Abstract
We explore the relative contributions of different structural elements to the stability of Aβ fibrils by molecular-dynamics simulations performed over a broad range of temperatures (298 K to 398 K). Our fibril structures are based on solid-state nuclear magnetic resonance experiments of Aβ(1–40) peptides, with sheets of parallel β-strands connected by loops and stabilized by interior salt bridges. We consider models with different interpeptide interfaces, and different staggering of the N- and C-terminal β-strands along the fibril axis. Multiple 10–20 ns molecular-dynamics simulations show that fibril segments with 12 peptides are stable at ambient temperature. The different models converge toward an interdigitated side-chain packing, and present water channels solvating the interior D23/K28 salt bridges. At elevated temperatures, we observe the early phases of fibril dissociation as a loss of order in the hydrophilic loops connecting the two β-strands, and in the solvent-exposed N-terminal β-sheets. As the most dramatic structural change, we observe collective sliding of the N- and C-terminal β-sheets on top of each other. The interior C-terminal β-sheets in the hydrophobic core remain largely intact, indicating that their formation and stability is crucial to the dissociation/elongation and stability of Aβ fibrils.
INTRODUCTION
Fibrils of Alzheimer's amyloid-beta (Aβ) peptides are the major component of Alzheimer's disease plaques (1,2). Amyloid fibrils are also associated with other neurodegenerative and prion diseases, type 2 diabetes (2–4), and insulin deficiency in the brain (5). In Alzheimer's disease, recent studies suggest a connection between Aβ plaques, oligomers, and neurotoxic effects (6), but its exact nature remains largely unknown (7). The detailed structural characterization of Aβ peptides both in solution (8–10) and in fibrils is a crucial step toward understanding the formation and stability of ordered fibrillar peptide aggregates (2,11–13). Knowledge of structural details of Aβ fibrils should in particular facilitate the design of inhibitors of fibril formation (14–16). Moreover, insights into the molecular self-assembly processes during fibril formation may also aid in the development of new soft nanomaterials (17,18).
Most computational investigations of the structure and dynamics of Aβ peptides have focused on monomers (10,19), dimers (20–22), and other low-order oligomers (20,23–25). By studying early aggregation events, molecular simulations using simplified, coarse models of various amyloid-forming molecules provided detailed mechanistic and structural insights in the formation of prefibrillar amyloid species (21,26–29). Larger systems have been studied to identify general aspects of amyloid assembly (30–35).
Here, taking advantage of structural information available from recent solid-state nuclear magnetic resonance (ssNMR) experiments (11,13,36,37) on the parallel cross-β structure of Aβ1–40 protofilaments, we perform all-atom/explicit solvent simulations of amyloid fibrils containing six two-peptide units, each with two U-shaped Aβ9–40 peptides in a plane roughly perpendicular to the fibril axis. Among the different structural topologies observed depending on experimental growth conditions (11,13,37), we focus on Aβ1–40 fibrils grown under gentle agitation. In a series of different models, we incorporate information from recent isotope-dilution ssNMR (11) suggesting that the N- and C-terminal β-strands within a given peptide may not be in contact, unlike previous models.
The objectives of our study are twofold: 1), to explore the contributions of the different structural elements of typical Aβ protofilaments to stability, conformational dynamics, and elongation/dissociation mechanisms; and 2), to investigate different possible protofilament models, including different β-sheet staggering and loop conformations, searching for common features that may be independent of structural details of specific models. Our simulations of both wild-type and mutated sequences cover a broad temperature range (298, 348, and 398 K), in which fibrils are experimentally stable (below ∼330 K) or fully dissociate (above ∼373 K) (38,39). We discuss structurally relevant fibril characteristics such as the secondary-to-quaternary structural elements (e.g., β-strands, intra- and intermolecular contacts), internal salt-bridges, the conformations of the amino acids in the loop region, and the interior hydration. We analyze the evolution of these elements at elevated temperatures, focusing in particular on the structure and dynamics of the Aβ monomers at the fibril ends to identify possible dissociation/elongation mechanisms.
The outline of the article is as follows. After introducing fibril terminology, we describe the different structural models and the simulation methodology. We then present simulation results for infinite periodic fibrils. From simulations of fully solvated finite fibrils, we extract information about secondary structure, side-chain packing, and interior salt bridges and hydration as well as fibril dynamics. We then interpret our results obtained at different temperatures in the context of fibril stability and elongation/dissociation mechanisms.
METHODS
In studies of Aβ amyloid aggregates, the term “fibril” is generally used for structures of aggregated peptides that have a rodlike appearance and low or unknown degree of molecular organization (12,40). The term “protofilament” refers more specifically to basic structural units of amyloid fibrils, consisting of Aβ peptide monomers arranged in two molecular layers along the fibril axis (12). Here, we study the molecular structure of Aβ peptides in such protofilaments and use the terms “fibril” and “protofilament” interchangeably. “Two-peptide” units refer to the two Aβ9–40 peptides in a plane roughly perpendicular to the fibril axis.
Structural models of Aβ protofilaments
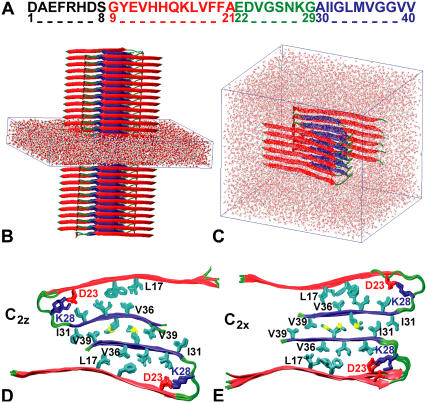
We use Aβ9–40 to model the full-length Aβ1–40 protofilaments (Fig. 1 A). N-terminal residues 1–8 are structurally disordered and not necessary for fibril growth (12,36,37). Structural models of Aβ protofilaments were built using peptide structures taken from the simulations of Buchete et al. (12) that were then stacked along the fibril axis (Fig. 2).
