Abstract
In most animals, longevity is achieved at the expense of fertility, but queen honey bees do not show this tradeoff. Queens are both long-lived and fertile, whereas workers, derived from the same genome, are both relatively short-lived and normally sterile. It has been suggested, on the basis of results from workers, that vitellogenin (Vg), best known as a yolk protein synthesized in the abdominal fat body, acts as an antioxidant to promote longevity in queen bees. We explored this hypothesis, as well as related roles of insulin–IGF-1 signaling and juvenile hormone. Vg was expressed in thorax and head fat body cells in an age-dependent manner, with old queens showing much higher expression than workers. In contrast, Vg expression in worker head was much lower. Queens also were more resistant to oxidative stress than workers. These results support the hypothesis that caste-specific differences in Vg expression are involved in queen longevity. Consistent with predictions from Drosophila, old queens had lower head expression of insulin-like peptide and its putative receptors than did old workers. Juvenile hormone affected the expression of Vg and insulin–IGF-1 signaling genes in opposite directions. These results suggest that conserved and species-specific mechanisms interact to regulate queen bee longevity without sacrificing fecundity.
Keywords: Apis mellifera, lifespan, social insect
Honey bees (Apis mellifera) provide an attractive model to identify the molecular mechanisms regulating variation in lifespan. Workers and queens develop from the same genome, but queen lifespan is ≈10-fold longer (1). Moreover, queen longevity is achieved without the typical tradeoff between longevity and reproduction. Queens lay up to 2,000 eggs per day (2) and live for 1–3 years. Workers, in contrast, have limited fecundity and live for 3–6 weeks during spring and summer in temperate climates (1).
Insulin–IGF-1 signaling (IIS) is a key integrative pathway regulating aging, fertility and other important biological processes in vertebrates and invertebrates. Down-regulation of IIS is associated with increased longevity and decreased fertility in Caenorhabditis elegans and Drosophila melanogaster (3). Although IIS functions are widely conserved, it is not known whether naturally occurring differences in longevity also are a result of variation in this pathway.
Recent advances in insect molecular endocrinology have revealed connections between IIS and juvenile hormone (JH), a major insect hormone with diverse influences on growth, reproduction, and longevity in many species (4). Studies with Drosophila point to a connection between IIS and JH (5, 6). Because queens are both long-lived and reproductively active, the unique relationship between JH and vitellogenin (Vg) in honey bees has attracted attention (7). Honey bee Vg is a 180-kDa glycolipoprotein (8) synthesized in fat body cells and released to the hemolymph. Vg is best known as a yolk protein and is taken up by developing oocytes (9). JH is a gonadotropin and regulates vitellogenesis in insects (10).
In most insects, JH and Vg titers are positively correlated (11). In honey bees, however, JH and Vg titers in general show an inverse pattern (12–15). In workers, the JH titer is low during the first 2–3 weeks of adult life when performing tasks in the hive such as brood care (“nursing”) and is high in foragers. Vg hemolymph levels follow an opposite pattern (12). In queens, both JH and Vg hemolymph titers are elevated in emerging virgin queens, but whereas JH drops and stays low thereafter (15), Vg remains high (12). Vg gene expression agrees with protein measurements (16–18) but has not been studied in old queens.
Recent results from studies using worker bees (7) suggest that the unique negative relationship between JH and Vg may be important to our understanding of queen longevity. Vg has antioxidant functions in workers, which led to the suggestion that Vg also operates to increase queen lifespan (7). We measured Vg mRNA levels in workers and queens of different ages and in different tissues and also compared their resistance to oxidative stress. Male (drone) bees also were studied for comparative purposes; drones show a more queen-like lifestyle, i.e., focused on reproduction, but they have a worker-like lifespan (2). We also explored relationships between IIS, JH, and Vg in queens and workers by comparing mRNA levels of three genes in the IIS pathway, AmILP-1, AmInR-1, and AmInR-2, and studying the effects of JH on the regulation of Vg and AmILP-1.
Results
Tissue-Specific Vg mRNA Levels in Queens, Workers, and Drones: Quantitative RT-PCR (qRT-PCR).
Knockdown of Vg expression in worker honey bees causes decreased resistance to oxidative stress (7), and Vg protein levels are higher in queens than workers (12). We determined whether queens also show higher levels of Vg expression than do workers (and drones) and whether old queens have higher Vg expression than do young queens. Such a pattern might be expected on the basis of findings showing that experimental up-regulation of antioxidant expression increases longevity in some (but not all) contexts (19, 20). We analyzed head, thorax, and abdomen samples separately; Vg synthesis in abdominal fat bodies has been well studied, but fat cells are also located in the head and thorax (21), and head fat cells play an important role in regulating Drosophila lifespan (22). We studied the following groups: queens, 1-day-old (QD), 1-week-old (QW), 1-month-old (QM), and 1-year-old (QY); workers, 1-day-old (WD), 1-week-old (WW), 1-month-old (WM); and drones: 1-day-old (DD) and 1-week-old (DW). QD and QW were virgins, QM and QY were mated, WW were nurse bees, and WM were foragers.
