Abstract
Mammalian thioredoxin reductases (TrxR) are homodimers, homologous to glutathione reductase (GR), with an essential selenocysteine (SeCys) residue in an extension containing the conserved C-terminal sequence -Gly-Cys-SeCys-Gly. In the oxidized enzyme, we demonstrated two nonflavin redox centers by chemical modification and peptide sequencing: one was a disulfide within the sequence -Cys59-Val-Asn-Val-Gly-Cys64, identical to the active site of GR; the other was a selenenylsulfide formed from Cys497-SeCys498 and confirmed by mass spectrometry. In the NADPH reduced enzyme, these centers were present as a dithiol and a selenolthiol, respectively. Based on the structure of GR, we propose that in TrxR, the C-terminal Cys497-SeCys498 residues of one monomer are adjacent to the Cys59 and Cys64 residues of the second monomer. The reductive half-reaction of TrxR is similar to that of GR followed by exchange from the nascent Cys59 and Cys64 dithiol to the selenenylsulfide of the other subunit to generate the active-site selenolthiol. Characterization of recombinant mutant rat TrxR with SeCys498 replaced by Cys having a 100-fold lower kcat for Trx reduction revealed the C-terminal redox center was present as a dithiol when the Cys59-Cys64 was a disulfide, demonstrating that the selenium atom with its larger radius is critical for formation of the unique selenenylsulfide. Spectroscopic redox titrations with dithionite or NADPH were consistent with the structure model. Mechanisms of TrxR in reduction of Trx and hydroperoxides have been postulated and are compatible with known enzyme activities and the effects of inhibitors, like goldthioglucose and 1-chloro-2,4-dinitrobenzene.
Keywords: evolution, thiols, disulfide, selenium, enzyme structure
Thioredoxin reductase (TrxR) is a member of the pyridine nucleotide-disulfide oxidoreductase family that includes glutathione reductase (GR), lipoamide dehydrogenase, and mercuric ion reductase (1). The members of this family are homodimeric flavoproteins containing one redox-active disulfide and one tightly bound FAD per subunit. TrxR catalyzes reduction of the redox-active disulfide in Trx (oxidized Trx) by NADPH. The dithiol form of Trx [Trx-(SH)2] is a major cellular reductant (2) involved in a number of thiol-dependent cellular reactions such as enzymatic synthesis of deoxyribonucleotides, defense against oxidative stress, redox regulation of gene expression, or signal transduction by thiol redox control (3, 4).
TrxR from mammalian species has long been known (5) to be very different from the well studied Escherichia coli TrxR (6), having subunits of 55 kDa instead of 35 kDa and showing a much broader substrate specificity (7). The mammalian TrxR directly reduces not only Trx from different species but also many nondisulfide substrates, such as selenite (8), lipid hydroperoxides (9), and H2O2 (10). Sequencing and cloning demonstrated that cytosolic human and rat TrxR are highly similar to GR (11, 12) and not to its E. coli counterpart (13). The sequence contains one active-site sequence motif—Cys59-Val-Asn-Val-Gly-Cys64—identical to the redox-active disulfide of GR (14). The C-terminal end contains an extension of 16 residues with a penultimate selenocysteine (SeCys) residue within the unique sequence, Gly-Cys-SeCys-Gly conserved in all mammalian TrxR reported to date (12, 15–19). The SeCys residue is essential for the catalytic activity of TrxR, because either its removal by carboxypeptidase digestion (12) or its modification by alkylation (12, 20) leads to inactivation. Also, the irreversible inhibitor 1-chloro-2,4-dinitrobenzene, which inactivates the mammalian enzyme as a TrxR but induces an NADPH oxidase activity in the enzyme (21), leads to alkylation of both the Cys497 and the SeCys498 residues in the enzyme (22). Furthermore, selenium deficiency leads to a major loss of TrxR activity (23–27).
We recently expressed and characterized mutant rat enzymes in E. coli, including SeCys498 replaced by Cys, and demonstrated that this folded protein contained FAD and was active. However, it had a 100-fold lower kcat and a 10-fold lower Km for Trx compared with the Se-containing wild-type rat enzyme (10). The truncated protein lacking the C-terminal SeCys-Gly dipeptide was also folded and contained FAD but was inactive although titration with NADPH yielded the characteristic thiolate-flavin charge transfer complex common for GR and mammalian TrxR (1, 28). In this paper, we have identified a selenenylsulfide as the active site of TrxR and propose a structure model and mechanisms for the enzyme.
Materials and Methods
Bovine cytosolic TrxR and the SeCys498-Cys rat TrxR were purified to homogeneity as described (5, 7, 10). Recombinant human Trx with the structural Cys63 and Cys72 residues replaced by Ser (C63S/C72S) was prepared as described previously (29). l-(1-tosylamido-2-phenyl)ethyl chloromethyl ketone-treated trypsin was purchased from Promega SDS, sodium dithionite from Merck, and NADPH and 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) from Sigma. [1-14C]iodoacetamide and [1-14C]iodoacetic acid were from American Radiolabeled Chemicals, St. Louis.