FIGURE 1.
MD simulations of Aβ9–40 protofilaments. (A) Aβ1–40 sequence and major structural elements: the unstructured N-terminal region (black), the N- (red) and C-terminal β-strands (blue), and the loop region (green). (B) Infinite-periodic fibril with solvent-filled simulation box. (C) Solvated 12-peptide fibril segment. (D,E) Top views of infinite fibrils with C2z (D) and C2x (E) topologies after 10 ns of MD.
FIGURE 2.
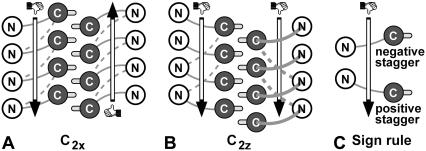
Schematic of Aβ9–40 protofilament β-strand staggering of fibrils with (A) C2x and (B) C2z topologies. The N- (open) and C-terminal β-strands (dark shaded circles) are roughly perpendicular to the plane. Thick and thin shaded connecting loops are located in front and behind, respectively. Solid loops correspond to the initial internal staggering of the S1 models (−0.5 for S1-C2x and +0.5 for S1-C2z), and dashed loops indicate −1.5 staggering of S2 and S3. M35 side chains (light shaded) indicate the 0.5 external staggering. Vertical arrows indicate the direction of the two fibril strands, defined by a right-hand rule for the N-to-C direction of the Aβ peptide.
We explore different staggerings (11,12,35) of the N- and C-terminal β-strands along the fibril axis to account for recent isotope-dilution experiments (11). We study three systems (S1, S2, and S3) that differ in the initial staggering of their N- and C-terminal β-strands (+0.5 for S1 and −1.5 for S2 and S3). Staggering is defined as the β-strand displacement along the oriented fibril axis in units of the ∼4.8 Å β-sheet interstrand spacing (11) with the sign determined by the intrinsic fibril direction (Fig. 2 C). A right-hand rule for the N-to-C direction of the peptide (Fig. 2 C) is used to define the staggering sign and the intrinsic fibril direction. Note that the staggering signs obtained with this rule are opposite to the ones depicted in Fig. 7 of Petkova et al. (11). To change the initial staggering from +0.5 (S1 system) to −1.5 (S2 and S3 systems), we reconnect the V24–N27 loops of stacked β-strands (Fig. 2). The S2 and S3 systems have different structures of the connecting loop, with V24–N27 being more extended in the S3 system. Additional simulations are performed for an S2 system with mutations V24A, S26A, and N27A.
FIGURE 7.
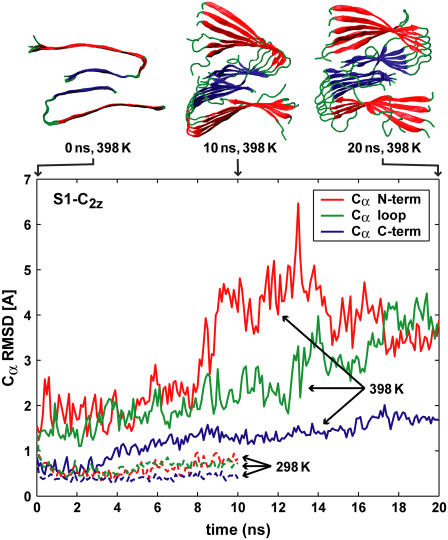
Cα RMSD values calculated for Aβ structural elements of the S1-C2z system, as a function of time, with respect to the average conformation of the 1–5 ns trajectory segment at 298 K. The solvent-exposed N-terminal β-strands (red) lose structural order at elevated temperatures faster than loop (green) and C-terminal (blue) regions.
For each Aβ fibril model, 10 ns of molecular dynamics (MD) simulations at 298 K were conducted in an infinite-periodic fibril setup (Fig. 1 B), followed by multiple simulations of finite fibril segments (Fig. 1 C) at 298, 348, and 398 K for up to 20 ns.
For S1 and S2 systems, we simulate both C2x and C2z topologies, which differ in the relative orientations of the Aβ peptides in a two-peptide unit normal to the fibril axis (see (12), and Fig. 1, D and E). For S3, we only consider the C2z topology that is more strongly supported by experiment (11). The two topologies differ in the relative orientation of the contacting C-terminal β-strands in the interface between the two U-shaped peptides within a two-peptide unit: the two strands are parallel in C2x structures, and antiparallel in C2z structures. As a consequence, the two stacks of U-shaped peptides along the fibril have either parallel (C2z) or antiparallel orientation (C2x), with equivalent fibril ends in the case of C2x fibrils (Fig. 2 A), but differing end structures in the case of C2z fibrils (Fig. 2 B). However, if the staggering sign or magnitude (Fig. 2 C) differ in the two molecular layers of the protofilament (as seen in some of our high-T simulations), the two fibril ends can differ even in the C2x case. We note that differences in the structure of the two ends may affect the relative fibril elongation kinetics. Similarly, symmetry differences between C2x and C2z topologies result in having the structurally disordered N-terminal residues (12,37) either close to each other (C2x, Fig. 1 E) or on the opposite sides of Aβ protofilaments (C2z, Fig. 1 D), with possible implications on their lateral aggregation properties.
