Abstract
Elucidating the origins of complex biological structures has been one of the major challenges of evolutionary studies. The bacterial flagellum is a primary example of a complex apparatus whose origins and evolutionary history have proven difficult to reconstruct. The gene clusters encoding the components of the flagellum can include >50 genes, but these clusters vary greatly in their numbers and contents among bacterial phyla. To investigate how this diversity arose, we identified all homologs of all flagellar proteins encoded in the complete genome sequences of 41 flagellated species from 11 bacterial phyla. Based on the phylogenetic occurrence and histories of each of these proteins, we could distinguish an ancient core set of 24 structural genes that were present in the common ancestor to all Bacteria. Within a genome, many of these core genes show sequence similarity only to other flagellar core genes, indicating that they were derived from one another, and the relationships among these genes suggest the probable order in which the structural components of the bacterial flagellum arose. These results show that core components of the bacterial flagellum originated through the successive duplication and modification of a few, or perhaps even a single, precursor gene.
Keywords: bacterial evolution, biological complexity, gene duplication
Bacterial flagella are complex and well honed organelles that provide swimming and swarming motilities and also play a central role in adhesion, biofilm formation, and host invasion (1). In the past several decades, extensive knowledge has accumulated about the structure, genetics, assembly, and regulation of flagella in widely diverse bacterial lineages (2–7). The typical bacterial flagellum consists of six components: a basal body (including MS ring, P ring, and L ring), a motor, a switch, a hook, a filament, and an export apparatus (2). In the best studied systems, those of Escherichia coli and Salmonella enterica sv. Typhimurium, >50 genes are involved in flagellar biosynthesis and function (3). Approximately half of these genes encode the structural components of the flagellum, and the rest are responsible for either the regulation of flagellar assembly or the detection and processing of environmental signals to which flagella respond.
Whereas E. coli and Salmonella have long served as the model organisms for studying flagellar assembly (2), there is extensive diversity among bacteria in the contents and organization of the gene complexes that specify flagella as well as structural variation in the flagellum itself (8, 9). For example, in Spirochaetes, flagella are located in the periplasm between the outer membrane sheath and cell cylinder (10); and, in accordance with their location, they have an enlarged C ring and rotor, and have a shape different from that seen in Salmonella (11). Furthermore, some bacteria, such as Vibrio parahaemolyticus, possess two flagellar systems (polar and lateral) that are encoded by distinct set of genes and use different motive forces (sodium and proton) but share a chemotaxis signal transduction system (12).
The bacterial flagellum has received attention as an exemplum of biological complexity; however, how this complexity and diversification have been achieved remains rather poorly understood. Although several scenarios have been posited to explain how this organelle might have been originated (13), the actual series of evolutionary events that have given rise to the flagellum, as might be inferred from the relationships of all genes that contribute to the formation and expression of this organelle across taxa, has never been accomplished.
Insights into the evolution of the bacterial flagellum have been gained from the homologies between flagellar proteins and those functioning in other systems (13). For example, the sequence similarity between flagellum-specific ATPase FliI and the β-subunit of ATP synthase led to the speculation that flagellum possibly evolved from this highly conserved, membrane-bound enzyme, whose subunits rotate during catalysis of ATP from ADP (14). Because the flagellar motor proteins MotA/B are homologous to the motor proteins in the Tol-pal and TonB systems (15), the flagellum was hypothesized to have originated as a simple proton-driven secretion system (16). Most significantly, there are well established sequence and structural homologies between bacterial flagella and the type III secretion system (TTSS) demonstrating that the two apparati derive from a common ancestor (17). Most evidence, including their much broader phylogenetic distribution, supports the view that the flagellum arose much earlier that the TTSS, which are largely limited to Proteobacteria (18–20).
