SUMMARY
Oxidation reactions represent an important degradation pathway of nucleic acid-based pharmaceuticals. To evaluate the role of metal contamination and chelating agents in the formation of reactive oxygen species (ROS) during lyophilization, ROS generation and the stability of lipid/DNA complexes were investigated. Trehalose-containing formulations were lyophilized with different levels of transition metals. ROS generation was examined by adding proxyl fluorescamine to the formulations prior to freeze-drying. Results show that ROS were generated during lyophilization, and both supercoil content and transfection rates decreased as the levels of metal-induced ROS increased. The experiments incorporating chelators demonstrated that some of these agents (e.g., DTPA, desferal) clearly suppress ROS generation, while others (e.g., EDTA) enhance ROS. Surprisingly, there was not a strong correlation of ROS generated in the presence of chelators with the maintenance of supercoil content. In this study, we demonstrated the adverse effects of the presence of metals (especially Fe2+) in nonviral vector formulations. While some chelators attenuate ROS generation and preserve DNA integrity, the effects of these additives on vector stability during lyophilization are difficult to predict. Further study is needed to develop potent formulation strategies that inhibit ROS generation and DNA degradation during lyophilization and storage.
Keywords: nonviral vectors, lyophilization, metal contamination, reactive oxygen species, chelating agents, gene delivery
INTRODUCTION
There is currently a growing interest in preserving nonviral vectors as dehydrated formulations because they offer the potential for prolonged stability at ambient temperature. The majority of lyophilization studies to date have focused on the acute stability of nonviral vectors and consistently reported that nonviral vectors can be stabilized in the presence of sugars [1–4]. Considering that nucleic acids are highly susceptible to oxidation [5, 6], formulation strategies must insure that DNA integrity is maintained during the freeze-drying process.
Free radical oxidation is often considered a major chemical degradation pathway for DNA-based pharmaceuticals [5, 7, 8]. A significant number of studies have demonstrated the high sensitivity of DNA to reactive oxygen species [ROS] (e.g., hydroxyl radical) that result in damaged bases and/or single and double-strand breaks [9–12]. Indeed, it is well established that exogenous factors such as the presence in excipients of trace amounts of transition metals (e.g., Cu2+ and Fe2+) can catalyze the generation of hydroxyl radicals via the Fenton reaction, which can extensively damage DNA [5–7, 13, 14]. In an effort to circumvent this problem, researchers have utilized strategies such as the addition of chelating agents to their formulations [7, 8, 15]. Importantly, while these findings suggest that trace metal contaminants can cause DNA degradation and that chelating agents can be used to prevent ROS in aqueous solution, to our knowledge no study has explored the role of the generation of reactive oxygen species (ROS) in the stability of lipid/DNA during drying. Thus, in the present study we evaluated the effects of metal contamination and chelating agents on the formation of ROS during acute lyophilization. Formulations containing the cationic lipid 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and L-α-dioleoylphosphatidyl-ethanolamine (DOPE) (1:1 molar ratio) complexed with plasmid DNA were lyophilized in trehalose fortified with different levels of transition metals (Cu2+, Fe2+, Fe3+). We investigated the role of metal ions in the generation of ROS during acute lyophilization by adding a fluorescent probe (proxyl fluorescamine) to the formulations prior to freeze-drying. We also explored the effect of four different chelators on metal-induced ROS generation and the acute stability of lipid/DNA complexes (as indicated by DNA integrity and biological activity upon rehydration). Here, we show that ROS are generated during lyophilization even in the absence of metals, although the addition of metals increased ROS levels. Generally, both supercoil content and transfection rates diminished as the levels of metal-induced ROS increased. The experiments incorporating chelators showed that some of these agents (e.g., diethylenetriaminepentaacetic acid [DTPA], desferrioxamine mesylate [desferal]) clearly suppress ROS generation, while others (e.g., ethylenediaminetetraacetic acid [EDTA]) enhance ROS levels. Surprisingly, there was not a strong correlation of ROS levels (as measured by proxyl fluorescamine) generated in the presence of chelators with the maintenance of supercoil content. The results obtained in this study demonstrate the adverse effects of the presence of metal contaminants, especially iron in the ferrous form, in DNA-containing formulations. In addition, this study underscores the importance of developing formulations strategies that minimize the generation of ROS.
MATERIALS AND METHODS
Chemicals
Trehalose was a gift from Ferro-Pfanstiehl Laboratories (Waukegan, IL). DOTAP and DOPE were obtained in a 1:1 weight ratio from Avanti Polar Lipids (Alabaster, AL). The DNA plasmid encoding the protein reporter luciferase (5.9 kb) was a generous gift from Valentis (Burlingame, CA). DNA was dissolved in sterile 2.5 mM Tris-HCl pH 8.5 and diluted to a concentration of 1 mg/mL prior to use. The luciferase assay kit was obtained from Promega (Madison, WI). Ethidium bromide solution (10 mg/mL) was purchased from Sigma (St. Louis, MO). Proxyl fluorescamine, 5-(2-carboxyphenyl)-5-hydroxy-1-[(2,2,5, 5-tetramethyl-1-oxypyrrolidin-3-yl) methyl]-3-phenyl-2-pyrrolin-4-one potassium salt, was obtained from Molecular Probes Inc. (Eugene, OR). Diethylenetriaminepentaacetic acid (DTPA), bathophenanthroline disulfonate (BPS) and ferrous chloride were obtained from Acros Organics (Fairlawn, NJ). Anhydrous cupric chloride, ferric chloride, ethylenediaminetetraacetic acid (EDTA), and desferrioxamine mesylate (desferal) were purchased from Sigma (St. Louis, MO). Sodium chloride (NaCl), potassium chloride (KCl), sodium bicarbonate (NaHCO3), calcium chloride (CaCl2), magnesium sulfate (MgSO4) and potassium phosphate monobasic (KH2PO4) were purchased from Sigma (St. Louis, MO). All chemicals were of analytical grade and used without further purification.
Preparation of liposomes
Liposomes containing DOTAP in a 1:1 (w/w) ratio with the zwitterionic lipid DOPE were prepared as previously described [16]. Briefly, the lipid mixture in chloroform was dried under a stream of nitrogen gas and placed under vacuum (10 mTorr) for 1 h to remove residual chloroform. The dried lipid was resuspended in sterile distilled water to a concentration of 2 mg/mL, sonicated to clarity with a Branson Sonifier 250, and stored at 4°C. The liposomes were freshly sonicated immediately before use.
Lipoplex preparation
The complexes were prepared with a 3:1 lipid:DNA weight ratio (480 μg DOTAP-DOPE and 160 μg DNA in 2.5 mM Tris-HCl pH 8.5) in polypropylene microcentrifuge tubes by gentle mixing, and incubated for 20 min at room temperature as previously described [17]. This method of preparation results in a heterogeneous suspension of particles with a calculated +/− charge ratio of 0.7 [16]. Depending on the experiment, freshly prepared metal ion solutions (1x106 ppb: CuCl2, FeCl2, FeCl3) were added to two milliliter aliquots of the resulting suspension of lipid/DNA complexes to achieve a range of final metal concentrations (0–1000 ppb). Accordingly, freshly prepared chelating agent solutions (DTPA, desferal, BPS and EDTA) were subsequently added to achieve final chelator concentrations ranging from 0–800 μM. Proxyl fluorescamine was introduced into these formulations at a final concentration of 0.45 μM. The resulting suspensions were then diluted with an equal volume of an 8% excipient solution (trehalose) in Tris buffer as previously described [18]. Aliquots of 400 μL containing 16 μg of DNA were transferred to clear 1-mL and amber 2-mL flat-bottomed borosilicate lyophilization vials (for samples with and without proxyl fluorescamine dye, respectively) (West Co., Litiz, PA). The stoppers were obtained from West Co. (Litiz, PA), washed with distilled water, and dried overnight in an oven (≈ 60°C) prior to use.
