Abstract
Recent studies suggest that the chromosome 16 inversion, associated with acute myeloid leukemia M4Eo, takes place in hematopoietic stem cells. If this is the case, it is of interest to know the effects of the resulting fusion gene, CBFB-MYH11, on other lineages. Here we studied T-cell development in mice expressing Cbfb-MYH11 and compared them with mice compound-heterozygous for a Cbfb null and a hypomorphic GFP knock-in allele (Cbfb−/GFP), which had severe Cbfb deficiency. We found a differentiation block at the DN1 stage of thymocyte development in Cbfb-MYH11 knock-in chimeras. In a conditional knock-in model in which Cbfb-MYH11 expression was activated by Lck-Cre, there was a 10-fold reduction in thymocyte numbers in adult thymus, resulting mainly from impaired survival of CD4+CD8+ thymocytes. Although Cbfb-MYH11 derepressed CD4 expression efficiently in reporter assays, such derepression was less pronounced in vivo. On the other hand, CD4 expression was derepressed and thymocyte development was blocked at DN1 and DN2 stages in E17.5 Cbfb−/GFP thymus, with a 20-fold reduction of total thymocyte numbers. Our data suggest that Cbfb-MYH11 suppressed Cbfb in several stages of T-cell development and provide a mechanism for CBFB-MYH11 association with myeloid but not lymphoid leukemia.
Introduction
The core-binding factor (CBF) family is a group of transcription factors composed of α and β subunits.1 There are 3 members of the α subunit in mammals, Cbfbα1 (Runx2), Cbfα2 (Runx1), and Cbfα3 (Runx3), encoded by 3 separate genes.2,3 There is only a single gene encoding the β subunit, Cbfb.4 Cbfβ binds Runx proteins and stabilizes their interactions with DNA.5–8 Cbfβ may also be able to prevent the ubiquitin-mediated degradation of Runx proteins.9 Runx1 and Cbfb null mice both have impaired definitive hematopoiesis and die at embryonic day (E) 12.5 from massive hemorrhage.10–12 They are required for the initial stages of hematopoiesis in the aorta-gonad-mesonephros (AGM) region13,14 and are critical for embryonic angiogenesis as well.15 Due to their phenotypic similarities, Cbfb is required for Runx1 functions in embryonic hematopoiesis.
In humans, acute myeloid leukemia (AML) subtype M4Eo is associated with a chromosome 16 inversion, inv(16)(p13; q22), in which CBFB fuses to MYH11, the gene encoding smooth muscle myosin heavy chain (SMMHC). The fusion gene, CBFB-MYH11, produces a chimeric protein CBFβ-SMMHC.4 CBFβ-SMMHC has increased binding affinity for Runx1 (compared to wild-type CBFβ) and can suppress its function through several potential mechanisms, including sequestration and active repression.16,17 We have previously created a knock-in mouse model expressing the Cbfb-MYH11 fusion gene.18 Heterozygous Cbfb-MYH11 knock-in mice exhibit a phenotype (a block of definitive hematopoiesis, hemorrhage, and embryonic lethality) nearly identical to that of the Cbfb12 and Runx1 null mice,10,11 suggesting that the fusion gene Cbfb-MYH11 dominantly suppresses Runx1/Cbfb function in vivo.18
We recently also generated a knock-in mouse model for a Cbfb-GFP fusion.14 Homozygous Cbfb-GFP (CbfbGFP/GFP) mice died at birth due to bone formation defects similar to those observed in Runx2-deficient mice, suggesting that Cbfb is also required for Runx2 function in bone formation.19 Interestingly, AGM hematopoiesis was relatively normal and there was no hemorrhage in the CbfbGFP/GFP embryos at E12.5.14 Our data show that the Cbfb-GFP allele is hypomorphic and that Runx2 is more sensitive to Cbfb dosage than Runx1 is. The hypomorphic nature of the Cbfb-GFP allele and Cbfb dosage sensitivity is supported by the observation that Cbfb−/GFP mice also die at birth with similar but more severe bone formation defects (L.Z. and P.P.L., unpublished results, December 2005).
Runx proteins have specific functions during T-cell development. T-lymphoid progenitor cells migrate from the fetal liver or bone marrow to the thymus,20 where they differentiate into mature T cells through a series of defined stages with characteristic gene rearrangements and expression of specific surface markers. The cells at the earliest stages of development in the thymus lack expression of both CD4 and CD8, and therefore are called double negative (DN) cells. DN cells can be subdivided into 4 stages based on cell surface expression of CD25 and CD44. For the predominant (αβ)T lineage, progression beyond the third DN stage (CD44loCD25+) requires TCRβ gene rearrangement. The developing thymocytes then start to express both CD4 and CD8 to become double positive (DP) cells, of which a subset is selected to become mature CD4+CD8− and CD4−CD8+ single-positive (SP) cells.21–23 Studies of Runx1 and Runx3 null mice indicate that Runx1 is required for active repression of CD4 in DN thymocytes, whereas Runx3 is required for establishing epigenetic silencing of CD4 in the CD8-lineage thymocytes.24,25 Runx1 is also required for the developmental progression of DN-stage thymocytes.24 Runx3-deficient cytotoxic CD8+ T cells, but not helper CD4+ cells, have defective responses to antigen, suggesting that Runx3 is important for both lineage specification and function of CD8-lineage T lymphocytes.24,25
Previous studies in our laboratories suggested the involvement of Cbfb in the development of lymphoid lineages.26,27 We hypothesized that Cbfb has a critical role in T-cell development and that Cbfb-MYH11 suppresses Cbfb and impairs T-cell development. In this study, we analyzed thymocytes in several Cbfb-deficient mouse models, including Cbfb knockout,12 Cbfb-GFP knock-in,19 conditional expression of Cbfb-MYH11,26 and Cbfb-MYH11 chimeras.18 This study demonstrates that Cbfb-MYH11 suppresses Cbfb in thymocyte development during DN stages and reduces the survival of thymocytes, but has limited effect on CD4 expression.
Materials and methods
Animals
All animals used and the procedures performed in this study were approved by the NHGRI Animal Care and Use Committee. Cbfb knockout, Cbfb-GFP knock-in, conventional, and conditional Cbfb-MYH11 knock-in (Cbfb+/56M) mice have been described previously.12,18,19,26 The Tg(Lck-Cre) and the Tg(Lck-hBcl2) transgenic mice were obtained from the Jackson Laboratories (Bar Harbor, ME), and the Tg(Tcrb) mice were purchased from Taconic Farms (Germantown, NY). Mice, 5 to 6 weeks old, were used in experiments unless indicated in the text.