Molecular dynamics simulations
Our molecular dynamics (MD) simulations of Aβ amyloid protofilaments use the methods described in Buchete et al. (12). Here, we study systems of 12 Aβ9–40 peptides (as compared to eight peptides in (12)), at both ambient (298 K) and elevated temperatures (348 K and 398 K). All simulations of solvated amyloid fibrils were performed using the NAMD2 program (41) with the CHARMM27 (42) force field parameters. The fibrils were explicitly solvated with TIP3P water molecules (43). All simulations were performed in the NPT ensemble. The Langevin piston method (41,44,45) was used to maintain a constant pressure of 1 atm. The temperature was controlled by using Langevin dynamics with a coupling coefficient of 1 ps (41). We used periodic boundary conditions and the particle-mesh Ewald method (46) with a real-space cutoff distance of 10 Å and a grid width smaller than 1 Å. The switching distance for nonbonded electrostatics and van der Waals interactions was 8.5 Å with a cutoff distance of 10 Å, and the integration time step was 1 fs (with the exception of the S3-C2z system where 2 fs time steps were used in conjunction with constrained bonds of hydrogen atoms (47)).
Initially, between 10 and 20 ns of MD were performed for four-layer infinite-periodic fibril systems hydrated with up to 7071 water molecules. From those systems, six-layer fibril segments were extracted and fully solvated with up to 21,913 water molecules leading to simulated systems of ∼70,000 atoms. After successive stages of minimization, heating and equilibration, up to 20 ns of MD runs were performed at 298 K. Additional runs of 10–20 ns were performed at 348 K and 398 K. The combined simulation time for all three systems (S1, S2, and S3) and temperatures is 320 ns (i.e., 80 ns for the infinitely-long systems and 240 ns for simulations of protofilament segments containing 12 Aβ9–40 peptide monomers).
RESULTS AND DISCUSSION
β-sheet staggering in infinite-periodic fibrils
After 10 ns of MD, the infinite-periodic fibrils S1, S2, and S3 maintain their secondary structure and overall topology despite a compaction in the side-chain packing (Fig. 1, D and E, and Fig. S1 that is published as Supplementary Material). In all cases, the side chains of the N- and C-terminal β-strands interdigitate during the equilibration, resulting in a well-packed structure. During the initial equilibration at ∼100 ps in the 298 K simulation of the infinite system with S1-C2x topology, the staggering changed from +0.5 to −0.5, indicating a cooperative sliding of both N-terminal β sheets relative to the two C-terminal sheets.
T-dependent secondary structure
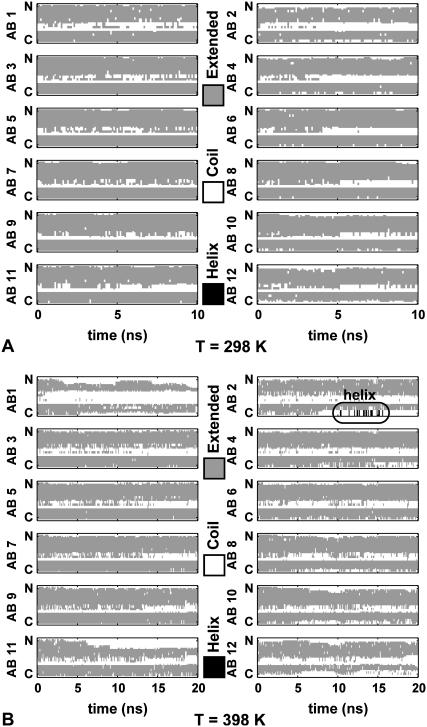
From the final structure of the infinite-periodic MD runs, stacks of six two-peptide units (i.e., 12 Aβ9–40 monomers) are separated and fully solvated. In subsequent NPT MD simulations, the resulting finite protofilament fragments are free to twist about the fibril axis, unlike the infinite-periodic fibrils. Fig. 3, A and B, illustrate the time evolution of the secondary structure content for the Aβ segments of the S1-C2z system along the 10 ns trajectory at 298 K and at 398 K, respectively.
FIGURE 3.
Secondary structure (60) of the S1-C2z system at (A) 298 K and (B) 398 K. Shaded representation indicates extended β-strand regions, and open representation indicates coil. Helical segments shown in solid representation are boxed. Peptides AB1/AB2 and AB11/AB12 form the top and bottom layers of the protofilaments, respectively.
For all three models, S1, S2, and S3, both N- and C-terminal β-sheet regions are stable at room temperature for the duration of the simulations (Fig. 3 A, and Figs. S2 A and S3 A). Even peptides located at the ends of six-layer fibril segments present stable β-strand regions. Occasional loss of β-sheet content is more common in the S2 and S3 systems, especially in the loop regions.
However, at elevated temperatures (Fig. 3 B, and Figs. S2 B and S3 B) there is a significant loss of β-sheet content in all cases. Consistent with the interpretation of calorimetry experiments (38,39), temperatures above 100°C are sufficient to overcome the hydrophobic interactions between the β-sheet residues, eventually dissociating the fibrils. The interior of the S1-C2z system, starting with +0.5 staggering, presents smaller changes than the S2 and S3 systems initiated with −1.5 staggering. Snapshots of initial and final conformations in MD trajectories of up to 20 ns, illustrating the structural elements of the Aβ protofilament systems, are shown in Figs. S4–S8.
“Surface melting” of N-terminal β-strands
At elevated temperatures, the loss of structure is initiated at the surface of the protofilaments (Fig. 3 B), as seen also in proteins (48). The solvent-exposed N-terminal β-strands are found to be most susceptible to such “surface melting.” In contrast, the interior C-terminal strands maintain their structure for up to 20 ns at all temperatures. Even at the fibril ends, the C-terminal strands appear more stable—an observation with possible implications on the potential fibril dissociation/elongation mechanism, as discussed below in more detail.
