Abstract
Background/Aim
The role of humoral immunity in hepatitis C virus (HCV) infection is poorly understood. Nevertheless, there is increasing interest in characterizing the neutralizing antibodies in the serum of HCV-infected patients. Focus reduction assays have been widely used to evaluate neutralizing antibody responses against a range of non-cytopathic viruses. Based on the recent development of a HCV cell culture system using the genotype 2 JFH-1-strain, we developed a focus reduction assay for HCV-neutralizing antibodies.
Methods
The focus reduction assay was based on a standard microneutralization assay in which immunostained foci on tissue culture plates are counted. The neutralizing anti-HCV antibodies titers of purified serum immunoglobulin samples from seventy-seven individuals were determined using a 50% focus reduction neutralization assay. Each titer was determined as the log value of the reciprocal antibody dilution that reduced the number of viral foci by 50%. IgG antibodies were first purified from each serum in order to avoid the facilitating effect of HDL on HCV entry.
Results
The assay's cut-off using an ELISA and RNA HCV-negative samples was found to be 1.25 log, corresponding to a dilution of 1:18. The assay was compared with a commercial HCV ELISA and exhibited specificity and sensitivity values of 100% and 96.5%, respectively, and good reproducibility (with intra-assay and inter-assay coefficients of variation of 6.7% and 12.6%, respectively). The assay did not show any cross-reactivity with anti-HIV, anti-HBs or heterophile antibody-positive samples. The neutralizing antibodies titers were 2.13 log (1:134) for homologous samples from HCV genotype 2 infected patients harboring the same genotype as JFH-1 and 1.93 log (1:85) for heterologous samples from patients infected by genotypes other than type 2. These results confirm the presence of broadly cross-neutralizing antibodies already reported using the HCV pseudoparticles system.
Conclusion
This study presents a simple, specific and reproducible cell culture-based assay for determination of HCV-neutralizing antibodies in human sera. The assay should be an important tool for gauging the relationship between the neutralizing antibodies response and viral load kinetics in acutely or chronically infected patients and for investigating the possible eradication or prevention of HCV infection by neutralizing antibodies.
Background
Hepatitis C virus (HCV, a member of the Flaviviridae family) is an enveloped, positive-stranded RNA virus that preferentially replicates in hepatocytes. At least 170 million people worldwide are persistently infected with hepatitis C virus. Chronic HCV infection is associated with a significant risk of progression to cirrhosis and hepatocellular carcinoma [1]. Antiviral therapy with pegylated alpha-interferon and ribavirin (the current best therapeutic regimen) is only successful in about 50% of all treated patients.
Better knowledge of the viral and host factors that determine HCV clearance or persistence during the acute stage of infection is needed in order to improve antiviral therapy and to develop efficient vaccines. Studies focusing on innate and cellular immune responses have shown that a sufficiently large HCV inoculum is able to evade, subvert or circumvent the host's defences. At present, the chimpanzee is the only reliable experimental animal model in which the initial post-HCV infection events and the efficacy of vaccine candidates can be evaluated [2]. It has been shown that HCV-specific T-cell immunity is important in the control of HCV infection [3,4]. Several studies have indicated a role for humoral immunity in the acute stage of HCV infection but this aspect remains poorly characterized. The E1 and E2 glycoproteins are thought to be the viral attachment proteins and thus the main targets for HCV-neutralizing antibodies; identification of protective epitopes conserved across different strains of HCV is therefore a major challenge in vaccine design. A number of antibodies capable of blocking E2 binding to cells or cell receptors have been described, [5-8] some of which neutralize HCV entry in animal or cellular models [9,10]. Cell entry has been shown to involve several surface molecules (notably including the tetraspanin CD81 and the SR-BI receptor [11,12]), although further studies are needed to better understand how viral entry occurs and how it might be neutralized. Detection of neutralizing antibodies in human blood had been problematical until an efficient and reliable cell culture system for HCV became available. Hence, the development of an in vitro neutralization assay for HCV could be extremely valuable for characterizing the humoral immune response to HCV and for evaluating the potential of passive and active immunization against hepatitis C. Recent studies using an in vitro neutralization assay system (based on infectious retroviral pseudoparticles (HCVpp) bearing HCV envelope glycoproteins) have confirmed that HCV-infected patient sera can indeed neutralize infection [13,14]. However, it has also been shown that the neutralizing activity of antibodies from HCV-infected patients is attenuated by a factor present in human serum, identified as the high-density lipoprotein (HDL) fraction [11,13,15]. HDL facilitation of HCVpp entry is a post-binding event [16], suggesting that HDLs favour internalization of virions and thus the latter's escape from neutralizing antibodies.