Freeze-drying protocol
Sample vials were placed on the shelf of an FTS Durastop lyophilizer (Stone Ridge, NY). The lyophilization cycle used was performed as follows: shelves were cooled to −40°C, and held for 2 h (sample temperature ≈ −37°C), the chamber pressure was then reduced to 60 mTorr, primary drying at −40°C for 30 h, secondary drying at 25°C for 6 h. After secondary drying, sample vials were stoppered under vaccum and stored at −80°C until rehydrated. For freezing studies, the same conditions were used to cool the samples in the lyophilizer. Samples were maintained on a −40°C shelf overnight, and rapidly thawed in a water bath (37°C) prior to analysis.
Rehydration protocol
Lyophilized samples containing the appropriate levels of plasmid, cationic agent and excipient (trehalose) were rehydrated to a final volume of 400 μL with filtered distilled water and incubated for 30 min at room temperature. Appropriate volumes of rehydrated samples were used for transfection (7.5 μl) and analysis of DNA structure (50 μl; described below).
Cell culture
African green monkey kidney cells (COS-7: ATCC No. CRL1651) were obtained from American Type Culture Collection (Rockville, MD). Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 50 units/mL penicillin G, and 50 μg/mL streptomycin sulfate, and were propagated by reseeding at 1−3 x 105 cells/100 mm dish every 2–3 days. In our experiments, cultures were freshly seeded at 2500 cells/well in 96-well plate 24 h before transfection.
Transfection assay
Aliquots of rehydrated and freshly prepared lipid/DNA complexes containing 0.3 μg DNA in the presence or absence of the metal ions Cu2+, Fe2+, and Fe3+, and the chelating agents DTPA, desferal, BPS and EDTA, were diluted to a final volume of 100 μL with serum-free, antibiotic-free DMEM and distributed into wells of a 96-well plate containing COS-7 cells freshly washed with phosphate buffered saline (PBS). Cells were subsequently incubated with lipoplexes for 4 h before the medium was replaced with 100 μL DMEM containing serum and antibiotics, and allowed to grow for approximately 40 h before the cell culture medium was discarded. Cells were washed and then lysed with 80 μL of lysis buffer (Promega, Madison, WI), as previously described [19]. Twenty microliters of lysate were used to assay luciferase expression according to the manufacturer’s protocol (Promega). Luciferase activity was quantified by using a TD-20e Los Alamos Diagnostics 535 Luminometer (Mountain View, CA). Protein concentrations were determined by the Bradford method using a Bio-Rad protein assay (Hercules, CA), according to the directions provided by the manufacturer. Absorbances were measured at 550 nm with a THERMOmax microplate reader (Molecular Devices, Sunnyvale, CA). Of note, the four chelating agents used in this study showed no cytotoxic effect as assessed by a transfection assay. Conversely, chelating agents such as phytic acid and dimercaprol (BAL) exhibited cytotoxicity and were not further studied.
Extraction of DNA from nonviral vectors
Separation of DNA from cationic lipids was achieved by modifying a previously described methodology [2]. Briefly, rehydrated or freshly prepared samples containing 2 μg of DNA were mixed with a sodium dodecyl sulfate (SDS) solution to a final SDS concentration of 25 mM. The resulting solutions were incubated for fifteen minutes at 75°C and allowed to cool to room temperature. Aliquots containing 400 ng of DNA were used to quantify DNA supercoil content.
Quantification of supercoil content
Loss of supercoil content was assessed by agarose gel electrophoresis. Samples were run in a 0.9% agarose gel which was soaked in a solution of EtBr after the run was terminated. Quantification of bands was assessed by fluorometric image analysis using a Fluor-S MultiImager and Quantity One software (Biorad; Hercules, CA). Fluorescence intensity for supercoiled DNA bands was corrected by multiplying by a factor of 1.4, as previously described [20].
Measurement of reactive oxygen species (ROS) generated in an aqueous solution
Generation of ROS was obtained by adding FeCl2 and hydrogen peroxide to an oxygenated Krebs-Henseleit buffer (KH), as previously described [21]. Briefly, KH buffer, which contains (in mM): NaCl 118; NaHCO3 23.8; KCl 4.7; KH2PO4 1.2; CaCl2 2.5; MgSO4 1.2, was bubbled with 95% O2-5% CO2 (pH 7.4) for 10 min prior to the experiments. This buffer system was chosen because ROS levels have been previously quantified under these conditions [21], thereby allowing comparisons with that detected by proxyl fluorescamine. The order of addition of reaction reagents is considered an important factor [22]. Therefore, in our experiments we mixed KH with proxyl fluorescamine (0.45 μM) and then with H2O2 (10 μM): metals were always the last component added. We tested media containing Cu2+ at a 10 μM concentration and Fe2+ concentrations of 1–20 μM. When desferal was utilized in these experiments, it was also added before introduction of Fe2+ at a final concentration of 50 μM. The reaction mixtures were incubated for 1 minute at room temperature. Then, aliquots of 200 μL were transferred onto a 96-well fluorescent plate and generation of ROS was followed for 60 min at 10 min intervals. Fluorescence was measured on SpectraMAX Gemini EM fluorescence microplate reader (Molecular Devices, Sunnyvale, CA); excitation wavelength: 385 nm and emission wavelength: 485 nm. Background levels of fluorescence in fresh samples were very consistent; relative levels of reactive oxygen species in formulations are reflected by fluorescence intensity values. If not otherwise stated, all experiments were performed at room temperature. Parallel experiments were performed in oxygenated Tris-HCl buffer containing trehalose (data not shown). Data from these latter experiments are very similar to that reported for KH buffer (i.e., linear with time), except that ROS formation was less dramatic.
Measurement of ROS in lyophilized samples
Lyophilized samples and fresh lipid/DNA complexes were assayed using the fluorogenic spin trap proxyl fluorescamine, which detects superoxide and free hydroxyl radicals, as described elsewhere [23]. Dried samples containing proxyl fluorescamine (16 μg of plasmid and 0.45 μM final dye concentration) were rehydrated with distilled water to assess the levels of ROS after rehydration. Vials were incubated for 30 min and then 200 μL of suspensions were transferred onto a 96-well fluorescent plate. Fluorescence was then measured on SpectraMAX Gemini EM fluorescence microplate reader as indicated previously. The background fluorescence in fresh controls lacking metals was very consistent, and was subtracted from each sample.
Statistical analysis
Formation of reactive oxygen species in lyophilized formulations was compared to that of fresh preparations. Comparisons of means having p values less than 0.05 were judged to be significantly different. Statistically significant differences were determined using a two-tailed student t test with Graphpad Prism Software (San Diego, CA).
RESULTS
In the present study we utilized proxyl fluorescamine, a water-soluble dye that is known to fluoresce upon reaction with ROS [23], to monitor ROS generation during acute freeze-drying. We first wanted to validate the applicability of this technique towards the detection of ROS. Accordingly, we chose solution conditions in which ROS formation has been extensively characterized [21]. The results in Figure 1 clearly show that this fluorogenic spin trap agent is capable of detecting ROS generation. Under aerobic conditions, we observed that the addition of micromolar concentrations of metals (Fe2+, Cu2+) resulted in a relative increase in fluorescence as compared with those observed in the presence of KH buffer alone. The effect of metals is evident by an initial burst of ROS upon mixing (prior to the initial measurement) followed by a time-dependent increase as previously described [21]. Moreover, the addition of hydrogen peroxide caused a further increase in fluorescence intensity in both metal-containing solutions. Fe2+ generated higher ROS levels when compared to an equimolar concentration of Cu2+, as revealed by the fluorescence measurements performed in the presence or absence of hydrogen peroxide. ROS formation mediated by Fe2+ was clearly lowered by desferal because this chelator completely prevented the time-dependent increase of ROS in solution (Figure 1A). As expected, greater ROS formation was observed in the presence of progressively higher Fe2+ concentrations (Fig. 1B). Furthermore, under conditions in which ROS generation has been shown to be linear with time [21], proxyl fluorescamine detected time-dependent linear increases in the levels of ROS generated in Fe2+-containing formulations (Figs. 1B and C).
Figure 1.