External and internal staggering of β-strands in Aβ fibril segments
For all systems studied here, the C-terminal β-strands of the two interior β-sheets are displaced along the fibril axis by ∼2.4 Å, corresponding to an external staggering of 0.5. The odd-numbered side chains I31, M35, and V39 adopt an interdigitated packing pattern along both the fibril axis and the β-strand direction. MD runs at elevated temperatures show that the C-terminal strands of different peptides form a highly stable protofilament core.
The internal staggering between the N and C-terminal β-strands of a peptide remains unchanged at 298 K (+0.5 for S1-C2z, −0.5 for S1-C2x, and −1.5 for S2 and S3). However, at elevated temperatures, we observe a tendency of the S2 and S3 systems to go from −1.5 to +0.5 staggering.
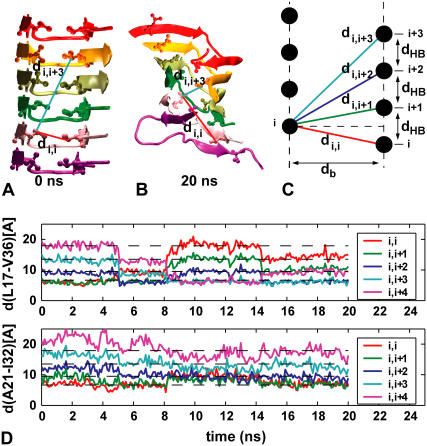
The most dramatic change in the staggering of N- and C-terminal strands occurs at 398 K in the S1-C2z system. Starting from +0.5 staggering, a whole N-terminal sheet is collectively displaced after ∼5 ns, and then again at 8 and 14 ns. As the N-terminal sheet slides on top of the C-terminal sheet, the staggering of the β-strands changes from +0.5 to +1.5, then +3.5, and back to +2.5. The concerted β-sheet sliding is evident in the time-dependent Euclidian distances di,i+k between terminal heavy atoms of facing side chains of N- and C-terminal β-strands of molecules i and i + k (Fig. 4). In our simulations, we note that the staggering changes less for residues close to the loop than for those at the tip of the strands (Fig. 4 D), with N- and C-terminal strands being at an angle to each other.
FIGURE 4.
Sliding of N- and C-terminal β-sheets in S1-C2z system at 398 K. (A) Starting configuration. (B) Configuration after 20 ns. (C) Schematic of di,i+k distances. (D) di,i+k as a function of time for residue pairs L17-V36 and A21-I32 far from the loop and close to it, respectively. Dashed lines indicate di,i+k distances expected for ideal β-sheets.
D23/K28 salt bridge
Consistent with the ssNMR data (11,13,36), all our starting fibril conformations have D23/K28 salt bridge contacts in the loop region. Other studies also showed that charged residues are important for the dynamics of protein aggregation and the stability of β-sheet structures (32,49–51). Consistent with previous results (12), our trajectories of both infinite and finite fibril segments show that the D23/K28 bridges are maintained at 298 K. Positive and negative charges alternate along the fibril axis as in a one-dimensional ionic crystal, with +0.5 staggering for the S1 system, and −1.5 for S2 and S3. We note that D23/K28 salt bridges maintain their staggering even in high-T simulations where the N- and C-terminal β-sheets slide on top of each other.
Water channels along fibril axis
In the infinite-periodic fibrils, small cavities form near the D23/K28 salt bridges but do not fill with water. However, water penetrates into finite fibril segments to form narrow water channels solvating the interior D23/K28 salt bridges. Indirect experimental evidence for interior hydration of Aβ fibrils comes from differential scanning calorimetry (38). However, ssNMR data do not indicate large structural differences between lyophilized and wet fibrils (11,37). Interestingly, experimental studies showed that both Aβ1–40 peptides (52,53) and Aβ1–42 peptides (54) can form ion channels through lipid membranes. However, such Aβ pores (55) likely occur at a larger scale between trans-membrane bundles of protofilaments. Recent simulations of fibrils with up to 32 Aβ16–22 peptides in antiparallel β-strands have also shown similar water channels hydrating the K16/E22 side chains (56).
Stability of connecting loops
A recent study suggested that the loops connecting the N- and C-terminal strands play an important role in aggregation (19,57). To explore the relative contributions of the loop and hydrophobic core to the overall stability and fibril dissociation, we mutated the residues V24–N27 in the salt-bridge region from VGSN to AGAA.
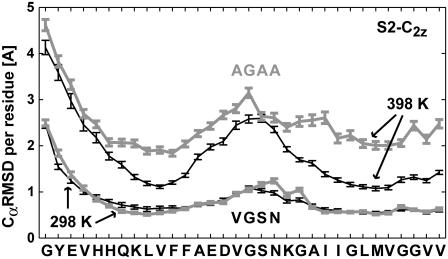
Fig. 5 shows a plot of the Cα root mean-square distance (RMSD) of individual residues at 298 K and 398 K. RMSD values were calculated for the interior eight Aβ9–40 peptides of finite S2-C2z protofilaments, and averaged over the 5–10 ns trajectory segments. At room temperature, we observe only a small increase in the flexibility of the loop region and no significant change in the β-strand segments. At elevated temperature (398 K), the mutations result not only in increased loop flexibility, but also produce large fluctuations in the β-strand regions. Figs. S6 and S7 show snapshots along the trajectories for both wild-type and mutated systems. Overall, the fibril does not appear to be strongly affected by these loop mutations at room temperature, but the enhanced flexibility of the mutated loops results in a significant loss of structure at 398 K.
FIGURE 5.
Cα RMSD per residue calculated for two-peptide units from the interior of S2-C2z fibril segments, at 298 K and 398 K, for the wild-type (solid, thin) and a mutated (shaded, thick) loop sequence: V24GSN27 → A24GAA27. Error bars indicate one estimated standard deviation of the average Cα-RMSD values.