Labeling of Redox-Active Cysteine and SeCys Groups and Tryptic Digestion.
All operations were performed with deaerated and N2 equilibrated buffers. To isolate peptides from the oxidized form of calf liver TrxR, 40 μM of subunits in 250 μl of 5 M of Gdn⋅HCl/0.1 M Tris⋅HCl, pH 8.0, 5 mM EDTA was alkylated with 2 mM of [1-14C]iodoacetic acid at room temperature overnight in the dark. The mixture was desalted on a Fast Desalting PC 3.2/10 column (Pharmacia Biotech) in 0.2 M ammonium bicarbonate with a Pharmacia Smart system and digested as described below.
To specifically label DTT-reducible bridges in the oxidized enzyme, a double alkylation procedure was used. The oxidized form of calf liver TrxR (20 μM of subunits) was denatured in 6 M Gdn⋅HCl/1 M Tris⋅HCl, pH 7.5, 2 mM EDTA, and alkylated by incubation with 2 mM of 4-vinylpyridine in a final volume of 250 μl at room temperature for 4 h in the dark. After desalting as described above, the protein solution (50 μl) was mixed with 150 μl of 8 M Gdn⋅HCl/0.1 M Tris⋅HCl, pH 7.5, 2 mM EDTA, and 0.23 mM of DTT under N2 to reduce disulfide or selenenylsulfide bridges. Nascent thiols or selenol groups were then alkylated with 0.69 mM of [1-14C]iodoacetamide for 6 h in the dark at room temperature. The resulting fully derivatized protein was desalted and then digested with 5 μg of l-(1-tosylamido-2-phenyl)ethyl chloromethyl ketone-treated trypsin. After overnight incubation at 37°C, another 5 μg of the trypsin was added and incubation continued for an additional 6 h. The digests were stored at −20°C until separation of the resulting peptides.
Separation of Peptides and Sequence Analysis.
Tryptic peptides were separated by reverse-phase chromatography on a Sephasil C2/C18 column by using a smart system (Pharmacia). A linear gradient from 0 to 80% acetonitrile in 0.1% trifluoroacetic acid was used at a flow rate 80 μl/min over 150 min. The selected peptides were sequenced by using automated Edman degradation on a Procise Protein Sequencing System as described previously (12).
Mass Spectrometric Analysis of the C-Terminal Tryptic Peptide.
Calf thymus TrxR (about 20 μM of subunits) was denatured in 6 M Gdn⋅HCl/0.1 M Tris⋅HCl, pH 7.5/2 mM EDTA. The denatured protein was then alkylated by incubation with 2 mM of 4-vinylpyridine at room temperature for 4 h. The protein desalting, digestion, and subsequent peptide separation procedures were as described above. The C-terminal peptide was identified by comparison with its retention time in the previously well-characterized tryptic peptide map and was analyzed with electrospray mass spectrometry by using an AutoSpec instrument (Micromass, Manchester, U.K.) at the Protein Analysis Center of the Karolinska Institute, Sweden.
Anaerobic Titration with Sodium Dithionite.
The TrxR solution was placed in a cuvette and sodium dithionite solution in a separate glass vial, both covered with a rubber septa and bubbled with argon through needles penetrating the rubber septa, while air was evacuated although a send pair of needles. This treatment lasted ≈30 min. To the enzyme solution was then added aliquots of the dithionite solution with a gas-tight Hamilton syringe. Spectra were recorded at 25°C by using a Shimadzu UV-visible spectrophotometer.
Quantitative Determination of Thiol Content.
Thiols were determined with DTNB in a total reaction volume of 200 μl containing 1 μM of enzyme subunit with a control containing only buffer. Enzyme was reduced by incubation with 4 mM NADPH in a total volume of 45 μl at room temperature for 2 min, after which the enzyme was denatured by adding 150 μl of 8 M guanidine hydrochloride (Gdn⋅HCl) in 50 mM Hepes and 2 mM EDTA, pH 7.6. The thiol (or selenol) content was determined after addition of 1.58 mM DTNB by absorbance at 412 nm recorded in a Shimadzu UV-visible spectrophotometer. Measurements were made anaerobically with 1-cm cuvettes covered with rubber septa. The absorbance at 412 nm of each sample was recorded against the relevant control solution in a Shimadzu UV-visible spectrophotometer. The thiol (or selenol) content was calculated by using an extinction coefficient of thionitrobenzoate at 412 nm [(14.15 mM−1⋅cm−1 in buffer and 13.7 mM−1⋅cm−1 in 6 M Gdn⋅HCl (30)].
Enzyme Concentration.