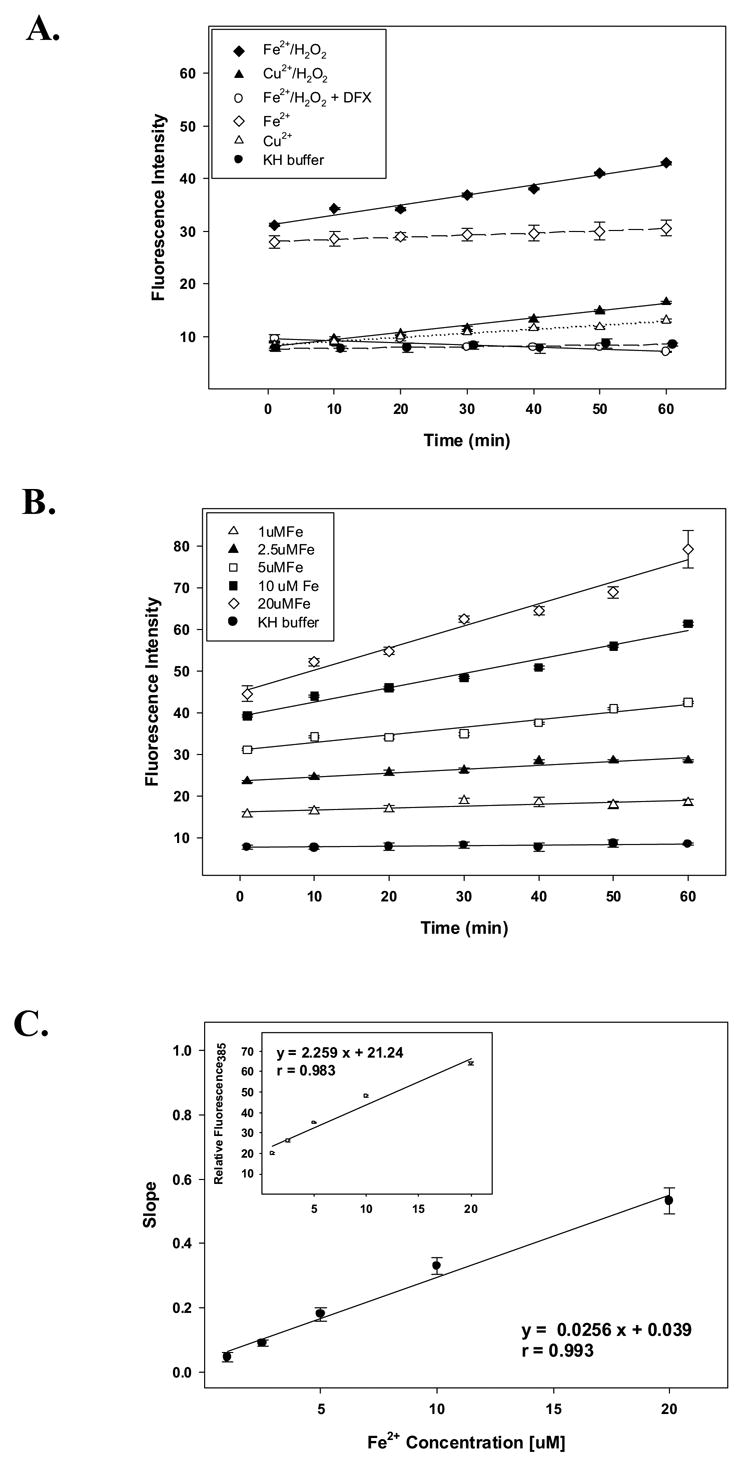
Levels of reactive oxygen species (ROS) assessed by monitoring the fluorescence intensity of proxyl fluorescamine at 385 nm in aqueous solutions. (A) Effect of Fe2+, Cu2+ (10 μM) and desferal (DFX, 50 μM) on the generation of ROS in the presence or absence of hydrogen peroxide (10 μM). (B) Time-dependent formation of ROS at different Fe2+ concentrations in the presence of 10 μM H2O2. (C) Effect of Fe2+ concentration on the rate of ROS generation. Inset shows relative ROS levels as a function of Fe2+ concentration after 30 min of reaction. The order of reagents was performed as described in the Methods section. Samples were incubated for 1 min, and reaction was followed for 60 min. The values represent the mean ± 1 S.D. of triplicate determinations.
We next wanted to assess the formation of ROS in lipid/DNA complexes during acute lyophilization. In order to accomplish this goal, proxyl fluorescamine was added to samples prior to freeze-drying, and fluorescence was assessed after thawing/rehydration. When complexes were freeze-thawed, we did not observe significant changes in ROS levels. However, the presence of lipid/DNA complexes significantly enhanced ROS levels after drying (p<0.0001; Fig. 2). It is worth noting that samples dried without added metals generated ROS levels comparable to metal-fortified liquid formulations. Thus, in order to evaluate the effect of metal ions on the generation of ROS during drying, lipid/DNA complexes formulated in trehalose were treated with increasing concentrations of copper and iron ions prior to freeze-drying. When formulations were fortified with Cu2+, progressive increases in ROS levels are observed after lyophilization/rehydration (Fig. 3A). As ROS levels increase, we observe a corresponding loss in transfection efficiency that is exacerbated at the highest metal concentration (i.e., 1000 ppb; Fig. 3A). We also observe a significant loss of supercoil content and the appearance of linear DNA at all metal concentrations (Fig. 3B).
Figure 2.
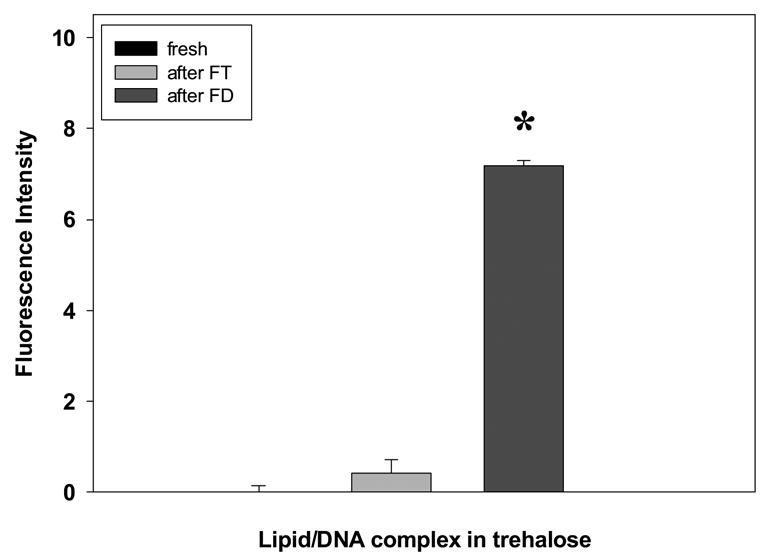
Effect of freeze-thawing and freeze-drying on the generation of reactive oxygen species (ROS). Levels of ROS assessed by monitoring the fluorescence intensity of proxyl fluorescamine at 385 nm following freeze-thawing (FT) and freeze-drying (FD) were assessed by using a fluorogenic spin trap probe, proxyl fluorescamine, that was added to the formulations prior to freezing or freeze-drying as described in Methods section. (*) Significant different sample with p < 0.0001. Reported values are represented as the mean ± 1 S.D. of triplicate determinations.
Figure 3.
