During meiosis, recombination between homologous chromosomes creates a physical connection that is necessary for proper chromosome segregation at the first meiotic division. For many years, the paradigm used to explain the molecular mechanism of recombination has been the double-strand break repair (DSBR) model (Fig. 1; Szostak et al. 1983). (The definitions of the acronyms used in this review, in order of their appearance, can be found in Table 1.) One key feature of this model is that recombination is initiated by the formation of programmed DSBs (double-strand breaks). The meiosis-specific protein responsible for catalyzing these breaks is a topoisomerase-like protein called Spo11 (Bergerat et al. 1997; Keeney et al. 1997). The identification and functional analysis of Spo11 homologs from a wide variety of organisms, including worms, fruit flies, fission yeast, and mammals, indicates that repair of programmed DSBs is a universal feature of meiotic recombination (Keeney 2001). A second key feature of the model is the generation of a recombination intermediate called a double Holliday junction (dHJ). Differential resolution of this intermediate was proposed to determine the formation of crossover (CO) versus noncrossover (NCO) chromosomes. Recent advances indicate, however, that the decision of whether a recombination event will result in a CO or NCO chromosome occurs much earlier, soon after DSB formation (Allers and Lichten 2001a; Hunter and Kleckner 2001; Clyne et al. 2003). In addition, studies of a newly discovered endonuclease, Mus81, indicate that, in addition to dHJ resolution, COs may be formed by the processing of non-dHJ intermediates (Heyer et al. 2003; Osman et al. 2003). The decision of which crossover pathway to use appears to vary between organisms and may, therefore, be evolutionarily significant (de los Santos et al. 2003).
Figure 1.
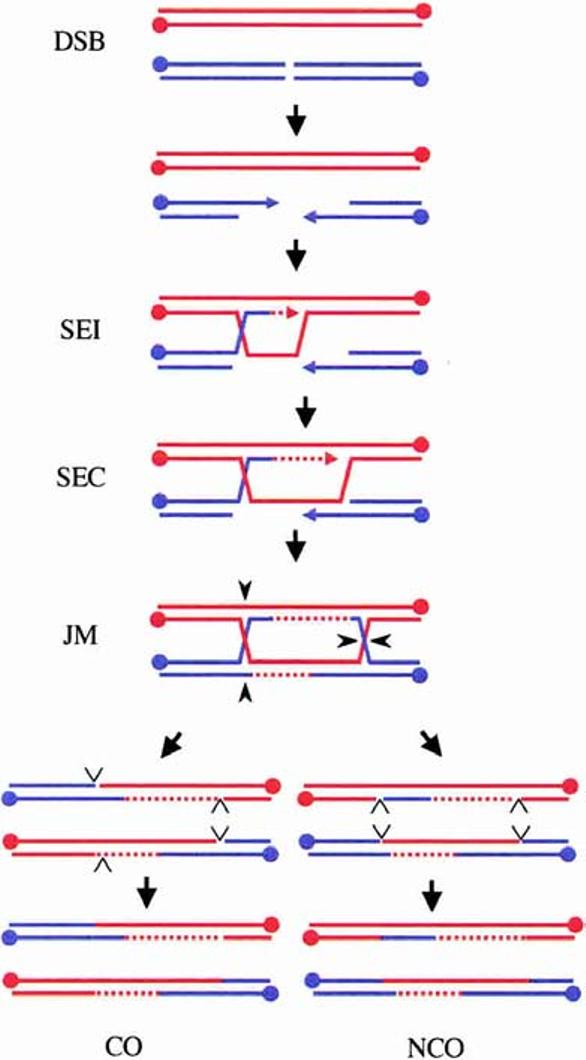
The DSBR model in meiosis. Shown are the duplex DNA strands of two non-sister chromatids from a pair of homologous chromosomes (Homolog 1 is red; Homolog 2 is blue). Meiotic DSBs are repaired following 5′-end resection and the formation of an SEI intermediate. Elongation of the invading strand extends the D-loop until SEC initiates a second round of DNA synthesis. Ligation of the free ends creates a joint molecule containing dHJs. Resolution of the dHJs in an opposite sense (e.g., black carets shown here) leads to the formation of a CO, whereas cleavage in the same sense (e.g., cleavage of the crossed strands) results in an NCO. Resolution consists of two steps: symmetric cleavage to produce duplex DNA with nicks (open carets) and ligation of the nicks. 5′-Ends of the DNA strands are represented by dots, 3′-ends are represented by arrows, and DNA synthesis is indicated by dashed lines.
Table 1.
Definition of acronyms in order of appearance
| Acronym | Definition |
|---|---|
| DSBR | Double-strand break repair |
| DSB | Double-strand break |
| dHJ | Double Holliday junction |
| CO | Crossover |
| NCO | Noncrossover |
| SEI | Single end invasion |
| SEC | Second end capture |
| HJ | Holliday junction |
| SDSA | Synthesis-dependent strand annealing |
| MMS | Methylmethane sulfonate |
| CPT | Camptothecin |
| pRFs | Pseudoreplication forks |
| RLS | Regressed lagging strand |
The double-strand break repair model for meiotic recombination
A modified version of the Szostak et al. (1983) DSBR model is shown in Figure 1. Many of the recombination intermediates predicted by this model have been directly observed in budding yeast, and the gene products necessary for processing of different recombination intermediates have been identified (Smith and Nicolas 1998). In addition to SPO11, a large number of additional genes, whose roles are not yet understood, are required for DSB formation (Keeney 2001). The 5′-ends on either side of the break are resected to produce single-stranded (ss) 3′-tails. Resection requires a trimeric complex of proteins containing Rad50, Xrs2, and Mre11 as well as Sae2/Com1 (Smith and Nicolas 1998). One of the 3′-tails invades the duplex of a homologous chromatid, thereby producing a D-loop or single end invasion (SEI) intermediate (Hunter and Kleckner 2001). Invasion of the homologous chromosome is promoted by two recA homologs, Rad51 and the meiosis-specific protein, Dmc1, as well as the presence of a barrier to sister-chromatid repair mediated by the meiosis-specific kinase Mek1 (Bishop et al. 1992; Wan et al. 2004). DNA synthesis extends the invading strand until the displaced strand in the D-loop overlaps sufficiently to anneal with the 3′-tail on the other side of the break to create a second end capture (SEC) intermediate. DNA synthesis and ligation create an intact dHJ-containing intermediate (Schwacha and Kleckner 1995). Resolution of both HJs in the same direction creates an NCO chromosome, whereas resolution of the junctions in opposite directions creates a CO chromosome. HJ resolvases from prokaryotes, such as RuvC and RusA, have been shown to make symmetrical nicks in strands of like polarity of HJs resulting in nicked duplex products (West 1997). Simple ligation of these nicked products restores intact duplexes. This version of the DSBR model assumes that eukaryotic HJ resolvases resolve HJs by a similar mechanism.