For spectral experiments, TrxR concentration was estimated from 11,300 M−1⋅cm−1 as the extinction coefficients of the enzyme-bound FAD at 463 nm assuming two FAD per dimer. The protein concentrations were also determined by absorbance at 280 nm with the value for 0.1% solution of 1.768 for calf thymus TrxR and 1.625 for SeCys498-Cys rat TrxR (10).
Results
Identification of a Disulfide Between Cys59 and Cys64 as Well as a Selenenylsulfide Between Cys497 and SeCys498.
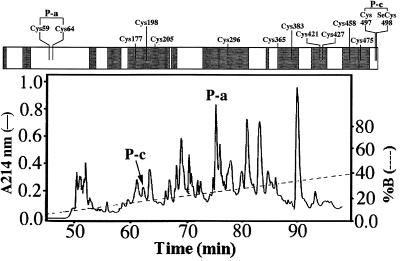
We previously sequenced large parts of bovine TrxR, which contains a total of 13 cysteine and one SeCys residue (12) [Terashima, H. (1998), GenBank accession no. AF053984]. Redox-active residues labeled after NADPH reduction were identified by treating the oxidized enzyme with iodoacetic acid before incubation with NADPH and subsequent alkylation of sulfhydryl or selenol groups with vinylpyridine. This procedure led to sequencing of two peptides, one of which contained the internal sequence CVNVGC, identical to the active site of GR and different from the C-terminal peptide RSGGNILQTGCUG (12). We now used oxidized enzyme in 5 M Gdn⋅HCl and blocked reactive groups by [14C]iodoacetic acid and removed the denaturant by chromatography in 0.2 M ammonium bicarbonate followed by digestion with trypsin. The resulting peptides were separated by reverse-phase chromatography (Fig. 1) and analyzed by Edman degradation. Although almost all Cys containing peptide were not present, suggesting a trypsin-resistant core, two peptides in high final yield denoted P-a and P-c (Fig. 1), corresponding to those containing Cys59 and Cys64 (P-a) and Cys-497 and SeCys498 (P-c). During Edman degradation, the phenylthiohydantoin (PTH) derivatives of Cys59,64,497, and PTH-SeCys498 were absent at the corresponding cycles, forming a characteristic gap in the peptide sequence (Table 1). These observations strongly support a notion of a disulfide connecting Cys59 and Cys64 and a bridge connecting Cys497 and SeCys498. The two bridges are intrasubunit linkages, because the native enzyme was detected only with a size corresponding to the monomeric form in a nonreduced SDS/PAGE (12), ruling out any intersubunit covalent linkages.
Figure 1.
Tryptic peptide map of the alkylated oxidized form of bovine TrxR. Solid line indicates absorbance at 214 nm; dashed line, the percent CH3CN in 0.1% TFA. The S-S bridge and Se-S bridge peptides are labeled P-a and P-c, respectively. The positions of the 13 Cys residues and the SeCys in bovine TrxR are shown (Upper), with peptides identified by sequencing indicated by white boxes and peptides not found in this digest in gray boxes.
Table 1.
Peptide sequence of bovine TrxR
| C-terminal peptide | |||||||||||||||
| Oxidized form (P-c in Fig. 1) | S | G | G | N | I | L | Q | T | G | ND‡ | ND‡ | G | |||
| Yield, pmol | 311 | 598 | 515 | 359 | 478 | 383 | 320 | 265 | 243 | 27 | |||||
| Reduced form (P-c in Fig. 2) | S | G | G | N | I | L | Q | T | G | C* | U† | G | |||
| Yield, pmol | 27 | 33 | 33 | 26 | 36 | 30 | 22 | 12 | 20 | 12 | ∼10 | 7 | |||
| Active-site peptide | |||||||||||||||
| Oxidized form (P-a in Fig. 1) | W | G | L | G | G | T | ND‡ | V | N | V | G | ND‡ | I | P | K |
| Yield, pmol | 196 | 177 | 225 | 171 | 163 | 161 | 153 | 120 | 143 | 143 | 108 | 45 | 26 | ||
| Reduced form (P-a in Fig. 2) | W | G | L | G | G | T | C* | V | N | V | G | C* | I | P | K |
| Yield, pmol | 21 | 27 | 64 | 25 | 25 | 24 | 12 | 25 | 19 | 22 | 22 | 9 | 17 | 8 | 4.5 |
When cysteine was labeled with iodoacetamide, the resulting PTH-Cys (Cam) eluted almost exactly with PTH-Glu.
†U indicates the PTH-SeCys peak eluting next to PTH-Pro.
‡ND indicates that in the corresponding cycle, no PTH peak was detected.
