Abstract
Apolipophorin III (apoLp-III) is a prototypical apolipoprotein used for structure-function studies. Besides its crucial role in lipid transport, apoLp-III is able to associate with fungal and bacterial membranes and stimulate cellular immune responses. We recently demonstrated binding interaction of apoLp-III of the greater wax moth, Galleria mellonella, with lipopolysaccharides (LPS). In the present study, the requirement of helix bundle opening for LPS binding interaction was investigated. Using site-directed mutagenesis, two cysteine residues were introduced in close spatial proximity (P5C/A135C). When the helix bundle was locked by disulfide bond formation, the tethered helix bundle failed to associate with LPS. In contrast, the mutant protein regained its ability to bind upon reduction with dithiothreitol. Thus, helix bundle opening is a critical event in apoLp-III binding interaction with LPS. This mechanism implies that the hydrophobic interior of the protein interacts directly with LPS, analogous to that observed for lipid interaction.
Keywords: apolipoprotein, apolipophorin, Galleria mellonella, innate immunity, lipopolysaccharide
Introduction
Apolipophorin is an abundant hemolymph protein involved in lipid transport processes [1]. It has served as an excellent model protein to study the structure-function relationship of exchangeable apolipoproteins [2–3]. ApoLp-III has been isolated and well characterized in several insect sources, most noticeably Locusta migratoria, Manduca sexta, and Galleria mellonella [3]. ApoLp-III is composed of a bundle of five amphipathic α-helices oriented in an antiparallel fashion [4–6]. The protein is normally present in this closed conformation, but can switch to an open conformation upon association with lipid and lipoprotein surfaces [7–8]. It has become increasingly clear that apoLp-III from the greater waxmoth, G. mellonella, plays a role in innate immunity [9]. Recent studies have shown that apoLp-III is able to bind to lipopolysaccharides (LPS), lipoteichoic acid and β-glucans [10–13]. In addition, apoLp-III is able to act as an immune activator of cellular reactions, i.e. stimulating phagocytosis, increase of antibacterial activity and nitric oxide production [10–12, 14–17].
It appears that the stimulus of cellular immune reactions is predominantly caused by apoLp-III in the lipid bound conformation. This immune activating role of apoLp-III was observed with the protein bound to dimyristoylphosphatidylcholine (DMPC), when the protein is present in an open conformation [10, 15, 17]. However, it seems likely that apoLp-III bound to a foreign membrane component is the relevant factor triggering an immune response. We and others have shown that insect apoLp-III binds to LPS [11, 13, 18]. LPS is an abundant membrane component of gram-negative bacteria, and is composed of a relatively small lipid A domain that anchors LPS to the outer membrane. The numerous oligosaccharides are located in the LPS core and the distal O-antigen repeat region [19]. Similar to apoLp-III, vertebrate apolipoproteins have been reported to associate and detoxify lipopolysaccharides [20–21]. ApoLp-III shares many structural and functional features with the mammalian protein apolipoprotein (apo) E [22], and the study of apoLp-III with lipopolysaccharides has the potential to provide important insights in the role apolipoproteins play in innate immunity. In previous studies we found no evidence that charge is a factor in the apoLp-III LPS binding interaction [18]. Instead, it is plausible that hydrophobic interactions are the driving force. Since most hydrophobic amino acid residues are buried in the protein interior, the helix bundle needs to open to allow such a binding interaction. In addition, apoLp-III helix bundle opening potentially allows a binding interaction with a large variety with of structures of foreign origin, thus apoLp-III can function as a pattern recognition protein.
To investigate if helix bundle opening and interaction of the hydrophobic interior is required for LPS binding, we tethered the helix bundle to prevent opening. Using site-directed mutagenesis two cysteine residues were introduced in apoLp-III which upon disulfide formation locks the protein in a closed state, preventing a conformational opening. We demonstrate that when the helix bundle is locked, apoLp-III is unable to interact with LPS.
Materials and methods
Site-directed mutagenesis
Site-directed mutagenesis was carried out with the pET-22b(+) vector that contains the coding region for G. mellonella apoLp-III [14]. To introduce the desired cysteine residues, the following primers were used (cysteine coding triplets underlined): 5’-ACGCGTCCACGTGCCTGCAGGACCTGG-3’ for P5C, and 5’-GACCAACAAGAAGCTGTGCCCGCAGATCAAGTCC-3’ for A135C (MWG Biotech, High Point, NC). Mutations were introduced using the QuickChange Multi-site-directed mutagenesis kit (Stratagene, La Jolla, CA). To verify the desired mutations, DNA sequencing of the mutated plasmids was carried out at the CSUPERB Microchemical Core Facility DNA Lab at California State University, San Diego.