Quantitative PCR
Thymocytes were sorted by flow cytometry and DNA was extracted with Qiagen DNAeasy kit (Qiagen, Valencia, CA) from the separated cell populations. Cybergreen quantitative polymerase chain reaction (PCR; ABI7500, cybergreen kit from Invitrogen, Carlsbad, CA) was used to determine the efficiency of Cre-mediated deletion and 2 sets of PCR primers were used simultaneously. Primers for Cbfb exons 5 and 6 are: forward (5′-CAGGAAGATGCATTAGCACAA) and reverse (5′-AGATCATCACCGCCACCTAA) and primers for the neomycin gene are neo forward 5′-ATCAGGATGATCTGGACGAAGA, and neo reverse 5′-CCACAGTCGATGAATCCAGAA. PCRs were run as follows: 50°C for 2 minutes, 95°C for 10 minutes, and 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute. By comparing Ct numbers of PCRs between these 2 sets of primers, the deletion efficiency can be calculated (according to manufacturer's suggestions). DNA from Cbfb+/56M mice was used to standardize the Ct numbers.
Genotype analysis
DNA isolated from tail snips was used for all genotyping analysis. The Cbfb56M allele was detected by PCR using primers specific for Cbfb exons 5 and 6 as described and Tg(Lck-Cre) allele was detected by primers Cre for 5′CGATGCAACGAGTGATGAGG3′, and Cre rev 5′GCATTGCTGTCACTTGGTCGT3′. The DNA samples were initially denatured at 94°C for 2 minutes, followed by 35 cycles of amplification (30 seconds each at 94°C, 53°C [for exon 5 and 6]/60°C [Cre primers], and 72°C), and a final 5-minute extension step at 72°C.
Flow cytometry
Cells from the thymus, spleen, and lymph node were washed with fluorescence-activated cell sorting (FACS) buffer (5% fetal calf serum in phosphate-buffered saline) and counted. Spleen and peripheral blood samples were incubated in ACK lysing buffer (Biowhittaker, Walkersville, MD) prior to staining with antibodies. Cells were stained with FITC, phycoerythrin (PE), PE-Cy5, APC, and APC-Cy7–conjugated antibodies to CD4, CD8, CD25, CD44, CD3, HSA, CD5, CD19, B220, TCRβ, TCRγδ, Ly9.1, and annexin V (BD PharMingen, San Diego, CA) for flow cytometry analysis. Appropriate isotype controls were used in each experiment. FACSCalibur and BDAria (BD PharMingen) were used to acquire data. FACS data were analyzed using Flowjo software (Tree Star, Ashland, OR).
Cell death and proliferation assays
Annexin-V and 7-AAD staining (BD PharMingen) were used to determine cell death and apoptosis. A BrdU-FITC or BrdU-APC kit (BD PharMingen) was used to measure proliferation in vivo following the manufacturer's instruction. In brief, 1 mg BrdU was injected intraperitoneally into mice and 16 hours later, the thymus tissue was harvested and stained following the manufacturer's protocol. FACS data were acquired using BDAria and LSRII (BD PharMingen) and analyzed with Flowjo software.
Histologic analysis and TUNEL assay
Tissues were fixed in 10% neutral-buffered formalin overnight at 4°C, dehydrated, and embedded in paraffin. Tissue blocks were sectioned at 5 μm and stained with hematoxylin and eosin. The TdT-mediated dUTP nick-end labeling (TUNEL) assay was performed using Apoptag peroxidase kit (S7100, Intergen, Purchase, NY) following the manufacturer's protocol.
In vitro reporter assay
CAT reporter constructs28,29 were transfected into the CD4− Jurkat cell clone (D1.1, from American Type Culture Collection, Manassas, VA) alone or with Runx1, Cbfb, and CBFB-MYH11 cDNA constructs16 by electroporation (BTX electroporation system, Electro Cell manipulator 600; BTX Instrument Division, Harvard Apparatus, Holliston, MA). CAT activity was measured following manufacturer's protocol (CAT ELISA, Roche Diagnostics, Basel, Switzerland). A luciferase vector was cotransfected to standardize transfection efficiency. The transfection was performed for 3 times with similar results.
Statistics
Data are shown as mean ± SE, and a Student t test was used to compare 2 groups of samples.
Results
Conditional expression of Cbfb-MYH11 in thymocytes
To express Cbfb-MYH11 in thymocytes, we crossed a mouse line (Cbfb+/56M) carrying a conditional Cbfb-MYH11 knock-in,26 which switches from expressing Cbfb to expressing Cbfb-MYH11 after Cre-mediated excision of an inserted exon 5/6 cDNA cassette, with Tg(Lck-Cre) mice that express Cre from the Lck promoter, which starts expressing in DN3 thymocytes30 (Figure 1A). The Tg(Lck-Cre)/Cbfb+/56M DP mice were viable and lived a similar life span (monitored for up to 1 year) compared with control littermates.
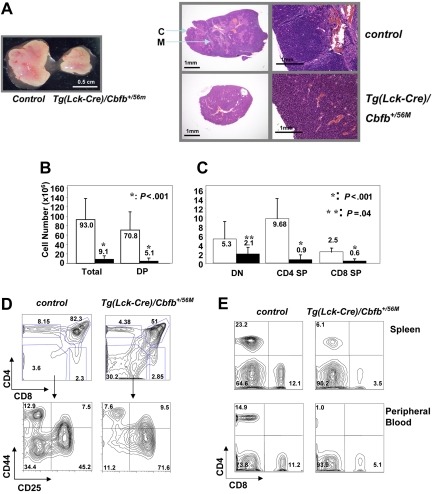
Figure 1.
Lck-Cre–mediated conditional Cbfb-MYH11 expression in adult thymus. (A) The Cbfb56M allele contains an insertion in intron 4 that includes exons 5 and 6 of Cbfb flanked by 2 lox sites. It also contains the knocked-in MYH11 cDNA in exon 5. Cre-mediated excision of the exons 5/6 cassette led to the restoration of the knock-in Cbfb-MYH11 fusion (Cbfb56M excised allele). PCR primers used for quantitative PCR to determine the efficiency of Cre-mediated deletion were: e5 and e6 primers for the Cbfb56M allele; neo primers for both the Cbfb56M and the Cbfb56M excised alleles. (B) Total thymocyte numbers from Tg(Lck-Cre) and Tg(Lck-Cre)Cbfb+/56M embryos at E17.5 (n = 4 for each genotype) and E18 (n = 3 for Tg(Lck-Cre) and n = 6 for Tg(Lck-Cre)Cbfb+/56M). (C) Representative contour plots of cell surface marker expression of thymocytes from Tg(Lck-Cre)/Cbfb+/56M and littermate control Tg(Lck-Cre) embryos at E18.0. The cells were stained with CD4, CD8, CD44, and CD25. The top panel shows CD4 and CD8 distribution, and the bottom panel shows CD44 and CD25 staining of DN cells.