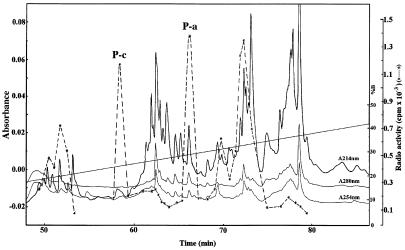
In the second experiment, free thiols in denatured oxidized enzyme (6 M Gdn⋅HCl) were labeled with 4-vinylpyridine followed by desalting to 0.2 M ammonium bicarbonate. The enzyme was denatured again in 6 M Gdn⋅HCl, reduced by DTT and alkylated with excess [1-14C]iodoacetamide. After desalting, the tryptic peptide map indicated complete digestion (Fig. 2), having two separate peaks with high specific radioactivity denoted as “P-c” and “P-a.” Analyses by Edman degradation revealed that “P-c” corresponded to the C-terminal Cys497 and SeCys498-containing peptide, and “P-a” was the Cys59 and Cys64-containing peptide (Table 1). These results showed that accessible Cys59, Cys64, Cys497, and SeCys498 had been formed after reduction with DTT.
Figure 2.
Tryptic peptide map of DTT-reduced and double alkylated bovine TrxR. Elution of peptides was monitored by absorbance at 214 nm, 280 nm, and 254 nm [for S-(4-pyridylethyl)-l-cysteine], as shown with solid lines. The dashed line shows the cpm from [14C]carboxymethylcysteine in each fraction.
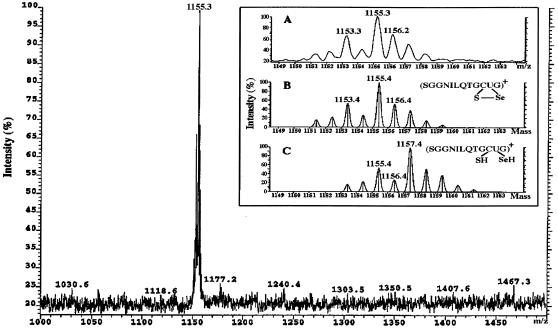
To further prove the existence of a Cys497-SeCys498 linkage, we analyzed the C-terminal peptide of the pyridylated nonreduced enzyme by mass spectrophotometry. The peptide gave a peak with maximum at m/z 1,155.3 (Fig. 3A) and a spectrum in perfect agreement with the theoretical mass spectrum of the peptide containing a selenenylsulfide (Fig. 3B), clearly shifted from that of the thiol- and selenol-containing peptide (Fig. 3C) and obviously different from that of a pyridylated peptide. These results proved that two redox-active bridges of mammalian TrxR are present as a disulfide linking Cys59 and Cys64 and a selenenylsulfide linking the penultimate SeCys with its adjacent Cys residue.
Figure 3.
Electrospray ionization (ESI) mass spectrum of the C-terminal tryptic peptide from oxidized bovine TrxR. (A Inset) shows the detailed mass spectrum of the peptide detected by ESI, in comparison to the simulated spectra of the peptide with (B) or without (C) a selenenylsulfide.
Thiols in the Bovine and Rat Mutant Enzymes.
In the oxidized bovine TrxR denatured by 6 M Gdn⋅HCl under anaerobic conditions, the number of titratable free SH groups was 10.4 ± 0.7 (n = 7) per subunit. When NADPH was used for reduction of the enzyme before denaturation, the number of free SH groups was 14.2 ± 0.5 (n = 5) and thus the newly formed groups that reacted with DTNB was 3.8, consistent with the total of 13 cysteines and 1 SeCys residue [Terashima, H. (1998) GenBank accession no. AF053984], of which four were joined in two NADPH-reducible bridges. The SeCys498-Cys rat mutant TrxR contains a total of 15 cysteine residues (12, 10). Previously we demonstrated that both Cys498 and Cys497 were alkylated by 4-vinylpyridine after denaturation of the oxidized rat enzyme, indicating that they were present as thiols. This result was also evident here from SH-determination with DTNB. Furthermore, preincubation with NADPH showed an increase of only two SH groups for the rat mutant SeCys498-Cys enzyme to 14.9 ± 1.1 (n = 7).
Characterization of the SeCys498-Cys Enzyme.
Alkylation of the rat SeCys498-Cys enzyme in 6 M Gdn⋅HCl with [14C]iodoacetic acid followed by desalting and tryptic digestion as described above for the bovine enzyme (Fig. 1) showed a peptide map (data not shown) where the active-site disulfide peptide P-a and the C-terminal peptide P-c were identified by Edman degradation. The P-c peptide was fully carboxymethylated demonstrating free Cys497 and Cys498 SH- groups. When an aliquot of this monoalkylated enzyme was denatured in 5 M Gdn⋅HCl and treated with DTT followed by carboxymethylation and tryptic digestion, the peptide map demonstrated that the P-a peptide now was eluted in a different position and was alkylated as well as the P-c peptide. This confirmed that the SeCys498-Cys enzyme as isolated has two thiols in the C terminus but a disulfide in the N-terminal active-site motif.
Redox Titrations.