Recent data from the Lichten and Kleckner labs have challenged the idea that both COs and NCO chromosomes are derived from dHJs. For example, mutant conditions exist in which HJs are unresolved and COs are greatly reduced. Nevertheless, the formation of NCO chromosomes is unaffected (Allers and Lichten 2001a; Clyne et al. 2003). These results support the argument that repair of DSBs can occur by two separate pathways during budding yeast meiosis—a CO pathway that involves the formation of fully mature dHJ intermediates and an NCO pathway that does not (Fig. 2). NCOs are proposed to arise by displacement of the extended invading strand followed by reannealing to the 3′-tail on the other side of the break (synthesis-dependent strand annealing or SDSA; for review, see Paques and Haber 1999). Kinetic studies of the formation of different recombination intermediates during meiosis in wild-type cells support this view (Allers and Lichten 2001a; Hunter and Kleckner 2001).
Figure 2.
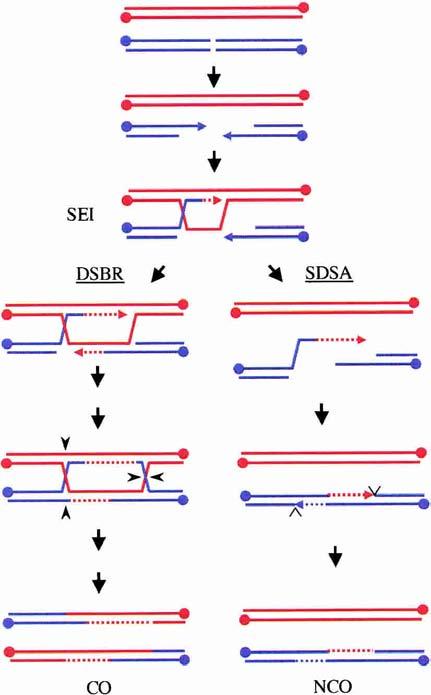
An alternative model for generating NCOs in budding yeast meiosis. Evidence at present suggests that SEI intermediates become committed to forming COs if they are repaired via the DSBR pathway, presumably through a biased resolution of dHJs in the direction of COs (left pathway). The SEI intermediate may be disrupted, however, to allow repair to proceed via the SDSA pathway, resulting in NCO products (right pathway).
The Mus81* endonuclease functions in DNA metabolism in mitotic and meiotic cells
Although many of the proteins necessary for mediating different steps of the DSBR pathway have been discovered, what has long remained elusive is the identification of a eukaryotic HJ resolvase. In the past few years, there has been much debate as to whether the Mus81 endonuclease may represent this Holy Grail of recombination (Haber and Heyer 2001; Heyer et al. 2003). The Mus81 protein shares homology with the XPF/Rad1 family of proteins involved in DNA nucleotide excision repair. Mus81 acts as a heterodimer with a second protein called Mms4 in the budding yeast, Saccharomyces cerevisiae, and Eme1 in the fission yeast, Schizosaccharomyces pombe (Boddy et al. 2001; Kaliraman et al. 2001). The human partner protein for Mus81 has been named both Mms4 and Eme1 (Ciccia et al. 2003; Ogrunc and Sancar 2003). For simplicity's sake, in this review Mus81/Mms4 and Mus81/Eme1 complexes will be indicated by Mus81*.
The MUS81/MMS4/EME1 genes were identified independently by several different labs, and mus81/mms4/eme1 mutant phenotypes are consistent with a role in DNA repair (Haber and Heyer 2001). In mitotic cells, mus81/mms4 mutants exhibit sensitivity to DNA damaging agents such as methylmethane sulfonate (MMS) and camptothecin (CPT), which are believed to stall replication forks and generate DSBs as a result of problems with DNA replication (Prakash and Prakash 1977; Interthal and Heyer 2000; Doe et al. 2002; Bastin-Shanower et al. 2003). Mus81* is not generally required for DSB repair in vegetative cells, however, as mus81/mms4 mutants are resistant to DSB-producing agents such as ionizing radiation and bleomycin (Prakash and Prakash 1977; Xiao et al. 1998; Boddy et al. 2000; Interthal and Heyer 2000). Mus81 physically interacts with the budding yeast recombination protein Rad54 and with the fission yeast DNA damage checkpoint kinase Cds1 (Boddy et al. 2000; Interthal and Heyer 2000). Mus81 is a nuclear protein and, in human cells, is recruited to sites of UV-induced damage specifically in S phase (Fu and Xiao 2003; Gao et al. 2003). These observations are consistent with the idea that Mus81* plays a role in rescuing stalled replication forks. In addition, mus81 and mms4 are synthetically lethal with mutants in SGS1 (RQH1 in fission yeast), a conserved 3′–5′ DNA helicase known to be required for genome stability (Boddy et al. 2000; Mullen et al. 2001). This lethality can be rescued by preventing homologous recombination, suggesting that Mus81* acts on some type of recombination intermediate in mitotic cells (Fabre et al. 2002; Bastin-Shanower et al. 2003).