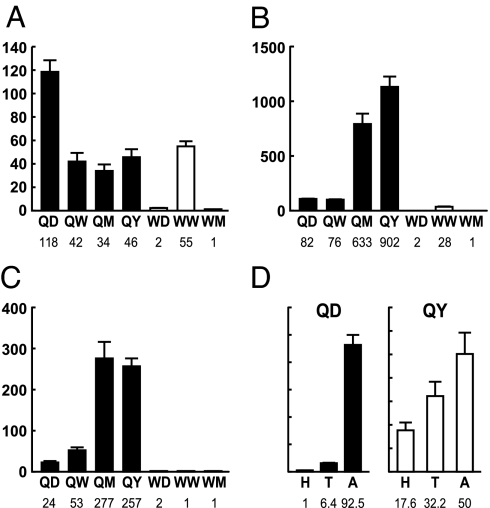
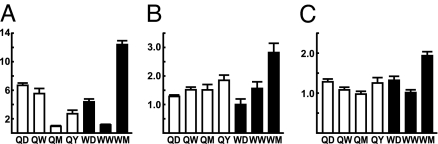
In queens, the concentration of abdominal Vg mRNA was highest in QD and then dropped to ≈1/3 that level in QW, QM, and QY (Fig. 1A). In workers, Vg mRNA abdominal concentration was very low in WD, increased ≈25-fold in WW, and dropped ≈50-fold to just above detection limits in WM. In drones, Vg mRNA abdominal concentration was low in DD and undetectable in DW [supporting information (SI) Fig. 9].
Fig. 1.
Tissue-specific analyses of Vg mRNA levels in queen and worker honey bees. (A–C) The y axis indicates the relative concentration of Vg mRNA in abdomen (A), thorax (B), and head (C). Queens are indicated by solid bars. Workers are indicated by open bars. Equal concentrations of RNA were used for these analyses. Values were calculated relative to the lowest group mean in each graph (WM) set as 1X. (D) Percentages of Vg mRNA per queen body part segment. QD are indicated by solid bars. QY are indicated by open bars. Equal volumes of RNA were used, without equalizing concentration, to obtain measures of total Vg mRNA per body part. Values depicted by each bar are given below each x axis. Results of statistical analysis in text: n = 7–12 individuals per group.
We also detected Vg expression in both thorax and head, especially in queens (Fig. 1 B and C), which is previously unreported. Queens showed an age-related increase in Vg thorax mRNA concentration. QY had ≈12-fold higher levels than QD and QW and ≈ 900-fold higher levels than WW. Queens also showed an age-related increase in Vg head mRNA concentration; unlike in thorax, head Vg peaked in QM and remained high in QY. QM and QY had ≈5- to 10-fold higher head concentrations than QD and QW and ≈125- to 300-fold higher head concentrations than workers. Workers had negligible Vg mRNA concentrations in head and thorax, and they decreased with age. In drones, Vg mRNA concentration in DD and QW head or thorax was very low (SI Fig. 9).
To further assess the possible significance of Vg expression in queen thorax and head, we calculated total amounts of Vg mRNA per body part. Queen abdomen contains more fat body cells than does thorax or head (21), and abdomen contained the most Vg RNA; however, head and thorax together contained almost as much Vg mRNA as in old queens (99.8%) (Fig. 1D). In contrast, head and thorax Vg RNA together amounted to only 7% of abdominal levels for young queens. Additional measurements across tissues and castes indicated that the proportion of Vg mRNA relative to total RNA was higher in queen head and thorax relative to abdomen (SI Fig. 9). We estimate that Vg transcription accounts for >25% of total transcriptional activity in queen head and thorax. These results indicate that the head and thorax contribute significantly to the total Vg mRNA pool in QM and QY.
Tissue-Specific Vg mRNA Levels in Queens, Workers, and Drones: In Situ Hybridization.
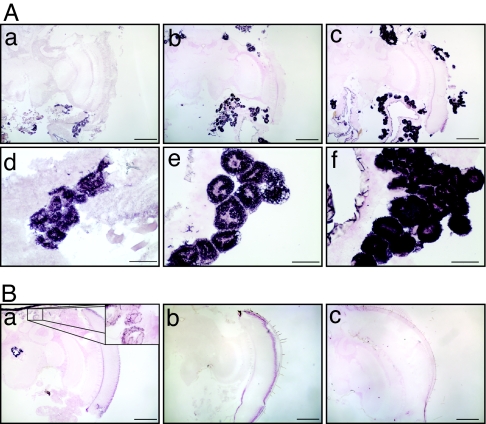
In situ hybridization confirmed the qRT-PCR results. Old queens had much higher head expression of Vg than young queens (Fig. 2A). Head fat cells were observed in queens, workers, and drones, with queen and drone more similar to one another in density and location; workers in particular had relatively few head fat cells (Fig. 2). In queen head, transcript localization was limited to fat cells. Vg expression was detected in head fat cells in young but not old workers and also was not detected in drone head (Fig. 2B). This finding shows that the high level of Vg mRNA found in the heads of old queens is not a property of head fat cells in general in honey bees but a specific feature of the queen. Results for thorax also were consistent with the above qRT-PCR analyses (SI Fig. 10). As expected, we also detected populations of abdominal fat cells expressing Vg in queen and worker (Fig. 3). These in situ results are consistent with qRT-PCR findings.
Fig. 2.
Localization of Vg mRNA in queen, worker, and drone head. (A) Queens. Shown are QD (a and d), QM (b and e), and QY (c and f). (a–c) Coronal sections show Vg mRNA in the fat cells of the dorsal and ventral sinus of the head capsule surrounding the brain (see also (SI Fig. 11). (d–f) High-magnification images of fat cells reveal an age-related increase in cell size and Vg mRNA signal intensity. In QM and QY, where staining was most intense, the fat cells were very large (up to 100 μm), exhibited a characteristic deformed nucleus (52), and showed clear cytoplasmic expression of Vg transcript. (B) Workers and drones. Shown are WW (a), WM (b), and DW (c). In WW (nurse bees), Vg mRNA is detected in a small cluster of fat cells (a Inset) but not in WM (b) or 2-week-old drones (c). (Scale bars: A a–c and B a–c, 400 μm; A d–f, 100 μm.)