Protein expression and purification
Escherichia coli BL21 (DE3) cells were transformed with the mutated plasmids, and cells were grown in Minimal Medium as described [23]. Since apoLp-III escaped the bacteria and appeared in the culture medium, cells were removed by centrifugation and the supernatant was collected and concentrated to 20 mL by tangential flow filtration with a 10K Minimate cassette membrane (Pall Life Sciences, Ann Arbor, MI). ApoLp-III was further purified by G-75 gel-filtration chromatography and reversed-phase HPLC (Beckman system gold, C8 Zorbax semi preparative column) using a linear gradient of acetonitrile in water containing 0.05% trifluoroacetic acid [23]. The protein was freeze-dried and stored at -20 °C. Before use, purified protein was dissolved in phosphate buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), and the protein concentration measured with the BCA method using bovine serum albumin as the reference protein (Pierce, Rockford, IL).
Protein analysis
Circular dichroism was carried out in a Jasco 810 polarimeter, using a protein concentration of 1.0 mg/mL and a circular cuvet with a 0.02 cm path length [24]. Far UV profiles were the average of five separate scans between 185 and 260 nm. Polyacrylamide gel electrophoresis (PAGE) was carried out with sodium dodecyl sulfate (SDS) or under non-denaturing conditions (native PAGE) using Novex Tris-glycine 4–20 % gradient gels (Invitrogen, Carlsbad, CA). Prior to electrophoresis, 10 μg (SDS-PAGE) or 25 μg protein (native PAGE) was incubated for 3 h in the absence or presence of 100-fold molar excess dithiothreitol (DTT). Gels were stained with Amido Black.
Phospholipid vesicle transformation
Unilamellar vesicles of DMPC (Avanti Polar Lipids, Birmingham, AL) were prepared in PBS by extrusion through 200 nm filters as described previously [23]. Phospholipid (250 μg) and protein (250 μg) were equilibrated at 24 °C, mixed (500 μl final volume), and the change in absorbance (325 nm) was measured with a Shimadzu spectrophotometer maintaining the incubation temperature with a Peltier controlled cell holder at 24 °C.
Fluorescence spectroscopy
ApoLp-III tyrosine fluorescence was measured in a Fluoromax-2 fluorescence spectrophotometer. ApoLp-III samples (50 μg) were excited at 278 nm and emission followed between 290 and 400 nm (5 nm slit width) in a 1 mL cuvet. After the emission spectrum of apoLp-III was obtained (under oxidizing or reducing conditions), 50 μg E. coli LPS (O55:B5, Sigma-Aldrich, Milwaukee, USA) was added and incubated for an additional 30 min. Subsequently, the tyrosine emission spectrum was obtained for the apoLp-III/LPS mixtures, and corrected for LPS background. To prevent inner filter effects and light scattering, the sample absorbance was kept below 0.1.
Results
Double cysteine mutant engineering
ApoLp-III from G. mellonella contains 163 amino acid residues (Mw 18 kDa) folded into a bundle of five antiparallel α-helices. To tether the helix bundle, two cysteines were introduced at strategic positions (the native protein lacks cysteine). Pro-5 and Ala-135 reside at the beginning of helix-1 or -5, respectively, and are in close spatial proximity according to our model structure that was based on the NMR solution structure of M. sexta apoLp-III [5, 18]. Disulfide bond formation at the introduced cysteine residues would potentially lock the protein in a closed conformation (Figure 1). Similar mutations in L. migratoria (T18C/A147C) and M. sexta apoLp-III (A8C/A138C) have been shown to effectively tether the helix bundle in a closed state. These mutants also lost their ability to transform bilayer vesicles into discoidal complexes, indicating the importance of helix-1 and -5 in the initial binding process [25, 26]. The ability to transform vesicles will be used as a control to verify the anticipated loss of lipid binding in G. mellonella apoLp-III (see lipid binding paragraph). Site-directed mutagenesis was used to introduce the codons for cysteine in the apoLp-III/pET vector, and the resulting mutant protein (P5C/A135C-apoLp-III) was expressed in E. coli and purified. Circular dichroism analysis in the far UV region of the mutant protein produced ellipticity scans that were essentially similar to that of wild-type apoLp-III (not shown). This indicated that the structural integrity of the disulfide mutant was maintained. SDS-PAGE analysis to verify disulfide bond formation (Figure 2) showed that the electrophoretic mobility of 5PC/A135C-apoLp-III was increased compared to the double cysteine mutant in the presence of DTT. The mobility of reduced P5C/A135C-apoLp-III was similar to that of wild-type apoLp-III, and no differences were observed for wild-type protein in the presence or absence of DTT. This indicated that the disulfide bond was formed under oxidizing conditions, while in the presence of DTT the protein existed in a reduced state. This allowed binding analysis of tethered and non-tethered P5C/A135C-apoLp-III to test if helix bundle opening is required for LPS binding interaction.
Figure 1.
