Abstract
In this paper, we describe the structure of chitinase B from Serratia marcescens, which consists of a catalytic domain with a TIM-barrel fold and a 49-residue C-terminal chitin-binding domain. This chitinase is the first structure of a bacterial exochitinase, and it represents one of only a few examples of a glycosyl hydrolase structure having interacting catalytic and substrate-binding domains. The chitin-binding domain has exposed aromatic residues that contribute to a 55-Å long continuous aromatic stretch extending into the active site. Binding of chitin oligomers is blocked beyond the −3 subsite, which explains why the enzyme has chitotriosidase activity and degrades the chitin chain from the nonreducing end. Comparison of the chitinase B structure with that of chitinase A explains why these enzymes act synergistically in the degradation of chitin.
The degradation of abundant insoluble carbohydrate polymers such as cellulose and chitin is achieved in nature with the help of batteries of glycosyl hydrolases with different substrate preferences and product specificities. For example, the degradation of chitin, a linear polysaccharide of β(1, 4)-linked N-acetylglucosamine (GlcNAc) residues, by the soil bacterium S. marcescens involves at least four enzymes (the exo- and endochitinases ChiA, ChiB, and ChiC, and an N-acetylglucosaminidase) (1–4). In addition to a catalytic domain, most enzymes involved in cellulose and chitin degradation usually contain one or more domains that are involved in substrate binding (refs. 5–7; see also http://afmb.cnrs-mrs.fr/pedro/DB/ncmCBM12.html). Removal of such domains often results in enzymes that are still active but display severely impaired binding to polymeric substrates (see examples in refs. 7 and 8). For cellulases, there is abundant structural information for a variety of catalytic domains and for isolated carbohydrate-binding domains (6, 9), but there is only one available crystal structure of a catalytic domain together with a (143-residue) CeBD (10).
Chitinases belong to families 18 and 19 of the glycosyl hydrolases (9). The catalytic domain of family 18 chitinases has a TIM-barrel fold (2, 11) and includes a conserved glutamate residue that presumably acts as an acid during catalysis (Glu144 in ChiB; Fig. 1; refs. 12–14). Catalysis proceeds with retention of the anomeric configuration, which is achieved by a mechanism in which the carbonyl oxygen of the N-acetyl group of the −1 sugar (nomenclature according to ref. 15) acts as nucleophile (12–14). Judged from their sequences, most family 18 chitinases, including ChiA, ChiB, and ChiC from S. marcescens, contain domains putatively involved in the interaction with chitin (5, 7, 8, 16). Perrakis et al. (2, 5) have determined the structure of complete ChiA, revealing the location of a 114-residue domain with a fibronectin III-like fold that most likely participates in chitin-binding (2, 5, 8). On the basis of sequence analyses, ChiB has been suggested to consist of a catalytic domain followed by a putative linker region and a small domain of approximately 45 residues sharing sequence homology with similarly short (partly putative) carbohydrate-binding domains found in chitinases, cellulases, and xylanases (7, 16). The chitin-binding domain (ChBD) has been classified (by sequence) as a family XII ChBD and exhibits sequence homology with family V cellulose-binding domains (CeBDs) (6). With the exception of an NMR structure for an isolated family V CeBD (17), nothing is known about the structure of these small domains and about how they are positioned with respect to their cognate catalytic domains.
Figure 1.
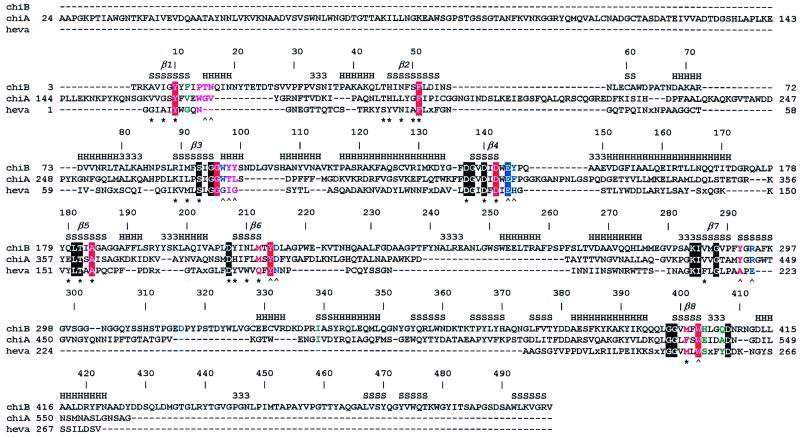
Sequence alignment of chitinases. ChiB, chitinase B from S. marcescens; ChiA, chitinase A from S. marcescens; heva, hevamine from Hevea brasiliensis. An x in the sequences indicates a small insertion in hevamine. The secondary structure of ChiB is indicated at the top of the sequence alignment by H, α-helix; S, β-strand; 3, 310-helix. The TIM barrel β-strands are indicated by β1–8. Every 10th ChiB residue is labeled with its corresponding sequence number. Boxes indicate conserved residues. The color coding indicates the subsites of the chitotetraose model (see also Fig. 3A). Purple, −3; green, −2; red, −1; blue, +1. A ∧ indicates hydrogen bonding to an atom on the chitotetraose model, and a ★ indicates involvement of the side chain in the hydrogen-bonding network inside the TIM barrel core of ChiB. The catalytic Glu144 is part of the family 18 DxxDxDxE motif. The following definitions of loops and domains are used throughout the text: the porch loop (residues 14–27), the support loop (residues 233–262), the flexible loop (residues 315–325), the α/β-domain (residues 295–373) and the linker (residues 425–450), and the ChBD (residues 451–498).