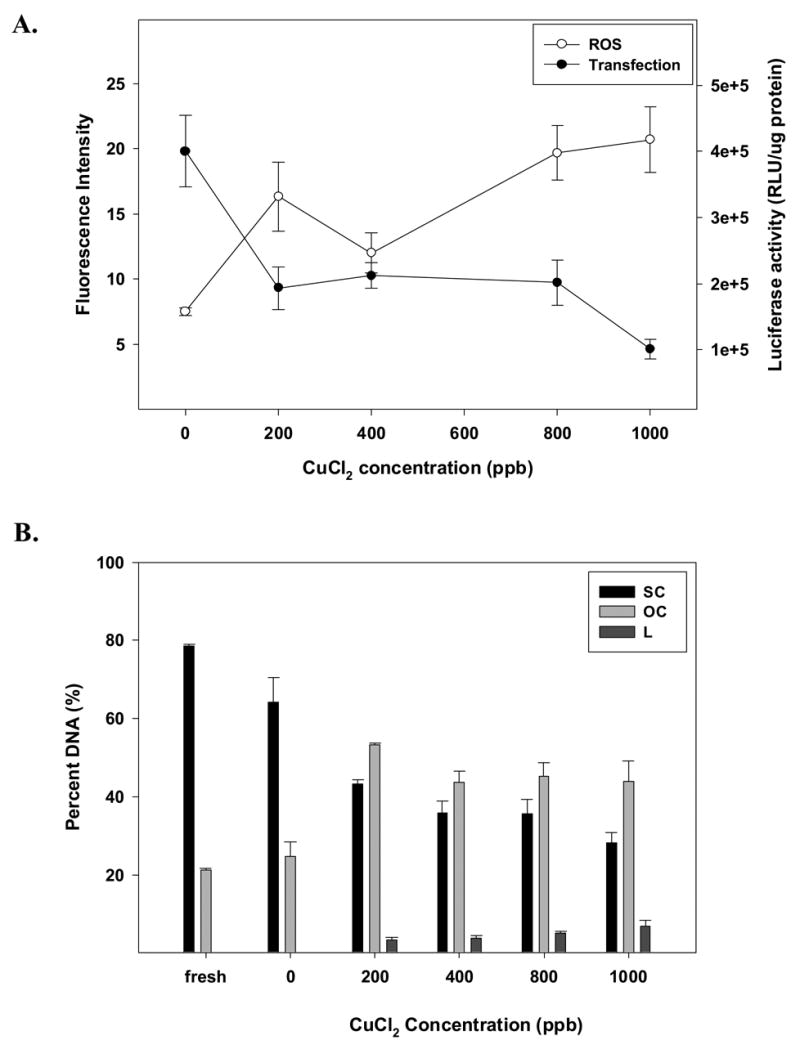
Effect of increasing amounts of Cu2+ on the stability of DOTAP:DOPE/DNA complexes formulated in trehalose during acute lyophilization, in terms of (A) ROS content and transfection rates, or (B) DNA integrity. Percent of DNA is shown as supercoil content (SC), open circle (OC) and linear form (L) remaining in samples after acute lyophilization. The results are expressed as mean values ± 1 S.D. of measurements on triplicate samples.
We also investigated the potential role of iron on chemical degradation and biological activity of lyophilized lipid/DNA vectors. We specifically studied lipoplexes containing both the reduced and oxidized forms of iron (Figs. 4 and 5). When vectors were treated with the reduced form of iron (Fe2+), we observed a progressive generation of ROS associated with reduced biological activity and supercoil content (Figs. 4A and B). It should be noted that reduction in supercoil content was much greater in formulations containing Fe2+ as compared to samples treated with Cu2+. The damaging effect of Fe3+ on dried complexes was smaller than that observed in formulations containing Fe2+ or Cu2+. In fact, the effect of the ferric ion concentrations up to 400 ppb did not significantly alter ROS levels, supercoil content, or biological activity (Fig. 5). While ROS levels and transfection rates were affected by higher Fe3+ concentrations, we did not observe a corresponding loss of supercoil content (Fig. 5B).
Figure 4.
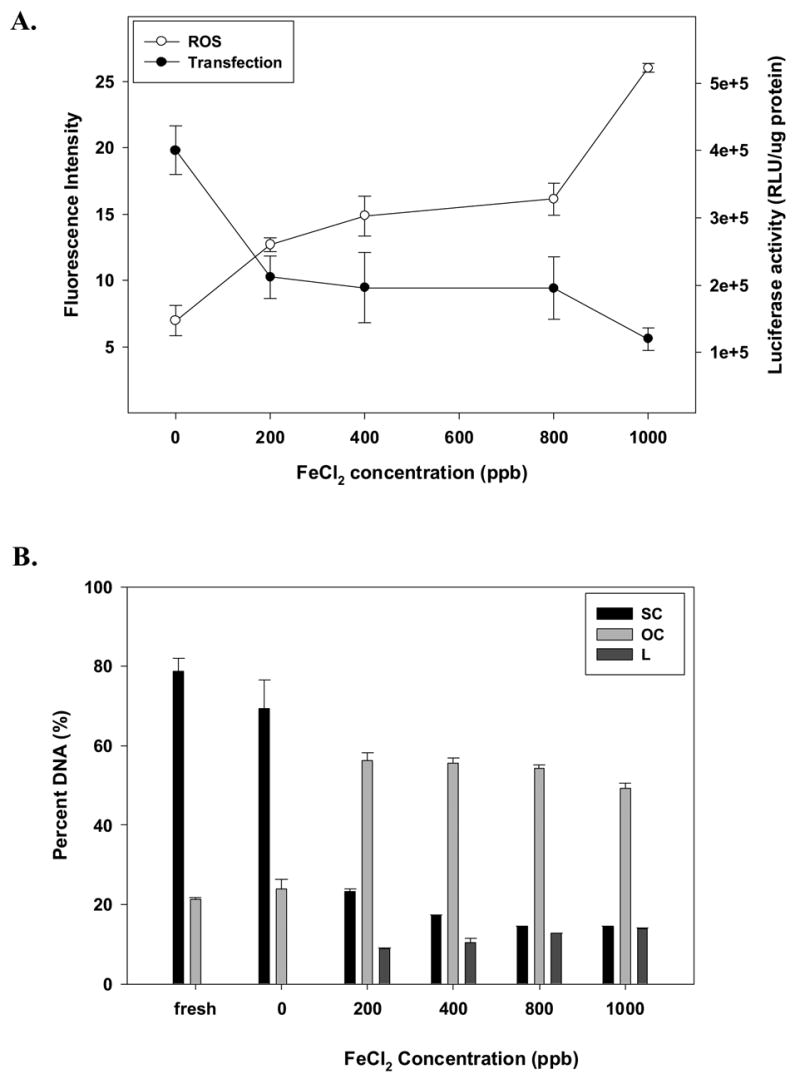
Effect of increasing amounts of Fe2+ on the stability of DOTAP:DOPE/DNA complexes formulated in trehalose during acute lyophilization, in terms of (A) ROS content and transfection rates, or (B) DNA integrity. Percent of DNA is shown as supercoil content (SC), open circle (OC) and linear form (L) remaining in samples after acute lyophilization. The results are expressed as mean values ± 1 S.D. of measurements on triplicate samples.
Figure 5.
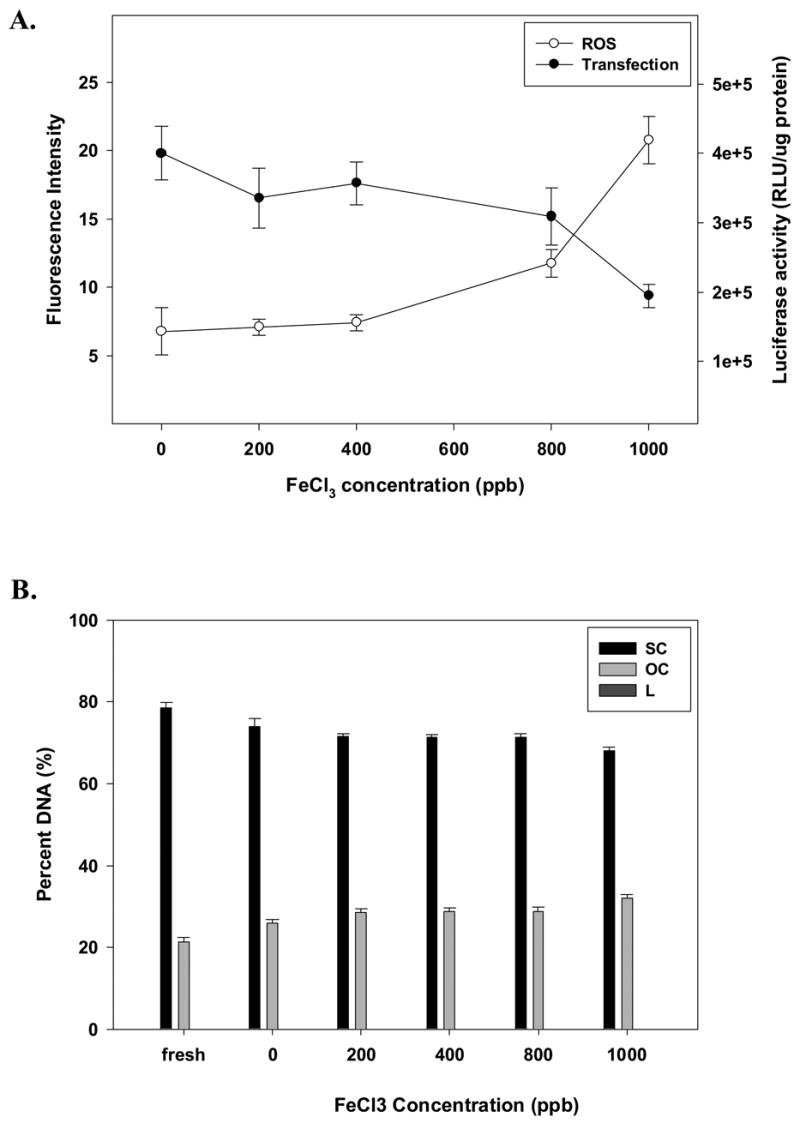
Effect of increasing amounts of Fe3+ on the stability of DOTAP:DOPE/DNA complexes formulated in trehalose during acute lyophilization, in terms of (A) ROS content and transfection rates, or (B) DNA integrity. Percent of DNA is shown as supercoil content (SC), open circle (OC) and linear form (L) remaining in samples after acute lyophilization. The results are expressed as mean values ± 1 S.D. of measurements on triplicate samples.