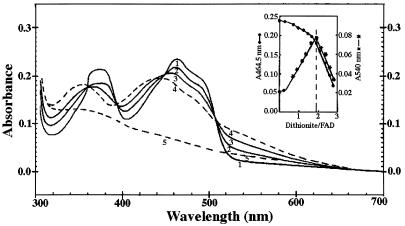
Titration of the SeCys498-Cys rat TrxR with dithionite resulted in highly similar spectral changes (Fig. 4) to those observed by Williams and coworkers analyzing human placenta TrxR (28). There were two spectral phases. First was the formation of a typical EH2 spectrum (28), in which two flavin absorbance bands were decreased and blue shifted, whereas a new long-wavelength flavin-thiolate charge transfer band was formed with a maximum at 540 nm, where neither oxidized enzyme nor fully reduced enzyme shows absorbance (28). The set of four isosbestic points seen in Fig. 4 indicated that with less than 1.9 equivalents of dithionite, the charge-transfer complex is the predominant species. In the second phase, addition of another equivalent of dithionite resulted in complete reduction of the enzyme with flavin bleaching and disappearance of the flavin-thiolate charge transfer absorbance band. To produce this fully reduced enzyme, about three equivalents of dithionite per enzyme subunit were required. This is one equivalent less than that of the human placenta (28) or the bovine enzyme (data not shown) and consistent with an already reduced C-terminal dithiol pair in this mutant enzyme.
Figure 4.
Sodium dithionite titration of rat SeCys498-Cys TrxR. Enzyme (21.2 nmol of subunits) in 0.6 ml of 0.1 M phosphate buffer/1 mM EDTA/pH 7.5 was titrated with 10 mM sodium dithionite dissolved in 50 mM sodium pyrophosphate buffer, pH 8.5, at 25°C in the presence of 150 nM methyl viologen. Spectra show oxidized enzyme (curve 1), the enzyme reduced with 0.94 equivalents (curve 2), 1.4 equivalents (curve 3), 1.9 equivalents (curve 4), and 2.83 equivalents of dithionite (curve 5). A slight artifact in the spectra at 360 nm is because of lamp change at this wavelength. Inset shows the effects of dithionite addition on the absorbances at 464 nm and at 540 nm.
Discussion
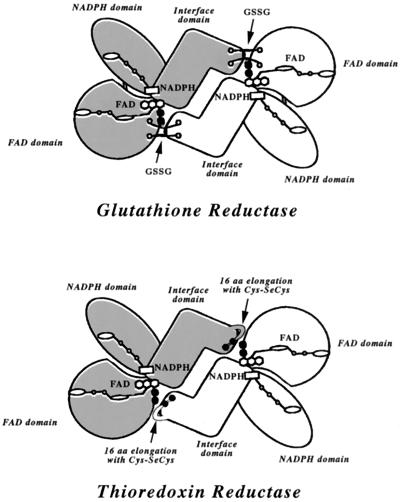
Williams and coworkers (28) demonstrated elegantly that the mechanism of TrxR from human placenta is similar to the mechanisms of lipoamide dehydrogenase and GR and distinct from the mechanism of TrxR from E. coli. All TrxR from mammalian cells, of which there are a growing number of isoenzymes (19, 31–33), have the conserved C-terminal sequence -Gly-Cys-SeCys-Gly and a subunit with homology to GR (10) including the identical conserved active site of the latter enzyme (19, 31–33). However, the detailed mechanism of mammalian TrxR has not been known. Before discussing our results, we will propose a structure of mammalian TrxR based on the homology to GR (Fig. 5). TrxR has a 16-residue C-terminal extension with the conserved SeCys residue. We propose that this is in a sense taking the place of glutathione disulfide, the substrate for GR, so that the Cys-SeCys residues of the one subunit are in close proximity to the redox-active disulfide/dithiol of the second subunit. Electrons can then be transferred out from the redox-active disulfide/dithiol to the SeCys and Cys residues, which constitute the active site of the enzyme. This will be in agreement with the highly unusual evolutionary scenario proposed by Williams et al. (28) for mammalian TrxR and the pyridine nucleotide-disulfide oxidoreductase family of dimeric flavoenzymes. A similar example of a structure in favor of our model is in mercuric reductase, where a Cys-Cys sequence near the C terminus can receive electrons from the active center dithiol (34). Also, the TrxR from Plasmodium falciparum, which is a homodimer of subunits with Mr 59,000, has two C-terminal Cys residues (Cys535 and Cys540), which interact with the active-site disulfide/dithiol (Cys88 and Cys93) from the other subunit (35).
Figure 5.
Proposed model of mammalian TrxR based on homology to GR. (Upper) Model of GR with FAD, NADPH, and interface domains (adapted from refs. 1, 6, 13, 14). Glutathione disulfide bound is indicated as well as the active-site cysteines (black circles). (Lower) Model of mammalian TrxR. The elongation with 16-residues is extending from the interface, positioning SeCys498 and Cys497 in one subunit adjacent to Cys59 and Cys64 of the other subunit.