In both fission and budding yeast, mus81/mms4/eme1 mutants have severe meiotic phenotypes. In fission yeast, spore viability is reduced to ≤1%, with high levels of chromosome missegregation (Boddy et al. 2001). In budding yeast, depending on temperature and strain background, up to 90%–100% of mus81/mms4 diploids arrest in prophase as a result of unprocessed recombination intermediates triggering the meiotic recombination checkpoint (de los Santos et al. 2001, 2003). In those rare tetrads that are formed, spore viability is reduced to ∼40% (Interthal and Heyer 2000; de los Santos et al. 2003). In both yeasts, these phenotypes are caused by a failure to repair meiosis-specific DSBs, as defects in meiotic progression and spore viability are rescued by mutations that prevent initiation of recombination (Boddy et al. 2001; de los Santos et al. 2001; Kaliraman et al. 2001). Clearly, Mus81* plays an essential role in meiotic recombination in both S. pombe and S. cerevisiae. The critical question then has been, what is the mechanism of its action?
Model I: Mus81* cleaves intact HJs
Mus81* was proposed to act at the HJ resolution step of the DSBR pathway, in part because partially purified Mus81* from fission yeast and human cells was observed to cleave intact HJs in vitro (Fig. 3D; Boddy et al. 2001; Chen et al. 2001). In addition, meiotic COs are reduced by mus81 in both fission and budding yeast (al-though to different extents), whereas the frequency of NCOs is unaffected or even elevated (de los Santos et al. 2003; Osman et al. 2003; Smith et al. 2003). This is the expected result for a resolvase mutant assuming that dHJs are the precursors of COs and that NCOs form via a non-dHJ mechanism such as SDSA. It is important to note, however, that although there is direct support for this idea in budding yeast, there are no physical data demonstrating that dHJs are the precursors to COs in fission yeast. The most compelling evidence in support of the idea that Mus81 is an HJ resolvase is the fact that many of the mus81/mms4/eme1 mutant phenotypes, including meiotic chromosome missegregation and spore inviability in fission yeast, sensitivity to DNA damaging agents, and synthetic lethality with rqh1− are efficiently suppressed by overexpression of rusA, a bacterial resolvase with high specificity for HJs (Boddy et al. 2001; Doe et al. 2002; Odagiri et al. 2003). This argument assumes that rusA is directly substituting for the activity of Mus81*. However, the suppression could be indirect. For example, overexpression of rusA also partially suppresses the defects exhibited by rqh1− cells that lack a 3′-to-5′ helicase, not an endonuclease (Doe et al. 2000).
Figure 3.
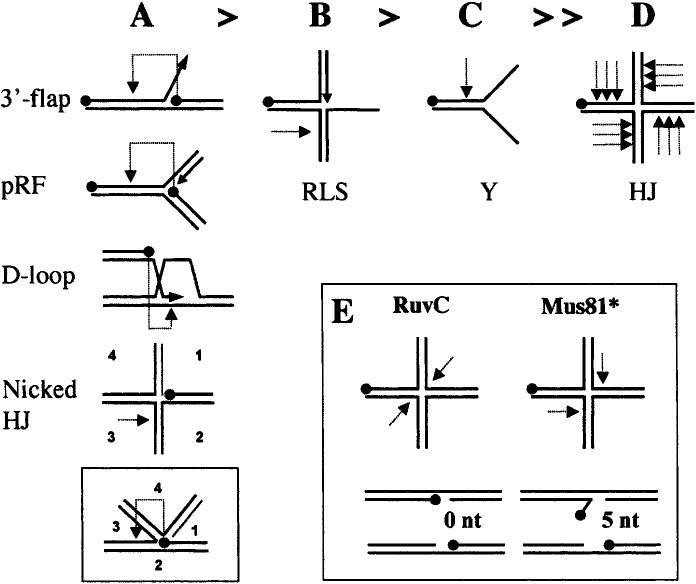
Substrate-specificity of Mus81* in vitro. (A–D) Summary of results from a variety of cleavage studies using rMus81* with substrates presented in order of cleavage efficiency. The most efficient substrates (A) are cleaved well owing to the presence of a 5′-end at their branch junctions (indicated by a black dot). Cleavage of these substrates (light dotted arrow) is directed one-half helical turn upstream of this 5′-end (pRF, pseudoreplication fork). (A, inset) This mechanism is consistent with the cleavage of nicked HJs in the strand opposing the nick. Efficient cleavage of the remaining substrates (B–D) requires an excess of enzyme over substrate. A regressed lagging-strand substrate (RLS, panel B) is cleaved in the opposing strand, while a Y substrate (C) is cleaved upstream of the branch junction. A further increase in enzyme concentration results in nicking of mobile HJs at multiple sites on the 5′-side of the mobile core. (E) Contrast of the HJ cleavage products obtained by symmetrical cleavage with RuvC (nicked duplexes) to the unligatable flap and gap products produced by Mus81*. These 5′-flap products may isomerize into 3′-flaps that are further processed by Mus81* to produce only gapped molecules.