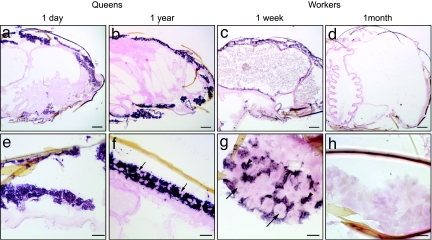
Fig. 3.
Localization of Vg mRNA in queen and worker abdomen. Shown are QD (a and e), QY (b and f), WW (c and g), and WM (d and h). (Lower) Higher magnification images of the corresponding (sagittal) section in the Upper images. QD (a) and QY (b) showed similar patterns of Vg mRNA localization in fat cells adjacent to cuticle. Vg mRNA was detected in fat cells of WW (c and g), but not in WM (d and h). These are the expected patterns of distribution on the basis of localization of Vg protein (12, 14) and serve to validate the more unexpected findings in Fig. 2. No hybridization signal was observed in control sections hybridized with a Vg sense strand probe for every group and body part analyzed. Abdominal fat body is organized as a layer of fat cells (trophocytes and oenocytes) adjacent to the cuticle, mostly lining the dorsal and ventral blood sinuses (51). Oenocytes were not present in QD fat bodies; they were present in QY, scattered among trophocytes (f, arrows). Vg mRNA was detected in trophocytes (g, arrowheads), but not in oenocytes (g, arrows) in WW. (Scale bars: a–d, 400 μm; e–f, 100 μm; g and h, 50 μm.
Vg Protein in Queen and Workers.
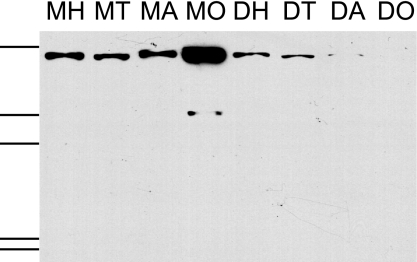
Western blot analysis was used to further confirm and extend the mRNA findings. A well characterized anti-Vg antibody (23) recognized a 180- to 190-kDa protein in samples of queen and worker abdominal, thorax, and head fat body. The tissue was washed in saline to remove hemolymph, because we were interested in tissue-associated Vg. The identity of the protein recognized by the antibody in the queen head samples was confirmed by mass spectrometry as Vg (SI Table 1). Vg protein was present in both worker and queen head, but Vg concentration was higher in QM than QD (Fig. 4). The total amount of Vg per tissue was highest in ovary, as expected (SI Fig. 12A). QM had higher total amounts of Vg in head and thorax than QD (SI Fig. 12A). In workers, the greatest amount of Vg was detected in abdomen, less in thorax, and even less in head (SI Fig. 12B).
Fig. 4.
Queen anti-Vg Western blot analysis. Two micrograms of total protein were loaded per lane onto a SDS/10% PAGE gel; results thus reflect Vg protein concentration. Additional experiments were made without normalizing protein amount, thus providing a measure of total Vg per tissue (SI Fig. 12). Shown for QM are head (MH), thorax (MT), abdominal fat body (MA), and ovary (MO); Shown for QD are head (DH), thorax (DT), abdominal fat body (DA), and ovary (DO). Size marker bands are 192, 117, 99, 54, 37, and 29 kDa, respectively.
Queen–Worker Differences in Resistance to Oxidative Stress.
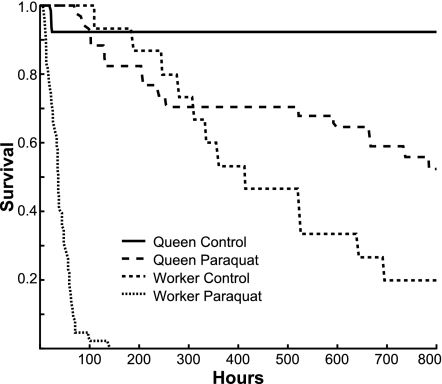
Queens have higher circulating Vg protein levels than workers (11), so if Vg acts as an antioxidant to promote longevity, then queens should be relatively more resistant to oxidative stress. Paraquat-injected queens had significantly higher survival than paraquat-injected workers (log-rank test: χ2 = 81.7, P < 0.0001) (Fig. 5). The effect of paraquat on queen survival was weaker than on worker survival (log-rank test: χ[1]2 = 138.1, P < 0.0001; proportional hazards: χ2 = 22.5, P < 0.0001). These results indicate that queens are more resistant to oxidative stress than workers.
Fig. 5.
Queen–worker differences in resistance to oxidative stress. Paraquat-injected queens had significantly higher survival than paraquat-injected workers. Paraquat reduced the median survival of workers from 359 to 35 h (log-rank test, χ2 = 43.2, P < 0.0001) and reduced the 800-h survival of queens from 92% to 52% (log-rank test, χ2 = 5.1, P = 0.02). Median survival time was not estimable because >50% of queens were alive at the time the experiment ended.
Queen–Worker Differences in Insulin Signaling.
