Abstract
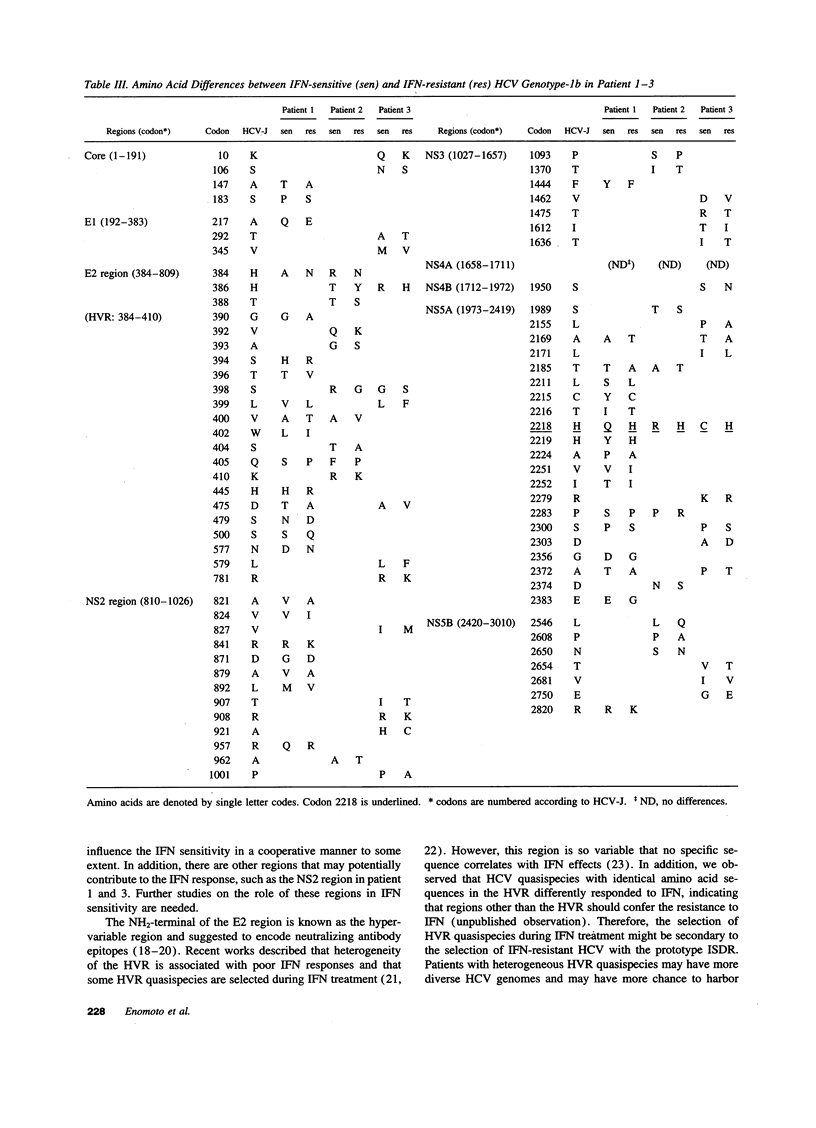
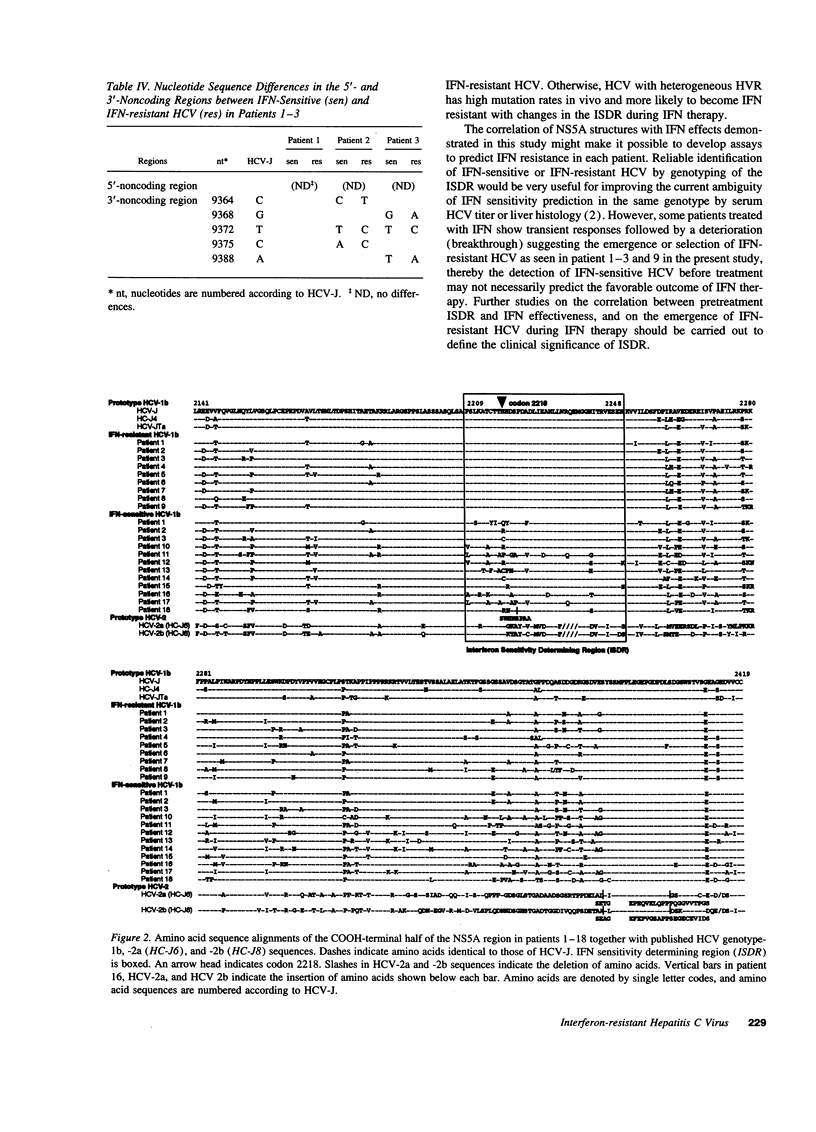
We have previously demonstrated that sensitivity to interferon is different among hepatitis C virus (HCV) quasispecies simultaneously detected in same individuals and that interferon-resistant HCV quasispecies are selected during the treatment. To determine the genetic basis of their resistance to interferon, HCV genotype-1b was obtained from serum of three patients before and during interferon therapy, and their full-length nucleotide and deduced amino acid sequences were determined. Comparison of the pairs of interferon-resistant and interferon-sensitive HCV isolates in respective individuals demonstrated clusters of amino acid differences in the COOH-terminal half of the NS5A region (codon 2154-2383), which contained a common unique amino acid difference at codon 2218. Additional sequence data of the COOH-terminal half of the NS5A region obtained from six interferon-resistant and nine interferon-sensitive HCV confirmed the exclusive existence of missense mutations in a 40 amino acid stretch of the NS5A region around codon 2218 (from codon 2209 to 2248) in interferon-sensitive HCV. On the other hand, this region of interferon-resistant HCV was identical to that of prototype HCV genotype-1b (HCV-J, HCV-JTa, or HC-J4). We designated this region as the interferon sensitivity determining region. Thus, HCV genotype-1b with the prototype interferon sensitivity determining region appears to be interferon-resistant strains. The specific nature of these mutations might make it possible to predict prognostic effects of interferon treatment.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Enomoto N., Kurosaki M., Tanaka Y., Marumo F., Sato C. Fluctuation of hepatitis C virus quasispecies in persistent infection and interferon treatment revealed by single-strand conformation polymorphism analysis. J Gen Virol. 1994 Jun;75(Pt 6):1361–1369. doi: 10.1099/0022-1317-75-6-1361. [DOI] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., Wychowski C., Lin C., Feinstone S. M., Rice C. M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993 Mar;67(3):1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Kato N., Ootsuyama Y., Nakagawa M., Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Mizushima H., Tanji Y., Komoda Y., Hirowatari Y., Akagi T., Kato N., Kimura K., Shimotohno K. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10773–10777. doi: 10.1073/pnas.90.22.10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino K., Sainokami S., Shimoda K., Iino S., Wang Y., Okamoto H., Miyakawa Y., Mayumi M. Genotypes and titers of hepatitis C virus for predicting response to interferon in patients with chronic hepatitis C. J Med Virol. 1994 Mar;42(3):299–305. doi: 10.1002/jmv.1890420318. [DOI] [PubMed] [Google Scholar]