Recently, an HCV cell culture model (HCVcc) has been developed [17-19], allowing the production of virus particles that can be efficiently propagated in cell culture. Some preliminary neutralization assays have been carried out by these authors. In this study, we describe how we set up a standardized focus reduction neutralization assay based on HCVcc.
Results
HCV focus reduction neutralization assay
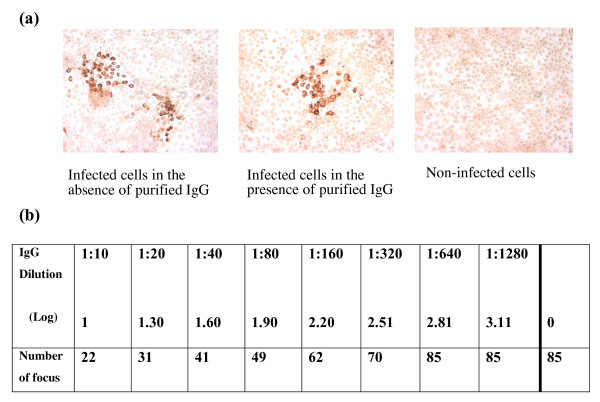
Focus reduction assays have been widely used to evaluate the neutralizing antibody responses to viruses that can form foci in infected cells. Following the recent development of the HCVcc model, the principle of the focus reduction assay has been applied to HCV-neutralizing antibodies detection. The JFH-1 HCV 2a viral strain was grown on a Huh-7 human hepatoma cell line. After three days of infection and cell permeabilization, detection of the HCV foci was carried out using an inactivated HCV-positive patient serum primary antibody and a peroxidase-coupled, Fc-specific anti-human IgG-antibody. The reaction was revealed with DAB peroxidase substrate. The viral foci were thus stained brown, making them easy to count (see Fig. 1a). It has been recently shown that the neutralizing activity of HCV antibodies is attenuated by a serum factor associated with the HDL fraction. Hence, HDLs were able to facilitate HCVpp and HCVcc entry via a mechanism which depended on the expression of the scavenger receptor BI (SR-BI) and its selective lipid-uptake function [11,15,16,20]. In view of the role of HDL in HCV entry, immunoglobulins were purified from each serum sample prior to determination of the neutralizing antibody titer (see Fig. 1b).
Figure 1.
(a) Typical pictures of HCVcc-infected Huh-7 cells observed under a light microscope (×40), showing the absence (on the right) and presence (on the left) of focus-forming units (FFU). (b) A sample-based neutralizing assay, showing the number of FFU in Huh-7 cells following serial dilutions of purified IgG from a HCV-positive serum sample. Neutralizing anti-HCV antibodies titers were expressed as the highest log dilution of IgG producing a 50% reduction in plaque count, as compared with controls in which the dose of virus was known.
Assay specificity and precision
The specificity of the HCV neutralization assay was assessed by testing 20 anti-HCV-ELISA-negative samples, including five positive for hepatitis B virus surface antibodies (anti-HBs) and five positive for heterophile antibodies. All samples tested negative with two commercial anti-HCV antibody detection assays (Axsym® HCV Version 3.0, Abbott, Wiesbaden, Germany; Vitros® Anti-HCV reagent pack, Ortho-Clinical Diagnostic, High Wycombe, United Kingdom) and HCV-RNA-negative with a qualitative, commercial assay (Cobas Amplicor HCV test Version 2.0, Roche Diagnostics, Meylan, France).
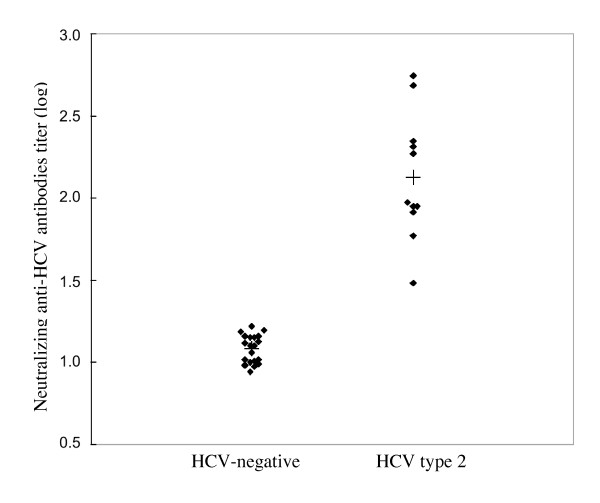
These anti-HCV-negative samples were compared with 11 samples from patients chronically infected with HCV genotype 2. The neutralization titers of anti-HCV-negative serum samples are shown in Fig. 2., with a mean value of 1.083 ± 0.083 (corresponding to a dilution of 1:12). The assay's cut-off (determined as the mean value for negative samples plus two standard deviations) corresponded to a dilution of 1:18. The assay exhibited specificity and sensibility values of 100% and 96.5%, respectively. The assay did not show any cross-reactivity with anti-HIV, anti-HBs or heterophile antibody-positive samples (data not shown). Conversely, the chronically HCV genotype 2-positive samples displayed strong reactions, with a mean value of 2.128 ± 0.365 (corresponding to a dilution of 1:134) (p < 0.001).