Old queens had lower head expression of AmILP-1 (insulin-like peptide) and its putative receptors (AmInR-1 and AmInR-2) than did old workers (Fig. 6 A–C). This result is consistent with predictions from Drosophila, in which experimental down-regulation of IIS extends lifespan (5, 24). AmILP-1 expression showed an age-dependent decrease in queens. In workers, by contrast, AmILP-1 was up-regulated in the older group. WM, the group with the shortest expected lifespan, had ≈10-fold higher AmILP-1 levels than WW and QM and 5-fold higher levels than QY. Expression of the genes encoding the two putative insulin receptors was not significantly different within queen age groups and young workers (WD, WW), but was lower than for WM (Fig. 6 B and C).
Fig. 6.
Caste-specific differences in the expression of AmILP-1 (A), AmInR-1 (B), and AmInR-2 (C) in queen and worker head. Sample notation is as in Fig. 1. Queens are indicated by open bars. Workers are indicated by closed bars. Results of statistical analysis in text: n = 7–12 individuals per group. The y axes are as in Fig. 1.
Effects of Methoprene on Vg and IIS Gene Expression.
Virgin QW were treated topically with 200 μg of the JH analog methoprene (in 5 μl of acetone) and sampled 24 h later. This dose is active in workers (25). Queens were either placed individually in small (≈1,000 adult bees) colonies (“colony queens”) or caged individually and held in the same otherwise queenless colony (“banked queens”). These manipulations were done to explore possible environmental influences on the impact of the hormone treatment; IIS is known to be nutritionally sensitive (26). Banked queens were expected to be fed more poorly than colony queens, and in workers, starvation induces higher rates of JH biosynthesis (27). Banked queens do have higher JH titers than colony queens (15), suggestive of a nutritional effect in banked queens, but this has not yet been measured directly.
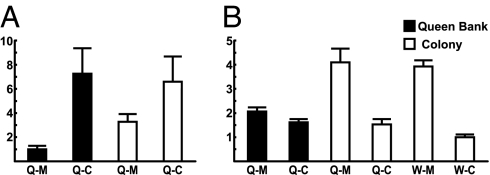
Methoprene caused a significant decrease in head Vg mRNA in both colony and banked queens (Fig. 7A), with a stronger effect on banked queens. In workers, where head Vg levels are just above background (Fig. 1), no differences were detected (data not shown). By contrast, methoprene caused an increase in AmILP-1 expression in both castes, although in queens this effect was only significant in colony queens (Fig. 7B). A similar effect was seen for treatment with JH III (data not shown), the endogenous hormone in honey bees. These results suggest that JH affects the expression of Vg and IIS genes in opposite directions. Effects vary by environment (colony type), which possibly reflects nutritional conditions.
Fig. 7.
Effects of methoprene on Vg (A) and AmILP-1 (B) expression in heads of QW and WW. Queens were either placed individually in small colonies (“colony”) or caged individually in a specially modified colony (“queen bank”). Queen and workers treated with methoprene or acetone vehicle (control) are indicated as Q-M and W-M or Q-C and W-C, respectively. Results are of statistical analysis in text. n = 7–12 individuals per group. The y axes are as in Fig. 1.
Discussion
There are striking differences in Vg expression between queen and worker honey bees. These findings, together with previous results showing that Vg acts as an antioxidant in workers (7) and the results of our queen–worker paraquat tests, support the hypothesis (7) that caste-specific differences in Vg are involved in queen longevity.
Abdominal Vg mRNA measurements were mostly consistent with previous results (16–18). Our in situ findings revealed that Vg expression occurs primarily or entirely in fat cells, and not in the ovaries, in contrast to previous Northern blot results (17); we suspect this discrepancy might be due to the presence of surrounding fat cells in their ovary samples (see Tissue Samples in Methods). Vg in ovarian follicle cells (9) likely is taken up from the hemolymph.
We found Vg expression in parts of the body not previously known to produce this yolk protein in insects (11); expression in worker head fat body also has recently been detected (28), consistent with our results. The age-related expression profile of Vg in queen head and thorax contrasted sharply with the abdomen profile. Abdomen Vg mRNA peaked early in adult life, but increased with age in head and thorax. Vg transcript in head and thorax accounts for a surprisingly large amount of the total Vg mRNA pool in old queens. These results suggest that the high Vg circulating titers observed throughout adult queen life (12) could be the result of the total and perhaps coordinated regulation of Vg expression throughout the body. Perhaps it is the total circulating pool of Vg throughout the body, independent of tissue of origin, that is important for longevity. Alternatively, perhaps tissue-specific expression is important; Vg expression was very high in queen head fat cells, cells that in Drosophila play an important role in regulating lifespan (22). There currently is insufficient information to discriminate between these possibilities.
Vg's antioxidant effect in honey bees has been attributed to its zinc-binding capacity (7), but other possibilities exist, including binding to iron (29, 30), a metal that catalyzes the generation of highly reactive hydroxyl radicals during the Fenton reaction (31). An earlier study (32) suggested that the Vg ortholog Vit-6 has antioxidant properties in C. elegans on the basis of its carbonylation activity in aged individuals, but Vit-2 and Vit-5 appear to promote aging in this species (33). Honey bees apparently lack genes encoding functional extracellular peroxidases (34), and queen longevity is not associated with higher expression of intracellular antioxidant genes (35). Perhaps Vg functionally replaces the traditional antioxidants to some extent, and queens have other mechanisms to minimize free radical damage, including decreased production of reactive oxygen species (35).