Here, we take advantage of complete genome sequence data to trace the history of each gene involved in the assembly and regulation of the bacterial flagellum. Our results show that flagellum originated very early, before the diversification of contemporary bacterial phyla, and evolved in a stepwise fashion through a series of gene duplication, loss and transfer events. In this article, we focus on the evolution of the core set of flagellar genes that is uniformly present in all flagellated bacteria. The later evolving and lineage-specific components of the flagellar gene complexes remain to be addressed.
Results
Defining the Core Set of Flagellar Genes.
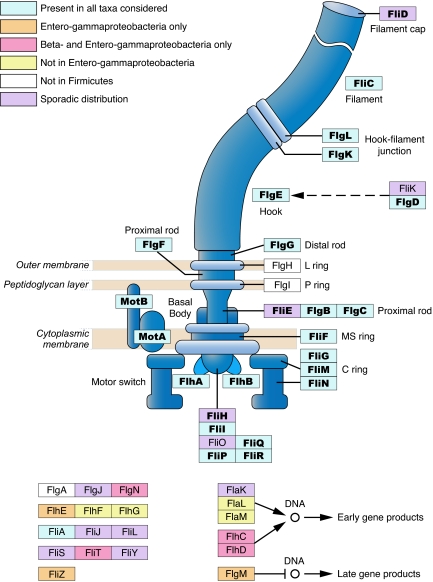
By querying the genomes of flagellated bacteria for which complete genome sequences are available, we obtained the phylogenetic distribution of every gene known to be involved in the biosynthesis and regulation of flagella. To investigate the origin and evolution of the bacterial flagellar system, we then applied a phylogenetic profiling method (21) to assort genes into functional groups based on their co-occurrence and shared distributions across genomes. Genes with different functional roles have distinct phylogenetic distributions and profiles; however, most of genes whose protein products constitute the structural components of the flagellum are present in all bacterial phyla considered (Fig. 1). This distribution suggests this core set of structural genes originated before the divergence of the major bacterial lineages and includes 21 genes that specify proteins that form the filament (fliC, which is often present in multiple copies), the hook-filament junction (flgK and flgL), the hook (flgE, which is present as a pseudogene in Thermotoga maritima), the rod (flgB, flgC, flgG, and flgF, which is missing only from Listeria innocua), the MS ring (fliF), the C ring (fliG, fliM, and fliN), the motor (motA and motB), and the export apparatus (flhA, flhB, fliI, fliP, fliR, and fliQ, which lacks a homolog in Clostridium tetani). In addition, flgD, encoding the hook-capping protein, which is required for flagellar assembly but does not contribute to the final structure, has homologs in all flagellated bacteria and thus was considered as part of the core set.
Fig. 1.
Distribution of flagellar proteins (excluding chemotaxis proteins) among flagellated bacterial species. Those proteins encoded by the core genes are designated in bold. This figure is redrawn with permission from that appearing in the KEGG pathway database (www.genome.jp/kegg/pathway/eco/eco02040.html).
Other flagellar structural genes that are broadly but not universally distributed across flagellated species include flgH, flgI, fliD, fliE, and fliH. The absence of certain of these genes from a genome is understandable once the characteristics of the particular bacteria are considered. For example, the L and P ring proteins FlgH and FlgI are not necessary in the Firmicutes because these bacteria lack the outer membrane in which these proteins are typically situated in Gram-negative bacteria. FlgH and FlgI are also not necessary in Spirochaetes, which have a periplasmic flagellum located inside of the outer membrane. The Firmicutes and Spirochaetes are viewed as two of the most basal bacterial lineages (22, 23), suggesting that flgH and flgI originated after the core set of structural proteins. In contrast, three other genes (fliD, fliE, and fliH) are present in all major groups but are missing sporadically from a few genomes, most noticeably in the Alphaproteobacteria. Because present-day distributions of these three genes are attributable to secondary loss, they too should be considered as part of the ancestral set of genes specifying the bacterial flagellum, bringing the total of core genes to 24.