The idea that Mus81* cleaves intact HJs was complicated by the discovery that recombinant budding yeast, fission yeast, and human Mus81* produced in Escherichia coli (designated rMus81*) have a significant preference for 3′-flap structures over intact HJs (Fig. 3, cf. A and D; Kaliraman et al. 2001; Doe et al. 2002; Ciccia et al. 2003). The primary determinant for optimal rMus81* activity in vitro appears to be the presence of a nick in the substrate. Mapping studies indicate that the free 5′-end directs cleavage to the upstream strand one-half helical turn 5′ of the nick (Bastin-Shanower et al. 2003; Osman et al. 2003). This rather nonspecific requirement results in efficient cleavage of several branched DNA substrates including 3′-flaps, pseudoreplication forks (pRFs), D-loops, and nicked HJs (Fig. 3A; Kaliraman et al. 2001; Doe et al. 2002; Gaillard et al. 2003; Osman et al. 2003; Whitby et al. 2003). When assayed in the presence of a molar excess of enzyme, however, a partial HJ substrate containing a single 3′-end at the junction, sometimes referred to as a regressed lagging-strand substrate (RLS; Fig. 3B), is efficiently cleaved on the same strand as a nicked HJ (Gaillard et al. 2003; Osman et al. 2003). In addition, a simple Y substrate (Fig. 3C), which is preferred by the related XPF–ERCC1 nuclease, is cleaved upstream of the branch point. Although rMus81* endonuclease activity on the RLS and Y substrates is suboptimal, their cleavage suggests that there may be additional determinants for rMus81* activity in vitro. All groups agree that intact HJs are very poor substrates for rMus81* (Fig. 3D).
In contrast to rMus81*, partially purified preparations of epitope-tagged Mus81* from S. pombe cell extracts (TEV-Mus81*) are active on intact HJs (Boddy et al. 2001; Gaillard et al. 2003). It has been suggested that the ability of TEV-Mus81* to cleave intact junctions is caused by the presence of a protein accessory factor or posttranslational modification (Gaillard et al. 2003). Mus81* partially purified from human cells, however, exhibits the same substrate specificity as the recombinant complexes, making it unlikely that the preference for 3′-flaps and pRFs over HJs exhibited by the bacterially produced enzymes is caused by a lack of a posttranslational modification (Constantinou et al. 2002). Based on the preference of Mus81* for nicked HJs, it has been proposed that Mus81* cleaves intact junctions by a “nick/counter-nick” mechanism (Gaillard et al. 2003). In this case, the first cut by Mus81* is slow and rate-limiting. This nick makes the junction more flexible and a better substrate for Mus81*, and the second cut is proposed therefore to be nearly simultaneous with the first. An alternative explanation for the differences observed between TEV-Mus81* and rMus81, however, could derive from impurities present in the crude TEV-Mus81* fraction. Resolving this paradox, therefore, will require comparison of rMus81/Eme1 from E. coli with highly purified Mus81* from S. pombe cells.
The HJ cleavage observed by Mus81* is nonsymmetrical and therefore leaves flaps and gaps on the HJs that cannot be repaired simply by ligation (Fig. 3E; Boddy et al. 2001; Constantinou et al. 2002). Therefore, if Mus81 resolves intact HJs, it does so by a non-RuvC/RusA-like mechanism. The idea that eukaryotic HJ resolution is mechanistically distinct from prokaryotic resolvases, while intriguing, is challenged by the discovery of a canonical RuvC-like activity in human cells. This activity, called Resolvase A, efficiently cuts intact HJs symmetrically to make ligatable, linear, nicked duplexes, is dependent on a branch migration activity, and is biochemically separable from Mus81* (Constantinou et al. 2002). Taken together, the biochemical data make it unlikely that the in vivo substrate of Mus81 is an intact HJ.
Further evidence that Mus81* is not required to cleave intact HJs comes from meiotic studies using budding yeast (de los Santos et al. 2001, 2003). In this organism, dHJs can be directly analyzed (Schwacha and Kleckner 1995). Whereas HJs would be predicted to accumulate in a resolvase mutant, mms4 exhibited a decrease in the frequency of dHJs during meiosis compared with wild type. In addition, the effect of mms4 on crossing over was, at most, twofold and appears to be dependent on chromosome size. Finally, overexpression of rusA had no effect on the meiotic defects of either mus81 or mms4 in budding yeast (de los Santos et al. 2003). Therefore Mus81* is not required to generate the bulk of meiotic COs in budding yeast. This fact, coupled with the observation that dHJs are the precursor to the majority of meiotic COs, provides compelling evidence that Mus81* is not resolving intact HJs in budding yeast.
Model II: Mus81* cleaves 3′-flaps generated by a partial strand displacement and annealing pathway of recombination
mus81/mms4 diploids are clearly generating unprocessed recombination intermediates during budding yeast meiosis, as evidenced by the prophase arrest triggered by the meiotic recombination checkpoint and the fact that significant numbers of unrepaired DSBs remain at late times in meiosis. Based on the in vitro preference of Mus81* for 3′-flaps, as well as the discovery of a class of joint molecules in which heteroduplex DNA is to one side of the dHJs (Fig. 4; Allers and Lichten 2001b), an alternative model was proposed for Mus81* function (de los Santos et al. 2001). In this model, a subset of dHJs was proposed to form by displacement of the 3′-end from the extended D-loop and its subsequent annealing to the other side of the break. If the newly synthesized DNA on this strand is longer than the resected 3′-tail on the other side of the break, a 3′-flap could result that would then be cleaved by Mus81* (Fig. 4). Similar models invoking 3′-flap cleavage by Mus81* in mitotic cells have also been proposed (Fabre et al. 2002; Bastin-Shanower et al. 2003).
Figure 4.