An insulin-like peptide and two insulin receptor genes were down-regulated in old queens relative to old workers. This finding parallels results from C. elegans and Drosophila, in which inactivation of InR genes enhances lifespan but, in contrast to queens, also decreases fertility (5, 36). However, effects of InR genes on longevity and fertility can be experimentally uncoupled (37, 38).
JH and Vg have been studied extensively in honey bees and other social insects, but experimental analysis linking them to IIS is thus far limited to Dipteran insects (4–6). We showed that JH affects the expression of at least one IIS gene. This result, plus those from Drosophila (5, 6), demonstrates reciprocal effects of these two hormone-signaling pathways. The finding that JH affects Vg and ILP-1 expression in opposite directions suggests the possibility of a regulatory feedback loop between these elements (as discussed below). A similar feedback loop, without IIS intermediates, was previously proposed to explain interactions between Vg and JH (39). We speculate that the unique negative relationship between JH and Vg in honey bees, shown in refs. 40 and 41 and extended in this study to the transcriptional level, might contribute to changes in IIS functioning in queens to allow for both high fertility and extended lifespan.
Studies with Drosophila also have revealed a connection between JH and resistance to oxidative stress. JH treatments decrease lifespan and increase sensitivity to oxidative stress (5, 42), suggesting that JH may down-regulate the expression of genes encoding antioxidant proteins in Drosophila. In honey bees, however, JH titers (12–15) and expression of antioxidant genes (35) are positively correlated; they are both low in queens and high in forager (worker) bees. These patterns, if functionally significant, would suggest another change in regulatory relationships involving JH besides the above-mentioned negative relationship between JH and Vg.
Interest in understanding how interactions among JH, Vg, and IIS affect a variety of different phenotypes in social insects is increasing (7, 43–45). We propose a verbal model to explain how Vg interacts with JH and IIS to regulate the striking differences in longevity between worker and queen honey bees (Fig. 8). This model differs from those in refs. 44 and 45 in highlighting how both conserved and species-specific regulatory mechanisms can interact to regulate longevity without sacrificing fecundity; roles of Vg and JH in this context have been considered earlier (7). Our model starts with the premise that queens have higher nutritional status than workers, which has been shown throughout the social insects (46). In honey bees, queens have greater abdominal lipid reserves and a richer diet: royal jelly, a proteinaceous exocrine secretion, instead of nectar and pollen (2).
Fig. 8.
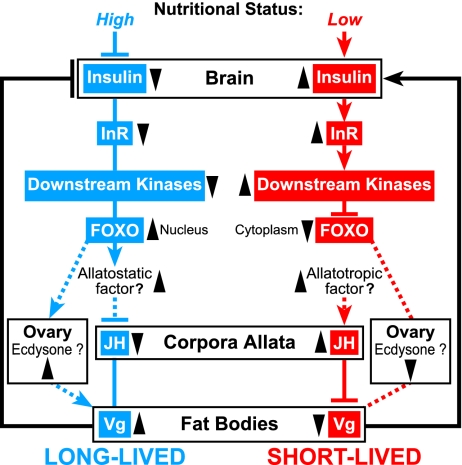
Model to explain how Vg, JH, and IIS interact to regulate longevity in the honey bee. Low nutritional status (red) up-regulates IIS (upward pointing arrowhead); insulin-like peptides (ILPs) bind to insulin-like receptors (InR) and signal by way of downstream kinases, resulting in phosphorylation of FOXO and cytoplasmic localization. In this state, FOXO does not repress JH synthesis, as proposed for Drosophila (4). High JH represses vitellogenesis, resulting in low Vg titers. Low Vg stimulates insulin secretion by a feedback loop mechanism. High nutritional status (blue) results in opposite effects, including decreased IIS activity, FOXO nuclear translocation, low JH, and high Vg. In addition to its antioxidant function, Vg might promote queen longevity by FOXO-mediated mechanisms, as in Drosophila and C. elegans (3, 4, 22, 33). The model includes both known and hypothesized elements from honey bee (39–40), and is supported by Fig. 7 results on the effects of JH on Vg and ILP gene expression, findings showing that knockdown of Vg by RNAi causes an increase in circulating JH titers (41) and AmILP-1 and 2 up-regulation by both Vg RNAi and starvation (M.C. and G.E.R., unpublished results). The model provides an explanation for differences in longevity between queens and workers and also can be used to explain differences in worker lifespan that relate to the in-hive and foraging phases of activity.
The key elements of this model are: (i) IIS is hypothesized to play a prominent role in the regulation of longevity in honey bees as a conserved mechanism, but the traditional relationship between nutrition and IIS is reversed. High nutritional status inhibits the secretion of insulin-like peptide, but the core of the IIS pathway in honey bees is hypothesized to be conserved. (ii) The traditional positive relationship between JH and Vg is inverted in honey bees, as noted above; JH inhibits Vg expression (40). (iii) The effects of JH on IIS are hypothesized to be mediated by Vg; this would render Vg a central signaling molecule as suggested in refs. 39 and 41.
This model raises several interesting questions, including the following: How were these traditional signaling relationships reversed, how might they relate to social evolution, and what are the functional consequences and tradeoffs? The JH system already is known to be involved in regulating two aspects of the honey bee's intricate social life, queen-worker caste determination, and division of labor among workers (10). The novel functions of Vg in honey bees may have evolved before the evolution of caste differentiation, because there are novel functions in both queen and worker honey bees (7). This suggests that it would be fruitful to study the functions of Vg in other members of the Hymenoptera, both social and nonsocial.