Therefore, the task of elucidating the evolution of the flagellum rests on establishing how this set of 24 structural genes originated. The remaining flagellar genes, including those that play regulatory or auxiliary roles in flagellar assembly and function (such as the master regulators flhC and flhD, and the hook length control gene fliK), have highly variable distributions and are excluded from the core set, even though some of the genes are known to be essential for proper functioning of the flagellar system in a particular species. (The evolutionary histories of these regulatory genes, along with that of a second bacterial flagellar system remain to be described.)
Phylogenetic Analysis of Flagellar Core Genes.
To ascertain whether the 24 genes that form the flagellar core set have congruent evolutionary histories with one another, we compared the phylogenetic tree inferred for each core gene to that based on concatenated alignments of proteins encoded by 14 of the core genes. (These 14 genes were selected because they were present in all species included in this study and encoded the proteins having a high proportion of alignable positions.) For each of the 24 genes, all branches with >75% bootstrap values agreed with those in the concatenated tree, indicating that no alternative branching orders show strong support and that each of these genes has followed a common history in bacteria since they originated.
Congruence of Flagellar Genes with Organismal Phylogeny of Bacteria.
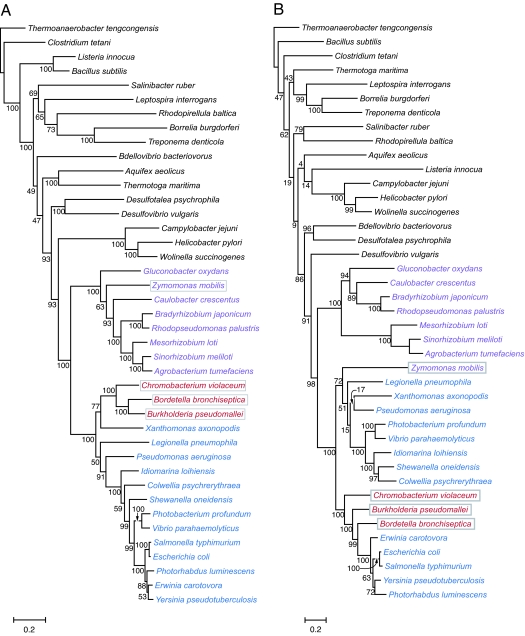
The distribution of the 24 core genes among divergent bacterial phyla is most consistent with an ancient origin, predating the shared ancestor of Bacteria. However, the distribution could have been achieved through later horizontal transfer. We tested these alternatives by comparing the phylogeny of the flagellar core proteins with the phylogeny of the corresponding bacterial phyla based on 25 universally distributed genes. The phylogenies are largely congruent on branches that have >75% bootstrap support; however, there are two inconsistencies between the core-gene and the organismal phylogenies; in the placement of both the alphaproteobacterial Zymomonas mobilis and a clade of three Betaproteobacteria within the Gammaproteobacteria (Fig. 2). Because individual flagellar genes within the core set show the same evolutionary history (see above), these incongruities have likely resulted from the transfer of the entire flagellar gene complexes between proteobacterial lineages after their separation from other major bacteria groups.
Fig. 2.
Congruence between species tree and flagellar protein tree. (A) Species tree based on concatenated protein alignment of 25 single-copy proteins. (B) Flagellar protein tree based on concatenated protein alignment of 14 flagellar core proteins. Bacterial groups are shaded to highlight incongruencies resulting from gene transfer events.
Core Flagellar Proteins Arose Through the Duplication and Diversification of a Single Precursor.