In this paper, we present a 1.9-Å resolution structure of the complete ChiB from S. marcescens. The structure is one of few in which both the catalytic core and the ChBD are observed. The complete structure provides insight into the catalytic function of ChiB and reveals how the ChBD relates to the catalytic domain and may contribute to substrate binding. Comparison of ChiB with ChiA provides an explanation for the chitotriosidase activity of ChiB and the synergism previously observed when incubating chitin with both enzymes (16).
Materials and Methods
Purification and Crystallization.
Recombinant S. marcescens ChiB was overexpressed in Escherichia coli and purified by a protocol consisting of a previously described hydrophobic interaction chromatography step (16), preceded by a standard ion-exchange chromatography step using Q-Sepharose Fast Flow (Amersham Pharmacia). The final protein material was collected in an ammonium carbonate buffer as previously described (16) and freeze-dried. The material was dissolved to 1 mg/ml in 50 mM Tris, pH 7.5, and dialyzed against the same buffer for 24 h. Subsequent concentration (with an Ultrafree-MC 30-kDa cutoff filter from Millipore) yielded a 10 mg/ml protein solution that was used directly for hanging drop-vapor diffusion crystallization experiments.
For crystallization, 2 ml of protein solution was mixed with an equal volume of well solution containing 50 mM citrate (pH 5.6), 0.5 M Li2SO4, and 0.25 M (NH4)2SO4. Crystals appeared within 1–2 weeks at room temperature and grew to a maximum size of 0.4 × 0.4 × 0.8 mm. Heavy atom derivatives were made by adding 4 ml of a 10–25 mM heavy atom solution directly to the vapor-diffusion drop containing the crystal at 8 h before data collection.
Data Collection.
Data were collected on a Nonius FR591 rotating anode with XRM-216 focusing mirrors (Prophysics) and a MAR345 image plate detector, as well as on beamlines X11/X31 at Deutsches Elektronen Synchrotron, Hamburg, with MAR345 and Mar 30-cm image plates, respectively. Because radiation damage was limited, all data were collected at room temperature. Images were processed using denzo, and reflections were merged using scalepack, of the hkl suite (18). Data collection statistics are summarized in Table 1.
Table 1.
Details of data collection and heavy atom refinement
| Datasets | Wavelength, Å | Resolution, Å | Redundancy | Completeness, % | Rmerge, % | Rdiff, % | No. of sites | PP, acen | PP, cen | RCullis, iso, acen | RCullis, iso, cen |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Native 1, 2 | 0.95, 0.91 | 40–2.5, 40–1.9 | 3.9, 3.7 | 99.6, 99.7 | 5.1, 6.9 | — | — | — | — | — | — |
| HgCl2 | 0.82 | 40–3.0 | 4.0 | 98.1 | 5.9 | 26.5 | 2 | 0.77 | 0.64 | 0.88 | 0.81 |
| PtCl4 | 0.88 | 40–2.5 | 3.4 | 95.7 | 8.7 | 19.9 | 10 | 1.97 | 1.35 | 0.65 | 0.65 |
| Au(CN)2 | 1.54 | 25–2.5 | 4.0 | 98.4 | 6.9 | 18.3 | 7 | 0.73 | 0.61 | 0.89 | 0.87 |
| cisPt 1, 2 | 0.88, 1.54 | 40–2.5, 25–2.2 | 3.7, 3.2 | 97.9, 99.3 | 6.7, 7.7 | 12.0, 14.9 | 10, 9 | 1.58, 1.72 | 1.11, 1.21 | 0.71, 0.68 | 0.73, 0.67 |
| Re 1, 2 | 0.98, 1.54 | 40–2.5, 25–2.2 | 3.7, 4.1 | 98.8, 98.9 | 7.1, 6.8 | 16.8, 21.0 | 10, 11 | 1.21, 1.24 | 0.98, 1.02 | 0.77, 0.76 | 0.77, 0.74 |
Re, potassium perrhenate; cisPt, cis-platinum(II)diaminedichloride; Rmerge=∑h∑i|Ii(hkl)−<I(hkl)>|/∑h∑i<I(hkl)>; Rdiff=∑h|Fp−Fph|/∑p|Fp|; PP, phasing power =∑hfc/∑hɛ; Rcullis=∑||FPH−|FP+fc||/∑||FPH−|FP||; fc is the calculated heavy atom factor, ɛ is the lack-of-closure error, I is the intensity, Fp is the native structure factor, and Fph is the heavy atom derivative structure factor; cen, centrics; acen, acentrics.
Structure Determination and Refinement.