We used quantitative PCR to determine the efficiency of Cre-mediated deletion. The efficiency of Lck-Cre–mediated deletion of the Cbfb exon 5/6 cassette varied in different stages of T-cell development, as well as among different mice. The deletion efficiency was 27% to 58% in DN cells, 72% to 91% in DP cells, and 27% to 55% in CD4+ SP and 40% to 58% in CD8+ SP cells (n = 5). In the spleen, the deletion efficiencies were 33% to 41% for CD4+ SP cells and 14% to 37% for CD8+ SP cells (n = 3). DP cells had the highest deletion efficiency, whereas mature CD4+ and CD8+ SP cells in the thymus and spleen had lower efficiencies, suggesting that cells with the exon 5/6 deletion did not efficiently develop from DP to SP stages.
Defective T-cell development in the Tg(Lck-Cre)/Cbfb+/56M mice
Analysis of embryonic thymi at E18 revealed that there were 2-fold fewer thymocytes in the Tg(Lck-Cre)/Cbfb+/56M mice compared with littermate controls and a milder decrease at E17.5 (Figure 1B). Surprisingly, we did not see an increased percentage of CD4+ SP cells nor decreased CD8+ SP populations (Figure 1C), as observed in Runx1 and Runx3 knockouts.
In the adult Tg(Lck-Cre)/Cbfb+/56M mice the thymi were reduced in size grossly (Figure 2A). Histologically the thymi had a near homogenous architecture, lacking the distinct cortical and medullary structures as observed in the wild-type thymi (Figure 2A). The total cellular numbers in the Tg(Lck-Cre)/Cbfb+/56M thymi were on average 10-fold lower than those of the control mice (Figure 2B). Further analysis showed that this reduced cellularity was primarily due to severe reduction of DP cells, since there were 14 times fewer DP cells in the Tg(Lck-Cre)/Cbfb+/56M thymi than in the control; in contrast the numbers of DN cells were only slightly reduced (Figure 2B-C). As a result, the Tg(Lck-Cre)/Cbfb+/56M thymi had proportionally more DN (38% ± 24% versus 6% ± 4%, n = 10, P < .01) and fewer DP (46% ± 23% versus 80% ± 5%, n = 10, P < .01) cells (Figure 2D). Within the DN population, the proportion of DN3 cells were significantly higher (64% ± 11% versus 38% ± 9% in controls, P = .01, n = 4), whereas that of DN4 cells was significantly lower (14% ± 2% versus 42% ± 4%, P < .001, n = 4) in the Tg(Lck-Cre)/Cbfb+/56M thymi, suggesting the existence of a developmental block between these 2 stages (Figure 2D). We also observed fewer CD4+ and CD8+ SP cells in the Tg(Lck-Cre)/Cbfb+/56M thymi (Figure 2C), and the spleen and peripheral blood contained lower percentages of CD4+ and CD8+ SP T cells as well (Figure 2E). Notably, there was a correlation between deletion efficiency and the severity of decreased cellularity: the higher the extent of deletion, the less the cellularity. Overall, conditional expression of Cbfβ-SMMHC in the thymus resulted in thymocyte developmental blockage and severely reduced production of mature T cells.
Figure 2.
T-cell developmental defects in the adult Tg(Lck-Cre)/Cbfb+/56M mice. (A) Tg(Lck-Cre)/Cbfb+/56M mice had smaller thymi (right) and their thymic architecture appeared to be homogeneous, as compared to the control mice (left). C indicates cortex; M: medulla. Left panel (gross view): pictures were taken with an Olympus SZ-40 (Tokyo, Japan) dissecting microscope camera with zoom setting at 2.5×. For the panel on the right, the 2 pictures on the left were taken with an Olympus SZ-40 dissecting microscope camera with zoom setting at 4×; the 2 pictures on the right were taken with a Nikon ECLIPSE E800 (Tokyo, Japan) microscope at 100× (10×/0.45 NA objective and 10× eyepiece). (B) Total and DP thymocyte numbers from adult thymi are graphed. (C) Numbers of DN, CD4+ SP, and CD8+ SP thymocytes are graphed. For panels B and C: □, littermate controls (n = 9); ■, Tg(Lck-Cre)/Cbfb+/56M mice (n = 10). Statistically significant P values (Student t test) are indicated. (D) Representative contour plots of cell surface marker expression of thymocytes from Tg(Lck-Cre)/Cbfb+/56M and littermate control mice. The cells were stained with CD4, CD8, CD44, and CD25. The top panels show CD4 and CD8 distribution, and the bottom panels show CD44 and CD25 staining of DN cells. (E) Representative contour plots of CD4 and CD8 expression of spleen and peripheral blood cells from Tg(Lck-Cre)/Cbfb+/56M and littermate control mice.
Decreased proliferation is not responsible for reduced thymocyte number in the Tg(Lck-Cre)/Cbfb+/56M mice
To determine the mechanism for the reduced thymocyte numbers in the Tg(Lck-Cre)/Cbfb+/56M mice, we examined the proliferation status of the thymocytes by measuring BrdU incorporation. The percentages of BrdU+ cells in total thymocytes and DN cells were comparable between the Tg(Lck-Cre)/Cbfb+/56M and the control mice (Figure 3A; n = 3 for control and n = 5 for Tg(Lck-Cre)/Cbfb+/56M mice). Interestingly the percentage of proliferating cells in the DP population was increased in the Tg(Lck-Cre)/Cbfb+/56M mice (34.2% versus 23.1% in the control mice, P = .05). Considering that the proportion of DP cells was reduced in the Tg(Lck-Cre)/Cbfb+/56M mice, the finding suggested that there might be increased turnover or reduced survival of DP cells in those mice. We also examined the distribution of BrdU+ cells among the differentiation stages of thymocytes and found that the distribution was comparable to that of the total thymocyte population (comparing the lower panels of Figure 3B with the upper panels of Figure 2D). Overall the results suggested that the reduction in cellularity in the Tg(Lck-Cre)/Cbfb+/56M thymi could not be entirely explained by proliferation defects.
Figure 3.
BrdU incorporation by thymocytes in the Tg(Lck-Cre)/Cbfb+/56M mice. (A) Percentages of BrdU+ cells among total thymocytes, DP cells, and DN cells in the littermate control (□, n = 3) and the Tg(Lck-Cre)/Cbfb+/56M mice (■, n = 6). *Only the difference in DP BrdU+ cells between the littermate control and the Tg(Lck-Cre)/Cbfb+/56M mice reached statistical significance (P = .05). (B) Distribution of BrdU+ cells relative to CD4 and CD8 expression.