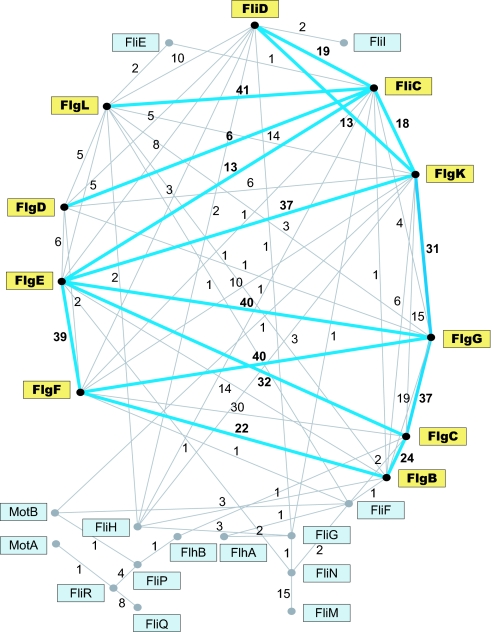
When each of the 24 core flagellar proteins of E. coli are compared (via BLAST) to all proteins encoded in the E. coli genome, their best and often only hits are to other core flagellar proteins. Pair-wise comparisons among these core proteins revealed that ten are homologous to other core proteins when applying an e-value cutoff of 10−4 (Fig. 3). This pattern indicates that the structural genes specifying the portion of flagellum residing outside of cytoplasmic membrane (i.e., the rod, hook, and filament) are paralogs and were derived from one another through duplications.
Fig. 3.
Network of relationships among flagellar core proteins. Above each link is the number of genomes for which homology between a particular protein pair was detected by pairwise comparison at a cutoff value of 10−4 or lower. Blue lines linking yellow-boxed proteins portray the homology network revealed when core proteins of E. coli were subjected to pairwise comparisons.
Aside from these matches to other core proteins, pairwise comparisons of these flagellar proteins to the >4,000 nonflagellar proteins encoded by the entire E. coli genome recovered cumulatively a total of only 24 hits that reached the same level of significance. Among these matches, half (including some with e-values as low as 3e−10 to the flagellar core proteins) are involved in other secretion systems, such as the P pilus and the Type V secretion system, which is consistent with the idea that the flagellum originated as a secretion system. An additional 10 of the 24 hits (with e-values ranging from 10−5 to 10−6) are membrane proteins, and the remaining two are prophage tail-fiber proteins. Thus, we conclude that despite their antiquity, the similarities among core proteins to one another are more common and, on average, stronger than to nonflagellar proteins.
Because the genes that constitute the core set are ancient and highly diverged, it is possible that some of the relationships among genes might not be recognized from analyses limited to the E. coli flagellar complex. We repeated this analysis and compared the core gene set of each other flagellated bacterium to all proteins encoded in the corresponding genomes and among themselves, and we obtained a similar result, i.e., the best (and often the only) hits of the flagellar core genes were to other flagellar core genes. However, by extending this analysis beyond E. coli, the similarity-relationships and links among several other core genes were resolved. For example, a highly significant match between fliM and fliN (that was not detected for E. coli homologs) was evident in 15 genomes from diverse bacterial subdivisions (Fig. 3). In addition, the interacting export components encoded by fliP, fliR, and fliQ are related based on their protein sequences within several taxa. And even among the 10 E. coli core genes that originally showed similarity to one another, there were several new interconnections (e.g., flgB to both flgE and flgG, and between flgL and flgK) revealed by performing the analysis on other genomes. Cumulatively, each of the 24 core genes shows significant similarly to one or more of the other core genes (Fig. 3), a pattern that would result from their successive origination from one another by independent gene duplications and/or gene fusions.
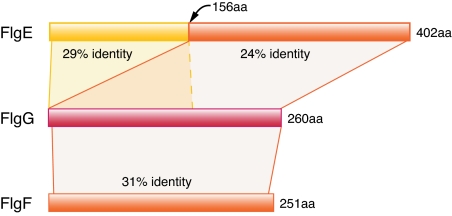
The similarity among the proximal rod protein FlgF, the distal rod protein FlgG, and the hook protein FlgE exemplifies the relationships among these flagellar proteins (Fig. 4). FlgF and FlgG are of similar size (251 aa vs. 260 aa in E. coli) and show 31% amino acid identity over their entire lengths. In contrast, the flgE gene is much longer and appears to have evolved from flgG through an intragenic duplication that added a 160-aa domain to the N terminus of its encoded protein. PSI-BLAST searches reveal two significant alignments between FlgE and FlgG in E. coli: one with 24% identity between whole length of FlgG and the C terminus of FlgE (156–401 aa), and the other with 29% identity between the N terminus of two proteins (≈160 aa). That flgE evolved by a duplication is also supported by the fact that there are two versions of flgE in the genus Bacillus: among sequenced genomes, four species (B. subtilis, B. clausii, B. licheniformis, and B. halodurans) contain a shorter version, which is similar in length to flgG, and three species (B. thuringiensis, B. cereus, and B. anthracis) have the longer version.