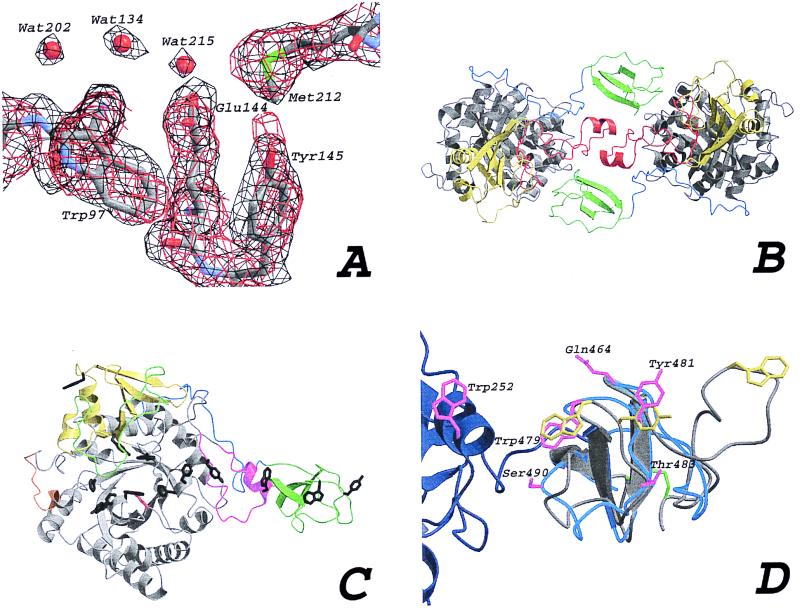
From the native data set, a Matthews coefficient of 2.5 Å3/D was obtained, assuming two ChiB molecules in the asymmetric unit. Two symmetry-related solutions of the self-rotation function [calculated with glrf (19)] were found at φ = 38°, ψ = 90°, κ = 180°, and φ = 52°, ψ = 90°, κ = 180°, with a peak height of 6.8 σ. Molecular replacement approaches using ChiA as a search model were inconclusive, thus we proceeded to solve the structure by using the multiple isomorphous replacement using anomalous scattering (MIRAS) approach, with multiple wavelengths for 2 of the 5 derivatives (see Table 1). Patterns of isomorphous and anomalous differences were calculated with the ccp4 package (20) and were consistent and straightforward to interpret. Initial sites were found and refined by using solve (21). Further refinement and incorporation of additional sites was performed with mlphare (22). Final experimental phases were calculated for data from 40–2.5 Å, with an overall figure of merit of 0.71 (see Table 1). Combination of the MIRAS phases with the native amplitudes resulted in a readily interpretable map (Fig. 2A). The program findncs (23) was applied to the list of heavy atom sites and was able to both confirm the orientation of the noncrystallographic 2-fold axis found from the self-rotation function and provide the translational component. The phases were then further improved by application of phase extension to 1.9 Å, 2-fold averaging and solvent flattening, in a single run with the program dm (24), using the automask option. The correlation coefficient between the masked map areas before averaging was 0.62, improving to 0.86 after the last cycle. This masking allowed for the calculation of better maps (Fig. 2A), which were used as a starting point for autobuilding with warpntrace (25). In 140 cycles (about 24 h on a Silicon Graphics 225 MHz R10000 chip), warpntrace was able to build 910 of 998 residues in 27 chains with a connectivity index of 0.94. The new warpntrace side-chain placement algorithm (A. Perrakis, personal communication) was applied, placing side chains according to a combination of a rotamer library and real space torsional refinement. The resulting model was then further improved with model building in o (26), including crosscopying of main-chain fragments that were built in one of the monomers, but absent in the other. This procedure resulted in a model containing 990 of 998 residues. Initial rigid body refinement in cns (27) of this model resulted in an R factor of 0.27. Further iterations of refinement with cns (including simulated annealing) and model building with o resulted in the final model as described in Table 2 and Fig. 2B. Throughout the refinement, structural integrity was monitored by using what if (28) and procheck (29). The structure has good geometry (Table 2), with two residues in nonallowed regions of the Ramachandran plot. No density was observed for residues 1, 2, and 499 for monomer A, whereas residues 1–3 were missing for monomer B. In addition, the loop extending over the active site (around residue 320; see Fig. 2C) is poorly defined, as indicated by the electron density maps and relatively high B factors.
Figure 2.
(A) Comparison of experimental and final maps. An area around the active site residue Glu144 is drawn in a stick representation. A 1 σ contoured Fo,φMLPHARE map is shown in black, calculated by using the phases at the end of heavy atom refinement with MLPHARE. A 2Fo-Fc,φcalc map is shown at the end of refinement with CNS, contoured at 1.4 σ (in red). (B) The two molecules in the asymmetric unit, color-coded to identify various regions. The TIM barrel (gray), the α/β-domain (yellow), the support loop (red), the linker (blue), and the ChBD (green). (C) ChiB, as in Fig. 2B, with the flexible loop covering the active site (green), the active site residue (red sticks), the porch loop (orange), and the exposed aromatic residues (black sticks). (D) Superposition of the ChBD of ChiB (blue ribbon) and the CeBD of endoglucanase Cel5 (gray ribbon). Most of the support loop of the catalytic domain of ChiB is shown as a dark-blue ribbon. Trp252 also is shown in magenta. The substrate-binding residues for the CeBD are shown in yellow, and the equivalent residues in the ChBD are shown in magenta. The disulfide bond between the termini of the CeBD is shown in green. Polar residues lining the path of aromatic residues in ChiB are shown in magenta. Labels correspond to the ChiB sequence. Note the almost exact overlap of the conserved β-strands.