Conformational dynamics at fibril ends
At elevated temperature, structure is lost most rapidly at the fibril ends, followed by the N-terminal sheets of the interior peptides. The core of the fibrils comprised of the C-terminal β-sheets remains largely intact. The loss of structure is more pronounced in the S2 and S3 systems with larger initial staggering, indicating that (at least at the finite lengths studied here) they are less thermo-stable than S1. The dissociation of peptides from the fibril ends seems to initiate at the N-terminal residues and in the loop region. At 398 K, transient helical conformations form in the loop and the N-terminal regions (emphasized by boxes in Fig. 3 B and in Fig. S2 B). Transient helices have also been detected during early fibril formation experiments and simulations (19,22,49,58).
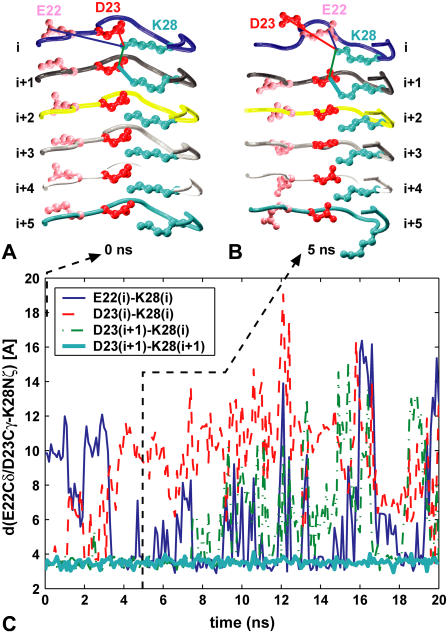
At 398 K, in several instances we observe an exchange in the salt bridge partners from the initial D23/K28 pair to E22/K28 as the end peptides begin to dissociate from the fibril (Fig. 6). The presence of E22/K28 salt bridges has been suggested both for the small peptide aggregates implicated in cytotoxicity and for Aβ monomers in solution (10,19,28,33), but not Aβ1–40 fibrils (36).
FIGURE 6.
Salt bridges (A) D23/K28 and (B) E22/K28 at fibril ends. (C) Time evolution of D23/K28 and E22/K28 distances in the S1 system at 398 K, color-coded as in panels A and B.
Fibril dissociation/elongation
Fig. 7 shows Cα RMSD values calculated for a typical trajectory (system S1-C2z) at room temperature (298 K) and at 398 K. The RMSD is calculated by using only the Cα atoms in the four interior two-peptide units in the Aβ9–40 fibril segments, and by aligning the simulation frames to the average obtained for the 1–5 ns trajectory segment. The sequence segments H5-F12, E14-G21, and I23-G30 (see Fig. 1 A) were used for the N-terminal sheet, loop, and C-terminal sheet, respectively. Noticeably, the C-terminal β-strands preserve their structural order longer than other regions of the Aβ peptides. Cα RMSD values calculated for either the N-terminal sheet or the loop, are always significantly higher, especially at elevated temperatures.
The higher stability of the C-terminal β-strand regions observed in the MD simulations suggests a possible mechanism for fibril elongation that follows the reversed steps of dissociation. In the resulting hypothetical scenario, the initial monomer addition at the end of a growing fibril is driven by strong hydrophobic interactions stabilizing the C-terminal β-strands. In a second stage, the less stable N-terminal β-strands would form. In a final step of this fibril-elongation scenario, the more flexible loop with its relatively hydrophilic residues would adopt the fibril conformation. During elongation, the E22/K28 ion pair suggested for the free monomer in solution (19,28) is replaced by D23/K28.
Overall, based on our qualitative observations the main driving force for fibril elongation appears to be the formation of C-terminal β-sheets. Their hydrophobic character and matching side-chain motif I31×G33×M35×G37 permits the interdigitation of large and small side chains that stabilizes the quaternary contacts between the two C-terminal β-sheets (16).
Recent experiments showed that perturbing the hydrogen bonds in N- and C-terminal β-sheets through selective N-methylation affects both fibril growth and structure (15). Disrupting the backbone hydrogen bonds of the N-terminal sheets resulted in relatively slow growth of fibrils with a “fuzzy” boundary; with hydrogen bonds in the C-terminal sheets disrupted, the growth was slightly less affected, and the fibrils had a sharply defined surface. These experimental results are consistent with our simulation observations. The C-terminal sheets are held together by strong hydrophobic interactions that would compensate for the partial loss of hydrogen bonds in the N-methylated system and produce wild-type-like fibrils. Increased fibril twisting could be caused by the loss of directional hydrogen bonding, and a gain in importance of packing interactions (59). In contrast, N-methylation at the N-termini disrupts the more fragile N-terminal sheets, with peptide ends sticking into the solvent to produce fibrils with a “fuzzy” boundary.
CONCLUSIONS
We explored the contributions of the different structural elements of Aβ protofilaments to the stability, conformational dynamics, and to the fibril elongation/dissociation mechanism. Using multiple 10–20 ns long MD simulations of fibril systems of ∼70,000 atoms, we studied protofilament models that differ 1), in the relative orientation of the C-terminal β-strands at the fibril core (C2x and C2z, Fig. 1, D and E); 2), the β-sheet staggering; and 3), connecting loop conformation (S1, S2, and S3 systems, Fig. 2) as well as sequence (VGSN versus AGAA). Our NPT simulations cover a broad range of temperatures (298, 348, and 398 K), for both wild-type and mutated sequences, allowing us to probe the structural stability and the early dissociation events occurring in Aβ protofilaments. We find that all models are stable at room temperature, and converge toward an interdigitated side-chain packing for intermolecular contacts within and between the two-peptide units of the protofilaments.