Figure 2.
Neutralization of HCVcc JFH-1 in infected Huh-7 cells for two distinct populations of samples, characterized by the presence (HCV genotype 2) or absence (HCV-negative) of anti-HCV antibodies.
Inter-assay variability was determined by testing one HCV genotype 2 sample in 10 consecutive experiments (n = 10), whereas intra-assay variability was evaluated by testing the same sample 10 times (n = 10) in the same experiment, whilst running the dilution series. The intra-assay and inter-assay coefficients of variation (CV) of the log neutralization titers were 6.7% and 12.6%, respectively.
Homologous and heterologous genotype reactivity
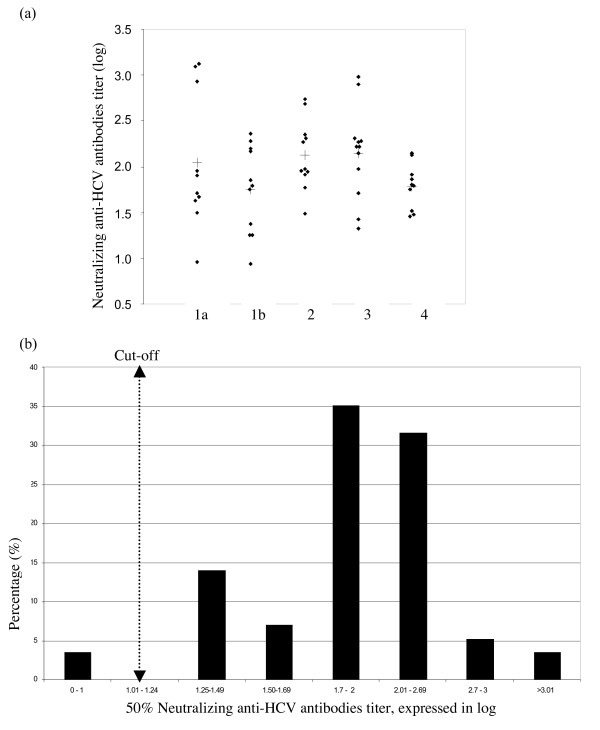
Fifty-seven HCV-positive antibodies samples were evaluated using the HCV focus reduction neutralization assay. The genotypes were distributed as follows; for types 1a, 1b, 2, 3, 4 and 5, we studied 11, 11, 11, 12, 10 and 2 samples, respectively. The mean values of the different genotypes is shown in Fig. 3. and Table 1. The mean log neutralization titers for genotypes 1a, 2 and 3 are very similar (2.046 ± 0.671 for genotype 1a, 2.128 ± 0.365 for genotype 2 and 2.148 ± 0.478 for genotype 3). The mean average values are lower for genotype 1b (1.747 ± 0.462) and genotype 4 (1.786 ± 0.236). Strikingly, very high heterologous titers were observed for five patients – three infected with HCV genotype 1a and two infected with HCV genotype 3 (see Fig. 3a). There were too few genotype 5 samples to compare with the other genotypes but the corresponding results nevertheless indicate that the neutralization assay is suitable for this genotype. The two determined log neutralization values for genotype 5 were 1.753 and 1.764, respectively. These results confirm the presence of broadly cross-neutralizing antibodies, as already reported using the previous HCV pseudoparticle system (HCVpp).
Figure 3.
(a) Neutralization results for genotypes 1a, 1b, 2, 3 and 4 with respect to HCVcc JFH-1 in infected Huh-7 cells. (b) Distribution of the neutralization anti-HCV antibodies titers (independently of the genotype).
Table 1.