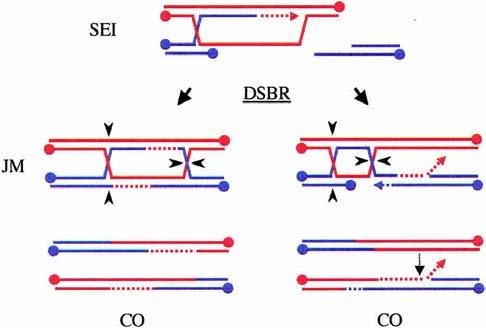
A role for Mus81* in cleaving 3′-flaps. Model II proposes that most meiotic SEI intermediates are processed into COs via the DSBR pathway (left). In a subset of SEI intermediates, however, the invading strand may be elongated excessively (right). In this case, the second strand is captured, not by the D-loop, but by the elongated strand following displacement. Any 3′-flaps that arise from strand annealing are substrates for Mus81* (light dotted arrow). In this pathway, heteroduplex DNA is found adjacent to the dHJs rather than between them.
Budding yeast has two genetically separable pathways for generating meiotic crossovers
The 3′-flap model (II) assumes that there are two pathways for generating COs, one that uses MUS81–MMS4 and one that does not. MSH4 and MSH5 encode meiosis-specific MutS homologs that have no role in mismatch repair, but instead are required to promote COs between homologous chromosomes (Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995). Similar to Mus81 and Mms4/Eme1, Msh4 and Msh5 function together as a heteroligomer (Pochart et al. 1997). Deletion of MSH4 and/or MSH5 reduces crossing over two- to threefold and spore viability to ∼40%. Interestingly, the COs that occur in msh4 mutants do not exhibit genetic interference (Novak et al. 2001). Interference is a phenomenon by which COs are distributed throughout the genome, perhaps to ensure that every chromosome receives at least one. Because COs are necessary for proper Meiosis I segregation, defects in interference may result in chromo-some missegregation, or nondisjunction, at the first meiotic division. Consistent with a loss of interference, the spore lethality observed in msh4/msh5 diploids is caused by chromosome nondisjunction (Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995). In contrast, the COs remaining in mms4 diploids exhibit interference, and spore lethality is not caused by nondisjunction (de los Santos et al. 2003). msh5 mms4 diploids exhibit a sixfold decrease in crossing over and only ∼20% viable spores, confirming that these genes define two independent pathways for generating crossovers, an MSH4–MSH5-dependent interference pathway (designated Class I) and an MUS81–MMS4-dependent noninterference pathway (Class II).
The discovery of two different CO pathways in budding yeast may explain why spore viability is decreased to a much lesser extent in budding yeast mus81 diploids compared with fission yeast mus81 mutants (40% vs. <1%). In fission yeast, none of the COs display interference and the gene products required for the Class I pathway (e.g., MSH4 and MSH5) are absent from the S. pombe genome (Munz 1994; Villeneuve and Hillers 2001). Therefore, although the MUS81–MMS4 pathway accounts for a relatively minor fraction of COs in budding yeast, it is responsible for most, if not all, of the COs in fission yeast (Fig. 5).
Figure 5.
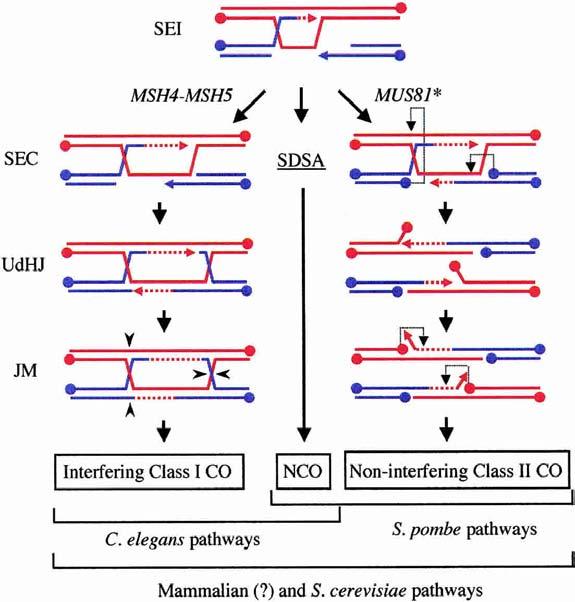
The role of Mus81* in creating COs without HJs. Model III proposes that meiotic SEI intermediates have three fates depending on the organism and pathway choice. All organisms use SDSA to produce NCOs (center). Class I interfering COs occur in organisms containing MSH4–MSH5 and arise from the DSBR pathway (left). Class II noninterfering COs are proposed to arise from the action of Mus81* on an SEC intermediate (right). The SEC intermediate provides targets for Mus81* cleavage at the D-loop and at the half-junction (light dotted arrows). Cleavage at these sites results in an obligatory CO product containing 5′-flaps and gaps. Here the flaps are shown to isomerize into 3′-flaps prior to cleavage by a second round of Mus81* action. Filling-in of the gaps and ligation completes the CO product (not shown).
Neither the HJ resolvase model nor the 3′-flap model for Mus81* function is satisfactory. As mentioned above, biochemical and genetic data do not support a role for Mus81* in the cleavage of intact HJs. A problem with the 3′-flap model is that it provides no way of easily discriminating between COs in the Class I and Class II pathways. Intact dHJs are postulated in the 3′-flap intermediates, and these must be resolved, presumably by the same resolvase that cleaves the junctions formed along the interference pathway (Fig. 3). Where does the MSH4–MSH5 dependence come into play? The most disturbing feature about these two models is that they are organism-specific. That is, Mus81* would have to behave like an HJ resolvase in fission yeast meiosis (Model I) and a 3′-flap endonuclease in budding yeast meiosis (Model II). The most plausible model would be one that is consistent with both the in vitro preference of the enzyme and the genetic data from both yeasts. Recently, Whitby and colleagues have proposed a model for Mus81 function that satisfies these criteria (Osman et al. 2003).