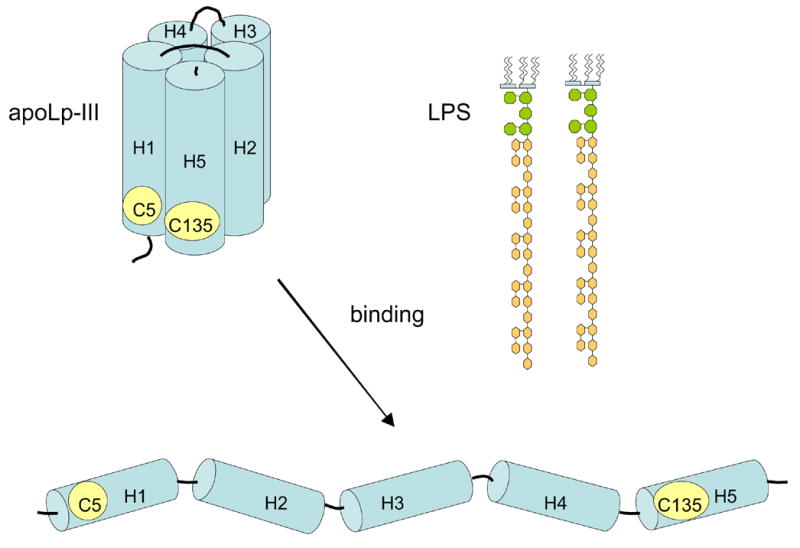
ApoLp-III mutant design. A schematic structure of apoLp-III is shown with the location of Cys-5 and Cys-135. Disulfide bond formation in the closed (unbound) state is expected to prevent helix bundle opening and therefore the protein may not be able to associate with LPS.
Figure 2.
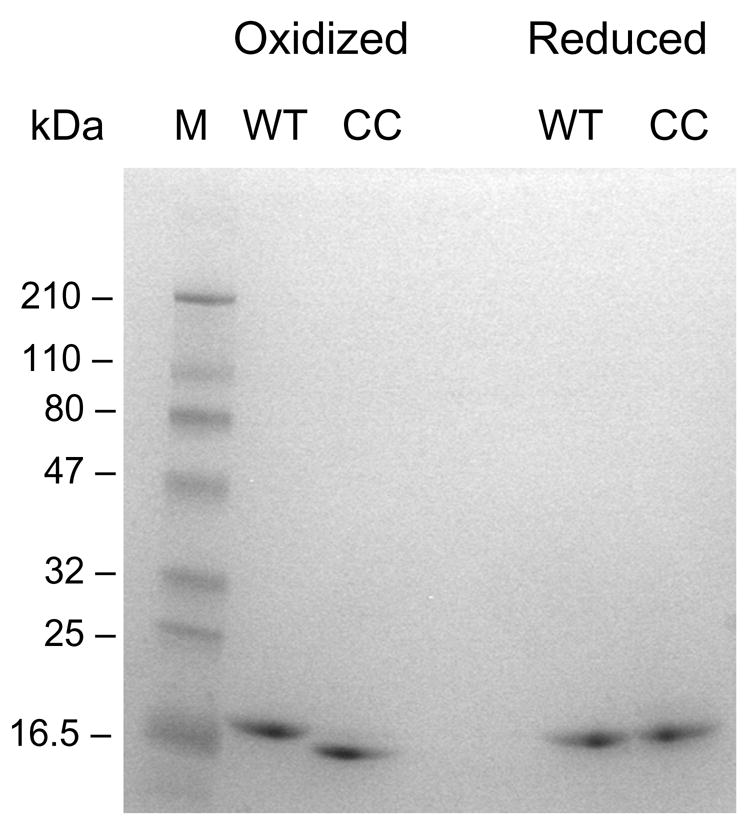
Disulfide bond formation between Cys-5 and Cys-135. Wild-type (WT) and the double cysteine mutant (CC) were analyzed by SDS-PAGE under oxidizing (- DTT, left lanes) and reducing conditions (+ DTT, right lanes).
Lipopolysaccharide binding
Previous studies have shown that apoLp-III-LPS association can be monitored by changes in mobility of apoLp-III or LPS when subjected to electrophoresis under non-denaturing conditions [13]. ApoLp-III association with LPS increases the electrophoretic mobility of micellar LPS, while at the same time apoLp-III disappears from its original position on the gel. As shown in Figure 3, the electrophoresis pattern of oxidized P5C/A135C-apoLp-III in the absence or presence of LPS is similar (left lanes). This indicates the absence of a binding interaction. In contrast, addition of LPS to reduced P5C/A135C-apoLp-III caused a major difference in the electrophoresis pattern, and apoLp-III virtually disappeared from its original position indicating LPS binding (right lanes).
Figure 3.