HCV JFH-1 neutralization assay
| Genotype | Nba | M/Fb ratio | Age (yr) | HCV RNAc (log IU/mL) | Mean ± SDd | Rangee |
| 1a | 11 | 7/4 | 43 ± 6 | 5.90 ± 0.6 | 2.046 ± 0.671 | 0.926 – 3.121 |
| 1b | 11 | 5/6 | 48 ± 16 | 6.16 ± 0.5 | 1.747 ± 0.462 | 0.934 – 2.356 |
| 2 | 11 | 9/2 | 63 ± 12 | 5.50 ± 0.9 | 2.128 ± 0.365 | 1.486 – 2.742 |
| 3 | 12 | 8/4 | 40 ± 11 | 5.66 ± 0.6 | 2.148 ± 0.478 | 1.326 – 2.977 |
| 4 | 10 | 4/6 | 45 ± 11 | 6.07 ± 0.7 | 1.786 ± 0.236 | 1.453 – 2.152 |
| 5 | 2 | 1/1 | 66 ± 14 | 6.06 ± 0.6 | 1.755 ± 0.005 | nd |
| all | 57 | 34/23 | 48 ± 15 | 5.83 ± 0.7 | 1.970 ± 0.491 | 0.934 – 3.121 |
a Number of samples tested.
b F, female; M, male.
c HCV RNA measured using the Amplicor HCV Monitor assay, version 2.0 (Roche Diagnostics, Meylan, France) and expressed in log equivalents of IU/mL.
d Neutralizing anti-HCV antibodies titers expressed in log units; mean ± SD
e Range of neutralizing anti-HCV antibodies titers, expressed in log units
The distribution of the log neutralization titers across all the HCV ELISA and RNA-positive samples as a function of the HCV genotype is shown in Fig. 3b. More than 60% of the neutralizing antibodies titers fell in the range from 1.7 to 2.69 log titers, corresponding to dilutions of 1:50 and 1:500, respectively. Overall, 3.5% of the samples displayed a titer greater than log 3.0 (1:1000) and, conversely, 3.5% displayed a titer below the cut-off value, i.e. log 1.25 (1:10). Thus, of 57 HCV-infected patients, only two did not test positive for neutralizing antibodies in this assay (the titers were 0.960 and 0.932, respectively).
Discussion
The role of neutralizing antibodies during acute and chronic viral infection remains an important question and has generated controversial results. Initially, the presence of neutralizing antibodies was shown to control the HCV load and to contribute to viral eradication in patients capable of clearing the infection [13]. In other studies, the appearance of neutralizing antibodies was delayed and restricted to IgG1 antibodies in patients who develop a chronic infection [2,21]. The chimpanzee model has been critical for the study of HCV transmission and host immune responses; however, neutralizing antibodies were not detected in some animals that resolved their infection – suggesting a minimal role in viral clearance, as also observed in human studies [14,15]. Experimentally infected chimpanzees and naturally infected humans can be re-infected with homologous and heterologous HCV strains, suggesting that the humoral immunity that develops after spontaneous resolution of acute hepatitis C is not sterilizing [22-24]. During chronic infection in humans, the presence and/or production of neutralizing antibodies do not suffice for curing the infection but could regulate the spread of the virus. Thus, it can be postulated that during chronic infection, viral mutants can continuously escape the renewed production of neutralizing antibodies.
Retroviral pseudoparticles have been used to develop a very interesting tool for measuring neutralizing antibodies in vitro [14]. The assay has demonstrated the presence of HCV-neutralizing antibodies in human sera with relatively high titers (>1:320) and broadly neutralizing activity against different HCV genotypes. However, this model does not represent genuine HCV virions; in particular, the budding of retroviral particles is thought to be very different and may involve a variety of cellular pathways. Characterization of infectious retroviral pseudotype particles bearing HCV glycoproteins have been shown to be very heterogeneous, and so it is possible that these pseudoparticles may not be as relevant as the native HCV virions [25].
The recent development of a cell culture model for HCV enables the production of native HCV virions that can be efficiently propagated in cell culture [17-19]. This cell culture system has allowed us to develop a neutralization assay for evaluating the level and the proportion of HCV-neutralizing antibodies in chronically infected HCV patients. We analysed a number of parameters (such as practicability, reproducibility and specificity) and tested the effect of a range of variables (viral inoculum size, incubation time, fixation and permeabilization methods, blocking and revelation reagents) on these parameters (data not shown). Overall, the neutralization assay described in this study performs similarly to standardized neutralization assays for many other viruses [26-28].
The assay relies on the ability of the specific JFH-1 genotype 2 viral strain to replicate and multiply on a Huh-7 human hepatoma cell line in a cell culture model, enabling the rapid detection of viral foci after 72 hours of infection. Moreover, no secondary foci were detectable at this time point. Fixation with paraformaldehyde and permeabilization with Triton X-100 were chosen in order to preserve antigenicity and prevent the cell monolayer from detaching during washes. Development with DAB peroxide substrate made it easy to count specifically coloured viral foci. The viral inoculum size is an important parameter; it has to be low enough to enable good assay sensitivity but high enough to produce a statistically significant number of foci, i.e. allowing the reduction in the number of foci (and thus the effect of neutralization) to be monitored. Thus, 100 FFUs were used as the inoculum in this neutralization assay.