Increased apoptosis in the Tg(Lck-Cre)/Cbfb+/56M thymi
We used annexin V staining to determine if there was increased apoptosis in the Tg(Lck-Cre)/Cbfb+/56M thymi. Annexin V staining of thymocytes showed an overall increase in apoptosis at the DN, DP, and SP stages in the Tg(Lck-Cre)/Cbfb+/56M mice (Figure 4A). The increases at all stages were statistically significant except for the CD8 SP cells (data not shown). Among DN cells, the increase of apoptosis was higher at DN2 and DN3 stages, but not at the DN4 stage (data not shown). TUNEL assay of thymic sections also revealed that there were more apoptotic cells in the Tg(Lck-Cre)/Cbfb+/56M thymi than in the controls (7.3% ± 1% (n = 4) versus 4% ± 1% (n = 3); P = .012).
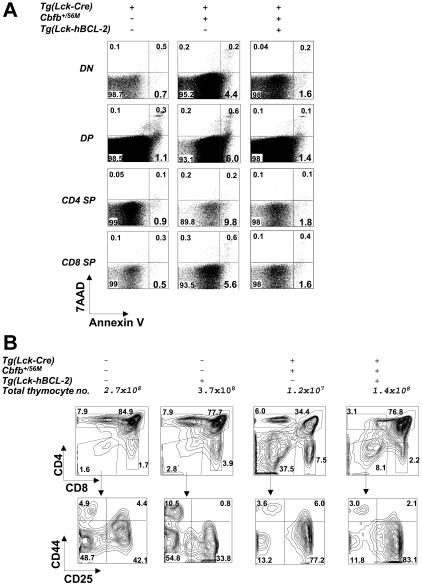
Figure 4.
Increased apoptosis of thymocytes in the Tg(Lck-Cre)/Cbfb+/56M mice. (A) Dot plots of annexin V and 7AAD staining in DN, DP, and CD4 and CD8 SP cells of the Tg(Lck-Cre), Tg(Lck-Cre)/Cbfb+/56M, and Tg(Lck-Cre)/Cbfb+/56M/Tg(Lck-hBcl-2) mice (all littermates). (B) Contour plots of cell surface marker expression on the thymocytes of mice with the genotypes as shown. The total thymocyte numbers for each genotype are also given.
If increased apoptosis was responsible for the decreased thymic cellularity, promoting thymocyte survival should increase thymocyte numbers. To test this possibility, we crossed the Tg(Lck-Cre)/Cbfb+/56M mice with the Tg(Lck-hBcl-2) mice, which carry a human BCL-2 transgene under the control of the Lck promoter.31 As shown in Figure 4A, Bcl-2 overexpression reduced apoptosis in almost every stage of T-cell development in Tg(Lck-Cre)/Cbfb+/56M mice to levels close to those of the wild-type controls. The Tg(Lck-Cre)/Cbfb+/56M/Tg(Lck-hBcl-2) mice had vastly increased total thymocyte numbers and increased DP population compared to the Tg(Lck-Cre)/Cbfb+/56M littermates (Figure 4B). However, the differentiation block in DN3 was not rescued (Figure 4B). These data indicate that there are at least 2 separate defects in the Tg(Lck-Cre)/Cbfb+/56M thymi: an increased apoptosis in the DP cells that is largely responsible for the reduced cellularity and a differentiation block between DN3 and DN4.
Transgenic expression of a rearranged Tcrb does not rescue the DN developmental block in the Tg(Lck-Cre)/Cbfb+/56M mice
The TCR genes undergo rearrangements during the DN2-DN3 stage. Successful rearrangement of the Tcrb locus and expression of TCRβ protein are required for the development of αβT cells beyond the DN3 stage.32 Given that the Tcrb enhancer contains Runx-binding sites,33 the partial developmental block in DN3 thymocytes in the Tg(Lck-Cre)/Cbfb+/56M mice raised the possibility that Cbfβ-SMMHC could impair Tcrb gene rearrangement or reduce Tcrb expression.
To address these possibilities, we first measured the cell surface TCRβ expression of DN, DP, and SP cells and found that it was comparable between the Tg(Lck-Cre)/Cbfb+/56M and the control mice (data not shown). In addition, we crossed the Tg(Lck-Cre)/Cbfb+/56M mice with the Tg(Tcrb) mice, which express a rearranged, functional Tcrb transgene that is capable of rescuing the DN stage block in Rag2-deficient mice.32 The Tg(Lck-Cre)/Cbfb+/56M/Tg(Tcrb) mice exhibited a similar phenotype as that in the Tg(Lck-Cre)/Cbfb+/56M mice, with severely reduced thymic cellularity and accumulation of DN thymocytes (Figure S1, available on the Blood website; see the Supplemental Figure link at the top of the online article), indicating that the defect caused by Cbfb-MYH11 was not rescued by the rearranged Tcrb gene. In addition, we measured the percentage and the total number of TCRγδ+ cells in the thymus, because TCRβ− cells may have developed into the TCRγδ lineage. We found that the total number of TCRγδ+ cells in the thymi was comparable to littermate controls (data not shown).
Cbfb-MYH11 derepresses CD4 expression in vitro but not in vivo
Runx1 and Runx3 play important roles in silencing CD4 expression, through a silencer element in intron 1 of the gene.28,34 Runx1 is required for CD4 silencer activity in DN thymocytes.24 Runx3 is required for CD4 silencing in CD8 SP cells and Runx3 deficiency results in the appearance of mature (HSAlo) DP thymocytes and peripheral DP T cells, both of which are made of CD8+ SP cells aberrantly expressing CD4.24 Analysis of Cbfb−/GFP and CbfbGFP/GFP embryos (Figure 7A,C), suggested that Cbfβ is required for the function of at least Runx1 in CD4 silencing. Therefore, if Cbfβ-SMMHC behaves as a dominant-negative suppressor of Cbfβ function, as is widely believed in the field, it should have generated a similar phenotype.
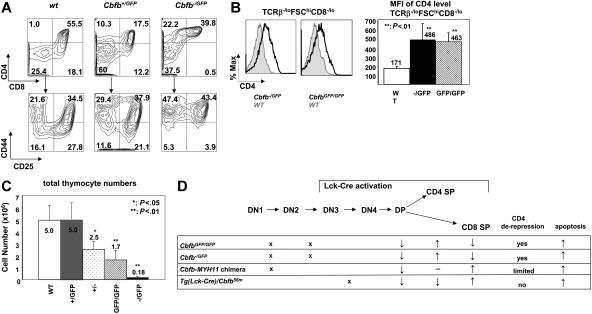
Figure 7.