Fig. 4.
Protein sequence similarity among the proximal rod protein FlgF, the distal rod protein FlgG, and the hook protein FlgE in E. coli. Whereas FlgF and FlgG are homologous over their entire lengths, FlgE contains an intragenic duplication at its N terminus.
From the matrix of relationships and protein sequence alignments of the flagellar core genes of E. coli, it is also possible to infer the order in which many of these genes and their corresponding structures originated. The low levels of protein identity among these paralogs, paralogous pairs are between 18% and 32% identical, required that we apply a method that combines the output of series of multiple alignment programs to derive a consensus alignment. The alignments on the terminal regions of the proteins, especially at the C terminus, offer the highest confidence. An unrooted neighbor-joining tree and a maximum-likelihood tree [supporting information (SI) Fig. 5] show that the rod proteins originated with either FlgB or FlgC, which are both short proteins, and then generated FlgF and FlgG (and hook protein FlgE) through a series of duplication events. The evolutionary relationships of these flagellar genes parallel the locations of their encoded proteins in contemporary flagella. The proximal, then distal, rod proteins precede (both evolutionarily and physically) the hook proteins, which preceded the hook-filament junction and filament proteins.
Discussion
Comparisons of the complete genome sequences of flagellated bacteria revealed that the flagellum is based on an ancestral set of 24 core genes for which homologs are present in genomes of all bacterial phyla. The most striking finding from our analysis is that these core genes originated from one another through a series of duplications, an inference based on the fact that they still retain significant sequence homology. The individual core genes show phylogenetic histories congruent with one another, and this core flagellar phylogeny is largely consistent in its deepest branches with the phylogenetic relationships as currently resolved for Bacteria. Taken together, these results indicate that the core set of flagellar genes arose and was assembled from a single or few ancestral sequences, and that the individual genes diversified, before the shared ancestor of Bacteria.
Although sequence similarities among some of the rod and hook proteins were noted in early analyses (24), the degree of paralogy for the ancestral set of flagellar genes, and its implications for the origins of the bacterial flagellum, have gone unrecognized. From a phylogeny of these core proteins, it is possible to reconstruct the order in which they appeared, which in turn, can help elucidate the progression by which the flagellum was originally formed. Based on their relationships and on the physical locations of proteins forming the flagellum, the rod, hook, and filament proteins originated in an order that mirrors the “inside-out” flagellar assembly process (2, 3). The earliest proteins are proximate to the cytoplasmic membrane with later proteins situated distally, first spanning the outer membrane and then giving rise to structures (i.e., the hook, junction, filament, and capping proteins) that extend outside of the bacterial cell. Thus, the flagellum represents a case whereby its order of assembly recapitulates its evolutionary history.
The structural features of the flagellum, along with the evidence of homology between FliI and ATP synthase subunits and between MotA/B and the secretion proteins TolQ-TolR, suggests that it originated as a primitive secretion system (16), first involving ATPase and then adding the rod, hook, and filament components by gene duplication and diversification. Its original role as a secretion apparatus is also supported by the clear links between the flagellum and the TTSS, a protein delivery system whose genetic architecture is similar to and derived from a flagellar gene complex (17, 20).