Model III: Mus81* cleaves D-loops and half-junctions
The most recent biochemical characterizations of rMus81* and TEV-Mus81* cleavage activity raise the possibility that D-loops and half-junctions are the in vivo substrates for the endonuclease, as opposed to 3′-flaps (Fig. 3; Gaillard et al. 2003; Osman et al. 2003). This idea is consistent with analyses that show that the presence of a flap is not as important as the presence of a 5′-nick (Bastin-Shanower et al. 2003). These findings led to the novel proposal that Mus81* generates COs without ever forming a dHJ intermediate (Fig. 5; Osman et al. 2003). In Model III, Mus81 cleaves a D-loop on one side of the break and the half-junction formed by SEC on the other side of the break (Fig. 5). This cleavage would result in gaps and/or 5′-flaps of ∼5 nt. Such flaps could isomerize to 3′-flaps and be cleaved by Mus81* or they could be processed by a 5′-flap endonuclease such as Rad27 in budding yeast or Rad2 in fission yeast (Alleva and Doetsch 1998; Kao et al. 2002). Interestingly, mus81 and rad27 are synthetically lethal in mitotic cells (Tong et al. 2001). Filling-in and ligation of the cleaved strands results in an obligatory CO without ever creating an intact dHJ intermediate (Osman et al. 2003). A variation of this model has been independently proposed by Heyer et al. (2003).
Model III beautifully reconciles the data between fission yeast and budding yeast. The assumption is that the SEI intermediate is the precursor to recombination in both organisms. In fission yeast, NCOs result from SDSA, whereas COs result almost exclusively from Mus81* cleavage (Fig. 5; Osman et al. 2003). In budding yeast, NCOs are also derived by SDSA, but COs can be generated by two independent pathways. The Class II, noninterfering COs result from Mus81* cleavage, similar to fission yeast. Whether D-loop cleavage by Mus81* occurs prior to SEC as drawn in Osman et al. (2003) or after SEC (Fig. 5) is not yet known. For the Class I, interference-dependent COs, the SEC intermediate is matured into a dHJ structure that is then resolved, presumably by an RuvC/RusA-like mechanism, to make COs. If Mus81* cleaves D-loops before SEC, then MSH4–MSH5 could act at the step promoting formation of the SEC intermediate. Alternatively, if Mus81* cleaves after SEC, MSH4–MSH5 might prevent this cleavage, thereby allowing maturation to dHJs. The MutS homolog, Msh2, is known to bind directly to HJs, so perhaps Msh4–Msh5 complexes bind directly to branched junctions as well (Alani et al. 1997; Marsischky et al. 1999). Msh4–Msh5 is part of the complex of proteins required for interference that includes Zip1, Zip2, and Zip3 (Agarwal and Roeder 2000). Binding of this complex to the junctions of developing recombination intermediates could create a physical barrier that prevents Mus81* from cutting.
In budding yeast, Class II COs comprise a minority class that is derived independently from Class I COs and NCOs. Therefore, mus81/mms4 mutants would be predicted to exhibit only a small decrease in COs and have no deleterious effects on gene conversion, which is exactly what is observed (de los Santos et al. 2003). In fission yeast, crossing over occurs primarily, if not entirely, by the Class II pathway. Therefore, by Model III, COs should be greatly reduced (Fig. 5). NCOs occur by SDSA after D-loop formation, and therefore should be unaffected. These predictions are consistent with the fission yeast recombination data (Osman et al. 2003; Smith et al. 2003). A strength of Model III, therefore, is that the differences observed in meiotic phenotypes between fission and budding yeast are explained, not by saying that Mus81* cleaves different substrates in the two yeasts, but rather that cleavage of the same substrate has a different relative contribution to the total number of meiotic COs in each yeast.
The rusA suppression of fission yeast mus81 meiotic phenotypes could be explained if SEC intermediates can spontaneously proceed to dHJs, thereby creating excellent substrates for RusA. If this occurs, then fission yeast has no way of normally resolving these dHJs, given the very low spore viability of mus81 mutants. The fact that rusA overexpression is unable to suppress budding yeast mus81 meiotic defects may mean that formation of dHJs cannot proceed spontaneously in S. cerevisiae, but requires the activity of proteins such as Msh4–Msh5. Alternatively, the failure of rusA to suppress mus81 budding yeast meiotic phenotypes could be due to the very significant differences in meiotic chromosome behavior exhibited by the two yeasts. During budding yeast meiosis, homologous chromosomes become physically associated by the formation of a meiosis-specific protein-aceous structure called the synaptonemal complex (SC; Roeder 1997). In contrast, in fission yeast, homologs do not synapse and no SC is formed (Bahler et al. 1993). The SC may, therefore, prevent RusA from accessing recombination intermediates during budding yeast meiosis. Consistent with this idea, rusA is able to suppress some mus81/mms4 phenotypes in vegetatively growing budding yeast cells where the SC is absent (Bastin-Shanower et al. 2003; Odagiri et al. 2003).
The semantics of resolution
Much of the controversy surrounding Mus81* has been the issue of whether it is an HJ resolvase or not. Both genetic and biochemical data support the argument against intact HJs being the target of Mus81*. Model III suggests that Mus81 cleavage of non-HJ intermediates results in the formation of COs after further DNA processing events. If this model is correct, Mus81 could be considered to “resolve” recombination intermediates. For many people, however, the term “resolvase” conjures up a specific paradigm—that of RuvC/RusA-like nucleases cleaving strands of like polarity in HJs to create ligatable, nicked, linear duplexes. For clarity, therefore, it might be most accurate to refer to Mus81*, not as a resolvase, but as a “nicked junction endonuclease.”