T-cell development alterations in Cbfb−/GFP and CbfbGFP/GFP embryos. (A) Representative contour plots of cell surface marker expression of thymocytes from Cbfb+/+, Cbfb+/GFP, and Cbfb−/GFP embryos at E17.5. The cells were stained with CD4, CD8, CD44, and CD25. The top panels show CD4 and CD8 distribution, and the bottom panels show CD44 and CD25 staining of DN cells. (B) CD4 expression in immature thymocytes from E17.5 embryos. The left panel shows the histogram overlay of CD4 expression in TCRβloFSChiCD8−/lo thymocytes of wild-type (filled gray line), Cbfb−/GFP, and Cbfb GFP/GFP mice (black line). The right panel shows the MFI of CD4 expression in TCRβloFSChiCD8−/lo cells from wild-type (n = 7), Cbfb−/GFP (n = 6), and CbfbGFP/GFP (n = 8). (C) Total thymocyte numbers from embryos at E17.5. The panel shows the means and SDs of wild-type (WT, n = 4), Cbfb+/GFP (+/GFP, n = 8), Cbfb+/−(+/−, n = 3), CbfbGFP/GFP (GFP/GFP, n = 6), and Cbfb−/GFP (−/GFP, n = 6) embryos. (D) Summary of T-cell phenotypes of CbfbGFP/GFP, Cbfb−/GFP, Cbfb-MYH11 chimera, and Tg(Lck-Cre)/Cbfb+/56M mice. The top panel shows the mouse thymocyte developmental diagram. The crosses in the lower panel indicate block of differentiation at the corresponding stages. The arrows in the lower panel indicate increase or decrease in percentage of the population, or apoptosis; − indicates no change.
To our surprise, CD4 expression was not derepressed in the Tg(Lck-Cre)/Cbfb+/56M mice. As can be seen in Figure 5A, the TCRβhiHSAlo mature thymocytes did not contain a CD4+CD8+ DP population as observed in Runx3-deficient thymi. In addition, the TCRβ-loFSChiCD8−/lo immature thymocytes did not show increases in CD4 expression either (Figure 5B). Moreover, the CD4+/CD8+ SP cell ratio in the spleen and peripheral blood was lower in the Tg(Lck-Cre)/Cbfb+/56M mice compared to that in the control mice (data not shown), and no DP population was detected (Figure 2E).
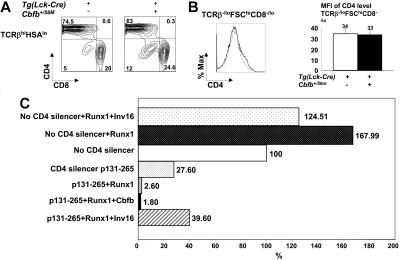
Figure 5.
Effects of Cbfb-MYH11 on CD4 expression in vivo and in vitro. (A) CD4 and CD8 distribution of TCRβhiHSAlo thymocytes from Tg(Lck-Cre) and Tg(Lck-Cre)/Cbfb+/56M mice. (B) CD4 expression in immature thymocytes. The left panel shows the histogram overlay of CD4 expression in TCRβloFSChiCD8−/lo thymocytes from Tg(Lck-Cre) (n = 10, gray line) and Tg(Lck-Cre)/Cbfb+/56M mice (n = 5, black line). The right panel shows the mean fluorescence intensity (MFI) of CD4 expression in TCRβloFSChiCD8−/lo cells. (C) CD4 silencer reporter assay. The indicated cDNA constructs were transfected into a CD4− Jurkat cell clone (D1.1) and CAT assays were performed. The CAT expression from the construct with no CD4 silencer was set at 100% and used to normalize the other results. No CD4 silencer: a construct with the CAT gene driven by CD4 enhancer/promoter. CD4 silencer p131-265: the above construct with 3 copies of the CD4 silencer core sequence. Runx1: Runx1 full-length cDNA. Cbfb: Cbfb full-length cDNA. Inv(16): CBFB-MYH11 cDNA.
Because Runx1 and Runx3 directly bind to the CD4 silencer and repress CD4 promoter expression in reporter assays,24 we determined the effect of Cbfβ-SMMHC in a similar reporter assay. The CAT reporter construct (p131-265), which contained 3 copies of the core CD4 silencer upstream of the CD4 enhancer/promoter,29 was transiently transfected into Jurkat cells, together with cDNA constructs for Runx1, Cbfb, or Cbfb-MYH11.16 As can be seen in Figure 5C, the CD4 core silencer reduced the transcriptional activity of the CD4 enhancer/promoter about 4-fold. Cotransfection with Runx1 alone, or with both Runx1 and Cbfb, further reduced the transcription activity by 10- to 15-fold. Cbfb-MYH11, on the other hand, restored transcription to above the level observed in the absence of Runx1. However, when the empty vector (without silencer) was cotransfected with Runx1 alone, or with both Runx1 and Cbfb-MYH11, no suppression effect was observed (Figure 5C). The result confirms that Cbfb-MYH11 is able to release Runx1 repression of the CD4 silencer and restore CD4 expression in reporter assays.
Early block of thymocyte development and limited CD4 derepression in Cbfb-MYH11 chimeras
To exclude the effect of Lck-Cre efficiency on these results, we also examined thymocyte development in the chimeric mice generated with embryonic stem (ES) cells containing conventional Cbfb-MYH11 knock-in.18 By using a mouse strain 129-specific marker Ly9.1, we could distinguish cells carrying Cbfb-MYH11 (derived from 129 ES cells) from wild-type B6 cells in the chimeras35 (Figure 6A). Thymocytes carrying Cbfb-MYH11 in the chimeras showed similar characteristics when compared to those in the Tg(Lck-Cre)/Cbfb+/56M mice and that there were increased DN and decreased DP populations (Figure 6B). Interestingly, there was a severe DN1 block in the Cbfb-MYH11 chimera (Figure 6B), which could not have been observed in the Tg(Lck-Cre)/Cbfb+/56M mice because Lck-Cre started to express at DN3. Thymocytes that carried Cbfb-MYH11 also had a significantly increased level of apoptosis (Figure 6C). In addition, the CD4/CD8 ratio was decreased, indicating no CD4 derepression by Cbfb-MYH11 at the level of SP cells. Further analysis of immature cells (FSChiTCRβlo/−CD8lo/−) revealed a small increase of CD4 expression (Figure 6D), which was not as strong as that in the Runx1-deficient mice24 or the Cbfb−/GFP mice (see next section). Cbfb-MYH11 therefore could suppress Runx1 function in CD4 repression in vivo, but at a very limited level.
Figure 6.
Developmental defects of thymocytes in Cbfb-MYH11 chimeras. (A) Identification of Ly9.1+ and Ly9.1− thymocytes from a Cbfb-MYH11 chimera. (B) Analysis of thymocyte development in the Cbfb-MYH11 chimeras by FACS. Top panels show CD4 and CD8 distribution of Ly9.1+ and Ly9.1− cells. Bottom panels show CD44 and CD25 distribution of DN cells from Ly9.1+ and Ly9.1− cells. (C) The annexin V and 7AAD stainings of Ly9.1+ and Ly9.1− cells are displayed. (D) CD4 expression in immature thymocytes. The left panel shows a representative histogram overlay of CD4 expression in TCRβloFSChiCD8−/lo thymocytes from Ly9.1+ and Ly9.1− cells. The right panel shows the MFI of CD4 expression in TCRβloFSChiCD8−/lo cells.