Methods
Bees.
Bees were from colonies maintained at the University of Illinois Bee Research Facility, Urbana, IL, and were derived from populations of a mixture of European subspecies of Apis mellifera. WD were obtained by removing frames of pupae from typical field colonies and placing them in an incubator (34°C and 80% relative humidity). Workers and queens of known age were obtained as in ref 34. Briefly, WW were collected while working in the hive; WM were collected while returning to the hive as foragers after ≈2 weeks of flight/foraging activity; QW were confined to the laboratory to prevent mating, and QM and QY mated naturally and headed colonies before sampling. DD were obtained as above; after emergence, they were kept in cages in the laboratory (25°C) and nourished by young workers that were fed with honey and pollen.
Tissue Samples.
Separate qRT-PCR analyses were performed on abdomen, thorax, and head/brain samples from each individual. Head samples for Vg analysis were initially prepared as dissected brains (35). However, results from in situ hybridization demonstrated that Vg expression is localized in fat cells surrounding the brain and not in the brain itself or anywhere else in the head. Subsequent analyses of Vg were performed by using whole heads. qRT-PCR analyses of dissected brains for Vg expression gave results (data not shown) that were not significantly different from what we present here (Fig. 1 and SI Fig. 9). Dissected brains were used for experiments in which AmILP-1 (GB17332-PA), AmInR-1 (GB15492-PA; XP_394771), and AmInR-2 (GB18331-PA; XP_001121597) mRNA was measured.
Real-Time qRT-PCR.
mRNA quantification was performed with an ABI Prism 7900 sequence detector, sequence-specific primers, and dual-labeled probes (SI Table 2). PCR assays were performed in two ways. We determined the mRNA concentration for each gene by using for each sample cDNA synthesized with 12 ng/μl RNA (Figs. 1 A–C, 6, and 7). We also determined the total amount of Vg mRNA per tissue by using cDNA synthesized with equal volumes of RNA (5 μl, 1/10 the total volume in which each RNA sample was eluted) (Fig. 1D). All PCR assays were performed in triplicate. qRT-PCR values of the focal genes were normalized by using an internal control gene (A. mellifera rp49; ref. 35) or two exogenous Arabidopsis thaliana RNA controls spiked during RNA extraction (XCP2, NM_101938) and cDNA synthesis (RCP1, NM_121758). XCP2 and RCP1 transcripts were synthesized in vitro by using the MEGAscript kit (Ambion, Austin, TX) and specific primers (SI Table 2). Transcript quantification calculations were performed by using the 2−ΔΔCt (47) or absolute quantification methods (48).
In Situ Hybridization.
Methods were as in ref. 49. Vg antisense and sense probes were prepared from PCR products generated by using Vg specific primers with T7 promoters attached to 5′ ends. We used 10-μm frozen sections.
Western Blot Analysis.
Tissues were dissected, washed three times with 1 ml of PBS, and homogenized in 400 μl of PBS plus protease inhibitor mixture set I (no. 539131; Calbiochem, San Diego, CA). Measurements were made in two different ways by using either 2 μg of protein (Fig. 4A) or 1 μl of each sample independent of protein concentration (Fig. 4B). Material was boiled in Laemmli buffer and analyzed by SDS/10% PAGE. Anti-Vg1 antibodies (23) were obtained from K. Hartfelder (Universidade de São Paulo, Ribeirao Preto, Brazil) and were used at 1:25,000. Anti-rabbit antibodies conjugated to peroxidase (A0545; Sigma, St. Louis, MO) were used at 1:10,000. Detection of protein was performed with Amersham Biosciences (Piscataway, NJ) ECL reagents (no. RPN2106).
Verification of Vg by Mass Spectrometry.
The band corresponding to Vg was excised from the gel, digested with trypsin (Promega, Madison, WI), and analyzed on a Waters (Milford, MA) Q-ToF equipped with an ultra performance liquid chromatography (UPLC) (column: Atlantis C-18 nanoACQUITY, 75 um × 20 cm; trap: Symmetry C-18, 200 μm × 2 cm). A linear gradient of water to 50% acetonitrile running at 250 nl/min was used to elute the peptides from the column. Data were analyzed with ProteinLynx Global Server 2.2.5 (Waters), Mascot (Matrix Sciences, London, U.K.) and PEAKS 4.0 (Bioinformatics Solutions, Ontario, Canada)
Paraquat Treatment.
Twenty-day-old workers were obtained by marking them with a paint dot and placing them in an indoor flight chamber until collection. Mated queens (≈1-month-old) were obtained from BeeWeaver (Navasota, TX) and maintained in an otherwise queenless colony with young workers (“queen bank”) for 1 week. Each queen was then placed in a Plexiglas cage with five attendant workers and maintained at 34°C on a 50% sucrose diet. Marked workers were placed in identical cages (n = 20). Workers were injected with paraquat (1,1′-dimethyl-4,4′-bipyridinium dichloride; N,N'-dimethyl-4,4′-bipyridinium dichloride) (Ultra Scientific, Hope, RI) to obtain a dose-response curve. We used a dose of 15 μg/μl (×10 higher than in ref. 7), which was the dose that resulted in greatest mortality over 3 days (Fig. 5). This protocol minimizes stress due to long-term confinement. To correct for differences in mass, queens were injected with a dose 70% > workers (25.5 μg/μl). Injections were made between the second and third abdominal tergites (10 μl of flexifil microsyringe, UltraMicroPump II, and Micro4 microsyringe pump controller; World Precision Instruments, Sarasota, FL). Controls were injected with saline. n = 34 queens and 45 workers (paraquat); n = 12 queens and 15 workers (control). Cages were censused every 4 h for the first 424 h and every 24 h thereafter. The experiment ended at 800 h.