To further investigate the role of Fe2+-induced ROS generation during lyophilization, we examined the effect of the addition of four iron chelators in lyophilized lipid/DNA complexes containing 400 ppb of Fe2+. Our results show that the presence of DTPA, desferal and BPS attenuated the formation of ROS (Fig. 6). Attenuation of ROS production by DTPA and BPS were significant at only one concentration (200 μM) as compared to dried vectors containing metal alone (p<0.05). While desferal significantly inhibited ROS formation at all concentrations studied (p<0.01), the presence of EDTA (at all levels tested) significantly enhanced the generation of reactive oxygen species in the dried cake (p<0.01; Fig. 6). We also measured supercoil content to assess the effectiveness of chelating agents on DNA integrity (Fig. 7). Our data show maintenance of supercoil content in the presence of DTPA and BPS (Fig. 7A and C). However, particularly surprising was the observation that desferal promoted the formation of double strand breaks and the emergence of linear DNA despite its ability to attenuate ROS production (Figs. 6 and 7B). A similar lack of correlation of ROS levels with supercoil content was observed with EDTA where supercoil content was maintained despite high ROS levels (Figs. 6 and 7D). Furthermore, DTPA, BPS, and EDTA were capable of maintaining biological activity when added at sufficient concentrations (Fig. 8). In contrast, low concentrations of desferal preserved transfection rates, but reduced biological activity was observed at high desferal concentrations (≥ 400 μM).
Figure 6.
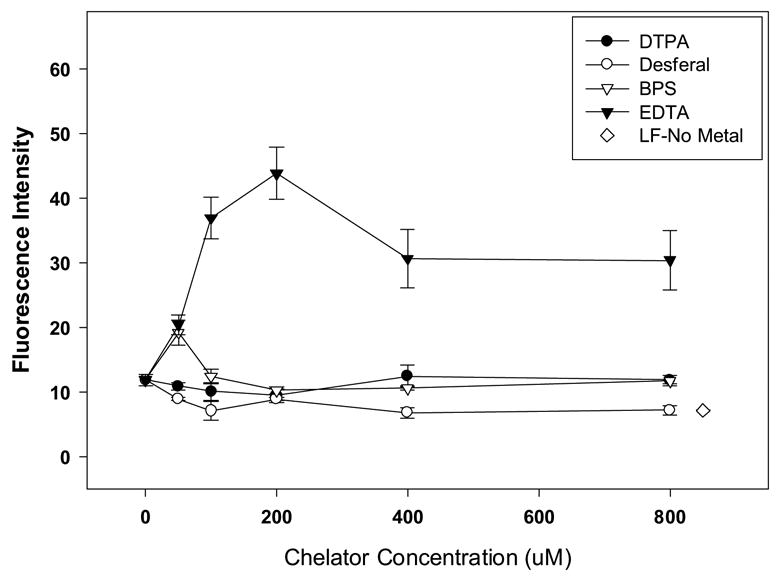
Effect of chelating agents on the ROS content of lyophilized vector formulations in the presence of 400 ppb Fe2+. The final concentrations of chelators were 0–800 μM. The results are expressed as mean values ± 1 S.D. of measurements on triplicate samples.
Figure 7.
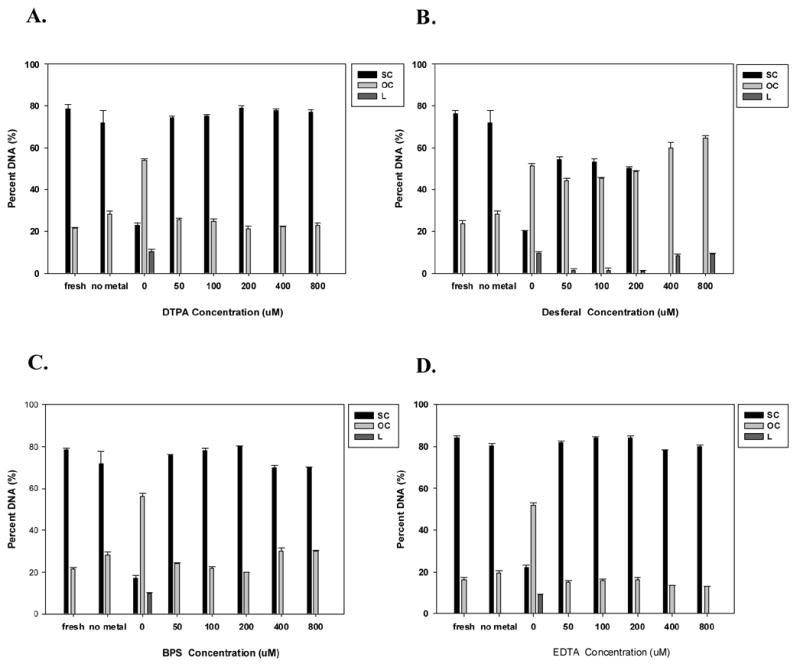
Effect of chelators on the DNA integrity of lyophilized vector formulations in the presence of 400 ppb Fe2+: (A) Diethylenetriaminepentaacetic acid (DTPA), (B) desferrioxamine (desferal), (C) bathophenanthroline disulfonate (BPS), and (D) ethylenediaminetetraacetic acid (EDTA). Percent of DNA is shown as supercoil content (SC), open circle (OC) and linear form (L) remaining in samples after acute lyophilization/rehydration. The results are expressed as mean values ± 1 S.D. of measurements on triplicate samples.
Figure 8.
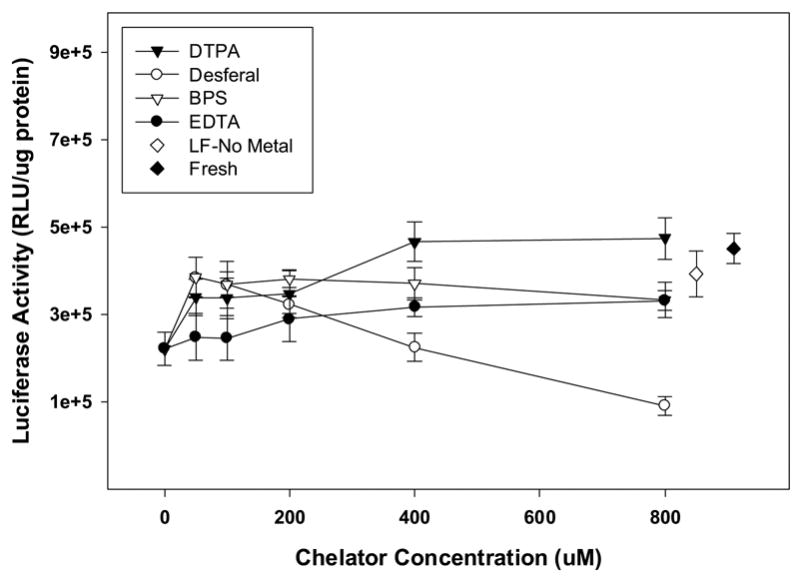
Effect of chelators on the in vitro transfection efficiency of lyophilized vector formulations in the presence of 400 ppb Fe2+. The results are expressed as mean values ± 1 S.D. of measurements on triplicate samples.