Table 2.
Refinement statistics
| Statistic | Value |
|---|---|
| R | 0.183 |
| Rfree | 0.213 |
| No. of molecules in the a.s.u. | 2 |
| No. of atoms | 8063 |
| No. of water molecules | 259 |
| Data/parameter ratio | 2.8 |
| Coordinate rmsd bonds | 0.016 Å |
| Coordinate rmsd angles | 1.87° |
| B-factor rmsd bonds (main chain) | 2.4 Å2 |
| B-factor rmsd bonds (side chain) | 3.8 Å2 |
| B-factor rmsd angles (main chain) | 3.0 Å2 |
| B-factor rmsd angles (side chain) | 5.0 Å2 |
| Average B-factor (main chain) | 23.4 Å2 |
| Average B-factor (side chain) | 26.9 Å2 |
| Average B-factor (water) | 29.6 Å2 |
a.s.u., asymmetric unit; rmsd, rms deviation.
Model of Chitotetraose in the ChiB Active Site.
A chitotetraose molecule was modeled into the ChiB active site by using an approach similar to that published by Tews and coworkers (13). The structures of hevamine in complex with the inhibitor allosamidin (occupying the −4, −3, −2, and −1 subsites) (30), and chitobiase in complex with chitobiose (occupying the −1 and +1 subsites) (3) both were superimposed on ChiB with the Ca atoms and a structural alignment obtained with dali (31). After a slight manual adjustment of the chitobiose, this method allowed the formation of a proper glycosidic link between the −2 sugar in allosamidin and the −1 sugar in chitobiose. The −4 sugar from the allosamidin structure was removed, because it had a large number of unfavorable contacts with a loop at the end of the substrate-binding cleft. After deletion of four active site waters that would overlap with the modeled chitotetraose, the complex was energy-minimized with programs from the gromos87 suite (32).
Results and Discussion
Overall Structure.
The structure of ChiB was solved by MIRAS and refined to 1.9-Å resolution. ChiB consists of a catalytic domain (residues 1–425; see Fig. 1) and a linker (residues 426–450), plus a small C-terminal domain (residues 451–499), referred to as the ChBD (Fig. 2). The catalytic domain has a fold similar to that of ChiA, including the existence of a tightly associated α/β-domain (residues 292–379; see Figs. 1 and 2C), which in ChiB provides a flexible loop (residues 310–325) near the active site. Another notable insertion in ChiB concerns residues 239–262. This insertion interacts with the ChBD and will be referred to as the support loop (Figs. 1 and 2). ChiB is folded into an elongated molecule with a largest dimension of 81 Å, attributable to the protruding ChBD (Fig. 2C). The active site is formed by a long, deep cleft (about 40 Å long, 15 Å wide, and 20 Å deep), which is characterized by a large number of exposed hydrophobic residues and harbors the catalytic residue Glu144 (Figs. 1 and 2). There are two outliers in the Ramachandran plot, both concerning the main-chain dihedrals of residues 317 and 320, which correspond to a poorly defined loop. There are three cis-peptide bonds (residues 50–51, 144–145, and 403–404) that also are present at equivalent positions in ChiA. The asymmetric unit contains two molecules of ChiB, related by a local twofold axis (Fig. 2B). The contacts between the two molecules are extensive (for example, the total buried surface per molecule is 810 Å2), but gel filtration and glutaraldehyde crosslinking experiments show that ChiB is a monomer in solution (B.S. and V.G.H.E., unpublished observations). The most notable features of the ChiB structure are the well-defined ChBD, a path of exposed aromatic residues, and a polar channel extending from the active site down into the TIM barrel core, which will be discussed in detail in Active Site and Polar Core.
ChBD.
A remarkable feature of the ChiB structure is the presence and location of the ChBD. This domain, formed by three antiparallel β-strands connected by long loops, is structurally homologous to the CeBD of endoglucanase Cel5 (Fig. 2D; ref. 17). The ChBD has two surface exposed aromatic residues (Trp479 and Tyr481), whose structural homologues in the CeBD (Trp43 and Tyr44) have been shown to be important for interaction with cellulose (33). The CeBD has an extra loop with an additional exposed tryptophan, which lies in line with the other exposed aromatic residues (Fig. 2D). One of the characteristic features of the family V CeBDs is a conserved disulfide bond between Cys4 and Cys61, which is not present in the ChBD structure. Bordering the path of aromatic residues lie a few polar residues that could serve to form specific hydrogen bonds with the N-acetyl groups of the chitin polymer (Fig. 2D). Starting from Trp479 and Tyr481 on the ChBD, a linear path of regularly spaced exposed aromatic residues leads into the active site, via Trp252 and Tyr240, both on the support loop (Fig. 2). The distances between subsequent exposed residues is 11 Å, on average, which is in the same range as the distance between equivalent atoms in the first and third GlcNAc of (GlcNAc)3 bound to hevamine (10.4 Å) (11). Thus, the incorporation of the support loop and the ChBD in ChiB extends the substrate-binding site considerably. The distance from the catalytic residue to the most remote of the exposed aromatic residues (Tyr481; Fig. 2D) is 55 Å, which corresponds to the length of a chitin chain with 10 subsites.