JH Analog Treatment.
One-week-old workers from a typical colony were treated topically on the dorsal abdomen with 200 μg of methoprene per 5 μl of acetone or 50 μg of JHIII per 5 μl of acetone and collected 24 later. This dose is known to be active in honey bees (25). One-week-old virgin queens were treated the same way and collected 24 h later. Queens were either placed individually in small (≈1,000 adult bees) colonies (“colony queens”) or caged individually and held together in a queen bank, both of which were in the field. All queens were derived from one queen inseminated with semen from one male.
Statistical Analyses.
qRT-PCR.
Analyses were performed on ΔΔCT values by using SAS version 8.2 for Linux (50).
Paraquat treatment.
Bees alive at 800 h were “right-censored” at that time. We compared Kaplan–Meier survival estimates among treatment groups with both log-rank tests and Wilcoxon tests (SAS PROC LIFETEST). Results of both tests were consistent, so we report only the P values given by the log-rank tests. Effects of paraquat on queen and workers were analyzed with contrast coding within the parametric regression procedure (SAS PROC LIFEREG) and the Cox proportional hazards procedure (PROC PHREG). For parametric models, we used the generalized gamma distribution, which was supported by likelihood-ratio tests that compared different survival distributions.
Supplementary Material
Acknowledgments
We thank K. Pruiett and T. Newman for expert assistance in the field and laboratory, respectively; D. B. Weaver (Weaver Apiariaries, Navasota, TX) for queens; Victor Chan for tissue preparation; K. Hartfelder for Vg antibody; Peter M. Yau (University of Illinois at Urbana–Champaign Proteomics Facility) for mass spectroscopic analysis; G. Amdam for advice; and C. Kenyon and members of the K.A.H. and G.E.R. Laboratories for reviewing the manuscript. This work was supported by National Institutes of Health/National Institute on Aging Grant AG022824 (to G.E.R. and K.A.H.).
Abbreviations
- DD
1-day-old drones
- IIS
insulin–IGF-1 signaling
- JH
juvenile hormone
- QD
1-day-old queens
- QW
1-week-old queens
- QM
1-month-old queens
- qRT-PCR
quantitative RT-PCR
- QY
1-year-old queens
- Vg
vitellogenin
- WD
1-day-old workers
- WW
1-week-old workers
- WM
1-month-old workers.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701909104/DC1.
References
- 1.Page RE, Jr, Peng CY. Exp Gerontol. 2001;36:695–711. doi: 10.1016/s0531-5565(00)00236-9. [DOI] [PubMed] [Google Scholar]
- 2.Winston ML. Biology of the Honey Bee. Cambridge, MA: Harvard Univ Press; 1987. [Google Scholar]
- 3.Finch CE, Ruvkun G. Annu Rev Genomics Hum Genet. 2001;2:435–462. doi: 10.1146/annurev.genom.2.1.435. [DOI] [PubMed] [Google Scholar]
- 4.Flatt T, Tu MP, Tatar M. BioEssays. 2005;27:999–1010. doi: 10.1002/bies.20290. [DOI] [PubMed] [Google Scholar]
- 5.Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 6.Tu MP, Yin CM, Tatar M. Gen Comp Endocrinol. 2005;142:347–356. doi: 10.1016/j.ygcen.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Seehuus SC, Norberg K, Gimsa U, Krekling T, Amdam GV. Proc Natl Acad Sci USA. 2006;103:962–967. doi: 10.1073/pnas.0502681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler DE, Kawooya JK. Arch Insect Biochem Physiol. 1990;14:253–267. doi: 10.1002/arch.940140405. [DOI] [PubMed] [Google Scholar]
- 9.Fleig R. Int J Insect Morphol Embryol. 1995;24:427–433. [Google Scholar]
- 10.Robinson GE, Vargo EL. Arch Insect Biochem Physiol. 1997;35:559–583. doi: 10.1002/(SICI)1520-6327(1997)35:4<559::AID-ARCH13>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 11.Raikhel AS, Dhadialla TS. Annu Rev Entomol. 1992;37:217–251. doi: 10.1146/annurev.en.37.010192.001245. [DOI] [PubMed] [Google Scholar]
- 12.Hartfelder K, Engels W. Curr Top Dev Biol. 1998;40:45–77. doi: 10.1016/s0070-2153(08)60364-6. [DOI] [PubMed] [Google Scholar]
- 13.Bloch G, Wheeler DE, Robinson GE. In: Hormones, Brain, and Behavior. Pfaff D, editor. Vol 3. New York: Academic; 2002. pp. 195–236. [Google Scholar]
- 14.Fluri P, Sabatini AG, Vecchi MA, Wille H. J Apic Res. 1981;20:221–225. [Google Scholar]
- 15.Fahrbach SE, Giray T, Robinson GE. Neurobiol Learn Mem. 1995;63:181–191. doi: 10.1006/nlme.1995.1019. [DOI] [PubMed] [Google Scholar]
- 16.Piulachs MD, Guidugli KR, Barchuk AR, Cruz J, Simoes ZL, Belles X. Insect Biochem Mol Biol. 2003;33:459–465. doi: 10.1016/s0965-1748(03)00021-3. [DOI] [PubMed] [Google Scholar]