The D23/K28 salt bridges maintain a stable and relatively rigid interdigitated structure. However, we find that during the initial stages of fibril dissociation the D23/K28 contacts in Aβ peptides at fibril ends can break to form the competing E22/K28 interaction. This observation suggests that the loss of E22/K28 contacts could be an important fingerprint of the transition experienced by the Aβ peptides from their solution structures toward the fibril conformations (19). As reported before (12), we find narrow water channels solvating the D23/K28 salt bridges interior to Aβ fibril segments.
Simulations at all temperatures reveal that β-strand staggering is a characteristic element of Aβ protofilaments, permitting a compact, interdigitated packing of side chains from neighboring β-sheets. We find that the type of staggering, as defined by using the intrinsic directionality of Aβ fibrils, can differ for different fibril models, and we observe a noticeable bias across several fibril models toward adopting positive staggerings (Fig. 2). As the most dramatic change in structure, we observed collective sliding of N- and C-terminal β-sheets on top of each other. Simulations show that Aβ peptides may adopt structural conformations with smaller β-sheet staggering for residues close to the loop than for those at the tip of the strands (Fig. 4 D), with N- and C-terminal strands of the same Aβ peptide being at an angle to each other. At elevated temperatures, simulations show features common to different models, most notably loss of order in the solvent-exposed N-terminal β-strands coupled to structural disorder in the loop regions. The loop regions and the N-terminal β-strands appear most sensitive to temperature increases. Mutations of loop residues are found to enhance the flexibility of the fibrils at elevated temperatures. Our simulations suggest that the hydrophobic fibril core comprising the C-terminal β-strands of the two molecular fibril layers is a major stabilizing element and its formation may constitute a crucial step in the aggregation and elongation of Aβ protofilaments.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Dr. Robert Tycko and Dr. Anant Paravastu for many helpful discussions. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, Maryland (http://biowulf.nih.gov).
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases.
References
- 1.Selkoe, D. J. 1994. Cell biology of the amyloid beta-protein precursor and the mechanism of Alzheimer's disease. Annu. Rev. Cell Biol. 10:373–403. [DOI] [PubMed] [Google Scholar]
- 2.Tycko, R. 2004. Progress towards a molecular-level structural understanding of amyloid fibrils. Curr. Opin. Struct. Biol. 14:96–103. [DOI] [PubMed] [Google Scholar]
- 3.Sunde, M., and C. C. F. Blake. 1998. From the globular to the fibrous state: protein structure and structural conversion in amyloid formation. Q. Rev. Biophys. 31:1–39. [DOI] [PubMed] [Google Scholar]
- 4.Caughey, B., and P. T. Lansbury. 2003. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 26:267–298. [DOI] [PubMed] [Google Scholar]
- 5.Lester-Coll, N., E. J. Rivera, S. J. Soscia, K. Doiron, J. R. Wands, and S. M. de la Monte. 2006. Intracerebral streptozotocin model of type 3 diabetes: relevance to sporadic Alzheimer's disease. J. Alzheimers Dis. 9:13–33. [DOI] [PubMed] [Google Scholar]
- 6.de Felice, F. G., M. N. N. Vieira, L. M. Saraiva, J. D. Figueroa-Villar, J. Garcia-Abreu, R. Liu, L. Chang, W. L. Klein, and S. T. Ferreira. 2004. Targeting the neurotoxic species in Alzheimer's disease: inhibitors of Aβ oligomerization. FASEB J. 18:1366–1372. [DOI] [PubMed] [Google Scholar]
- 7.Carulla, N., G. L. Caddy, D. R. Hall, J. Zurdo, M. Gairi, M. Feliz, E. Giralt, C. V. Robinson, and C. M. Dobson. 2005. Molecular recycling within amyloid fibrils. Nature. 436:554–558. [DOI] [PubMed] [Google Scholar]
- 8.Zhang, S., K. Iwata, M. J. Lachenmann, J. W. Peng, S. Li, E. R. Stimson, Y. Lu, A. M. Felix, J. E. Maggio, and J. P. Lee. 2000. The Alzheimer's peptide Aβ adopts a collapsed coil structure in water. J. Struct. Biol. 130:130–141. [DOI] [PubMed] [Google Scholar]
- 9.Shao, H. Y., S. C. Jao, K. Ma, and M. G. Zagorski. 1999. Solution structures of micelle-bound amyloid β-(1–40) and β-(1–42) peptides of Alzheimer's disease. J. Mol. Biol. 285:755–773. [DOI] [PubMed] [Google Scholar]
- 10.Lazo, N. D., M. A. Grant, M. C. Condron, A. C. Rigby, and D. B. Teplow. 2005. On the nucleation of amyloid β-protein monomer folding. Protein Sci. 14:1581–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petkova, A. T., W. M. Yau, and R. Tycko. 2006. Experimental constraints on quaternary structure in Alzheimer's β-amyloid fibrils. Biochemistry. 45:498–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchete, N. V., R. Tycko, and G. Hummer. 2005. Molecular dynamics simulations of Alzheimer's β-amyloid protofilaments. J. Mol. Biol. 353:804–821. [DOI] [PubMed] [Google Scholar]
- 13.Petkova, A. T., R. D. Leapman, Z. H. Guo, W. M. Yau, M. P. Mattson, and R. Tycko. 2005. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science. 307:262–265. [DOI] [PubMed] [Google Scholar]