- Kanazawa Y., Hayashi N., Mita E., Li T., Hagiwara H., Kasahara A., Fusamoto H., Kamada T. Influence of viral quasispecies on effectiveness of interferon therapy in chronic hepatitis C patients. Hepatology. 1994 Nov;20(5):1121–1130. [PubMed] [Google Scholar]
- Kato N., Hijikata M., Ootsuyama Y., Nakagawa M., Ohkoshi S., Sugimura T., Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N., Sekiya H., Ootsuyama Y., Nakazawa T., Hijikata M., Ohkoshi S., Shimotohno K. Humoral immune response to hypervariable region 1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J Virol. 1993 Jul;67(7):3923–3930. doi: 10.1128/jvi.67.7.3923-3930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N., Yokosuka O., Hosoda K., Ito Y., Ohto M., Omata M. Quantification of hepatitis C virus by competitive reverse transcription-polymerase chain reaction: increase of the virus in advanced liver disease. Hepatology. 1993 Jul;18(1):16–20. [PubMed] [Google Scholar]
- Lau J. Y., Davis G. L., Kniffen J., Qian K. P., Urdea M. S., Chan C. S., Mizokami M., Neuwald P. D., Wilber J. C. Significance of serum hepatitis C virus RNA levels in chronic hepatitis C. Lancet. 1993 Jun 12;341(8859):1501–1504. doi: 10.1016/0140-6736(93)90635-t. [DOI] [PubMed] [Google Scholar]
- Martell M., Esteban J. I., Quer J., Genescà J., Weiner A., Esteban R., Guardia J., Gómez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992 May;66(5):3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T., Kato N., Ohkoshi S., Shibuya A., Shimotohno K. Characterization of the 5' noncoding and structural region of the hepatitis C virus genome from patients with non-A, non-B hepatitis responding differently to interferon treatment. J Hepatol. 1994 May;20(5):623–629. doi: 10.1016/s0168-8278(05)80350-2. [DOI] [PubMed] [Google Scholar]
- Okada S., Akahane Y., Suzuki H., Okamoto H., Mishiro S. The degree of variability in the amino terminal region of the E2/NS1 protein of hepatitis C virus correlates with responsiveness to interferon therapy in viremic patients. Hepatology. 1992 Sep;16(3):619–624. doi: 10.1002/hep.1840160302. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Kojima M., Okada S., Yoshizawa H., Iizuka H., Tanaka T., Muchmore E. E., Peterson D. A., Ito Y., Mishiro S. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology. 1992 Oct;190(2):894–899. doi: 10.1016/0042-6822(92)90933-g. