in the function of this light-driven ion pump. Biochim Biophys Acta. 2000;1460:133–156. doi: 10.1016/s0005-2728(00)00135-3. [DOI] [PubMed] [Google Scholar]
- 43.Betancourt FM, Glaeser RM. Chemical and physical evidence for multiple functional steps comprising the M state of the bacteriorhodopsin photocycle. Biochim Biophys Acta. 2000;1460:106–118. doi: 10.1016/s0005-2728(00)00133-x. [DOI] [PubMed] [Google Scholar]
- 44.Facciotti MT, Rouhani S, Glaeser RM. Crystal structures of bR(D85S) favor a model of bacteriorhodopsin as a hydroxyl-ion pump. FEBS Lett. 2004;564:301–306. doi: 10.1016/S0014-5793(04)00208-X. [DOI] [PubMed] [Google Scholar]
- 45.Hatcher ME, Hu JG, Belenky M, Verdegem P, Lugtenburg J, Griffin RG, Herzfeld J. Control of the pump cycle in bacteriorhodopsin: mechanisms elucidated by solid-state NMR of the D85N mutant. Biophys J. 2002;82:1017–1029. doi: 10.1016/S0006-3495(02)75461-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rothschild KJ, Roepe P, Lugtenburg J, Pardoen JA. Fourier transform infrared evidence for Schiff base alteration in the first step of the bacteriorhodopsin photocycle. Biochemistry. 1984;23:6103–6109. doi: 10.1021/bi00320a031. [DOI] [PubMed] [Google Scholar]
- 47.Maeda A, Sasaki J, Pfefferle JM, Shichida Y, Yoshizawa T. Fourier transform infrared spectral studies on the Schiff base mode of all-trans bacteriorhodopsin and its photointermediates, K and L. Photochem Photobiol. 1991;54:911–921. [Google Scholar]
- 48.Lanyi JK. What is the real crystallographic structure of the L photointermediate of bacteriorhodopsin? Biochim Biophys Acta. 2004;1658:14–22. doi: 10.1016/j.bbabio.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 49.Balashov SP, Ebrey TG. Trapping and spectroscopic identification of the photointermediates of bacteriorhodopsin at low temperatures. Photochem Photobiol. 2001;73:453–462. doi: 10.1562/0031-8655(2001)073<0453:tasiot>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 50.Maeda A, Balashov SP, Lugtenburg J, Verhoeven MA, Herzfeld J, Belenky M, Gennis RB, Tomson FL, Ebrey TG. Interaction of internal water molecules with the schiff base in the L intermediate of the bacteriorhodopsin photocycle. Biochemistry. 2002;41:3803–3809. doi: 10.1021/bi011923p. [DOI] [PubMed] [Google Scholar]
- 51.Maeda A, Tomson FL, Gennis RB, Balashov SP, Ebrey TG. Water molecule rearrangements around Leu93 and Trp182 in the formation of the L intermediate in bacteriorhodopsin’s photocycle. Biochemistry. 2003;42:2535–2541. doi: 10.1021/bi020532n. [DOI] [PubMed] [Google Scholar]
- 52.Maeda A, Herzfeld J, Belenky M, Needleman R, Gennis RB, Balashov SP, Ebrey TG. Water-mediated hydrogen-bonded network on the cytoplasmic side of the schiff base of the L photointermediate of bacteriorhodopsin. Biochemistry. 2003;42:14122–14129. doi: 10.1021/bi0301542. [DOI] [PubMed] [Google Scholar]
- 53.Lanyi JK. A structural view of proton transport in bacteriorhodopsin. In: Wikstrom M, editor. In Biophysical and Structural Aspects of Bioenergetics. Royal Soc Chem; Cambridge, U.K: 2005. [Google Scholar]
- 54.Henderson R, Moffat JK. The difference Fourier technique in protein crystallography: errors and their treatment. Acta Crystallographica Section B. 1971;27:1414–1420. [Google Scholar]
- 55.Gerwert K, Souvignier G, Hess B. Simultaneous monitoring of light-induced changes in protein side- group protonation, chromophore isomerization, and backbone motion of bacteriorhodopsin by time-resolved Fourier-transform infrared spectroscopy. Proc Natl Acad Sci U S A. 1990;87:9774–9778. doi: 10.1073/pnas.87.24.9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kandori H, Yamazaki Y, Shichida Y, Raap J, Lugtenburg J, Belenky M, Herzfeld J. Tight Asp-85-Thr-89 association during the pump switch of bacteriorhodopsin. Proc Natl Acad Sci U S A. 2001;98:1571–1576. doi: 10.1073/pnas.98.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown LS, Gat Y, Sheves M, Yamazaki Y, Maeda A, Needleman R, Lanyi JK. The retinal Schiff base-counterion complex of bacteriorhodopsin: changed geometry during the photocycle Is a cause of proton transfer to aspartate 85. Biochemistry. 1994;33:12001–12011. doi: 10.1021/bi00206a001. [DOI] [PubMed] [Google Scholar]