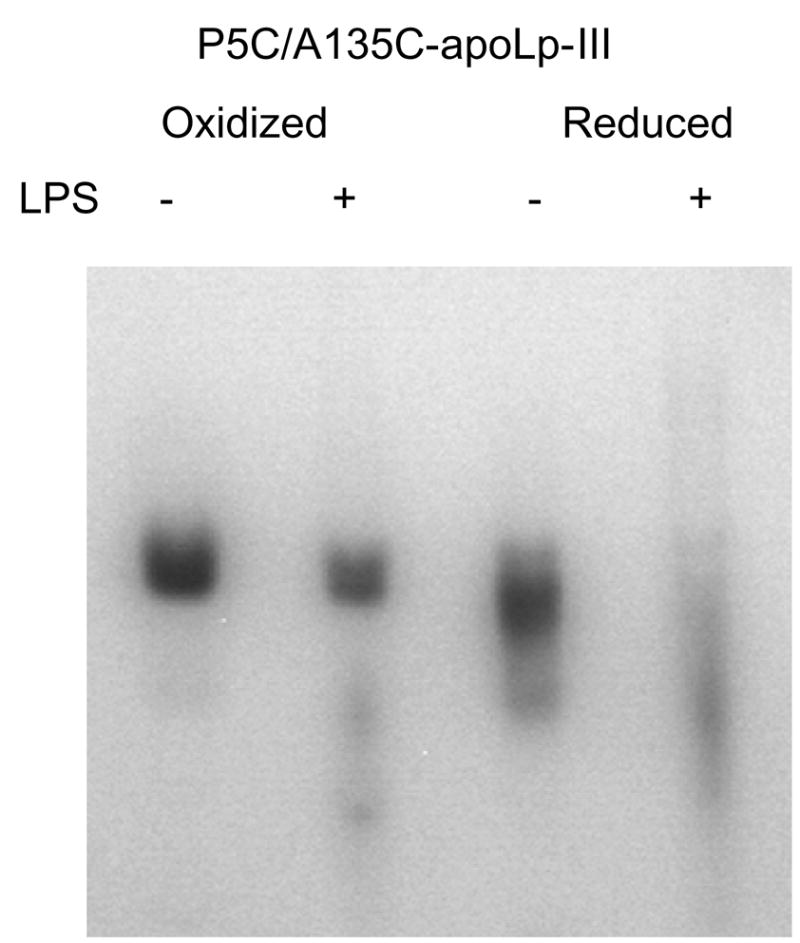
Native PAGE analysis of apoLp-III LPS binding. ApoLp-III was incubated in the presence or absence of LPS, and analyzed by PAGE under non-denaturing conditions. P5C/A135C-apoLp-III was incubated in the absence (left lanes) or presence of DTT (right lanes) prior to electrophoresis.
A second assay was employed to evaluate LPS binding of the disulfide-bonded mutant protein. We have developed a sensitive tyrosine fluorescence method, in which the change in intrinsic fluorescence intensity is measured as a function of the apoLp-III:LPS ratio. G. mellonella apoLp-III contains a lone tyrosine residue buried in the helix interior, while tryptophan is noticeably absent. In unbound protein Tyr-142 is in a quenched state with a weak emission peak at ~ 304 nm. In contrast, a prominent tyrosine fluorescence peak appears in LPS-bound apoLp-III, as the quenched state is abolished [13, 18]. As shown in Figure 4, the fluorescence intensity of oxidized P5C/A135C-apoLp-III did not change upon LPS addition, suggesting that LPS binding did not occur. However, the fluorescence intensity of the reduced mutant (in the presence of excess DTT) increased substantially, reaching a plateau at higher LPS concentrations. At LPS concentrations of > 100 μg/mL, the majority of apoLp-III appeared to be in an LPS-bound form, while no free apoLp-III was present in solution. This result indicates that P5C/A135C-apoLp-III regained its ability to associate with LPS after reduction of the disulfide bond by DTT.
Figure 4.
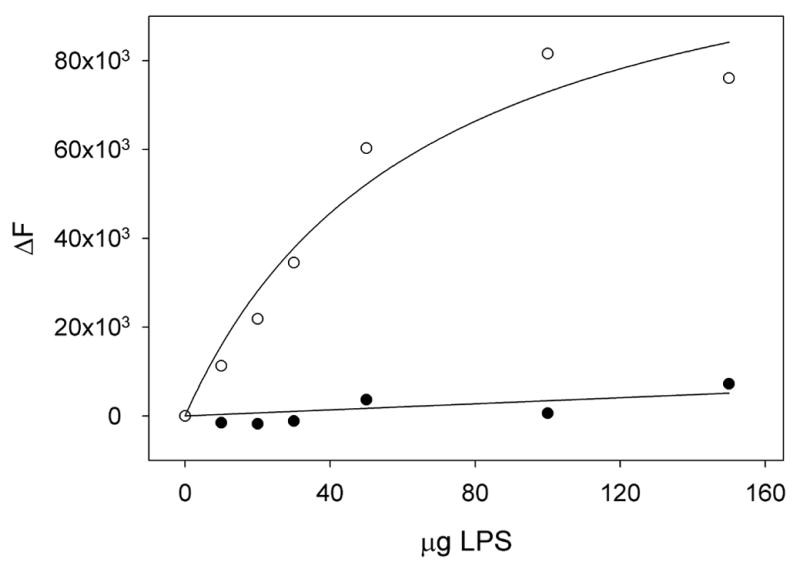
Tyrosine fluorescence analysis of LPS binding. Shown is the change in tyrosine fluorescence emission intensity (∆F) at 304 nm upon addition of increasing amounts of LPS. The change in fluorescence intensity is shown for P5C/A35C-apoLp-III in the oxidized state (closed circles) or in the reduced state (open circles).