- 14.Gordon, D. J., and S. C. Meredith. 2003. Probing the role of backbone hydrogen bonding in beta-amyloid fibrils with inhibitor peptides containing ester bonds at alternate positions. Biochemistry. 42:475–485. [DOI] [PubMed] [Google Scholar]
- 15.Sciarretta, K. L., A. Boire, D. J. Gordon, and S. C. Meredith. 2006. Spatial separation of β-sheet domains of β-amyloid: disruption of each β-sheet by N-methyl amino acids. Biochemistry. 45:9485–9495. [DOI] [PubMed] [Google Scholar]
- 16.Sato, T., P. Kienlen-Campard, M. Ahmed, W. Liu, H. L. Li, J. I. Elliott, S. Aimoto, S. N. Constantinescu, J. N. Octave, and S. O. Smith. 2006. Inhibitors of amyloid toxicity based on β-sheet packing of Aβ40 and Aβ42. Biochemistry. 45:5503–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheibel, T., R. Parthasarathy, G. Sawicki, X.-M. Lin, H. Jaeger, and S. L. Lindquist. 2003. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc. Natl. Acad. Sci. USA. 100:4527–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reches, M., and E. Gazit. 2006. Molecular self-assembly of peptide nanostructures: mechanism of association and potential uses. Curr. Nanosci. 2:105–111. [Google Scholar]
- 19.Baumketner, A., S. L. Bernstein, T. Wyttenbach, N. D. Lazo, D. B. Teplow, M. T. Bowers, and J.-E. Shea. 2006. Structure of the 21–30 fragment of the amyloid β-protein. Protein Sci. 15:1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang, W., S. Zhang, R. D. Kamm, and M. Karplus. 2004. Kinetic control of dimer structure formation in amyloid fibrillogenesis. Proc. Natl. Acad. Sci. USA. 101:12916–12921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urbanc, B., L. Cruz, F. Ding, D. Sammond, S. Khare, S. V. Buldyrev, H. E. Stanley, and N. V. Dokholyan. 2004. Molecular dynamics simulation of amyloid β dimer formation. Biophys. J. 87:2310–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarus, B., J. E. Straub, and D. Thirumalai. 2005. Probing the initial stage of aggregation of the Aβ10–35-protein: assessing the propensity for peptide dimerization. J. Mol. Biol. 345:1141–1156. [DOI] [PubMed] [Google Scholar]
- 23.Klimov, D. K., and D. Thirumalai. 2003. Dissecting the assembly of Aβ(16–22) amyloid peptides into antiparallel β sheets. Structure. 11:295–307. [DOI] [PubMed] [Google Scholar]
- 24.Gnanakaran, S., R. Nussinov, and A. E. García. 2006. Atomic-level description of amyloid beta-dimer formation. J. Am. Chem. Soc. 128:2158–2159. [DOI] [PubMed] [Google Scholar]
- 25.Ma, B. Y., and R. Nussinov. 2006. The stability of monomeric intermediates controls amyloid formation: Aβ25-35 and its N27Q mutant. Biophys. J. 90:3365–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fawzi, N. L., V. Chubukov, L. A. Clark, S. Brown, and T. Head-Gordon. 2005. Influence of denatured and intermediate states of folding on protein aggregation. Protein Sci. 14:993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dima, R. I., and D. Thirumalai. 2002. Exploring protein aggregation and self-propagation using lattice models: phase diagram and kinetics. Protein Sci. 11:1036–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borreguero, J. M., B. Urbanc, N. D. Lazo, S. V. Buldyrev, D. B. Teplow, and H. E. Stanley. 2005. Folding events in the 21–30 region of amyloid β-protein (Aβ) studied in silico. Proc. Natl. Acad. Sci. USA. 102:6015–6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen, H. D., and C. K. Hall. 2004. Molecular dynamics simulations of spontaneous fibril formation by random-coil peptides. Proc. Natl. Acad. Sci. USA. 101:16180–16185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, L., T. A. Darden, L. Bartolotti, D. Kominos, and L. G. Pedersen. 1999. An atomic model for the pleated β-sheet structure of Aβ amyloid protofilaments. Biophys. J. 76:2871–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cecchini, M., R. Curcio, M. Pappalardo, R. Melki, and A. Caflisch. 2006. A molecular dynamics approach to the structural characterization of amyloid aggregation. J. Mol. Biol. 357:1306–1321. [DOI] [PubMed] [Google Scholar]
- 32.Ma, B. Y., and R. Nussinov. 2002. Stabilities and conformations of Alzheimer's β-amyloid peptide oligomers (Aβ(16–22), Aβ(16–35) and Aβ(10–35)): sequence effects. Proc. Natl. Acad. Sci. USA. 99:14126–14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cruz, L., B. Urbanc, J. M. Borreguero, N. D. Lazo, D. B. Teplow, and H. E. Stanley. 2005. Solvent and mutation effects on the nucleation of amyloid β-protein folding. Proc. Natl. Acad. Sci. USA. 102:18258–18263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gsponer, J., and M. Vendruscolo. 2006. Theoretical approaches to protein aggregation. Protein Pept. Lett. 13:287–293. [DOI] [PubMed] [Google Scholar]
- 35.Fawzi, N. L., Y. Okabe, E.-H. Yap, and T. Head-Gordon. 2007. Determining the critical nucleus and mechanism of fibril elongation of the Alzheimer's Abetal–40 peptide. J. Mol. Biol. 365:535–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petkova, A. T., Y. Ishii, J. J. Balbach, O. N. Antzutkin, R. D. Leapman, F. Delaglio, and R. Tycko. 2002. A structural model for Alzheimer's beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. USA. 99:16742–16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paravastu, A. K., A. T. Petkova, and R. Tycko. 2006. Polymorphic fibril formation by residues 10–40 of the Alzheimer's β-amyloid peptide. Biophys. J. 90:4618–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasahara, K., H. Naiki, and Y. Goto. 2005. Kinetically controlled thermal response of β2-microglobulin amyloid fibrils. J. Mol. Biol. 352:700–711. [DOI] [PubMed] [Google Scholar]