In this study, we have proved that the C-terminal SeCys residue of oxidized bovine mammalian TrxR forms a selenenylsulfide with the adjacent Cys residue by using both Edman degradation and mass spectrometry. A number of arguments support the conclusion that the selenolthiol form of the selenenylsulfide is the actual active site of the enzyme for reduction of Trx and the many other substrates known (5, 7–10). The position of the active site at the C-terminal end is compatible with an open active site capable of reducing all kinds of molecules including DTNB, which is used to assay the enzyme (5, 7). The truncated enzyme lacking the SeCys-Gly dipeptide is inactive (10). Furthermore, replacement of the SeCys residue by a similar but redox-inactive Ser results in an inactive enzyme (10). However, both these mutant enzymes contain FAD and showed the same spectral changes on redox titration with NADPH, consistent with an intact reductive half reaction very similar to GR (10). The SeCys to Cys mutant rat enzyme shows activity with Trx having a 100-fold lower kcat and 10-fold lower apparent Km indicating an increased binding of Trx. Most importantly, the pH optimum of the mutant enzyme was 9 as opposed to 7 for the wild-type selenium-containing enzyme compatible with the involvement of a low pKa (5.25) selenol in the catalytic mechanism (10). Accumulated evidence showed that the native unreduced enzyme was resistant to either carboxypeptidase (12) or trypsin digestion (36) as well as to chemical modification by 1-chloro-2,4-dinitrobenzene (22), aurothioglucose (37), bromo [1-14C] acetic acid (20), and 5-iodoacetamidofluorescein (32). However, the enzyme was modified by all of the above treatments only after NADPH reduction. The selenenylsulfide between adjacent residues may assume a particular hairpin β-turn structure critically dependent on the larger atomic radius of the Se atom compared with an S atom. Our finding of a Cys-Cys dithiol in the mutant enzyme as isolated is consistent with an unfavorable structure for forming a disulfide.
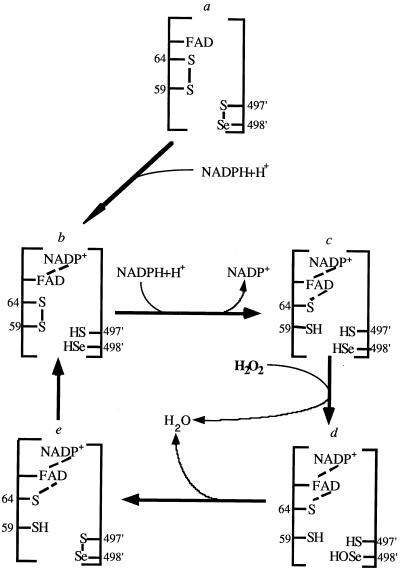
The mammalian Se-containing TrxR reduces lipid hydroperoxides (9) and H2O2 (10), and this activity is lost in the Cys mutant enzyme. So far, two redox states of the SeCys, a selenol and a selenenylsulfide oxidized form, have been unambiguously identified. This allows us to propose a mechanism for H2O2 reduction by mammalian TrxR (Fig. 6). The catalytic intermediates consist of a selenol, a putative selenenic acid, and the selenenylsulfide enzyme forms. The initial reaction is that the selenenylsulfide receives electrons from NADPH via the FAD and redox-active dithiol of the first subunit to produce a sulfhydryl and a selenol (-SeH) in the second subunit. Because of the low pKa value of the selenol, selenolate should be a predominant form under physiologic conditions. Because it is a strong nucleophilic, selenolate is more susceptible to oxidation by H2O2 than thiols, yielding selenenic acid (-SeOH). One cysteine thiol (most likely Cys497) reacts with the selenenic acid to produce water and to reform the selenenylsulfide. A second thiol (most likely Cys59 from the other subunit) would attack the bridge to regenerate the selenol. Therefore, the selenenylsulfide serves as either a catalytically essential redox center or transient intermediate during peroxide reduction. According to its high apparent Km value (2.5 mM) for H2O2 (10) and kcat of 100 × min−1, this antioxidant defense function of mammalian TrxR may be expected to play a role only with elevated H2O2 concentration.
Figure 6.
Simplified postulated mechanism for hydrogen peroxide reduction by mammalian TrxR. See Discussion.
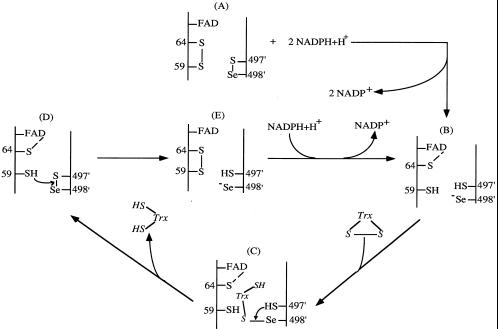
A mechanism for mammalian TrxR to react with Trx is proposed in Fig. 7. The catalytic reaction starts by reduction of the selenenylsulfide to the selenolate anion as for H2O2 reduction. The selenolate anion (-Se−) attacks the disulfide of Trx. The resulting enzyme–Trx-mixed selenenylsulfide (Fig. 7C) is attacked by Cys497 to regenerate the selenenylsulfide, which will be reduced by the active-site thiolate from the other subunit again (Fig. 7D). During the reaction, the active-site dithiol maintains the selenol in the reduced state. As the selenolate anion is both a better nucleophilic and a better leaving group than the thiolate anion, this explains why the reduction rate of Trx by the SeCys498-Cys mutant TrxR is much slower, and the dithiol should have a higher redox potential (38).