T-cell developmental defects in Cbfb−/GFP and CbfbGFP/GFP embryos
To investigate whether the phenotype described so far was due to suppression of Cbfb function by Cbfb-MYH11, we studied mice with a Cbfb null allele and a hypomorphic Cbfb-GFP allele. Cbfb−/− embryos die on E12.5 due to blockage of definitive hematopoiesis and multiple hemorrhages,12 precluding studies of T-cell development in those embryos. Cbfb−/GFP and CbfbGFP/GFP mice die at birth, with delayed bone formation and clavicle hypotrophy19 (L.Z. and P.P.L., unpublished data, December 2005). We therefore studied the thymi of embryos from Cbfb+/− × Cbfb+/GFP and Cbfb+/GFP × Cbfb+/GFP crosses at E17.5 and E18. The Cbfb−/GFP embryos had the most severe bone defects (and presumably most severe Cbfb deficiency) among all possible genotypes (Cbfb+/+, Cbfb+/GFP, Cbfb+/−, CbfbGFP/GFP, and Cbfb−/GFP). In the Cbfb−/GFP thymi, the CD8+ SP T-cell population was absent with elevated percentage of CD4+ SP cells (Figure 7A). There was a similar but less severe increase of the CD4 SP population and decrease of the CD8 SP population in the Cbfb+/GFP mice (Figure 7A). In addition, the average expression level of CD4 in the immature thymocytes in the Cbfb−/GFP and CbfbGFP/GFP embryos was greatly increased (Figure 7B). The CD4 expression increase in the immature thymocytes seemed to be dependent on Cbfb dosage because the CD4 expression level was unchanged in Cbfb+/GFP and Cbfb+/− embryos (data not shown). In addition, there were developmental blocks at stages DN1 and DN2 in the Cbfb−/GFP thymi (Figure 7A). These defects were also observed in the CbfbGFP/GFP and Cbfb+/−embryos at reduced severity (data not shown). The thymocyte numbers were more than 20-fold lower in the Cbfb−/GFP thymi compared to wild-type littermates, whereas CbfbGFP/GFP and Cbfb+/− embryos had about 2-fold fewer thymocytes (Figure 7C). The reduced cellularity in these Cbfb-deficient embryos was probably also due to increased apoptosis because we observed increased annexin V staining of DP cells in the Cbfb−/GFP thymi, whereas the incorporation of BrdU by the thymocytes was unchanged (data not shown).
These observations indicate that Cbfb is required for DN cell development, repression of CD4 expression in DN cells, and survival of maturing thymocytes. The less severe phenotype of cellularity and CD4 derepression in Cbfb+/−and Cbfb+/GFP mice suggests a dose effect of Cbfβ. Most of the alterations in T-cell development observed in the Cbfb-deficient mice were similarly observed in the Cbfb-MYH11 mice, except for CD4 derepression as summarized in Figure 7D.
Discussion
The function of Runx1 and Runx3 in T-cell development has been studied recently, and the role of Cbfb is emerging. In this report we provide evidence that Cbfb function is required for CD4 derepression, thymocyte development through DN stages, and the survival of thymocytes at several stages. More importantly, we demonstrated that CBFB-MYH11, the fusion gene associated with AML-M4Eo, suppresses most functions of Runx/Cbfb in thymocyte development (Figure 7D).
The developmental block at DN1 and DN2 in the Cbfb-MYH11 knock-in chimeras and Cbfb−/GFP mice is strong evidence that Cbfb is required at the earliest stages of thymocyte development. This block and the CD4 derepression in Cbfb−/GFP mice appeared to be similar to the reported Tg(Mx1-Cre)/Runx1f/rd mice36 and other Cbfb deficiency models,37 suggesting that Cbfb regulates the function of Runx1 in DN cells. Lck-Cre is activated around DN2-DN3; accordingly a developmental block at DN3 was observed in the Tg(Lck-Cre)/Cbfb+/56M mice, which appeared to be similar to what has been reported in the Tg(Lck-Cre)/Runx1f/f mice.24 These observations suggest that Cbfb and Runx1 are required at several DN stages.
The progression of T-cell development at the DN3 stage is regulated through multiple pathways. The most important signal comes from TCR gene rearrangement and expression of functional TCRβ chains.21,23 The resulting production of the pre-TCR proteins is a major driving force for thymocyte expansion and further differentiation.38 In the conditional Cbfb-MYH11 knock-in mice, the DN3 block and the associated apoptosis increase indicate difficulties of cells to go through this developmental stage. However, neither crossing with the Tg(Tcrb) mice to express a functional TCRβ protein nor crossing with the Bcl-2 transgenic mice to reduce apoptosis could rescue this DN3 stage block, suggesting that the rearrangement of TCR genes and the expression of functional β chains were not affected in the Tg(Lck-Cre)/Cbfb+/56M mice. It remains possible, however, that other events of the pre-TCR signaling pathway were affected.
Previous studies suggested that Runx/Cbfβ proteins regulate genes involved in cell cycle control,39 and that Cbfβ-SMMHC can slow proliferation of cultured cells.40 Our in vivo studies, on the other hand, suggest that Cbfb-MYH11 does not induce significant defects in cell cycle (data not shown) or proliferation. In addition, mice deficient in Ccnd3 (encoding cyclin D3) share a similar phenotype of post/pre-TCR blockage,41 and Runx/Cbfβ can regulate the expression of the Ccnd3 gene directly.40 However, we found normal expression of Ccnd3 in the Tg(Lck-Cre)/Cbfb+/56M mice by quantitative reverse transcription-PCR (data not shown).
Instead, we observed significant increases in apoptosis at most stages of T-cell development, which was likely responsible for the severely reduced thymocyte numbers in the Tg(Lck-Cre)/Cbfb+/56M mice. This observation was supported by our results that transgenic expression of Bcl-2 restored the thymocyte population and increased the percentage of DP cells. Interestingly the DN3 block was not released and the CD4/CD8 ratio remained the same in the Tg(Lck-Cre)Cbfb+/56m/Tg(Lck-hBcl2) mice. A direct link between Cbfβ-SMMHC and apoptosis pathways has not been reported before. Therefore, it will be highly interesting to understand the underlying mechanism. The cytokine IL-2 regulates both T-cell proliferation and apoptosis,42,43 and its high-affinity receptor, CD25 (IL-2Rα) is responsible for activation-induced cell death in vivo as shown in CD25 null mice.44 We observed increased production of cell surface CD25 in Cbfb-MYH11-expressing cells (data not shown). It is possible that the IL-2 signaling pathway is playing a role here to induce cell death by down-regulating Bcl-2, which is a surviving factor for T cells during development. It will be interesting to determine if apoptosis is similarly induced in the myeloid lineage and, if so, whether such changes are important for leukemogenesis. We have previously demonstrated that CBFB-MYH11 by itself was not sufficient for leukemogenesis and that additional mutations are needed for leukemia development.35,45 Antiapoptotic genetic changes may be one class of cooperating oncogenes for CBFB-MYH11.