Lipid binding
Since similar mutations in L. migratoria and M. sexta apoLp-III had an adverse effect on lipid binding, it is important to test the lipid binding properties of this G. mellonella apoLp-III mutant. In this way, a direct comparison between LPS and lipid interaction binding can be made. To verify that the mutant protein is defect in lipid binding, control experiments were carried out with model phospholipid bilayer vesicles. ApoLp-III has the ability to transform DMPC vesicles into much smaller discoidal complexes, a characteristic shared with other exchangeable apolipoproteins [8]. The transformation of unilamellar vesicles (200 nm in size) into discs (18 nm diameter, estimated by electron microscopy) is a spontaneous process at the lipid transition temperature, which is 24 °C for DMPC [23]. The transformation can be monitored spectrophotometrically as the turbid phospholipid suspension becomes transparent during formation of the much smaller discs. As shown in Figure 5, in the absence of apoLp-III the phospholipid vesicle suspension remained turbid. Addition of wild-type apoLp-III caused a rapid decrease in absorbance, indicating the appearance of small apoLp-III/DMPC discoidal complexes. In contrast, addition of P5C/A135C-apoLp-III to the phospholipid vesicles did not result in decreased sample absorbance. This indicated that the oxidized mutant was unable to transform the phospholipid vesicles. However, in the presence of DTT the cysteine double mutant regained its ability to transform vesicles into discoidal complexes, as indicated by the rapid absorbance decrease. Thus, disulfide bond formation prevented the transformation of phospholipid vesicles into discs. The non-tethered state of reduced P5C/A135C-apoLp-III showed that it was able to open its interior to allow protein lipid interactions and transform phospholipid bilayer vesicles. Thus, the decreased lipid binding of apoLp-III was a result of the introduction of two cysteine residues, and their oxidation into a disulfide-bonded state.
Figure 5.
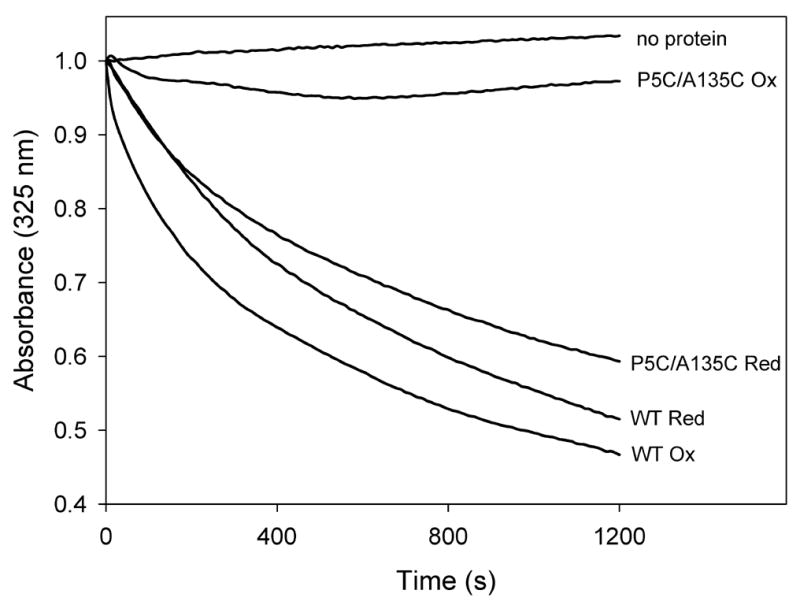
ApoLp-III induced DMPC transformation. The ability to transform DMPC vesicles into discoidal complexes was analyzed for wild-type (WT) and P5C/A135C-apoLp-III under oxidized (-DTT) or reduced (+ DTT) conditions. Transformation of vesicles into discs was monitored by the decrease in sample absorbance at 325 nm.