DISCUSSION
The potential for metal contaminants to reduce the stability of DNA-based therapeutics needs to be addressed before such formulations are employed as pharmaceutical products. However, most of the studies on oxidative damage to DNA have been performed in aqueous solution [7, 9, 10, 21]. Since proxyl fluorescamine has been successfully used to measure ROS (hydroxyl and superoxide radicals) in biological systems [23, 24], we first evaluated the applicability of this fluorescent probe to detect ROS generated in formulations fortified with metal ions. Our observations in solution demonstrate that this probe can be utilized to detect ROS formation induced by metal contaminants. Proxyl fluorescamine was then used as a probe to assess ROS generated during lyophilization. It is important to note that although this water-soluble probe does not allow identification of the specific oxygen radical, it provides valuable information on the relative levels of reactive oxygen species that are generated in our samples. Our results indicate that ROS are formed during the drying step of the lyophilization process. While the removal of water appears to facilitate radical formation, the presence of multivalent metal contaminants in DNA-based formulations is known to cause oxidation [7]. Moreover, sugars that are commonly employed for lyophilization can contain relatively high levels of metal contaminants (e.g., 20 ppm, unpublished observations). To gain a better understanding of the ability of metal ions to facilitate ROS formation and their subsequent effect on the stability of lipid/DNA complexes, we investigated the ability of three common metal contaminants to promote damage to nonviral vectors during lyophilization. The observed increase of ROS levels in formulations dried in the presence of increasing metal ion concentrations (Fe2+, Cu2+, Fe3+) strongly suggests that the generation of these species is promoted by the presence of trace amounts of metal ions. The results are consistent with the fact that transition metal ions are capable of binding to biomolecules and catalyzing free radical oxidation via the Fenton reaction [9, 14, 25]. Detection of ROS formation even in the absence of added metal ions may be due to metal contaminants in our samples. In fact, we have previously determined that even highly purified DNA used in clinical trials can possess ≈ 30–40 ppb iron (unpublished observations).
In good agreement with earlier studies, our results showed that the effect of Fe3+ was less damaging than its reduced form [7, 21]. This is likely explained by the fact that reduction of Fe3+ to Fe2+ is required for the formation of hydroxyl radicals via the Fenton reaction [9]. Similarly, cupric ions must also be reduced to cuprous ions (Cu+) in order to generate hydroxyl radicals via the Fenton reaction [14]. Thus, both Cu2+ and Fe3+ need to be reduced to exert their oxidative effect on DNA. In our hands, the addition of Cu2+ resulted in increased ROS levels, showing that this metal promotes ROS formation during lyophilization. The observation that the damaging effect of Cu2+ was more extensive than that produced by equimolar amounts of Fe3+ ions has been previously reported [26, 27]. As both multivalent ions possess high affinity for DNA [28–30], and similar binding affinities for guanine bases [28, 29, 31], one possible explanation for our observations may involve the greater efficiency of Cu2+, as compared to Fe3+, to produce hydroxyl radicals via redox cycling (k ≈ 5–8 x 109 M−1s−1 and 1.5x108 M−1s−1, respectively) [32].
Our results clearly demonstrate that trace amounts of transition metals (Cu2+, Fe2+) increase formation of single and double strand breaks, presumably through ROS production [11, 33, 34]. Our data also show that both the type of metal and its concentration influence the extent of DNA damage during acute lyophilization. We propose that the removal of water during lyophilization facilitates degradation by placing metal ions in close proximity to lipid/DNA complexes. We also observe that the metal-mediated strand breakage has a detrimental effect on biological activity (i.e., loss in transfection efficiency), which is consistent with a cause-effect relationship. This is in good agreement with other studies suggesting that metal-catalyzed Fenton reactions may play a prominent role in biological processes such as lipid-mediated transfection and inactivation in macrophages [35]. Furthermore, it is important to point out that the lipid moiety may also play a role in the metal-induced complex damage and the observed loss in biological activity. In fact, it has been reported that unsaturation renders lipids more prone to oxidation [14]. By analogy, the unsaturated lipids used in our studies (DOTAP and DOPE) can be susceptible to oxidation, and hence, the radicals generated from these oxidizing lipids (e.g., lipo-peroxyl radicals, lipid hydroperoxides) could react and ultimately damage nucleic acids compromising gene delivery efficiency [9, 14].
It is generally acknowledged that metal chelation is an effective strategy to prevent metal-mediated ROS generation in DNA-based formulations [5, 8]. To address the role of Fe2+-mediated degradation of lipid/DNA complexes during lyophilization, we examined the effect of chelating agents. As already noted, our data revealed that Fe2+ is the most damaging metal ion contaminant in the absence of chelating agents. When four different iron chelators were tested in the presence of an added excess of iron, only DTPA and BPS were able to reduce ROS levels, and maintain supercoil content and biological activity. These results may reflect the ability of DTPA to remove metal ions located in the proximity of DNA due to the fact that its affinity for iron (Log Ka of Fe2+ = 16) [36] is larger than the affinity of iron for double-stranded DNA (Log Ka for Fe2+ = 4.73) [29]. Although DTPA is considered a limited iron chelator [37, 38], it has also been reported that DTPA can alter the rate at which chelatable iron is reduced, thus inhibiting hydroxyl radical production from superoxide reaction with iron [13, 37, 39]. While there are no reports with information about the binding constants for the BPS-metal complex, it has been shown that BPS is a specific chelator for the ferrous iron, forming a redox-inactive complex that eventually prevents Fe2+-mediated hydroxyl radical formation [40, 41]. In contrast to DTPA and BPS, our findings showed that EDTA was not efficient in reducing the formation of reactive oxygen species. This is consistent with reports showing that the Fe2+/3+ –EDTA complex is a Fenton reaction active complex that can generate free and chelator:metal-bound hydroxyl (HO•) radicals [42, 43]. Surprisingly, we observed maintenance of supercoil content in the presence of EDTA despite higher levels of ROS. This apparent contradiction might be explained by the fact that steric and ionic repulsion between the EDTA-Fe complex and DNA minimizes their co-localization [44]. The effect of this physical separation on oxidative damage to DNA could be exacerbated by the reduced molecular motion within the glassy sugar matrix [45] which might further inhibit access of HO• radicals to the lipid/DNA complex [43].
In sharp contrast to the aforementioned chelators, our results showed that desferal, a well-characterized HO• radical scavenger [46], attenuated ROS formation but did not confer enhanced stability to lipid/DNA complexes during lyophilization, as indicated by the observed loss in DNA integrity and biological activity. We suggest four possible factors that may contribute to these observations. First, it has been suggested that desferal can be used to inhibit only reactions that are dependent on Fe3+ reduction (e.g., the Haber-Weiss cycle) but not in those which depend on Fe2+ oxidation (Fenton reaction) [47]. Second, although it is generally accepted that Fe3+-desferal is resistant to reduction [48], it has also been demonstrated that a Fe3+-desferal complex has the potential to autoreduce its chelated iron to the ferrous state, and thereby generate ROS [49]. Third, it is possible that other radicals that are not detected by proxyl fluorescamine might contribute to the observed degradation (e.g., ferryl or perferryl radicals [22, 43]). Fourth, access of the Fe2+-desferal chelate to the lipid/DNA complex may be facilitated by its greater lipid solubility (as compared to DTPA- or EDTA- chelates), thereby providing greater access to the DNA in lipid/DNA complexes [50]. According to this scenario, a water-soluble indicator of ROS levels (e.g., proxyl fluorescamine) may not accurately measure the specific oxidative environment of DNA complexed with lipids, resulting in a poor correlation of measured ROS levels with DNA damage. Further studies are needed to identify the relative contributions of each of these potential factors.