In order to test different human samples, we had to take into account the ability of HDL to facilitate HCVcc entry via a mechanism which depends on expression of the scavenger receptor BI [11,15,16,20]. Given HDL's role in HCV entry, immunoglobulins were purified from each serum sample prior to determination of the neutralizing antibodies titer; this frees the assay of the risk of non-specific neutralization activity of the serum via the effects of HDL, the complement system and/or serum amyloid A protein (SAA) [29].
The HCV neutralization assay exhibited good reproducibility, for both duplicate assays and independent tests. As expected, the intra-assay coefficient of variation (CV) was lower than the interassay CV. The test also showed good specificity, since there was no interaction with anti-HIV, anti-HBV or heterophile antibodies. Very low titers were found with HCV ELISA and RNA-negative samples, and the assay's cut-off was determined as the mean titer for negative samples plus two standard deviations (1.25 log, corresponding to a dilution of 1:18).
Given that only the JFH-1 strain of HCV genotype 2a was available for the assay, we evaluated the neutralization titer of sera from patients chronically infected with other HCV genotypes, i.e. 1, 2, 3, 4 and 5. Most of these sera were detected as positive by the neutralization assay, except for two sera from HCV genotype 1-infected patients. These two samples presented a high specific antibody ratio according to the ELISA but only very low inhibition by neutralization assay (far below the cut-off, in fact). We conclude that either the samples lacked neutralizing antibodies or that any such antibodies that were present did not cross-neutralize with HCV genotype 2a. The sensitivity was 100% – not only for genotype 2 (the genotype of the strain used for the assay) but also for other HCV genotypes (except genotype 1). HCV genotype 5 antibodies were also measured but there were too few samples for accurate testing. Moreover, the positive sera (96.5%) had comparable and significantly high titers (1.99 ± 0.63), whatever the genotype. This finding suggests that most neutralizing antibodies are cross-reactive. Another possibility is that most of the patients had been previously infected by a genotype 2 strain. However, this is unlikely because few genotype 2 strains are circulating in France [30]. As expected for a neutralization test, the assay presented in the present study appeared to be very specific (independently of the genotype) and usable in most circumstances. For most viral infections, neutralization assays such as that described in this study are used as reference assays. Thus, we are confident that as other HCVcc genotypes become available, these assays will replace the pseudoparticle assay in the near future because they are probably more relevant. Our assay is somewhat time-consuming and could be simplified by using one dilution to count the foci; however, this type of "short cut" would make it difficult to extrapolate to the dilution neutralizing 50% of the inoculum. Another approach would consist in using recombinant HCV capable of expressing reporter genes (such as luciferase) in order to use a single dilution and obtain a quantitative result [31]. However, further neutralization studies using other genotypes are needed in order to complete our observations and to characterize the homologous and heterologous potencies of polyclonal and monoclonal neutralizing antibodies.
Conclusion
A simple, specific and reproducible cell culture-based neutralization assay was developed for the determination of neutralizing anti-HCV antibodies in human sera. This test should be an important tool for gauging the relationship between the neutralizing response and viral load kinetics in acutely and chronically infected patients.
Methods
Cell culture and HCV production
The Huh-7 human hepatoma cells [32] were grown in Dulbecco's minimum essential medium (Invitrogen) supplemented with 10% fetal bovine serum. All cell cultures were maintained in 5% CO2 at 37°C.
The plasmid pJFH-1 containing the full-length cDNA of the JFH-1 isolate (which belongs to subtype 2a (GenBank accession no. AB047639)), was a gift from Dr Wakita (Department of Microbiology, Tokyo Metropolitan Institute for Neuroscience, Tokyo, Japan) and has been described previously [17]. To generate genomic HCV RNA, the plasmid pJFH-1 was linearized at the 3' end of the HCV cDNA and used as a template for in vitro transcription, as described previously [33]. Viral stocks were obtained by harvesting cell culture supernatants and freezing them at -80°C. Virus titration was performed on Huh-7 cells with 6-well microtiter plates (Corning, NY) 72 hours after incubation, by immunostaining the cells with antibodies from a HCV-positive patient serum that had previously been inactivated at 56°C (see the section on the virus neutralization assay). The viral titer was determined in triplicate from the mean number of foci and expressed as focus forming units/mL (FFU/mL).