Discussion
It is generally accepted that the helical structure of apoLp-III has the ability to open its hydrophobic interior upon binding to a lipid, coined the conformational switch [8, 25]. Helix-helix interactions are replaced by helix-lipid interactions, allowing formation of a stable apoLp-III lipid complex. This conformational change has been studied in great detail with DMPC bilayer vesicles. Disulfide bond engineering has been successfully applied to M. sexta and L. migratoria apoLp-III to demonstrate the requirement for helix repositioning upon lipid or lipoprotein association [27, 28]. Pyrene excimer fluorescence and fluorescence resonance energy transfer studies indicated that upon DMPC binding the protein becomes an extended α-helix, wrapping around the periphery of the disc [25, 29]. Similar models have been proposed for apoE and apoA-I [30–32].
To test whether a similar conformational opening of the helix bundle is required for LPS binding, introduction of cysteine residues was required. The selection of amino acid residues was based on our model structure [18], and similar mutations in the homologous apoLp-IIIs from M. sexta and L. migratoria were previously successful in prevention of phospholipid vesicle transformation. M. sexta apoLp-III shares a 71 % amino acid identity with G. mellonella, while there is little sequence homology shared with L. migratoria apoLp-III although the proteins are very similar in their tertiary fold [33]. Disulfide bond formation between Cys-5 and Cys-135 in G. mellonella apoLp-III locked the protein in a closed state. This tethered protein lost its ability to associate with LPS as evidenced by PAGE and tyrosine fluorescence analysis. Control experiments showed that the protein also failed to transform phospholipid bilayer vesicles. Thus, a conformational change of apoLp-III is essential for LPS binding and appears to be mechanistically similar compared to lipid binding. Other support for a conformational change is the observed LPS-induced increase in fluorescence emission intensity of Tyr-142 [13, 18]. The quenched state is abolished, which indicates relocation of the buried tyrosine residue into a new microenvironment, facilitated by helix repositioning. Similarly, tyrosine fluorescence intensity was substantially enhanced in M. sexta apoLp-III upon DMPC association [7]. The requirement of apoLp-III to open its interior to undergo a LPS binding interaction indicates an important role for the amphipathic α-helix. This result also explains why truncated mutants of G. mellonella apoLp-III retained their ability to associate with LPS, as these mutants were comprised of three complete amphipathic α-helices [23].
ApoLp-III is an abundant hemolymph protein normally found in an unbound state. It seems likely that when apoLp-III encounters LPS in vivo, superficial binding is followed by helix bundle opening and a stable binding interaction to neutralize LPS. The structural flexibility of apoLp-III may provide a good system for binding interaction with a wide variety of foreign structures composed of carbohydrates and or lipids. This allows it to function as an effective pattern recognition protein [34]. Since it was pointed out that apoLp-III in the open conformation (as discoidal complexes of apoLp-III and DMPC) acts as an immune activator, the protein conformational switch may trigger secondary cellular immune responses [11]. Our studies support this concept, and for the first time we have demonstrated a conformational opening of apoLp-III upon binding with an immunological relevant substrate. It remains unclear how cells will be able to discriminate between LPS-bound apoLp-III and diacylglycerol-enriched lipophorins which appear during lipid mobilization [1]. However, apoLp-III immune responses have been shown in larvae, and the apoLp-III associated state with lipophorin occurs during insect flight. In addition, there may be differences in the open conformation depending on the nature of the substrate surface. To date, high resolution structural data of apolipoprotein in their biologically active forms are not available, although at lower resolution significant progress has been made [35].
The traditional role of apolipoproteins in lipid transport processes is currently expanding into innate immunity [36–37]. However, further study is crucial to understand the underlying mechanisms of apolipoprotein function in innate immunity [32–33]. ApoLp-III has served as an excellent model system for structural studies of lipid transport. The protein may also be useful to investigate apolipoprotein function in innate immunity. Given the structural similarity between apoLp-III and vertebrate apoE it is possible that these proteins undergo similar conformational changes upon interaction with LPS. In conclusion, the present study presents a mechanism of apolipoprotein association with foreign membrane components, and provides a molecular basis for the physiological observations carried out by others.