The ChBD is connected to the catalytic domain by a linker. This linker starts at residue 424, where the last helix of the TIM barrel core ends, and ends at approximately residue 451, the first residue of the ChBD. The structure (Fig. 2C) reveals that this linker runs along the surface of the catalytic domain and has no defined secondary structure. Linkers between catalytic and substrate-binding domains in glycosyl hydrolases have been suggested as being flexible, but this seems not to be the case in ChiB. First, the electron density for the linker is well defined (with an average B-factor of 24.8 Å2 for all atoms), which would not be expected if it were highly mobile. Second, the linker has many contacts with the catalytic core (nine hydrogen bonds and nine apolar side chain–side chain contacts). The ChBD itself has a few additional contacts with the support loop (Fig. 2C), involving residues Leu493 (apolar contact) and Lys494 (hydrogen bond) on the ChBD. The general picture that emerges is that the ChBD is intimately linked to the rest of the protein. This finding is in agreement with preliminary microcalorimetry and limited proteolysis studies, which indicate that unfolding of ChiB is highly cooperative (B.S., B. Kjellesvik, and A. Filimonov, unpublished observations).
Active Site and Polar Core.
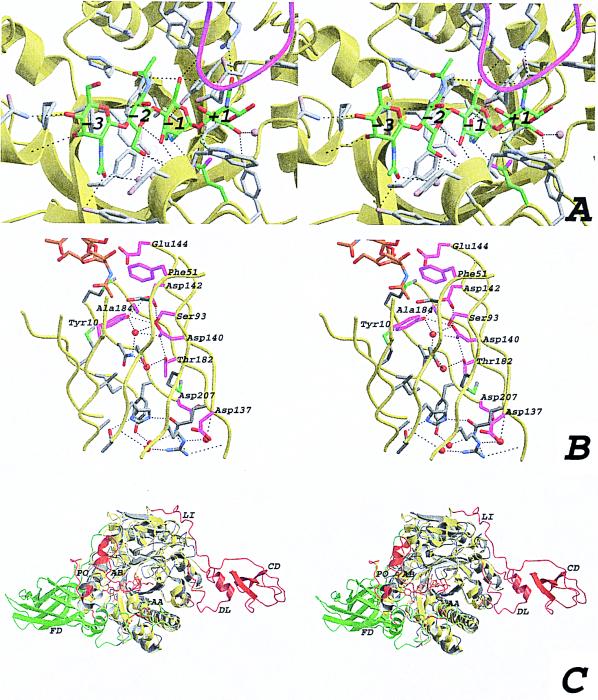
Soaking experiments with substrate (GlcNAc4) so far have not yielded different Fourier maps with significant signal. Given the long soaking times (several days) and the fact that data collection was at room temperature, it is possible that the ChiB molecules in the crystal hydrolyzed all available substrate. However, the tight packing between the monomers in the asymmetric unit could also prevent binding of the substrate. To test this theory, we modeled the structure of chitotetraose in the active site by using the hevamine–allosamidin and chitobiase–chitobiose complexes (13). Careful manual adjustments and energy minimization resulted in a plausible model (Fig. 3A). The oligosaccharide interacts with the protein through both hydrophobic and polar contacts, involving the residues marked in a structure-based alignment of three family 18 chitinases (Fig. 1). The substrate model fits the active site well, without any steric clashes with atoms from other ChiB monomers in the crystal, suggesting that the packing would allow ChiB still to be active. Four well-defined water molecules in the ChiB active site are replaced with carbonyl or hydroxyl groups from the chitin fragment. The oxygen of the scissile bond is within 3.3 Å of Oɛ2 of the catalytic Glu144. The −3 subsite is located near the end of the cleft, where a barrier is formed by the short helix and loop between residues 14 and 26 (the porch loop), which prevents binding of substrates extending longer than 3 units from the scissile glycosidic bond. If an additional GlcNAc would bind to the −4 subsite, based on the position seen in the hevamine–GlcNAc3 complex (11), several clashes would exist. Defining a clash as van der Waals radii overlapping for more than 0.25 Å, the residues Pro14, Thr15, Asn16, and Gln17 in ChiB would have a total of 61 steric clashes with atoms C1, C2, N2, C3, O3, C4, O4, C5, O5, C6, O6, C7, O7, and C8 on the predicted position of the GlcNAc at position −4. In addition, there are no exposed hydrophobic residues on the porch loop. Given the steric clashes and this nature of the porch loop, we think it is unlikely that the substrate extends beyond −3 in ChiB.