Figure 7.
Postulated mechanism for Trx reduction by mammalian TrxR. See Discussion.
The mechanism (Fig. 7) is also compatible with the effects of the irreversible inhibitor 1-chloro-2,4-dinitrobenzene, which we showed previously modified both the SeCys498 and the adjacent Cys497 by alkylation leading to a total loss of Trx reduction. However, the enzyme was still active because it had a 30-fold increase in NADPH oxidase activity compared with the wild-type enzyme, producing superoxide (22). This demonstrates that the first reductive half-reaction is intact in the modified enzyme. These results also suggest that if the truncated enzyme is produced during selenium starvation, an increased NADPH oxidase activity may be producing mutagenic superoxide or other species of free radicals. Further studies will be required to test this hypothesis.
Acknowledgments
This study was supported by the Swedish Cancer Society (project 961), the Swedish Medical Research Council (projects 13X-3529 and 03XS-013005–01A), the K. A. Wallenberg Foundation, and the I.-B. and A. Lundberg Foundation. We thank Dr. William Griffiths for the mass spectrum analysis and Mr. Geng Chang for the purification of human Trx.
Abbreviations
- Trx
thioredoxin
- TrxR
Trx reductase
- SeCys
selenocysteine
- Gdn⋅HCl
guanidine hydrochloride
- DTNB
5,5′-dithio(bis 2-nitrobenzoic acid)
- GR
glutathione reductase
- PTH
phenylthiohydantoin
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100114897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100114897
References
- 1.Williams C H., Jr . In: Chemistry and Biochemistry of Flavoenzymes. Müller F, editor. III. Boca Raton, FL: CRC; 1992. pp. 121–211. [Google Scholar]
- 2.Holmgren A. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 3.Holmgren A. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 4.Nakamura H, Nakamura K, Yodoi J. Annu Rev Immunol. 1997;15:351–369. doi: 10.1146/annurev.immunol.15.1.351. [DOI] [PubMed] [Google Scholar]
- 5.Luthman M, Holmgren A. Biochemistry. 1982;21:6628–6633. doi: 10.1021/bi00269a003. [DOI] [PubMed] [Google Scholar]
- 6.Williams C H., Jr FASEB J. 1995;9:1267–1276. doi: 10.1096/fasebj.9.13.7557016. [DOI] [PubMed] [Google Scholar]
- 7.Arnér E S J, Zhong L, Holmgren A. Methods Enzymol. 1999;300:226–239. doi: 10.1016/s0076-6879(99)00129-9. [DOI] [PubMed] [Google Scholar]
- 8.Kumar S, Björnstedt M, Holmgren A. Eur J Biochem. 1992;207:435–439. doi: 10.1111/j.1432-1033.1992.tb17068.x. [DOI] [PubMed] [Google Scholar]
- 9.Björnstedt M, Hamberg M, Kumar S, Xue J, Holmgren A. J Biol Chem. 1995;270:11761–11764. doi: 10.1074/jbc.270.20.11761. [DOI] [PubMed] [Google Scholar]
- 10.Zhong, L. & Holmgren, A. (2000) J. Biol. Chem., in press. [DOI] [PubMed]
- 11.Gasdaska P Y, Gasdaska J R, Cochran S, Powis G. FEBS Lett. 1995;373:5–9. doi: 10.1016/0014-5793(95)01003-w. [DOI] [PubMed] [Google Scholar]
- 12.Zhong L, Arnér E S J, Ljung J, Åslund F, Holmgren A. J Biol Chem. 1998;273:8581–8591. doi: 10.1074/jbc.273.15.8581. [DOI] [PubMed] [Google Scholar]
- 13.Kuriyan J, Krishna T S R, Wong L, Guenther B, Pahler A, Williams C H., Jr Nature (London) 1991;352:172–174. doi: 10.1038/352172a0. [DOI] [PubMed] [Google Scholar]
- 14.Karplus P A, Schulz G E. J Mol Biol. 1987;195:701–729. doi: 10.1016/0022-2836(87)90191-4. [DOI] [PubMed] [Google Scholar]