Acknowledgments
This study was supported by the National Institutes of Health: Minority Biomedical Research Support, Support of Continuous Research Excellence (GM 063119-05), and student support was provided by the Howard Hughes Medical Institute # 52002663.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Van der Horst DJ, Van Hoof D, Van Marrewijk WJA, Rodenburg KW. Alternative lipid mobilization: the insect shuttle system. Mol Cell Biochem. 2002;239:113–119. [PubMed] [Google Scholar]
- 2.Narayanaswami V, Ryan RO. Molecular basis of exchangeable apolipoprotein function. Biochim Biophys Acta. 2000;1483:15–36. doi: 10.1016/s1388-1981(99)00176-6. [DOI] [PubMed] [Google Scholar]
- 3.Weers PMM, Ryan RO. Apolipophorin III: role model apolipoprotein. Insect Biochem Mol Biol. 2006;36:231–240. doi: 10.1016/j.ibmb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Breiter DR, Kanost MR, Benning MM, Wesenberg G, Law JH, Wells MA, Rayment I, Holden HM. Molecular structure of an apolipoprotein determined at 2.5-Å resolution. Biochemistry. 1991;30:603–608. doi: 10.1021/bi00217a002. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Sykes BD, Ryan RO. Structural basis for the conformational adaptability of apolipophorin III, a helix bundle exchangeable apolipoprotein. Proc Natl Acad Sci USA. 2002;99:1188–1193. doi: 10.1073/pnas.032565999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan D, Zheng Y, Yang D, Wang J. NMR solution structure and dynamics of an exchangeable apolipoprotein, Locusta migratoria apolipophorin III. J Biol Chem. 2003;278:21212–21220. doi: 10.1074/jbc.M208486200. [DOI] [PubMed] [Google Scholar]
- 7.Wientzek M, Kay CM, Oikawa K, Ryan RO. Binding of insect apolipophorin III to dimyristoylphosphatidylcholine vesicles. Evidence for a conformational change. J Biol Chem. 1994;269:4605–4612. [PubMed] [Google Scholar]
- 8.Weers PMM, Ryan RO. Apolipophorin III: a lipid-triggered molecular switch. Insect Biochem Mol Biol. 2003;33:1249–1260. doi: 10.1016/j.ibmb.2003.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Wiesner A, Losen S, Kopáek P, Weise C, Götz P. Isolated apolipophorin III from Galleria mellonella stimulates the immune reactions of this insect. J Insect Physiol. 1997;43:383–391. doi: 10.1016/s0022-1910(96)00113-8. [DOI] [PubMed] [Google Scholar]
- 10.Halwani AE, Niven DF, Dunphy GB. Apolipophorin-III and the interactions of lipoteichoic acids with the immediate immune responses of Galleria mellonella. J Invertebr Pathol. 2000;76:233–241. doi: 10.1006/jipa.2000.4978. [DOI] [PubMed] [Google Scholar]
- 11.Dunphy G, Halwani A. Haemolymph proteins of larvae of Galleria mellonella detoxify endotoxins of the insect pathogenic bacteria Xenorhabdus nematophilus (enterobacteriaceae) J Insect Physiol. 1997;43:1023–1029. doi: 10.1016/s0022-1910(97)00072-3. [DOI] [PubMed] [Google Scholar]
- 12.Whitten MMA, Tew IF, Lee BL, Ratcliffe NA. A novel role for an insect apolipoprotein (apolipophorin III) in beta-1,3-glucan pattern recognition and cellular encapsulation reactions. J Immunol. 2004;172:2177–2185. doi: 10.4049/jimmunol.172.4.2177. [DOI] [PubMed] [Google Scholar]
- 13.Pratt CC, Weers PMM. Lipopolysaccharide binding of an exchangeable apolipoprotein, apolipophorin III, from Galleria mellonella. Biol Chem. 2004;385:1113–1119. doi: 10.1515/BC.2004.145. [DOI] [PubMed] [Google Scholar]
- 14.Niere M, Meisslitzer C, Dettloff M, Weise C, Ziegler M, Wiesner A. Insect immune activation by recombinant Galleria mellonella apolipophorin III. Biochim Biophys Acta. 1999;1433:16–26. doi: 10.1016/s0167-4838(99)00148-x. [DOI] [PubMed] [Google Scholar]
- 15.Dettloff M, Wiesner A. Immune stimulation by lipid-bound apolipophorin III. In: Wiesner A, Dunphy GB, Marmaras VJ, Morishima I, Sugumaran M, Yamakawa M, editors. Techniques in Insect Immunology. SOS Publications; Fair Haven: 1999. pp. 243–251. [Google Scholar]
- 16.Dettloff M, Kaiser B, Wiesner A. Localization of injected apolipophorin III in vivo – new insights into the immune activation process directed by this protein. J Insect Physiol. 2001;47:789–797. doi: 10.1016/s0022-1910(01)00069-5. [DOI] [PubMed] [Google Scholar]
- 17.Niere M, Dettloff M, Maier T, Ziegler M, Wiesner A. Insect immune activation by apolipophorin III is correlated with the lipid-binding properties of the protein. Biochemistry. 2001;40:11502–11508. doi: 10.1021/bi010117f. [DOI] [PubMed] [Google Scholar]