Figure 3.
(A) Stereo view of the active site with the modeled chitotetraose (same view as in Fig. 1C). The ChiB backbone is shown as a yellow ribbon. The modeled chitotetraose is shown in a stick representation, with the carbons colored green. Side chains within 5 Å of the chitotetraose are depicted by gray sticks, and also are indicated in Fig. 1. Possible hydrogen bonds are drawn as black dashed lines, and the residues involved are indicated in Fig. 1. The four water molecules that are predicted to be replaced by the substrate are shown as blue transparent spheres. The GlcNAc residues are labeled from −3 to +1, corresponding to their location with respect to the active site residue (15). The loop around residue 316, partially covering the active site, is shown in magenta. (B) Stereo view of the interior of the ChiB TIM barrel. The strands forming the TIM barrel are shown as a yellow ribbon. Side chains of residues lining the inside of the barrel are shown as sticks. Side chains conserved in ChiA, ChiB, and hevamine are colored magenta. Water molecules in the structure are shown as red spheres. Hydrogen bonds are shown as black dashed lines. Conserved residues are labeled according to the ChiB sequence. Part of the chitotetraose model is shown as sticks, with carbon atoms colored orange. (C) Stereo view of a superposition of ChiA and ChiB. Both structures are shown in a ribbon representation. ChiB is colored yellow, except for residues that correspond to insertions in ChiB with respect to ChiA, which are colored red. ChiA is colored gray except for residues that correspond to insertions in ChiA with respect to ChiB, which are colored green. Some insertions are indicated with two-letter labels. AA, active site covering loop in ChiA; AB, active site covering loop in ChiB; CD, ChBD in ChiB; DL, ChBD support loop in ChiB; FD, fibronectin domain in ChiA; LI, linker in ChiB; PO, porch loop in ChiB.
The sugar at the −3 subsite interacts with the protein through four hydrogen bonds and one apolar contact (see Fig. 1). None of the side chains involved in these contacts is conserved. The −2 subsite is characterized by relatively few interacting side chains (4), although some of the apolar residues interacting with the −1 sugar have a few additional contacts with the −2 sugar. The −1 GlcNAc has by far the most protein contacts. A total of 10 side chains interact with the −1 sugar, most of which are apolar. Interestingly, almost all residues that are both conserved and interact with the chitotetraose are part of the −1 subsite (Fig. 1). This subsite, with its many interacting side chains, has been shown to distort the −1 sugar that plays a role in catalysis (13, 14). The +1 subsite is characterized by the Oɛ1 atom of the catalytic Glu144 and by five additional residues, none conserved, which form a total of five hydrogen bonds. Starting with the +1 subsite, the active site curves upward toward the support loop and the ChBD.
The catalytic Glu144 is located at the end of β-strand 4 of the TIM barrel (Fig. 1). Below it, moving away from the active site and down into the TIM barrel core, lies a string of conserved aspartic acid residues (Asp142, Asp140, and Asp137), reaching to the backside of the TIM barrel (Figs. 1 and 3B). These residues form the DxxDxDxE motif observed in all family 18 chitinases (Fig. 1). Together with additional negatively charged residues around the active site and a cluster of positively charged residues at the backside of the TIM barrel, they generate a dipole, pointing from the active site residue down through the center of the barrel. Inspection of the inside of the barrel (Fig. 3B) reveals an extensive network of hydrogen bonds in the core of the TIM barrel. Many of these hydrogen bonds are formed to two well-ordered (B-factors of 18.3 and 18.9 Å2) water molecules that are buried in the center of the core (Fig. 3B). The inside of TIM barrels in other proteins is usually formed by tightly packed apolar residues. There are exceptions, however. For instance, the structure of enolase reveals a similar polar network through the TIM barrel core (34), and in methylmalonyl-CoA mutase, short polar residues line the inside of the barrel, which is used for supplying the active site with substrate (35). Most of the residues lining the inside of the TIM barrel core in ChiB, including some polar ones, are conserved in ChiA and hevamine (Fig. 1). Unlike ChiB, however, neither ordered water molecules nor the continuous string of hydrogen bonds are observed on the inside of the barrel in either ChiA or hevamine. The precise function of the polar network inside the TIM barrel is unclear. However, mutagenesis studies have shown that acidic residues in the DxxDxDxE motif are important for catalytic activity (B.S., B. Gaseidnes, and V.G.H.E., unpublished data; refs. 36, 37).
Comparison with ChiA and Other Glycosidic Hydrolases.