Conclusion
For years, recombination models have assumed that COs in eukaryotic cells would follow the paradigm from bacteria of resolution of intact HJs by an RuvC/RusA-like resolvase. The discovery of Mus81* has raised the possibility that cells have evolved at least two ways for generating COs, the Class I pathway, which follows the paradigm of intact HJ resolution, and the Class II pathway, which may not. During evolution, the choice of which pathway(s) to use has varied with different organisms (Fig. 5). Fission yeast appears to depend exclusively on the Class II pathway, which may be because of the failure of homologous chromosomes to synapse. In contrast, the majority of COs in budding yeast occur by the Class I pathway, and the Class II pathway may exist primarily as a backup to ensure that any recombination intermediates that fail to be converted into intact dHJs be repaired. In worms, deletion of MSH4 or MSH5 eliminates all COs, suggesting that meiotic COs in this organism rely exclusively on the Class I pathway (Fig. 5; Zalevsky et al. 1999; Kelly et al. 2000).
One outcome in having two independent ways of generating COs is the ability to modulate interference. Interestingly, mms4 exhibits a stronger phenotype on a small chromosome that displays a lower level of interference than on a large chromosome on which interference is stronger (Kaback et al. 1999; de los Santos et al. 2003). Therefore, the degree of interference may be regulated by altering the frequency with which the different CO pathways are used. The two extreme cases are fission yeast, where there is no interference and COs are generated primarily, if not exclusively, by the Class II pathway, and worms, where interference is strong (Hillers and Villeneuve 2003) and the Class I pathway apparently generates all COs. It is possible that mammals, like budding yeast, use both CO pathways during meiosis. Mammalian chromosomes exhibit interference, and MSH4 and MSH5 are meiosis-specific genes required for the proper execution of meiosis; Mus81* activity has been detected in somatic cells (de Vries et al. 1999; Broman and Weber 2000; Kneitz et al. 2000). However, whether mammals follow the worm paradigm, in which all COs are MSH4/MSH5-dependent, or the budding yeast paradigm, in which a minority of COs are generated by Mus81*, is as yet unknown. The study of this very interesting enzyme has therefore suggested a novel mechanism for generating meiotic COs and may ultimately shed light on how the distribution of COs is regulated in different organisms.
Acknowledgments
We thank Neil Hunter and Michael Lichten for helpful comments on the manuscript. We are grateful to W. Heyer, G. Smith, C. McGowan, P. Russell, and M. Whitby for communicating ideas/results prior to publication. Research in N.M.H.'s laboratory was supported by NIH grant GM50717 and March of Dimes grant 1-FY02-754, and research in S.J.B.'s laboratory is supported by NIH grant GM67956.
References
- Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. doi: 10.1016/s0092-8674(00)00029-5. [DOI] [PubMed] [Google Scholar]
- Alani E, Lee S, Kane MF, Griffith J, Kolodner RD. Saccharomyces cerevisiae MSH2, a mispaired base recognition protein, also recognizes Holliday junctions in DNA. J. Mol. Biol. 1997;265:289–301. doi: 10.1006/jmbi.1996.0743. [DOI] [PubMed] [Google Scholar]
- Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001a;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Allers T, Lichten M. Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol. Cell. 2001b;8:225–231. doi: 10.1016/s1097-2765(01)00280-5. [DOI] [PubMed] [Google Scholar]
- Alleva JL, Doetsch PW. Characterization of Schizosaccharomyces pombe Rad2 protein, a FEN-1 homolog. Nucleic Acids Res. 1998;26:3645–3650. doi: 10.1093/nar/26.16.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wyler T, Loidl J, Kohli J. Unusual nuclear structures in meiotic prophase of fission yeast: A cytological analysis. J. Cell Biol. 1993;121:241–256. doi: 10.1083/jcb.121.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin-Shanower SA, Fricke WM, Mullen JR, Brill SJ. The mechanism of Mus81–Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1–Rad10. Mol. Cell. Biol. 2003;23:3487–3496. doi: 10.1128/MCB.23.10.3487-3496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P. Damage tolerance protein mus81 associates with the FHA1 domain of checkpoint kinase cds1 [In Process Citation] Mol. Cell. Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Gaillard P-HL, McDonald WH, Shanahan P, Yates JR, III, Russell P. Mus81–Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Broman KW, Weber JL. Characterization of human crossover interference. Am. J. Hum. Genet. 2000;66:1911–1926. doi: 10.1086/302923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X-B, Melchionna R, Denis C-M, Gaillard P-HL, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Constantinou A, West SC. Identification and purification of the human Mus81/Eme1 endonuclease. J. Biol. Chem. 2003;278:25172–25178. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, Nasmyth K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 2003;5:480–485. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- Constantinou A, Chen XB, McGowan CH, West SC. Holliday junction resolution in human cells: Two junction endonucleases with distinct substrate specificities. EMBO J. 2002;21:5577–5585. doi: 10.1093/emboj/cdf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T, Loidl J, Larkin B, Hollingsworth NM. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics. 2001;159:1511–1525. doi: 10.1093/genetics/159.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries SS, Baart EB, Dekker M, Siezen A, de Rooij DG, de Boer P, te Riele H. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes & Dev. 1999;13:523–531. doi: 10.1101/gad.13.5.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Dixon J, Osman F, Whitby MC. Partial suppression of the fission yeast rqh1− phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 2000;19:2751–2762. doi: 10.1093/emboj/19.11.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Ahn JS, Dixon J, Whitby MC. Mus81–Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- Fabre F, Chan A, Heyer WD, Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Xiao W. Functional domains required for the Saccharomyces cerevisiae Mus81–Mms4 endonuclease complex formation and nuclear localization. DNA Repair. 