- 18.Leon JL, Pratt CC, Vasquez LJ, Weers PMM. Tyrosine fluorescence analysis of apolipophorin III - lipopolysaccharide interaction. Arch Biochem Biophys. 2006 doi: 10.1016/j.abb.2006.05.009. in press. [DOI] [PubMed] [Google Scholar]
- 19.Raetz CRH. Biochemistry of endotoxins. Annu Rev Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- 20.Van Oosten M, Rensen PCN, Van Amersfoort ES, Van Eck M, Van Dam A-M, Brevé JJP, Vogel JJPT, Panet A, Van Berkel TJC. Apolipoprotein E protects against bacterial lipopolysaccharide-induced lethality. A new therapeutic approach to treat gram-negative sepsis. J Biol Chem. 2001;276:8820–8824. doi: 10.1074/jbc.M009915200. [DOI] [PubMed] [Google Scholar]
- 21.Van Amersfoort ES, Van Berkel TJC, Kuiper J. Receptors, mediators, and mechanisms involved in bacterial sepsis and septic shock. Clin Microbiol Rev. 2003;16:379–414. doi: 10.1128/CMR.16.3.379-414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 23.Dettloff M, Niere M, Ryan RO, Kay CM, Wiesner A, Weers PMM. Differential lipid binding of truncation mutants of Galleria mellonella apolipophorin III. Biochemistry. 2002;41:9688–9695. doi: 10.1021/bi0200108. [DOI] [PubMed] [Google Scholar]
- 24.Weers PMM, Abdullahi WE, Cabrera JM, Hsu TC. Role of buried polar residues in helix bundle stability and lipid binding of apolipophorin III: destabilization by threonine 31. Biochemistry. 2005;44:8810–8816. doi: 10.1021/bi050502v. [DOI] [PubMed] [Google Scholar]
- 25.Sahoo D, Weers PMM, Ryan RO, Narayanaswami V. Lipid triggered molecular switch of apoLp-III helix bundle to an extended helix conformation. J Mol Biol. 2002;321:201–214. doi: 10.1016/s0022-2836(02)00618-6. [DOI] [PubMed] [Google Scholar]
- 26.Soulages JL, Arrese EL, Chetty PS, Rodriguez V. Essential role of the conformational flexibility of helices 1 and 5 on the lipid binding activity of apolipophorin III. J Biol Chem. 2001;276:34162–34166. doi: 10.1074/jbc.M105836200. [DOI] [PubMed] [Google Scholar]
- 27.Narayanaswami V, Wang J, Kay CM, Scraba DG, Ryan RO. Disulfide bond engineering to monitor conformational opening of apolipophorin III during lipid binding. J Biol Chem. 1996;271:26855–26862. doi: 10.1074/jbc.271.43.26855. [DOI] [PubMed] [Google Scholar]
- 28.Chetty PS, Arrese EL, Rodriguez V, Soulages JL. Role of helices and loops in the ability of apolipophorin-III to interact with native lipoproteins and form discoidal lipoprotein complexes. Biochemistry. 2003;42:15061–15067. doi: 10.1021/bi035456i. [DOI] [PubMed] [Google Scholar]
- 29.Garda HA, Arrese EL, Soulages JL. Structure of apolipophorin III in discoidal lipoproteins. Interhelical distances in the lipid-bound state and conformational change upon binding to lipid. J Biol Chem. 2002;277:19773–19782. doi: 10.1074/jbc.M110089200. [DOI] [PubMed] [Google Scholar]
- 30.Lu B, Morrow JA, Weisgraber KH. Conformational reorganization of the four-helix bundle of human apolipoprotein E in binding to phospholipid. J Biol Chem. 2000;275:20775–20781. doi: 10.1074/jbc.M003508200. [DOI] [PubMed] [Google Scholar]
- 31.Narayanaswami V, Maiorano JN, Dhanasekaran P, Ryan RO, Phillips MC, Lund-Katz S, Davidson WS. Helix orientation of the functional domains in apolipoprotein E in discoidal high density lipoprotein particles. J Biol Chem. 2004;279:14273–14279. doi: 10.1074/jbc.M313318200. [DOI] [PubMed] [Google Scholar]
- 32.Marcel YL, Kiss RS. Structure function relationships of apolipoprotein A-I: a flexible protein with dynamic lipid associations. Curr Opin Lipidol. 2003;14:151–157. doi: 10.1097/00041433-200304000-00006. [DOI] [PubMed] [Google Scholar]
- 33.Weise C, Franke P, Kopáek P, Wiesner A. Primary structure of apolipophorin-III from the greater wax moth, Galleria mellonella. J Prot Chem. 1998;17:633–641. doi: 10.1007/BF02780964. [DOI] [PubMed] [Google Scholar]
- 34.Kanost MR, Jiang H, Yu XQ. Innate immune responses of a lepidopteran insect. Manduca sexta Immunol Rev. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- 35.Peters-Libeu CA, Newhouse Y, Hatters DM, Weisgraber KH. Model of biologically active apolipoprotein E bound to dipalmitoylphosphatidylcholine. J Biol Chem. 2006;281:1073–1079. doi: 10.1074/jbc.M510851200. [DOI] [PubMed] [Google Scholar]
- 36.Mahley RW, Rall SC., Jr Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 37.Berbée JFP, Havekes LM, Rensen PCN. Apolipoproteins modulate the inflammatory response to lipopolysaccharide. J Endotoxin Res. 2005;11:97–103. doi: 10.1179/096805105X35215. [DOI] [PubMed] [Google Scholar]