After the structure of ChiA (2) and the endo/exocellulase E4 from Thermomonospora fusca (10), the structure of ChiB is the third structure of a cellulase/chitinase that contains both the catalytic domain and a sugar-binding domain. These domains are all dominated by β-strands, but have different sizes (114, 143, and 49 residues, respectively) and topologies. In contrast to the ChBDs in ChiA and ChiB, the CeBD in cellulase E4 does not have aromatic residues that are as conspicuously exposed as the aromatic residues in the ChBDs. ChiB resembles cellulase E4 in that the sugar-binding domain is located at the reducing side of the active site and in that the sugar binding cleft is blocked at the nonreducing side of subsites −3 and −4, respectively. These findings provide a structural explanation for the exoactivity of both enzymes and show that the enzymes degrade their polymeric substrates from the nonreducing end. The fact that the porch loop prevents sugar binding in the −4 position, whereas there is a proper −3 subsite, suggests that ChiB acts as a chitotriosidase. Analysis of degradation of oligomeric chitin with ChiB has shown that the enzyme converts GlcNAc6 predominantly into GlcNAc3 (16). This finding supports the idea that ChiB has a chitotriosidase activity, with the structural comparison suggesting cleavage from the nonreducing end. In addition, the alignment of the ChBD exposed aromatics with those in the catalytic core could support a mechanism for processivity, as suggested previously for cellulase E4 (10), where the polymer is allowed to slide into the active side while retaining its interaction with the ChBD/CeBD. If ChiB acts exclusively as a chitotriosidase, this would not be possible, because processivity with an odd number of GlcNAcs being cleaved off implies a subsequent 180° rotation of the polymer along its long axis. Mutations suggest that the native CeBD in endoglucanase Cel5 is important for binding to polymeric substrates (33), and we aim to investigate the function of the ChBD in ChiB by deletion mutants.
ChiA and ChiB both convert chitin into monomers and dimers of GlcNAc. Still, the two enzymes play different roles in the degradation of chitin, as illustrated by the fact that synergistic effects on the degradation rate are obtained upon combining the two enzymes (16). Differences between ChiA and ChiB can readily be explained by a structural comparison of the two enzymes (Figs. 1 and 2C). Whereas the support loop and the ChBD (both absent in ChiA; Fig. 1) extend the substrate-binding cleft of ChiB on the reducing side of the active site, the fibronectin III-like ChBD of ChiA (absent in ChiB; Fig. 1) extends the substrate-binding cleft at the nonreducing side. Thus, assuming that the extra domains play a key role in chitin binding, any chitobiosidase activity displayed by ChiA is likely to result in degradation of the chitin chain from the reducing end. Consequently, ChiA and ChiB should at least have additive effects. The synergy between ChiA and ChiB likely is to be attributable to the endo-activity displayed by the former enzyme only. The porch loop is absent in ChiA, meaning that the substrate-binding cleft is open on both sides of the active site. Interestingly, ChiB contains a 17-residue insertion between residues 314 and 336 (Fig. 1). This insertion is part of a mobile loop that extends well over the substrate-binding cleft, positioning Asp316 such that it could interact with bound substrate (Fig. 2A). ChiA has a similar loop on the other side of the substrate-binding cleft (inserted between residues 56 and 57 in ChiB; Fig. 1), which, however, is much shorter and does not extend over the substrate-binding cleft. Thus, the cleft in ChiA has a more groove-like character, typical for endo-enzymes, whereas the substrate-binding cleft of ChiB has a more tunnel-like character, which is often observed in exo-enzymes (38–40).
The combined crystal structures of multidomain ChiA and ChiB offer a detailed and exciting view on the machinery that S. marcescens employs for degrading chitin, and they add significantly to our general understanding of the mechanisms involved in the degradation of insoluble biopolymers such as chitin and cellulose. Interestingly, the structure of the N-acetylglucosaminidase of S. marcescens is also known (3), whereas work on the ChiC (both by homology modeling and x-ray crystallography; D.M.F.v.A. and V.G.H.E., unpublished observations) has yet to be initiated. This investigation will open the perspective of structural knowledge of a complete bacterial chitinolytic machinery to becoming available in the near future.
Acknowledgments
We thank Peter Everitt (European Molecular Biology Laboratory) and Simone Frank (Prophysics) for installing the mirrors on our rotating anode. Anastassis Perrakis is gratefully acknowledged for the use of the latest warpntrace algorithms. We thank the European Molecular Biology Laboratory-Hamburg outstation for use of beamlines X11 and X31 at Deutsches Elektronen Synchrotron. This work was supported by European Union Grant BIO4-CT-960670.
Abbreviations
- ChBD
chitin-binding domain
- CeBD
cellulose-binding domain
- ChiX
chitinase X
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1E15).