Our in vitro reporter assays demonstrated that Cbfβ-SMMHC could suppress Runx1/Cbfβ function and derepress CD4 expression. However, we did not observe any CD4 derepression in the Tg(Lck-Cre)/Cbfb+/56M mice, and only saw marginal increase of CD4 expression in DN cells (Figure 6D) in the Cbfb-MYH11 knock-in chimera mouse. There could be several reasons to explain the reason Cbfb-MYH11 did not derepress CD4 in vivo. Our data in Cbfb+/GFP, Cbfb+/−, CbfbGFP/GFP, and Cbfb−/GFP mice indicate that CD4 derepression is Cbfb dose-dependent. In the presence of one wild-type Cbfb allele, such as in Cbfb+/− and Cbfb+/GFP mice, CD4 derepression was almost undetectable. Cbfb-MYH11 may not be a very strong Cbfb repressor as previously believed, especially when expressed at the endogenous level in vivo. Further, previous studies showed that different domains of Runx1 are required for different functions. For example, the C-terminal VWRPY motif is important for thymic cellularity, but not for definitive hematopoiesis.46 Additional studies on the C-terminal domains by Kawazu et al demonstrated that VWRPY motif is necessary for CD4 repression, whereas the activation domain is critical for regulation of Tcrb expression.47 It is possible that Cbfb-MYH11 suppresses Runx1 domains at different levels. Finally, functions of Runx family members may have different sensitivity to Cbfb-MYH11 inhibition. The phenotype we observed in the Cbfb-MYH11 mice is more similar to mice with Runx1 deficiency,36 than those with Runx3 deficiency, suggesting that Runx1 may be more sensitive to Cbfb dosage change than Runx3.
Chromosome 16 inversion, which generates the CBFB-MYH11 fusion gene, is associated with AML in human patients.4 Clonality studies have provided evidence that the inversion occurs in hematopoietic stem cells.48,49 It is interesting then why the inversion is only associated with myeloid leukemias. In one recent study, it was shown that inv(16) can be detected in the B cells, in addition to the myeloid lineages, but not in the mature T cells.50 By crossing the conditional Cbfb-MYH11 knock-in mice with transgenic mice expressing the Cre gene under the control of the Mx1 promoter, which was inducible in hematopoietic stem cells, we showed previously that Cbfb-MYH11+ bone marrow cells did not contribute efficiently to the T-cell lineage after competitive transplantation.26 The data presented here provide strong evidence that Cbfb-MYH11 blocks T-cell development at several stages and reduces thymocyte survival by inducing apoptosis. The data could help to explain why CBFB-MYH11+ cells cannot be detected in the T-cell lineage in patients with AML with this fusion gene.
Supplementary Material
Acknowledgments
This work was supported in part by the Intramural Research Programs of the National Human Genome Research Institute and National Cancer Institute, National Institutes of Health.
The authors thank the outstanding support of the transgenic mouse core of the Intramural Research Program, National Human Genome Research Institute.
The authors also thank Dan Littman for CD4 reporter plasmids and Jan Melenhorst for helpful discussions.
Footnotes
The online version of this manuscript contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: L.Z., J.L.C., P.L.S., R.B., and P.P.L. designed research, analyzed data, and wrote the paper; L.Z., J.L.C., S.A., M.K., and L.X. performed experiments; L.H.C. contributed the conditional CBFB-MYH11 mice and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: P. Paul Liu, Genetics and Molecular Biology Branch, National Human Genome Research Institute, National Institutes of Health, 49 Convent Dr, Bldg 49, Rm 3A26, Bethesda, MD 20892; e-mail: pliu@mail.nih.gov.
References
- 1.Ogawa E, Maruyama M, Kagoshima H, et al. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci U S A. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogawa E, Inuzuka M, Maruyama M, et al. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology. 1993;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 3.Wang S, Wang Q, Crute BE, Melnikova IN, Keller SR, Speck NA. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu P, Tarle SA, Hajra A, et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 5.Huang X, Peng JW, Speck NA, Bushweller JH. Solution structure of core binding factor beta and map of the CBF alpha binding site. Nat Struct Biol. 1999;6:624–627. doi: 10.1038/10670. [DOI] [PubMed] [Google Scholar]
- 6.Goger M, Gupta V, Kim WY, Shigesada K, Ito Y, Werner MH. Molecular insights into PEBP2/CBF beta-SMMHC associated acute leukemia revealed from the structure of PEBP2/CBF beta. Nat Struct Biol. 1999;6:620–623. doi: 10.1038/10664. [DOI] [PubMed] [Google Scholar]
- 7.Warren AJ, Bravo J, Williams RL, Rabbitts TH. Structural basis for the heterodimeric interaction between the acute leukaemia-associated transcription factors AML1 and CBFbeta. EMBO J. 2000;19:3004–3015. doi: 10.1093/emboj/19.12.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tahirov TH, Inoue-Bungo T, Morii H, et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell. 2001;104:755–767. doi: 10.1016/s0092-8674(01)00271-9. [DOI] [PubMed] [Google Scholar]
- 9.Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, Ito Y. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 2001;20:723–733. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q, Stacy T, Miller JD, et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 13.North T, Gu TL, Stacy T, et al. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- 14.Kundu M, Chen A, Anderson S, et al. Role of Cbfb in hematopoiesis and perturbations resulting from expression of the leukemogenic fusion gene Cbfb-MYH11. Blood. 2002;100:2449–2456. doi: 10.1182/blood-2002-04-1064. [DOI] [PubMed] [Google Scholar]
- 15.Namba K, Abe M, Saito S, et al. Indispensable role of the transcription factor PEBP2/CBF in angiogenic activity of a murine endothelial cell MSS31. Oncogene. 2000;19:106–114. doi: 10.1038/sj.onc.1203257. [DOI] [PubMed] [Google Scholar]
- 16.Adya N, Stacy T, Speck NA, Liu PP. The leukemic protein core binding factor beta (CBFbeta)-smooth-muscle myosin heavy chain sequesters CBFalpha2 into cytoskeletal filaments and aggregates. Mol Cell Biol. 1998;18:7432–7443. doi: 10.1128/mcb.18.12.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shigesada K, van de Sluis B, Liu PP. Mechanism of leukemogenesis by the inv(16) chimeric gene CBFB/PEBP2B-MHY11. Oncogene. 2004;23:4297–4307. doi: 10.1038/sj.onc.1207748. [DOI] [PubMed] [Google Scholar]