It is worth noting that although we used the cleanest sugar available for our studies (copper and iron levels below detection limits of 2 ppb and 42 ppb, respectively), it is difficult to completely eliminate metal contamination [7]. While the removal of metal contaminants may be achievable at laboratory scale [7, 11, 12, 51], it has been suggested that demetalation is impracticable for large-scale purification and production [52]. Nonetheless, the present study indicates that in the absence of added metal, lipid/DNA complexes do not degrade significantly during acute lyophilization when very pure excipients are used, however even very low (e.g., undetectable) metal levels may induce damage during prolonged storage [53]. Therefore, demetalation may be a potentially useful tool to prevent Fenton reaction-mediated vector degradation during prolonged storage.
In conclusion, we have observed that trace amounts of metal ions significantly affected the stability of lipid/DNA complexes during acute lyophilization. Our results demonstrate that contaminating ferrous and cupric ions induce damage, and that their detrimental effect is concentration-dependent. We have also shown that DTPA and BPS can inhibit iron-induced damage by reducing ROS formation and preserving DNA integrity and biological activity during lyophilization. Future studies should focus on the development of effective strategies (e.g., chelator and/or antioxidant) to prevent generation of ROS during acute lyophilization and storage. Furthermore, identification of the specific chemical mechanism responsible for ROS generation will be a critical step in developing dried nonviral vector formulations that are stable during prolonged storage.
Acknowledgments
We thank Valentis Inc. for providing plasmid DNA encoding the luciferase gene. We also acknowledge Ferro-Pfanstiehl Laboratories for providing highly purified trehalose. This work was supported by Ferro-Pfanstiehl and the National Institutes of Health through NIBIB grant #1 RO1 EB005476-01. The authors thank Dr. Pablo Castello, Department of Pharmaceutical Sciences, School of Pharmacy at the University of Colorado, for his suggestions and helpful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Allison SD, Anchordoquy TJ. Mechanisms of protection of cationic lipid-DNA complexes during lyophilization. J Pharm Sci. 2000;89:682–691. doi: 10.1002/(SICI)1520-6017(200005)89:5<682::AID-JPS14>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 2.Anchordoquy TJ, Carpenter JF, Kroll DJ. Maintenance of transfection rates and physical characterization of lipid/DNA complexes after freeze-drying and rehydration. Arch Biochem Biophys. 1997;348:199–206. doi: 10.1006/abbi.1997.0385. [DOI] [PubMed] [Google Scholar]
- 3.Cherng J-Y, Wetering Pvd, Talsma H, Crommelin DJA, Hennink WE. Stabilization of polymer-based gene delivery systems. Int J Pharm. 1999;183:25–28. doi: 10.1016/s0378-5173(99)00037-x. [DOI] [PubMed] [Google Scholar]
- 4.Talsma H, Cherng JY, Lehrmann H, Kursa M, Ogris W, Hennink WE, Cotten M, Wagner E. Stabilization of gene delivery systems by freeze-drying. Int J Pharm. 1997;157:233–238. doi: 10.1016/s0378-5173(97)00244-5. [DOI] [PubMed] [Google Scholar]
- 5.Middaugh CR, Evans RK, Montgomery DL, Casimiro DR. Analysis of plasmid DNA from a pharmaceutical perspective. J Pharm Sci. 1998;87:130–146. doi: 10.1021/js970367a. [DOI] [PubMed] [Google Scholar]
- 6.Pogocki D, Schoneich C. Chemical stability of nucleic acid-derived drugs. J Pharm Sci. 2000;89:443–456. doi: 10.1002/(SICI)1520-6017(200004)89:4<443::AID-JPS2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 7.Evans RK, Xu Z, Bohannon KE, Wang B, Brunner MW, Volkin DB. Evaluation of degradation pathways for plasmid DNA in pharmaceutical formulations via accelerated stability studies. J Pharm Sci. 2000;89:76–87. doi: 10.1002/(SICI)1520-6017(200001)89:1<76::AID-JPS8>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 8.Waterman KC, Adami RC, Alsante KM, Hong J, Landis MS, Lombardo F, Roberts CJ. Stabilization of pharmaceuticals to oxidative degradation. Pharm Dev Technol. 2002;7:1–32. doi: 10.1081/pdt-120002237. [DOI] [PubMed] [Google Scholar]
- 9.Breen AP, Murphy JA. Reactions of oxyl radicals with DNA. Free Radic Biol Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 10.Imlay JA, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 11.Kukielka E, Cederbaum AI. DNA strand cleavage as a sensitive assay for the production of hydroxyl radicals by microsomes: role of cytochrome P4502E1 in the increased activity after ethanol treatment. Biochem J. 1994;302:773–779. doi: 10.1042/bj3020773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marrot L, Giacomoni PU. Enhancement of oxidative DNA degradation by histidine: the role of stereochemical parameters. Mut Res. 1992;275:69–79. doi: 10.1016/0921-8734(92)90010-m. [DOI] [PubMed] [Google Scholar]
- 13.Buettner GR, Jurkiewicz BA. Catalytic metals, ascorbate and free radicals: combinations to avoid. Radiat Res. 1996;145:532–541. [PubMed] [Google Scholar]
- 14.Valko M, Morris H, Cronin MTD. Metals, toxicity and oxydative stress. Curr Med Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 15.Adami RC, Collard WT, Gupta SA, Kwok KY, Bonadio J, Rice KG. Stability of peptide-condensed plasmid DNA formulations. J Pharm Sci. 1998;87:678–683. doi: 10.1021/js9800477. [DOI] [PubMed] [Google Scholar]
- 16.Molina MdC, Allison DS, Anchordoquy TJ. Maintenance of particle size during the freezing step of the lyophilization process is insufficient for preservation of activity: insight from other structural indicators. J Pharm Sci. 2001;90:1445–1455. doi: 10.1002/jps.1096. [DOI] [PubMed] [Google Scholar]
- 17.Anchordoquy TJ, Girouard LG, Carpenter JF, Kroll DJ. Stability of lipid/DNA complexes during agitation and freeze-thawing. J Pharm Sci. 1998;87:1046–1051. doi: 10.1021/js9801891. [DOI] [PubMed] [Google Scholar]
- 18.Allison SD, Molina MdC, Anchordoquy TJ. Stabilization of lipid/DNA complexes during the freezing step of the lyophilization process: The particle isolation hypothesis. Biochim Biophys Acta. 2000;1468:127–138. doi: 10.1016/s0005-2736(00)00251-0. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Anchordoquy TJ. The role of lipid charge density in the serum stability of cationic lipid/DNA complexes. Biochim Biophys Acta. 2004;1663:143–157. doi: 10.1016/j.bbamem.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Lloyd RS, Haidle CW, Robberson DL. Bleomycin-specific fragmentation of double-stranded DNA. Biochemistry. 1978;17:1890–1896. doi: 10.1021/bi00603a014. [DOI] [PubMed] [Google Scholar]
- 21.Urbanski NK, Beresewicz A. Generation of OH initiated by interaction of Fe+2 and Cu+ with dioxygen; comparison with the Fenton chemistry. Acta Biochimica Polonica. 2000;47:951–962. [PubMed] [Google Scholar]
- 22.Qian SY, Buettner GR. Iron and dioxygen chemistry is an important route to initiation of biological free radical oxidations: an electron paramagnetic resonance spin trapping study. Free Radic Biol Med. 1999;26:1447–1456. doi: 10.1016/s0891-5849(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 23.Pou S, Huang YI, Bhan A, Bhadti VS, Hosmane RS, Wu SY, Cao GL, Rosen GM. A fluorophore-containing nitroxide as a probe to detect superoxide and hydroxyl radical generated by stimulated neutrofils. Anal Biochem. 1993;212:85–90. doi: 10.1006/abio.1993.1295. [DOI] [PubMed] [Google Scholar]