- 15.Gladyshev V N, Jeang K T, Stadtman T C. Proc Natl Acad Sci USA. 1996;93:6146–6151. doi: 10.1073/pnas.93.12.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura T, Stadtman T C. Proc Natl Acad Sci USA. 1996;93:1006–1011. doi: 10.1073/pnas.93.3.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasdaska P Y, Berggren M M, Berry M J, Powis G. FEBS Lett. 1999;442:105–111. doi: 10.1016/s0014-5793(98)01638-x. [DOI] [PubMed] [Google Scholar]
- 18.Lee S R, Kim J R, Kwon K S, Yoon H W, Levine R L, Ginsburg A, Rhee S G. J Biol Chem. 1999;274:4722–4734. doi: 10.1074/jbc.274.8.4722. [DOI] [PubMed] [Google Scholar]
- 19.Miranda-Vizuete A, Damdimopoulos A E, Pedrajas J R, Gustafsson J A, Spyrou G. Eur J Biochem. 1999;261:405–412. doi: 10.1046/j.1432-1327.1999.00286.x. [DOI] [PubMed] [Google Scholar]
- 20.Gorlatov S N, Stadtman T C. Proc Natl Acad Sci USA. 1998;95:8520–8525. doi: 10.1073/pnas.95.15.8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnér E S J, Björnstedt M, Holmgren A. J Biol Chem. 1995;270:3479–3482. doi: 10.1074/jbc.270.8.3479. [DOI] [PubMed] [Google Scholar]
- 22.Nordberg J, Zhong L, Holmgren A, Arnér E S J. J Biol Chem. 1998;273:10835–10842. doi: 10.1074/jbc.273.18.10835. [DOI] [PubMed] [Google Scholar]
- 23.Berggren M, Gallegos A, Gasdaska J, Powis G. Anticancer Res. 1997;17:3377–3380. [PubMed] [Google Scholar]
- 24.Hill K E, McCollum G W, Boegli M E, Burk R F. Biochem Biophys Res Commun. 1997;234:293–295. doi: 10.1006/bbrc.1997.6618. [DOI] [PubMed] [Google Scholar]
- 25.Gladyshev V N, Factor V M, Housseau F, Hatfield D L. Biochem Biophys Res Commun. 1998;251:488–493. doi: 10.1006/bbrc.1998.9495. [DOI] [PubMed] [Google Scholar]
- 26.Gasdaska J R, Harney J W, Gasdaska P Y, Powis G, Berry M J. J Biol Chem. 1999;274:25379–25385. doi: 10.1074/jbc.274.36.25379. [DOI] [PubMed] [Google Scholar]
- 27.Fujiwara N, Fujii T, Fujii J, Taniguchi N. Biochem J. 1999;340:439–444. [PMC free article] [PubMed] [Google Scholar]
- 28.Arscott L D, Gromer S, Schirmer R H, Becker K, Williams C H., Jr Proc Natl Acad Sci USA. 1997;94:3621–3626. doi: 10.1073/pnas.94.8.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren X, Björnstedt M, Shen B, Ericson M L, Holmgren A. Biochemistry. 1993;32:9701–9708. doi: 10.1021/bi00088a023. [DOI] [PubMed] [Google Scholar]
- 30.Riddles P W, Blakeley R L, Zerner B. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- 31.Rigobello M P, Callegaro M T, Barzon E, Benetti M, Bindoli A. Free Radical Biol Med. 1998;24:370–376. doi: 10.1016/s0891-5849(97)00216-5. [DOI] [PubMed] [Google Scholar]
- 32.Sun Q A, Wu Y, Zappacosta F, Jeang K T, Lee B J, Hatfield D L, Gladyshev V N. J Biol Chem. 1999;274:24522–24530. doi: 10.1074/jbc.274.35.24522. [DOI] [PubMed] [Google Scholar]
- 33.Watabe S, Makino Y, Ogawa K, Hiroi T, Yamamoto Y, Takahashi S Y. Eur J Biochem. 1999;264:74–84. doi: 10.1046/j.1432-1327.1999.00578.x. [DOI] [PubMed] [Google Scholar]
- 34.Miller S M, Moore M J, Massey V, Williams C H., Jr Biochemistry. 1989;28:1194–1205. doi: 10.1021/bi00429a037. [DOI] [PubMed] [Google Scholar]
- 35.Wang P F, Arscott L D, Gilberger T W, Muller S, Williams C H., Jr Biochemstry. 1999;38:3187–3196. doi: 10.1021/bi982674g. [DOI] [PubMed] [Google Scholar]
- 36.Gromer S, Wissing J, Behne D, Ashman K, Schirmer R H, Flohe L, Becker K. Biochem J. 1998;332:591–592. doi: 10.1042/bj3320591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gromer S, Arscott L D, Williams C H, Jr, Schirmer R H, Becker K. J Biol Chem. 1998;273:20096–20101. doi: 10.1074/jbc.273.32.20096. [DOI] [PubMed] [Google Scholar]
- 38.Besse D, Siedler F, Dierks T, Kessler H, Moroder L. Angew Chem Int Ed Engl. 1997;36:883–885. [Google Scholar]