2003;2:1435–1447. doi: 10.1016/j.dnarep.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Gaillard P-HL, Noguchi E, Shanahan P, Russell P. The endogenous Mus81–Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell. 2003;12:747–759. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- Gao H, Chen X-B, McGowan CH. Mus81 endonuclease localizes to nucleoli and to regions of DNA damage in human S-phase cells. Mol. Biol. Cell. 2003;14:4826–4834. doi: 10.1091/mbc.E03-05-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE, Heyer WD. The fuss about Mus81. Cell. 2001;107:551–554. doi: 10.1016/s0092-8674(01)00593-1. [DOI] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Solinger JA. Holliday junctions in the eukaryotic nucleus: Resolution in sight? Trends Biochem. Sci. 2003;28:548–557. doi: 10.1016/j.tibs.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Hillers KJ, Villeneuve AM. Chromosome-wide control of meiotic crossing over in C. elegans. Curr. Biol. 2003;13:1641–1647. doi: 10.1016/j.cub.2003.08.026. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Ponte L, Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes & Dev. 1995;9:1728–1739. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- Hunter N, Kleckner N. The single-end invasion: An asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Interthal H, Heyer WD. MUS81 encodes a novel helix–hairpin–helix protein involved in the response to UV-and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000;263:812–827. doi: 10.1007/s004380000241. [DOI] [PubMed] [Google Scholar]
- Kaback DB, Barber D, Mahon J, Lamb J, You J. Chromosome size-dependent control of meiotic reciprocal recombination in Saccharomyces cerevisiae: The role of crossover interference. Genetics. 1999;152:1475–1486. doi: 10.1093/genetics/152.4.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman V, Mullen JR, Fricke WM, Bastin-Shanower SA, Brill SJ. Functional overlap between the Sgs1–Top3 and the fork endonuclease Mms4–Mus81: Potential mechanisms for re-establishing replication forks. Genes & Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HI, Henricksen LA, Liu Y, Bambara RA. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 2002;277:14379–14389. doi: 10.1074/jbc.M110662200. [DOI] [PubMed] [Google Scholar]
- Keeney S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Kelly KO, Dernberg AF, Stanfield GM, Villeneuve AM. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics. 2000;156:617–630. doi: 10.1093/genetics/156.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneitz B, Cohen PE, Avdievich E, Zhu L, Kane MF, Hou H, Jr., Kolodner RD, Kucherlapati R, Pollard JW, Edelmann W. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes & Dev. 2000;14:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Marsischky GT, Lee S, Griffith J, Kolodner RD. Saccharomyces cerevisiae MSH2/6 complex interacts with Holliday junctions and facilitates their cleavage by phage resolution enzymes. J. Biol. Chem. 1999;274:7200–7206. doi: 10.1074/jbc.274.11.7200. [DOI] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics. 1994;137:701–707. doi: 10.1093/genetics/137.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak JE, Ross-Macdonald PB, Roeder GS. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics. 2001;158:1013–1025. doi: 10.1093/genetics/158.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odagiri N, Seki M, Onoda F, Yoshimura A, Watanabe S, Enomoto T. Budding yeast mms4 is epistatic with rad52 and the function of Mms4 can be replaced by a bacterial Holliday junction resolvase. DNA Repair (Amst) 2003;2:347–358. doi: 10.1016/s1568-7864(02)00234-3. [DOI] [PubMed] [Google Scholar]
- Ogrunc M, Sancar A. Identification and characterization of human MUS81–MMS4 structure-specific endonuclease. J. Biol. Chem. 2003;278:21715–21720. doi: 10.1074/jbc.M302484200. [DOI] [PubMed] [Google Scholar]
- Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: A role for Mus81–Eme1 in meiosis. Mol. Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochart P, Woltering D, Hollingsworth NM. Conserved properties between functionally distinct MutS homologs in yeast. J. Biol. Chem. 1997;48:30345–30349. doi: 10.1074/jbc.272.48.30345. [DOI] [PubMed] [Google Scholar]
- Prakash L, Prakash S. Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics. 1977;86:33–55. doi: 10.1093/genetics/86.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder GS. Meiotic chromosomes: It takes two to tango. Genes & Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P, Roeder GS. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell. 1994;79:1069–1080. doi: 10.1016/0092-8674(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Smith KN, Nicolas A. Recombination at work for meiosis. Curr. Opin. Genet. Dev. 1998;8:200–211. doi: 10.1016/s0959-437x(98)80142-1. [DOI] [PubMed] [Google Scholar]
- Smith GR, Boddy MN, Russell P. Fission yeast Mus81–Eme1 Holliday junction resolvase is required for meiotic crossing-over but not for gene conversion. Genetics. 2003 doi: 10.1093/genetics/165.4.2289. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double strand-break model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Villeneuve AM, Hillers KJ. Whence meiosis? Cell. 2001;106:647–650. doi: 10.1016/s0092-8674(01)00500-1. [DOI] [PubMed] [Google Scholar]
- Wan L, de los Santos T, Zhang C, Shokat K, Hollingsworth NM. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic DSB repair in budding yeast. Mol. Biol. Cell. 2004;15:11–23. doi: 10.1091/mbc.E03-07-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SC. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- Whitby MC, Osman F, Dixon J. Cleavage of model replication forks by fission yeast Mus81–Eme1 and budding yeast Mus81–Mms4. J. Biol. Chem. 2003;278:6928–6935. doi: 10.1074/jbc.M210006200. [DOI] [PubMed] [Google Scholar]
- Xiao W, Chow BL, Milo CN. Mms4, a putative transcriptional (co)activator, protects Saccharomyces cerevisiae cells from endogenous and environmental DNA damage. Mol. Gen. Genet. 1998;257:614–623. doi: 10.1007/s004380050689. [DOI] [PubMed] [Google Scholar]
- Zalevsky J, MacQueen AJ, Duffy JB, Kemphues KJ, Villeneuve AM. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics. 1999;153:1271–1283. doi: 10.1093/genetics/153.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]