Although some bacterial genomes contain recent paralogs of particular flagellar genes, most flagellar genes originated very early and are highly divergent, which occasionally hampers the recognition of orthologs, or the similarity between core proteins, in some of the genomes that we considered. Although additional flagellar proteins can be recognized by adopting more sensitive search programs (25), virtually all of the flagellar gene homologs that we identified were confirmed by examining their genomic context and enabling us to define the set of core genes that are ancestral to all bacterial lineages. Those few core genes that are absent from a few genomes (fliD, fliE, and fliH) are likely to represent cases of gene loss and it has been shown, at least for fliH, that this gene is not always essential for flagellar assembly (26).
To ascertain the ancestry of the flagellar core genes, we searched initially for homologs of each gene within the E. coli genome, which has the highest proportion of functionally annotated genes. The resulting network, involving only 10 of the 24 core genes, provided a very conservative view of the relationships and paralogy among the core genes but showed that flagellar genes were derived largely from other flagellar genes with apparently little input from other coding sequences. Extending these analyses to include other genomes uncovered additional links among flagellar proteins and revealed that the entire set of core genes could be formed through the duplication and divergence of previously existing flagellar genes. That the analysis of the E. coli did not resolve all of the links among core genes is not surprising given that these genes are ancient and have followed independent histories within bacterial lineages. It was originally hypothesized that biological pathways and structures might expand through the successive addition and modification of their preceding components (27). Although there is diminishing evidence that the recruitment of new enzymes into metabolic pathways occurs by this process (28), it is apparently the manner by which the bacterial flagellum arose.
The origins of complex organs and organelles, such as the bacterial flagellum and the metazoan eye, have often been subjects of conjecture and speculation because each such structure requires the interaction and integration of numerous components for its proper function, and intermediate forms are seldom operative or observed. However, the analysis of biological complexity has changed with the application both of genetic procedures that serve to identify the contribution of individual genes to a phenotype and of comparative sequence analyses that can elucidate the evolutionary and functional relationships among genes that occur in all life-forms. As with the evolution of other complex structures and processes (29–32), we have shown the bacterial flagellum too originated from “so simple a beginning,” in this case, a single gene that underwent successive duplications and subsequent diversification during the early evolution of Bacteria.
Materials and Methods
Genome Sequences.
Protein and DNA sequences from 249 complete bacterial genomes were downloaded from the National Center for Biotechnology Information (NCBI) (ftp.ncbi.nih.gov/genomes/Bacteria) on December 28, 2005. E. coli flagellar genes were retrieved from the Kyoto Encyclopedia or Genes and Genomes (KEGG) database (www.genome.jp/kegg/pathway/eco/eco02040.html) and curated manually. In addition, the five flagellar genes that are absent from E. coli but present in Vibrio parahaemolyticus were retrieved from GenBank. Only flagellated bacterial species with published genome sequences were chosen for phylogenetic analysis. Species that possess flagellar gene homologs but are not known to produce operational flagella because of pseudogenes or missing components were excluded from the analysis because these flagellar systems are in derived or degenerated states. A given bacterial species was considered to be flagellated if listed as such in Bergey's Manual (33). Because most species within a genus have very similar flagellar systems, we selected only one genome from each genus for further analysis. This resulted in a total of 41 species representing 11 bacterial phyla/divisions (SI Table 1).
Phylogenetic Profiling.
E. coli flagellar proteins are used as queries to search annotated proteins from complete genomes with BLASTP. Reciprocal best hits recovered at a cutoff of 10−5 among proteins in the E. coli and the queried genome were considered to be orthologs. If no ortholog was recovered, we first used TBLASTN to query the complete genome to confirm that its absence was not attributable to annotation errors. As a secondary check, PSI-BLAST was also used to find potential homologs. Homologs recognized from either the TBLASTN or PSI-BLAST searches were then examined for gene context. In cases where genes were in a colinear region or had at least one of the same neighboring genes as in E. coli, we regarded the sequences as orthologous to the E. coli gene. If an ortholog was still not found, we considered that particular flagellar gene as absent from a genome. To search for any flagellar genes or proteins that might be highly diverged from their E. coli homologs and go undetected in previous searches, we repeated the entire analysis using the flagellar gene/proteins of Bacillus subtilis as queries. In cases where there were multiple copies of homologs of a particular gene, paralogy was established if they had the original E. coli query gene as their best hit when searching against all E. coli proteins. TTSS proteins, although homologous to flagellar proteins, are easily identified because they generally have lower similarity values and are not contained within the flagellar gene neighborhood.