Patients and clinical samples
Seventy-seven human serum samples were tested. Collection of the sera was approved by the local Ethics Committee and informed consent had been obtained from the donors. Fifty-seven of these samples were obtained from chronically infected HCV patients. The presence of HCV antibodies was determined and confirmed using two third-generation HCV EIA assays (Axsym® HCV Version 3.0, Abbott, Wiesbaden, Germany and Vitros® Anti-HCV reagent pack, Ortho-Clinical Diagnostic, High Wycombe, United Kingdom). HCV RNA was determined with a qualitative commercial assay (Cobas Amplicor HCV test Version 2.0, Roche Diagnostics, Meylan, France) and HCV genotyping was performed by direct sequencing, as described elsewhere [34]. The genotypes were distributed as follows: 11, 11, 11, 12, 10 and 2 samples of types 1a, 1b, 2, 3, 4 and 5, respectively. A set of 20 anti-HCV-negative serum samples was used to evaluate the assay's specificity, including five serum samples with positive hepatitis B virus surface antibody (anti-HBs) status and five sera from Epstein-Barr virus-infected patients that had tested positive for heterophile antibodies. All serum samples had been stored at -80°C upon collection and had not been thawed until the time of assay.
Serum immunoglobulins purification
Serum immunoglobulins G (IgG) fraction was purified using protein G-Sepharose (GE Healthcare, Orsay, France). 400 μL of heat-inactivated serum was mixed with 200 μL of protein G-Sepharose immunobeads for 30 min at 25°C and then centrifuged for 1 min at 3800 g. The supernatant was discarded and the immunobeads were then washed 3 times with 400 μL of Immunopure IgG binding buffer (Pierce, Rockford, USA). 400 μL Immunopure IgG Elution buffer (Pierce, Rockford, USA) were added to the immunobeads, which were mixed thoroughly and then centrifuged for 1 min at 5000 g. The supernatant was pooled and neutralized with 40 μL Tris-HCL 1 M pH 8.0. The IgG concentration was determined using a Bradford assay (Bio-Rad Protein Assay, Bio-Rad, Marnes-la-Coquette, France). Purified IgG was stored at -80°C.
HCV focus reduction neutralization assay
The HCV focus reduction neutralization assay was performed in 96-well microtiter plates. Serial dilutions of purified IgG (10 μg) ranging from 1:10 to 1:1,280 were established. Each dilution was tested twice. 25 μL of each sample was mixed with 25 μL of virus (100 FFU) in 96-well microtiter plates and incubated for 1 hour at 37°C, 5% CO2. A volume of 100 μL of Huh-7 cell suspension (10,000 cells/well) in culture medium was added and incubated for 5 hours at 37°C, 5% CO2. After 5 hours, the supernatants were removed and 100 μL of culture medium were added to the monolayers. After 72 hours, the cells were fixed with paraformaldehyde and permeabilized with 0.5% Triton X-100. Primary antibody (a HCV-positive patient serum inactivated at 56°C) was diluted to 1:500 prior to use and then incubated for 1 h at room temperature. A peroxidase-coupled, Fc-specific anti-human IgG antibody (Sigma, Saint Quentin Fallavier, France) diluted to 1:200 was dispensed onto the cell monolayer and incubated for 30 min at room temperature. The reaction was developed with DAB peroxidase substrate (Sigma, Saint Quentin Fallavier, France) and stopped after 10 min of incubation with distilled water. The number of HCV foci in each dilution was determined. Controls were included in each assay (non-neutralized virus, purified IgG from each patient at a 1:10 dilution). The dilution that neutralized 50% of the virus was calculated by curvilinear regression analysis using XLSTAT 2006 software (Addinsoft SARL, Paris, France) [35]. Each titer was determined as the log value of the reciprocal antibody dilution that reduced the number of viral foci by 50%.
Statistical analysis
Titers were expressed as logarithmic values and means ± standard deviation were calculated. Student's t-test was used to compare data between groups. p values below 0.05 were considered to be significant.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
CFo, SC and GD conceived, designed and performed the analysis. CFo, SD, EB, AS carried out the bioassay. SC and CFr performed the statistical analysis. DC, CW and VT have given final approval for the version to be published. SC and GD wrote the paper.
All authors have read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work was supported by grants from the Programme Hospitalier de Recherche Clinique de Picardie (Appel d'Offres Local, 2006) and the Agence Nationale de la Recherche sur le SIDA (ANRS, 2006).
We are grateful to Agnès Baron and Véronique Descamps for their expert technical assistance.
Contributor Information
Carole Fournier, Email: carole.fournier64@wanadoo.fr.