- 58.Delaney JK, Subramaniam S. The residues Leu 93 and Asp 96 act independently in the bacteriorhodopsin photocycle: studies with the Leu 93→Ala, Asp 96→Asn double mutant. Biophys J. 1996;70:2366–2372. doi: 10.1016/S0006-3495(96)79803-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayashi S, Ohmine I. Proton transfer in bacteriorhodopsin: structure, excitation, IR spectra, and potential energy surface analyses by an ab initio QM/MM method. J Phys Chem B. 2000;104:10678–10691. [Google Scholar]
- 60.Smith SO, Lugtenburg J, Mathies RA. Determination of retinal chromophore structure in bacteriorhodopsin with resonance Raman spectroscopy. J Membr Biol. 1985;85:95–109. doi: 10.1007/BF01871263. [DOI] [PubMed] [Google Scholar]
- 61.Lanyi JK, Schobert B. Propagating structural perturbation inside bacteriorhodopsin: Crystal structures of the M state and the D96A and T46V mutants. Biochemistry. 2006;45:12003–12010. doi: 10.1021/bi061310i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamazaki Y, Sasaki J, Hatanaka M, Maeda A, Kandori H, Needleman R, Shinada T, Yoshihara K, Brown LS, Lanyi JK. Interaction of tryptophan 182 with the 9-methyl group of the retinal in the L intermediate of bacteriorhodopsin. Biochemistry. 1995;34:577–582. doi: 10.1021/bi00002a024. [DOI] [PubMed] [Google Scholar]
- 63.Yamazaki Y, Tuzi S, Saito H, Kandori H, Needleman R, Lanyi JK, Maeda A. Hydrogen bonds of water and C==O groups coordinate long-range structural changes in the L photointermediate of bacteriorhodopsin. Biochemistry. 1996;35:4063–4068. doi: 10.1021/bi9524530. [DOI] [PubMed] [Google Scholar]
- 64.Brown LS, Dioumaev AK, Needleman R, Lanyi JK. Connectivity of the retinal Schiff base to Asp85 and Asp96 during the bacteriorhodopsin photocycle: the local-access model. Biophys J. 1998;75:1455–1465. doi: 10.1016/S0006-3495(98)74064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown LS, Dioumaev AK, Needleman R, Lanyi JK. Local-access model for proton transfer in bacteriorhodopsin. Biochemistry. 1998;37:3982–3993. doi: 10.1021/bi9728396. [DOI] [PubMed] [Google Scholar]
- 66.Dioumaev AK, Richter HT, Brown LS, Tanio M, Tuzi S, Saito H, Kimura Y, Needleman R, Lanyi JK. Existence of a proton transfer chain in bacteriorhodopsin: participation of Glu-194 in the release of protons to the extracellular surface. Biochemistry. 1998;37:2496–2506. doi: 10.1021/bi971842m. [DOI] [PubMed] [Google Scholar]
- 67.Dioumaev AK, Brown LS, Needleman R, Lanyi JK. Partitioning of free energy gain between the photoisomerized retinal and the protein in bacteriorhodopsin. Biochemistry. 1998;37:9889–9893. doi: 10.1021/bi980934o. [DOI] [PubMed] [Google Scholar]
- 68.Rummel G, Hardmeyer A, Widmer C, Chiu ML, Nollert P, Locher KP, Pedruzzi I, Landau EM, Rosenbusch JP. Lipidic cubic phases: New matrices for the three-dimensional crystallization of membrane proteins. J Struct Biol. 1998;121:82–91. doi: 10.1006/jsbi.1997.3952. [DOI] [PubMed] [Google Scholar]
- 69.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 70.Sheldrick GMST. High resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 71.Luecke H, Richter HT, Lanyi JK. Proton transfer pathways in bacteriorhodopsin at 2.3 Angstrom resolution. Science. 1998;280:1934–1937. doi: 10.1126/science.280.5371.1934. [DOI] [PubMed] [Google Scholar]
- 72.Kleywegt GJ, Jones TA. xdlMAPMAN and xdlDATAMAN - programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Cryst D. 1996;52:826–828. doi: 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- 73.Evans SV. SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J Mol Graph. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 74.Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