To evaluate whether the identification and distribution of particular flagellar proteins might be confounded by enhanced rates of protein evolution, we computed the overall percent protein identity for each flagellar protein in E. coli to its ortholog in Salmonella enterica sv. Typhimurium. (Salmonella and E. coli are used because orthology is easily ascertained, and alignments cover the entire lengths of proteins.) Most pairs of orthologs were >70% identical, and those with lower values (i.e., fast evolving) were FliC (54%), FliD (51%), FliK (47%), FliS (62%), and FliT (35%). Despite their relatively fast rates of evolution, FliC and FliD orthologs were detected in the genomes of all major bacterial groups and were included in the universally distributed core set. Moreover, this analysis also showed that “noncore” proteins are, by and large, not fast evolving and that their orthologs will be detected when present in a genome.
Similarity Among Core Proteins.
To detect similarities among core proteins in each of 41 flagellated bacteria, each protein was compared with each of the other core proteins in the same genome by using the Bl2seq program (with default options) in the NCBI BLAST package, applying an e-value cutoff of 10−4.
Phylogenetic Analysis of Flagellar Proteins.
Protein sequence alignments were generated in Muscle (34) with option (-maxiters 100), and poorly conserved regions were trimmed by using Gblocks method (35) with parameters (−b4 = 2 −b5 = n). PHYML (36) was used to construct maximum likelihood-based phylogenetic trees with 100 bootstrap replicates (with options 1 i 1 100 JTT e 4 e BIONJ y y). When multiple proteins from each genome were used to build a phylogenetic tree, individual protein alignments were first concatenated to form a single large alignment before phylogenetic analysis. To assess the evolutionary histories of individual members of the core set of flagellar genes, we compared the branching order inferred for alignments of each protein to that inferred for a concatenation of 14 well aligned proteins present in all flagellar systems. To construct a multiple sequence alignment of the highly divergent paralogous core proteins in E. coli, we used the mcoffee option of the T-Coffee program (37) (http://igs-server.cnrs-mrs.fr/Tcoffee/tcoffee_cgi).
Reconstruction of the Bacterial Species Tree.
Based on the concatenated alignments of 31 single-copy genes (predominantly ribosomal proteins), Ciccarelli et al. (23) reconstructed the relationships for sequenced representatives of three domains of life. Of these 31 genes, 25 are present in all of the bacterial genomes that we considered, and we used the concatenated alignment of their encoded proteins to build the bacterial species tree.
Supplementary Material
Acknowledgments
We thank Nancy Moran for originally suggesting this project and for input on the manuscript; Eduardo Rocha, Vincent Daubin, and Emmanuelle Lerat for numerous helpful comments about the analyses; and Becky Nankivell for assistance in making figures. This work was supported by National Institutes of Health Grants GM56120 and GM74738 (to H.O.).
Abbreviation
- TTSS
type III secretion system.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700266104/DC1.