Gilles Duverlie, Email: gilles.duverlie@sa.u-picardie.fr.
Catherine François, Email: catherine.francois@u-picardie.fr.
Aurelie Schnuriger, Email: schnurig@chups.jussieu.fr.
Sarah Dedeurwaerder, Email: Sarah02120@aol.com.
Etienne Brochot, Email: ebrochot@etud.u-picardie.fr.
Dominique Capron, Email: capron.dominique@chu-amiens.fr.
Czeslaw Wychowski, Email: czeslaw.wychowski@ibl.fr.
Vincent Thibault, Email: vincent.thibault@psl.ap-hop-paris.fr.
Sandrine Castelain, Email: sandrine.castelain@u-picardie.fr.
References
- Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- Logvinoff C, Major ME, Oldach D, Heyward S, Talal A, Balfe P, Feinstone SM, Alter H, Rice CM, McKeating JA. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc Natl Acad Sci USA. 2004;101:10149–10154. doi: 10.1073/pnas.0403519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollier C, Depla E, Drexhage JA, Verschoor EJ, Verstrepen BE, Fatmi A, Brinster C, Fournillier A, Whelan JA, Whelan M, et al. Control of heterologous hepatitis C virus infection in chimpanzees is associated with the quality of vaccine-induced peripheral Thelper immune response. J Virol. 2004;78:187–196. doi: 10.1128/JVI.78.1.187-196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechmann M, Liang TJ. Vaccine development for hepatitis C. Semin Liver Dis. 2000;20:211–226. doi: 10.1055/s-2000-9947. [DOI] [PubMed] [Google Scholar]
- Owsianka A, Clayton RF, Loomis-Price LD, McKeating JA, Patel AH. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J Gen Virol. 2001;82:1877–1883. doi: 10.1099/0022-1317-82-8-1877. [DOI] [PubMed] [Google Scholar]
- Flint M, Maidens C, Loomis-Price LD, Shotton C, Dubuisson J, Monk P, Higginbottom A, Levy S, McKeating JA. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol. 1999;73:6235–6244. doi: 10.1128/jvi.73.8.6235-6244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadlock KG, Lanford RE, Perkins S, Rowe J, Yang Q, Levy S, Pileri P, Abrignani S, Foung SK. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J Virol. 2000;74:10407–10416. doi: 10.1128/JVI.74.22.10407-10416.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allander T, Drakenberg K, Beyene A, Rosa D, Abrignani S, Houghton M, Widell A, Grillner L, Persson MA. Recombinant human monoclonal antibodies against different conformational epitopes of the E2 envelope glycoprotein of hepatitis C virus that inhibit its interaction with CD81. J Gen Virol. 2000;81:2451–2459. doi: 10.1099/0022-1317-81-10-2451. [DOI] [PubMed] [Google Scholar]
- Farci P, Alter HJ, Wong DC, Miller RH, Govindarajan S, Engle R, Shapiro M, Purcell RH. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu YK, Hijikata M, Iwamoto A, Alter HJ, Purcell RH, Yoshikura H. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J Virol. 1994;68:1494–1500. doi: 10.1128/jvi.68.3.1494-1500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosch B, Verney G, Dreux M, Donot P, Morice Y, Penin F, Pawlotsky JM, Lavillette D, Cosset FL. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J Virol. 2005;79:8217–8229. doi: 10.1128/JVI.79.13.8217-8229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- Lavillette D, Morice Y, Germanidis G, Donot P, Soulier A, Pagkalos E, Sakellariou G, Intrator L, Bartosch B, Pawlotsky JM, Cosset FL. Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J Virol. 2005;79:6023–6034. doi: 10.1128/JVI.79.10.6023-6034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosch B, Bukh J, Meunier JC, Granier C, Engle RE, Blackwelder WC, Emerson SU, Cosset FL, Purcell RH. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc Natl Acad Sci USA. 2003;100:14199–14204. doi: 10.1073/pnas.2335981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier JC, Engle RE, Faulk K, Zhao M, Bartosch B, Alter H, Emerson SU, Cosset FL, Purcell RH, Bukh J. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc Natl Acad Sci USA. 2005;102:4560–4565. doi: 10.1073/pnas.0501275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisset C, Callens N, Blanchard E, Op De Beeck A, Dubuisson J, Vu-Dac N. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J Biol Chem. 2005;280:7793–7799. doi: 10.1074/jbc.M411600200. [DOI] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisset C, de Beeck AO, Horellou P, Dreux M, Gustot T, Duverlie G, Cosset FL, Vu-Dac N, Dubuisson J. High-density lipoproteins reduce the neutralizing effect of hepatitis C virus (HCV)-infected patient antibodies by promoting HCV entry. J Gen Virol. 2006;87:2577–2581. doi: 10.1099/vir.0.81932-0. [DOI] [PubMed] [Google Scholar]