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Kurai K., Okada S., Yamamoto K., Lizuka H., Tanaka T., Fukuda S., Tsuda F., Mishiro S. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology. 1992 May;188(1):331–341. doi: 10.1016/0042-6822(92)90762-e. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Okada S., Sugiyama Y., Kurai K., Iizuka H., Machida A., Miyakawa Y., Mayumi M. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J Gen Virol. 1991 Nov;72(Pt 11):2697–2704. doi: 10.1099/0022-1317-72-11-2697. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Sugiyama Y., Okada S., Kurai K., Akahane Y., Sugai Y., Tanaka T., Sato K., Tsuda F., Miyakawa Y. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol. 1992 Mar;73(Pt 3):673–679. doi: 10.1099/0022-1317-73-3-673. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Alberti A., Alter H. J., Bonino F., Bradley D. W., Brechot C., Brouwer J. T., Chan S. W., Chayama K., Chen D. S. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology. 1994 May;19(5):1321–1324. [PubMed] [Google Scholar]
- Tanaka T., Kato N., Nakagawa M., Ootsuyama Y., Cho M. J., Nakazawa T., Hijikata M., Ishimura Y., Shimotohno K. Molecular cloning of hepatitis C virus genome from a single Japanese carrier: sequence variation within the same individual and among infected individuals. Virus Res. 1992 Apr;23(1-2):39–53. doi: 10.1016/0168-1702(92)90066-i. [DOI] [PubMed] [Google Scholar]
- Taniguchi S., Okamoto H., Sakamoto M., Kojima M., Tsuda F., Tanaka T., Munekata E., Muchmore E. E., Peterson D. A., Mishiro S. A structurally flexible and antigenically variable N-terminal domain of the hepatitis C virus E2/NS1 protein: implication for an escape from antibody. Virology. 1993 Jul;195(1):297–301. doi: 10.1006/viro.1993.1378. [DOI] [PubMed] [Google Scholar]
- Tsubota A., Chayama K., Ikeda K., Yasuji A., Koida I., Saitoh S., Hashimoto M., Iwasaki S., Kobayashi M., Hiromitsu K. Factors predictive of response to interferon-alpha therapy in hepatitis C virus infection. Hepatology. 1994 May;19(5):1088–1094. [PubMed] [Google Scholar]
- Weiner A. J., Geysen H. M., Christopherson C., Hall J. E., Mason T. J., Saracco G., Bonino F., Crawford K., Marion C. D., Crawford K. A. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada N., Tanihara K., Mizokami M., Ohba K., Takada A., Tsutsumi M., Date T. Full-length sequence of the genome of hepatitis C virus type 3a: comparative study with different genotypes. J Gen Virol. 1994 Nov;75(Pt 11):3279–3284. doi: 10.1099/0022-1317-75-11-3279. [DOI] [PubMed] [Google Scholar]