- 39.Meersman, F., and C. M. Dobson. 2006. Probing the pressure-temperature stability of amyloid fibrils provides new insights into their molecular properties. BBA Proteins Proteom. 1764:452–460. [DOI] [PubMed] [Google Scholar]
- 40.Zanuy, D., K. Gunasekaran, B. Y. Ma, H. H. Tsai, C. J. Tsai, and R. Nussinov. 2004. Insights into amyloid structural formation and assembly through computational approaches. Amyloid J. Protein Folding Disord. 11:143–161. [DOI] [PubMed] [Google Scholar]
- 41.Kale, L., R. Skeel, M. Bhandarkar, R. G. Brunner, A. N. Krawetz, J. Phillips, A. Shinozaki, K. Varadarajan, and K. Schulten. 1999. NAMD2: greater scalability for parallel molecular dynamics. J. Comput. Phys. 151:283–312. [Google Scholar]
- 42.MacKerell, A. D., D. Bashford, M. Bellott, R. L. Dunbrack, J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph-McCarthy, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, W. E. Reiher, B. Roux, M. Schlenkrich, J. C. Smith, R. Stote, J. Straub, W. Watanabe, J. Wiorkiewicz-Kuczera, D. Yin, and M. Karplus. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102:3586–3616. [DOI] [PubMed] [Google Scholar]
- 43.Jorgensen, W. L., J. Chandrasekhar, J. D. Madura, R. W. Impey, and M. L. Klein. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935. [Google Scholar]
- 44.Martyna, G. J., D. J. Tobias, and M. L. Klein. 1994. Constant-pressure molecular-dynamics algorithms. J. Chem. Phys. 101:4177–4189. [Google Scholar]
- 45.Feller, S. E., Y. H. Zhang, R. W. Pastor, and B. R. Brooks. 1995. Constant-pressure molecular-dynamics simulation—the Langevin piston method. J. Chem. Phys. 103:4613–4621. [Google Scholar]
- 46.Darden, T., D. York, and L. Pedersen. 1993. Particle-mesh Ewald—An Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- 47.Ryckaert, J. P., G. Ciccotti, and H. J. C. Berendsen. 1977. Numerical integration of Cartesian equations of motion of a system with constraints—molecular dynamics of n-alkanes. J. Comput. Phys. 23:327–341. [Google Scholar]
- 48.Li, A. J., and V. Daggett. 1996. Identification and characterization of the unfolding transition state of chymotrypsin inhibitor 2 by molecular dynamics simulations. J. Mol. Biol. 257:412–429. [DOI] [PubMed] [Google Scholar]
- 49.Massi, F., D. Klimov, D. Thirumalai, and J. E. Straub. 2002. Charge states rather than propensity for beta-structure determine enhanced fibrillogenesis in wild-type Alzheimer's beta-amyloid peptide compared to E22Q Dutch mutant. Protein Sci. 11:1639–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thirumalai, D., D. K. Klimov, and R. I. Dima. 2003. Emerging ideas on the molecular basis of protein and peptide aggregation. Curr. Opin. Struct. Biol. 13:146–159. [DOI] [PubMed] [Google Scholar]
- 51.Dima, R. I., and D. Thirumalai. 2004. Proteins associated with diseases show enhanced sequence correlation between charged residues. Bioinformatics. 20:2345–2354. [DOI] [PubMed] [Google Scholar]
- 52.Arispe, N. 2004. Architecture of the Alzheimer's AβP ion channel pore. J. Membr. Biol. 197:33–48. [DOI] [PubMed] [Google Scholar]
- 53.Quist, A., L. Doudevski, H. Lin, R. Azimova, D. Ng, B. Frangione, B. Kagan, J. Ghiso, and R. Lal. 2005. Amyloid ion channels: a common structural link for protein-misfolding disease. Proc. Natl. Acad. Sci. USA. 102:10427–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ambroggio, E. E., D. H. Kim, F. Separovic, C. J. Barrow, K. J. Barnham, L. A. Bagatolli, and G. D. Fidelio. 2005. Surface behavior and lipid interaction of Alzheimer β-amyloid peptide 1–42: a membrane-disrupting peptide. Biophys. J. 88:2706–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lashuel, H. A., D. Hartley, B. M. Petre, J. S. Wall, M. N. Simon, T. Walz, and P. T. Lansbury. 2003. Mixtures of wild-type and a pathogenic (E22G) form of Aβ40 in vitro accumulate protofibrils, including amyloid pores. J. Mol. Biol. 332:795–808. [DOI] [PubMed] [Google Scholar]
- 56.Rohrig, U. F., A. Laio, N. Tantalo, M. Parrinello, and R. Petronzio. 2006. Stability and structure of oligomers of the Alzheimer peptide Aβ16–22: from the dimer to the 32-mer. Biophys. J. 91:3217–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sciarretta, K. L., D. J. Gordon, A. T. Petkova, R. Tycko, and S. C. Meredith. 2005. Aβ40-Lactam(D23/K28) models a conformation highly favorable for nucleation of amyloid. Biochemistry. 44:6003–6014. [DOI] [PubMed] [Google Scholar]
- 58.Kirkitadze, M. D., M. M. Condron, and D. B. Teplow. 2001. Identification and characterization of key kinetic intermediates in amyloid β-protein fibrillogenesis. J. Mol. Biol. 312:1103–1119. [DOI] [PubMed] [Google Scholar]
- 59.Chothia, C., and J. Janin. 1982. Orthogonal packing of β-pleated sheets in proteins. Biochemistry. 21:3955–3965. [DOI] [PubMed] [Google Scholar]
- 60.Frishman, D., and P. Argos. 1995. Knowledge-based protein secondary structure assignment. Proteins. 23:566–579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.