- 18.Castilla LH, Wijmenga C, Wang Q, et al. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87:687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 19.Kundu M, Javed A, Jeon JP, et al. Cbfbeta interacts with Runx2 and has a critical role in bone development. Nat Genet. 2002;32:639–644. doi: 10.1038/ng1050. [DOI] [PubMed] [Google Scholar]
- 20.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 21.Rothenberg EV, Taghon T. Molecular genetics of T cell development. Annu Rev Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- 22.Staal FJ, Clevers HC. Regulation of lineage commitment during lymphocyte development. Int Rev Immunol. 2001;20:45–64. doi: 10.3109/08830180109056722. [DOI] [PubMed] [Google Scholar]
- 23.von Boehmer H. Selection of the T-cell repertoire: receptor-controlled checkpoints in T-cell development. Adv Immunol. 2004;84:201–238. doi: 10.1016/S0065-2776(04)84006-9. [DOI] [PubMed] [Google Scholar]
- 24.Taniuchi I, Osato M, Egawa T, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 25.Woolf E, Xiao C, Fainaru O, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A. 2003;100:7731–7736. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo YH, Landrette SF, Heilman SA, et al. Cbf beta-SMMHC induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer Cell. 2006;9:57–68. doi: 10.1016/j.ccr.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 27.Kundu M, Liu PP. Cbf beta is involved in maturation of all lineages of hematopoietic cells during embryogenesis except erythroid. Blood Cells Mol Dis. 2003;30:164–169. doi: 10.1016/s1079-9796(03)00030-5. [DOI] [PubMed] [Google Scholar]
- 28.Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 29.Taniuchi I, Sunshine MJ, Festenstein R, Littman DR. Evidence for distinct CD4 silencer functions at different stages of thymocyte differentiation. Mol Cell. 2002;10:1083–1096. doi: 10.1016/s1097-2765(02)00735-9. [DOI] [PubMed] [Google Scholar]
- 30.Lee PP, Fitzpatrick DR, Beard C, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 31.Sentman CL, Shutter JR, Hockenbery D, Kanagawa O, Korsmeyer SJ. bcl-2 inhibits multiple forms of apoptosis but not negative selection in thymocytes. Cell. 1991;67:879–888. doi: 10.1016/0092-8674(91)90361-2. [DOI] [PubMed] [Google Scholar]
- 32.Shinkai Y, Koyasu S, Nakayama K, et al. Restoration of T cell development in RAG-2-deficient mice by functional TCR transgenes. Science. 1993;259:822–825. doi: 10.1126/science.8430336. [DOI] [PubMed] [Google Scholar]
- 33.Levanon D, Goldstein RE, Bernstein Y, et al. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci U S A. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siu G, Wurster AL, Duncan DD, Soliman TM, Hedrick SM. A transcriptional silencer controls the developmental expression of the CD4 gene. EMBO J. 1994;13:3570–3579. doi: 10.1002/j.1460-2075.1994.tb06664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castilla LH, Garrett L, Adya N, et al. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat Genet. 1999;23:144–146. doi: 10.1038/13776. [DOI] [PubMed] [Google Scholar]
- 36.Growney JD, Shigematsu H, Li Z, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talebian L, Li Z, Guo Y, et al. T lymphoid, megakaryocyte, and granulocyte development are sensitive to decreases in CBFβ dosage. Blood. 2007;109:11–21. doi: 10.1182/blood-2006-05-021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Boehmer H. Unique features of the pre-T-cell receptor alpha-chain: not just a surrogate. Nat Rev Immunol. 2005;5:571–577. doi: 10.1038/nri1636. [DOI] [PubMed] [Google Scholar]
- 39.Peterson LF, Boyapati A, Ranganathan V, et al. The hematopoietic transcription factor AML1 (RUNX1) is negatively regulated by the cell cycle protein cyclin D3. Mol Cell Biol. 2005;25:10205–10219. doi: 10.1128/MCB.25.23.10205-10219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernardin-Fried F, Kummalue T, Leijen S, Collector MI, Ravid K, Friedman AD. AML1/RUNX1 increases during G1 to S cell cycle progression independent of cytokine-dependent phosphorylation and induces cyclin D3 gene expression. J Biol Chem. 2004;279:15678–15687. doi: 10.1074/jbc.M310023200. [DOI] [PubMed] [Google Scholar]
- 41.Sicinska E, Aifantis I, Le Cam L, et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell. 2003;4:451–461. doi: 10.1016/s1535-6108(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 42.Shi Y, Wang R, Sharma A, et al. Dissociation of cytokine signals for proliferation and apoptosis. J Immunol. 1997;159:5318–5328. [PubMed] [Google Scholar]
- 43.Van Parijs L, Refaeli Y, Lord JD, Nelson BH, Abbas AK, Baltimore D. Uncoupling IL-2 signals that regulate T cell proliferation, survival, and Fas-mediated activation-induced cell death. Immunity. 1999;11:281–288. doi: 10.1016/s1074-7613(00)80103-x. [DOI] [PubMed] [Google Scholar]
- 44.Willerford DM, Chen J, Ferry JA, Davidson L, Ma A, Alt FW. Interleukin-2 receptor alpha chain regulates the size and content of the peripheral lymphoid compartment. Immunity. 1995;3:521–530. doi: 10.1016/1074-7613(95)90180-9. [DOI] [PubMed] [Google Scholar]
- 45.Castilla LH, Perrat P, Martinez NJ, et al. Identification of genes that synergize with Cbfb-MYH11 in the pathogenesis of acute myeloid leukemia. Proc Natl Acad Sci U S A. 2004;101:4924–4929. doi: 10.1073/pnas.0400930101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishimura M, Fukushima-Nakase Y, Fujita Y, et al. VWRPY motif-dependent and -independent roles of AML1/Runx1 transcription factor in murine hematopoietic development. Blood. 2004;103:562–570. doi: 10.1182/blood-2003-06-2109. [DOI] [PubMed] [Google Scholar]
- 47.Kawazu M, Asai T, Ichikawa M, et al. Functional domains of Runx1 are differentially required for CD4 repression, TCRbeta expression, and CD4/8 double-negative to CD4/8 double-positive transition in thymocyte development. J Immunol. 2005;174:3526–3533. doi: 10.4049/jimmunol.174.6.3526. [DOI] [PubMed] [Google Scholar]
- 48.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 49.Haase D, Feuring-Buske M, Konemann S, et al. Evidence for malignant transformation in acute myeloid leukemia at the level of early hematopoietic stem cells by cytogenetic analysis of CD34+ subpopulations. Blood. 1995;86:2906–2912. [PubMed] [Google Scholar]
- 50.Chang H, Nayar R, Li D, Sutherland DR. Clonality analysis of cell lineages in acute myeloid leukemia with inversion 16. Cancer Genet Cytogenet. 2005;156:175–178. doi: 10.1016/j.cancergencyto.2004.03.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.