- 24.Li B, Gutierrez PL, Blough NV. Trace determination of hydroxyl radical in biological systems. Anal Chem. 1997;69:4295–4302. doi: 10.1021/ac970622b. [DOI] [PubMed] [Google Scholar]
- 25.Henle ES, Luo Y, Linn S. Fe+2, Fe+3, and oxygen react with DNA-derived radicals formed during iron-mediated Fenton reaction. Biochemistry. 1996;35:12212–12219. doi: 10.1021/bi961235j. [DOI] [PubMed] [Google Scholar]
- 26.Aruoma OI, Halliwell B, Gajewski E, Dizdaroglu M. Copper-ion-dependent damage to the bases in DNA in the presence of hydrogen peroxide. Biochem J. 1991;273:601–604. doi: 10.1042/bj2730601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoewe R, Prutz WA. Copper-catalyzed DNA damage by ascorbate and hydrogen peroxide: kinetics and yield. Free Radic Biol Med. 1987;3:97–105. doi: 10.1016/s0891-5849(87)80003-5. [DOI] [PubMed] [Google Scholar]
- 28.Izatt RM, Christensen JJ, Rytting JH. Sites and thermodynamic quantities associated with proton and metal ion interaction with ribonucleic acid, deoxyribonucleic acid, and their constituent bases, nucleosides, and nucleotides. Chem Rev. 1971;71:439–481. doi: 10.1021/cr60273a002. [DOI] [PubMed] [Google Scholar]
- 29.Ouameur AA, Arakawa H, Ahmad R, Naoui M, Tajmir-Riahi HA. A comparative study of Fe(II) and Fe(III) interactions with DNA duplex: major and minor grooves bindings. DNA Cell Biol. 2005;24:394–401. doi: 10.1089/dna.2005.24.394. [DOI] [PubMed] [Google Scholar]
- 30.Pezzano H, Podo F. Structure of binary complexes of mono- and polynucleotides with metal ions of the first transition group. Chem Rev. 1980;80:366–401. [Google Scholar]
- 31.Sagripanti JL, Kraemer KH. Site-specific oxidative DNA damage at polyguanosines produced by copper plus hydrogen peroxide. J Biol Chem. 1989;264:1729–1734. [PubMed] [Google Scholar]
- 32.Halliwell B, Gutteridge JMC. Free radicals in biology and medicine. 3. Oxford University Press; Oxford: 1999. [Google Scholar]
- 33.Pedreno E, Lopez-Contreras AJ, Cremades A, Penafiel R. Protecting or promoting effects of spermine on DNA strand breakage induced by iron or copper ions as a function of metal concentration. J Inorg Biochem. 2005;99:2074–2080. doi: 10.1016/j.jinorgbio.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 34.Srinivasan A, Lehmler HJ, Robertson LW, Ludewig G. Production of DNA strand breaks in vitro and reactive oxygen species in vitro and HL-60 cells by PCB metabolites. Toxicol Sci. 2001;60:92–102. doi: 10.1093/toxsci/60.1.92. [DOI] [PubMed] [Google Scholar]
- 35.Dokka S, Toledo D, Shi X, Castranova V, Rojanasakul Y. Oxygen radical-mediated pulmonary toxicity induced by some cationic liposomes. Pharm Res. 2000;17:521–525. doi: 10.1023/a:1007504613351. [DOI] [PubMed] [Google Scholar]
- 36.Graf E, Mahoney JR, Bryant RG, Eaton JW. Iron-catalyzed hydroxyl radical formation. Stringent requirement for free iron coordination site. J Biol Chem. 1984;259:3620–3624. [PubMed] [Google Scholar]
- 37.Buettner GR, Doherty TP, Patterson LK. The kinetics of the reaction of superoxide radical with Fe(III) complexes of EDTA, DETAPAC, and HEDTA. FEBS Lett. 1983;158:143–146. doi: 10.1016/0014-5793(83)80695-4. [DOI] [PubMed] [Google Scholar]
- 38.Miller DM, Buettner GR, Aust SD. Transition metals as catalysts of “autoxidation” reactions. Free Radic Biol Med. 1990;8:95–108. doi: 10.1016/0891-5849(90)90148-c. [DOI] [PubMed] [Google Scholar]
- 39.Buettner GR, Oberley LW, Leuthauser SWHC. The effect of iron on the distribution of superoxide and hydroxyl radicals as seen by spin trapping and on the superoxide dismutase assay. Photochem Photobiol. 1978;28:693–695. doi: 10.1111/j.1751-1097.1978.tb07001.x. [DOI] [PubMed] [Google Scholar]
- 40.Dean RT, Nicholson P. The action of nine chelators on iron-dependent radical damage. Free Radic Res. 1994;20:83–101. doi: 10.3109/10715769409147506. [DOI] [PubMed] [Google Scholar]
- 41.Zhu BZ, Kitrossky N, Chevion M. Evidence for production of hydroxyl radicals by pentachlorophenol metabolites and hydrogen peroxide: A metal-independent organic Fenton reaction. Biochem Biophys Res Commun. 2000;270:942–946. doi: 10.1006/bbrc.2000.2539. [DOI] [PubMed] [Google Scholar]
- 42.Engelmann MD, Bobier RT, Hiatt T, Cheng IF. Variability of the Fenton reaction characteristics of the EDTA, DTPA, and citrate complexes of iron. Biometals. 2003;16:519–527. doi: 10.1023/a:1023480617038. [DOI] [PubMed] [Google Scholar]
- 43.Yamazaki I, Piette LH. EPR spin-traping study on the oxidizing species formed in the reaction of the ferrous ion with hydrogen peroxide. J Am Chem Soc. 1991;113:7588–7593. [Google Scholar]
- 44.Lloyd DR, Phillips DH. Oxidative DNA damage mediated by copper(II), iron(II) and nickel(II) Fenton reactions: evidence for site-specific mechanisms in theformation of double-strands breaks, 8-hydroxydeoxyguanosine and putative intrastrand cross-links. Mutat Res. 1999;424:23–36. doi: 10.1016/s0027-5107(99)00005-6. [DOI] [PubMed] [Google Scholar]
- 45.Hancock BC, Zografi G. Characteristics and significance of the amorphous state in pharmaceutical systems. J Pharm Sci. 1997;86:1–12. doi: 10.1021/js9601896. [DOI] [PubMed] [Google Scholar]
- 46.Halliwell B. Use of desferrioxamine as a ‘probe’ for iron-dependent formation of hydroxyl radicals. Evidence for a direct reaction between desferal and the superoxide radical. Biochem Pharmacol. 1985;34:229–233. doi: 10.1016/0006-2952(85)90129-7. [DOI] [PubMed] [Google Scholar]
- 47.Burkitt MJ, Mason RP. Direct evidence for in vivo hydroxyl-radical generation in experimental iron overload: an ESR spin-trapping investigation. Proc Natl Acad Sci USA. 1991;88:8440–8444. doi: 10.1073/pnas.88.19.8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halliwell B. Protection against tissue damage in vivo by desferrioxamine: what is its mechanism of action? Free Radic Biol Med. 1989;7:645–651. doi: 10.1016/0891-5849(89)90145-7. [DOI] [PubMed] [Google Scholar]
- 49.Gutteridge JMC, Quinlan GJ, Swain J, Cox J. Ferrous ion formation by ferrioxamine prepared from aged desferrioxamine: a potential prooxidant property. Free Radic Biol Med. 1994;16:733–739. doi: 10.1016/0891-5849(94)90188-0. [DOI] [PubMed] [Google Scholar]
- 50.Schaich KM, Borg DC. Fenton reactions in lipid phases. Lipids. 1988;23:570–579. doi: 10.1007/BF02535600. [DOI] [PubMed] [Google Scholar]
- 51.Toyokuni S, Sagripanti JL. Association between 8-hydroxy-2′-deoxyguanosine formation and DNA strand breaks mediated by copper and iron. Free Radic Biol Med. 1996;20:859–864. doi: 10.1016/0891-5849(95)02184-1. [DOI] [PubMed] [Google Scholar]
- 52.Anchordoquy TJ, Armstrong TK, Molina MC, Allison DS, Zhang Y, Patel MM, Lentz YK, Koe GS. Formulation considerations for DNA-based therapeutics. In: Lu DR, Oie S, editors. Cellular drug delivery: principles and practice. Humana Press; New Jersey: 2004. pp. 237–264. [Google Scholar]
- 53.Molina MC, Armstrong TK, Zhang Y, Patel MM, Lentz YK, Anchordoquy TJ. The stability of lyophilized lipid/DNA complexes during prolonged storage. J Pharm Sci. 2004;93:2259–2273. doi: 10.1002/jps.20138. [DOI] [PubMed] [Google Scholar]