- 17.Guidugli KR, Piulachs MD, Belles X, Lourenco AP, Simoes ZL. Arch Insect Biochem Physiol. 2005;59:211–218. doi: 10.1002/arch.20061. [DOI] [PubMed] [Google Scholar]
- 18.Amdam GV, Simoes ZL, Guidugli KR, Norberg K, Omholt SW. BMC Biotechnol. 2003;3:1. doi: 10.1186/1472-6750-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun J, Tower J. Mol Cell Biol. 1999;19:216–228. doi: 10.1128/mcb.19.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orr WC, Mockett RJ, Benes JJ, Sohal RS. J Biol Chem. 2003;278:26418–26422. doi: 10.1074/jbc.M303095200. [DOI] [PubMed] [Google Scholar]
- 21.Snodgrass RE. Anatomy of the Honey Bee. Ithaca, NY: Cornell Univ Press; 1956. [Google Scholar]
- 22.Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 23.Bitondi MM, Simoes ZL. J Apic Res. 1996;35:27–36. [Google Scholar]
- 24.Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, Partridge L. Proc Natl Acad Sci USA. 2005;22:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan JP, Jassim O, Fahrbach SE, Robinson GE. Horm Behav. 2000;37:1–14. doi: 10.1006/hbeh.1999.1552. [DOI] [PubMed] [Google Scholar]
- 26.Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Dev Cell. 2002;2:239–249. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
- 27.Kaatz H, Eichmuller S, Kreissl S. J Insect Physiol. 1994;40:865–872. [Google Scholar]
- 28.Seehuus SC, Norberg K, Krekling T, Fondrk MK, Amdam GV. J Insect Sci. 2007 doi: 10.1673/031.007.5201. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Heusden MC, Fogarty S, Porath J, Law JH. Protein Expr Purif. 1991;2:24–28. doi: 10.1016/1046-5928(91)90004-3. [DOI] [PubMed] [Google Scholar]
- 30.Ishikawa S, Yano Y, Arihara K, Itoh M. Biosci Biotechnol Biochem. 2004;68:1324–1331. doi: 10.1271/bbb.68.1324. [DOI] [PubMed] [Google Scholar]
- 31.Halliwell B, Gutteridge JM. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura A, Yasuda K, Adachi H, Sakurai Y, Ishii N, Goto S. Biochem Biophys Res Commun. 1999;264:580–583. doi: 10.1006/bbrc.1999.1549. [DOI] [PubMed] [Google Scholar]
- 33.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 34.Corona M, Robinson GE. Insect Mol Biol. 2006;15:687–701. doi: 10.1111/j.1365-2583.2006.00695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corona M, Hughes KA, Weaver DB, Robinson GE. Mech Ageing Dev. 2005;126:1230–1238. doi: 10.1016/j.mad.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang RA. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 37.Dillin A, Crawford DK, Kenyon C. Science. 2002;298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- 38.Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amdam GV, Omholt SW. J Theor Biol. 2003;223:451–464. doi: 10.1016/s0022-5193(03)00121-8. [DOI] [PubMed] [Google Scholar]
- 40.Pinto LZ, Bitondi MM, Simoes ZL. J Insect Physiol. 2000;46:153–160. doi: 10.1016/s0022-1910(99)00111-0. [DOI] [PubMed] [Google Scholar]
- 41.Guidugli KR, Nascimento AM, Amdam GV, Barchuk AR, Omholt S, Simoes ZL, Hartfelder K. FEBS Lett. 2005;579:4961–4965. doi: 10.1016/j.febslet.2005.07.085. [DOI] [PubMed] [Google Scholar]
- 42.Salmon AB, Marx DB, Harshman LG. Evolution Int J Org Evolution. 2001;55:1600–1608. doi: 10.1111/j.0014-3820.2001.tb00679.x. [DOI] [PubMed] [Google Scholar]
- 43.Wheeler DE, Buck N, Evans JD. Insect Mol Biol. 2006;15:597–602. doi: 10.1111/j.1365-2583.2006.00681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunt GJ, Amdam GV, Schlipalius D, Emore C, Sardesai N, Williams CE, Rueppell O, Guzman-Novoa E, Arechavaleta-Velasco M, Chandra S, et al. Naturwissenschaften. 2007;94:247–267. doi: 10.1007/s00114-006-0183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Page RE, Jr, Amdam GV. BioEssays. 2007;29:334–343. doi: 10.1002/bies.20549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunt JH, Nalepa CA, editors. Nourishment and Evolution in Insect Societies. Boulder, CO: Westview; 1994. p. 449. [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Applied Biosytems. User Bulletin #2, ABI PRISM 7700 Sequence Detection System. Foster City, CA: Applied Biosystems; 2001. [Google Scholar]
- 49.Velarde RA, Sauer CD, Walden KK, Fahrbach SE, Robertson HM. Insect Biochem Mol Biol. 2005;35:1367–1377. doi: 10.1016/j.ibmb.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 50.Littell RC, Stroup WW, Freund RJ. SAS for Linear Models. Cary, NC: SAS Institute; 2002. [Google Scholar]
- 51.Cruz-Landim C. Rev Brasil Biol. 1985;45:221–232. [Google Scholar]
- 52.Bishop GP. J Exp Zool. 1958;137:501–525. doi: 10.1002/jez.1401370308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.