References
- 1.Kirov SM. FEMS Microbiol Lett. 2003;224:151–159. doi: 10.1016/S0378-1097(03)00445-2. [DOI] [PubMed] [Google Scholar]
- 2.Macnab RM. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 3.Macnab RM. Biochim Biophys Acta. 2004;1694:207–217. doi: 10.1016/j.bbamcr.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Berg HC. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 5.McCarter LL. Curr Opin Microbiol. 2006;9:180–186. doi: 10.1016/j.mib.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Aldridge P, Hughes KT. Curr Opin Microbiol. 2002;5:160–165. doi: 10.1016/s1369-5274(02)00302-8. [DOI] [PubMed] [Google Scholar]
- 7.Soutourina OA, Bertin PN. FEMS Microbiol Rev. 2003;27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 8.Penn CW, Luke CJ. FEMS Microbiol Lett. 1992;100:331–336. doi: 10.1111/j.1574-6968.1992.tb14060.x. [DOI] [PubMed] [Google Scholar]
- 9.Bardy SL, Ng SYM, Jarrell KF. Microbiology. 2003;149:295–304. doi: 10.1099/mic.0.25948-0. [DOI] [PubMed] [Google Scholar]
- 10.Charon NW, Goldstein SF. Annu Rev Genet. 2002;36:47–73. doi: 10.1146/annurev.genet.36.041602.134359. [DOI] [PubMed] [Google Scholar]
- 11.Murphy GE, Leadbetter JR, Jensen GJ. Nature. 2006;442:1062–1064. doi: 10.1038/nature05015. [DOI] [PubMed] [Google Scholar]
- 12.McCarter LL. J Mol Microb Biotech. 2004;7:18–29. doi: 10.1159/000077866. [DOI] [PubMed] [Google Scholar]
- 13.Pallen MJ, Matzke NJ. Nat Rev Microbiol. 2006;4:784–790. doi: 10.1038/nrmicro1493. [DOI] [PubMed] [Google Scholar]
- 14.Rizzotti M. Early Evolution: From the Appearance of the First Cell to the First Modern Organisms. Boston: Birkhauser; 2000. [Google Scholar]
- 15.Cascales E, Lloubes R, Sturgis JN. Mol Microbiol. 2001;42:795–807. doi: 10.1046/j.1365-2958.2001.02673.x. [DOI] [PubMed] [Google Scholar]
- 16.Musgrave I. In: Why Intelligent Design Fails: A Scientific Critique of the New Creationism. Young M, Edis T, editors. New Brunswick, NJ: Rutgers Univ Press; 2004. pp. 72–84. [Google Scholar]
- 17.Blocker A, Komoriya K, Aizawa S. Proc Natl Acad Sci USA. 2003;100:3027–3030. doi: 10.1073/pnas.0535335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hueck CJ. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen L, Paulsen IT, Tchieu J, Hueck CJ, Saier MH. J Mol Microb Biotech. 2000;2:125–144. [PubMed] [Google Scholar]
- 20.Saier MH. Trends Microbiol. 2004;12:113–115. doi: 10.1016/j.tim.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daubin V, Gouy M, Perriere G. Genome Res. 2002;12:1080–1090. doi: 10.1101/gr.187002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 24.Homma M, Derosier DJ, Macnab RM. J Mol Biol. 1990;213:819–832. doi: 10.1016/S0022-2836(05)80266-9. [DOI] [PubMed] [Google Scholar]
- 25.Pallen MJ, Penn CW, Chaudhuri RR. Trends Microbiol. 2005;13:143–149. doi: 10.1016/j.tim.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Minamino T, Gonzalez-Pedrajo B, Kihara M, Namba K, Macnab RM. J Bacteriol. 2003;185:3983–3988. doi: 10.1128/JB.185.13.3983-3988.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horowitz NH. Proc Natl Acad Sci USA. 1945;31:153–157. doi: 10.1073/pnas.31.6.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen RA. Annu Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 29.Fernald RD. Science. 2006;313:1914–1918. doi: 10.1126/science.1127889. [DOI] [PubMed] [Google Scholar]
- 30.Trezise AEO, Collin SP. Curr Biol. 2005;15:R794–R796. doi: 10.1016/j.cub.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 31.Olson EN. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang Y, Doolittle RF. Proc Natl Acad Sci USA. 2003;100:7527–7532. doi: 10.1073/pnas.0932632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boone DR, Castenholz RW, Garrity GM. Bergey's Manual of Systematic Bacteriology. New York: Springer; 2001. [Google Scholar]
- 34.Edgar RC. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castresana J. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 36.Guindon S, Gascuel O. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 37.Notredame C, Higgins DG, Heringa J. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.