References
- 1.Fuchs R L, McPherson S A, Drahos D J. Appl Environ Microbiol. 1986;51:504–509. doi: 10.1128/aem.51.3.504-509.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perrakis A, Tews I, Dauter Z, Oppenheim A B, Chet I, Wilson K S, Vorgias C E. Structure (London) 1994;2:1169–1180. doi: 10.1016/s0969-2126(94)00119-7. [DOI] [PubMed] [Google Scholar]
- 3.Tews I, Perrakis A, Oppenheim A, Dauter Z, Wilson K S, Vorgias C E. Nat Struct Biol. 1996;3:638–648. doi: 10.1038/nsb0796-638. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe T, Kimura K, Sumiya T, Nikaidou M, Suzuki K, Suzuki M, Taiyoji M, Ferrer S, Regue M. J Bacteriol. 1997;179:7111–7117. doi: 10.1128/jb.179.22.7111-7117.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perrakis A, Ouzonis C, Wilson K S. Folding Des. 1997;2:291–294. doi: 10.1016/S1359-0278(97)00040-0. [DOI] [PubMed] [Google Scholar]
- 6.Bayer E A, Chanzy H, Lamed R, Shoham Y. Curr Opin Struct Biol. 1998;8:548–557. doi: 10.1016/s0959-440x(98)80143-7. [DOI] [PubMed] [Google Scholar]
- 7.Svitil A L, Kirchman D L. Microbiology. 1998;144:1299–1308. doi: 10.1099/00221287-144-5-1299. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe T, Ito Y, Yamada T, Hashimoto M, Sekine S, Tanaka H. J Bacteriol. 1994;176:4465–4472. doi: 10.1128/jb.176.15.4465-4472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herissat B, Davies G. Curr Opin Struct Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 10.Sakon J, Irwin D, Wilson D B, Karplus P A. Nat Struct Biol. 1997;4:810–818. doi: 10.1038/nsb1097-810. [DOI] [PubMed] [Google Scholar]
- 11.Terwisscha van Scheltinga A C, Hennig M, Dijkstra B W. J Mol Biol. 1996;262:243–257. doi: 10.1006/jmbi.1996.0510. [DOI] [PubMed] [Google Scholar]
- 12.McCarter J D, Withers S G. Curr Opin Struct Biol. 1994;4:885–892. doi: 10.1016/0959-440x(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 13.Tews I, Terwisscha van Scheltinga A C, Perrakis A, Wilson K S, Dijkstra B W. J Am Chem Soc. 1997;119:7954–7959. [Google Scholar]
- 14.Brameld K A, Goddard W A., III J Am Chem Soc. 1998;120:3571–3580. [Google Scholar]
- 15.Davies G J, Wilson K S, Henrissat B. Biochem J. 1997;321:557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brurberg M B, Nes I F, Eijsink V G H. Microbiology. 1996;142:1581–1589. doi: 10.1099/13500872-142-7-1581. [DOI] [PubMed] [Google Scholar]
- 17.Brun E, Moriaud F, Gans P, Blackledge M J, Barras F, Marion D. Biochemistry. 1997;36:16074–16086. doi: 10.1021/bi9718494. [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 19.Tong L, Rossmann M G. Methods Enzymol. 1997;276:594–611. [PubMed] [Google Scholar]
- 20.ccp4, Collaborative Computational Project, No. 4. (1994) Acta Crystallogr. D50, 760–763. [DOI] [PubMed]
- 21.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otwinowski Z. Daresbury Study Weekend Proceedings. Daresbury Laboratory, Daresbury, U.K.: Science and Engineering Research Council; 1991. [Google Scholar]
- 23.Lu G. J Appl Crystallogr. 1999;32:365–368. [Google Scholar]
- 24.Cowtan K. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography No. 31. Warrington, U.K.: SERC Daresbury Laboratory; 1994. pp. 34–38. [Google Scholar]
- 25.Perrakis A, Morris R, Lamzin V S. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 26.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 27.Brünger A T, Adams P D, Clore G M, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, Read R J, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Vriend G. J Mol Graphics. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski R A, McArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 30.Terwisscha van Scheltinga A C, Armand S, Kalk K H, Isogai A, Henrissat B, Dijkstra B W. Biochemistry. 1995;34:15619–15623. doi: 10.1021/bi00048a003. [DOI] [PubMed] [Google Scholar]
- 31.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 32.van Gunsteren W F, Berendsen H C J. gromos, BIOMOS, Biomolecular Software. Univ. of Groningen, Groningen, The Netherlands: Laboratory of Physical Chemistry; 1987. [Google Scholar]
- 33.Simpson H D, Barras F. J Bacteriol. 1996;181:4611–4616. doi: 10.1128/jb.181.15.4611-4616.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stec B, Lebioda L. J Mol Biol. 1990;211:235–248. doi: 10.1016/0022-2836(90)90023-F. [DOI] [PubMed] [Google Scholar]
- 35.Mancia F, Smith G A, Evans P R. Biochemistry. 1999;38:7999–8005. doi: 10.1021/bi9903852. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe T, Kobori K, Miyashita K, Fujii T, Sakai H, Uchida M, Tanaka H. J Biol Chem. 1993;268:18567–18572. [PubMed] [Google Scholar]
- 37.Watanabe T, Uchida M, Kobori K, Tanaka H. Biosci Biotechnol Biochem. 1994;58:2283–2285. doi: 10.1271/bbb.58.2283. [DOI] [PubMed] [Google Scholar]
- 38.Davies G, Henrissat B. Structure (London) 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 39.Spezio M, Wilson D B, Karplus P A. Biochemistry. 1993;32:9906–9916. doi: 10.1021/bi00089a006. [DOI] [PubMed] [Google Scholar]
- 40.Rouvinen J, Bergfors T, Teeri T, Knowles J K C, Jones T A. Science. 1990;249:380–386. doi: 10.1126/science.2377893. [DOI] [PubMed] [Google Scholar]