- Netski DM, Mosbruger T, Depla E, Maertens G, Ray SC, Hamilton RG, Roundtree S, Thomas DL, McKeating J, Cox A. Humoral immune response in acute hepatitis C virus infection. Clin Infect Dis. 2005;41:667–675. doi: 10.1086/432478. [DOI] [PubMed] [Google Scholar]
- Prince AM, Brotman B, Huima T, Pascual D, Jaffery M, Inchauspe G. Immunity in hepatitis C infection. J Infect Dis. 1992;165:438–443. doi: 10.1093/infdis/165.3.438. [DOI] [PubMed] [Google Scholar]
- Lai ME, Mazzoleni AP, Argiolu F, De Virgilis S, Balestrieri A, Purcell RH, Cao A, Farci P. Hepatitis C virus in multiple episodes of acute hepatitis in polytransfused thalassaemic children. Lancet. 1994;343:388–390. doi: 10.1016/S0140-6736(94)91224-6. [DOI] [PubMed] [Google Scholar]
- Farci P, Alter HJ, Govindarajan S, Wong DC, Engle R, Lesniewski RR, Mushahwar IK, Desai SM, Miller RH, Ogata N, et al. Lack of protective immunity against reinfection with hepatitis C virus. Science. 1992;258:135–140. doi: 10.1126/science.1279801. [DOI] [PubMed] [Google Scholar]
- Flint M, Logvinoff C, Rice CM, McKeating JA. Characterization of infectious retroviral pseudotype particles bearing hepatitis C virus glycoproteins. J Virol. 2004;78:6875–6882. doi: 10.1128/JVI.78.13.6875-6882.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal O, Olshevsky U, Lustig S, Paran N, Halevy M, Schneider P, Zomber G, Fuchs P. Development of a tissue-culture-based enzyme-immunoassay method for the quantitation of anti-vaccinia-neutralizing antibodies in human sera. J Virol Methods. 2005;130:15–21. doi: 10.1016/j.jviromet.2005.05.027. [DOI] [PubMed] [Google Scholar]
- Wang S, Sakhatskyy P, Chou TH, Lu S. Assays for the assessment of neutralizing antibody activities against Severe Acute Respiratory Syndrome (SARS) associated coronavirus (SCV) J Immunol Methods. 2005;301:21–30. doi: 10.1016/j.jim.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska E, Liu D, Wu HY, Quiroz J, Rappaport R, Yang DP. Development of an improved microneutralization assay for respiratory syncytial virus by automated plaque counting using imaging analysis. Virol J. 2005;2:84. doi: 10.1186/1743-422X-2-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parren PW, Burton DR. The antiviral activity of antibodies in vitro and in vivo. Adv Immunol. 2001;77:195–262. doi: 10.1016/S0065-2776(01)77018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payan C, Roudot-Thoraval F, Marcellin P, Bled N, Duverlie G, Fouchard-Hubert I, Trimoulet P, Couzigou P, Cointe D, Chaput C, et al. Changing of hepatitis C virus genotype patterns in France at the beginning of the third millenium: The GEMHEP GenoCII Study. J Viral Hepat. 2005;12:405–413. doi: 10.1111/j.1365-2893.2005.00605.x. [DOI] [PubMed] [Google Scholar]
- Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol. 2006;80:5308–5320. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- Rouille Y, Helle F, Delgrange D, Roingeard P, Voisset C, Blanchard E, Belouzard S, McKeating J, Patel AH, Maertens G, et al. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J Virol. 2006;80:2832–2841. doi: 10.1128/JVI.80.6.2832-2841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelain S, Khorsi H, Zawadzki P, Sueur JM, Capron JP, Eb F, Duverlie G. Direct blotting electrophoresis for sequencing and genotyping hepatitis C virus. J Virol Methods. 1997;65:237–243. doi: 10.1016/S0166-0934(97)02184-8. [DOI] [PubMed] [Google Scholar]
- Martin NC, Pardo J, Simmons M, Tjaden JA, Widjaja S, Marovich MA, Sun W, Porter KR, Burgess TH. An immunocytometric assay based on dengue infection via DC-SIGN permits rapid measurement of anti-dengue neutralizing antibodies. J Virol Methods. 2006;134:74–85. doi: 10.1016/j.jviromet.2005.12.002. [DOI] [PubMed] [Google Scholar]